Open Access Article
Open Access ArticleTarget-induced activation of DNAzyme for highly sensitive colorimetric detection of bleomycin via DNA scission†
Xiaofei Liaoa,
Mengyan Lia and
Li Zou *ab
*ab
aSchool of Pharmacy, Guangdong Pharmaceutical University, Guangzhou 510006, PR China. E-mail: lizou@gdpu.edu.cn
bKey Specialty of Clinical Pharmacy, The First Affiliated Hospital of Guangdong Pharmaceutical University, Guangzhou 510699, PR China
First published on 22nd June 2022
Abstract
In this work, a label-free and sensitive colorimetric sensing strategy for the detection of bleomycin (BLM) was developed on the basis of BLM-mediated activation of G-quadruplex DNAzyme via DNA strand scission. A G-quadruplex based hairpin probe (G4HP) containing the scission site (5′-GT-3′) of BLM at the loop region and guanine (G)-rich sequences at its 5′-end was employed in this protocol. In the presence of BLM, it may cleave the 5′-GT-3′ site of the hairpin probe with Fe(II) as a cofactor, releasing the G-tetrads DNA fragment, which may further bind hemin to form a catalytic G-quadruplex-hemin DNAzyme. The resultant G-quadruplex DNAzyme has notable peroxidase-like activity, which effectively catalyzes the oxidation of 2,2′-azino-bis(3-ethylbenzothiozoline-6-sulfonic acid) (ABTS) by H2O2 to produce the blue-green-colored free-radical cation (ABTS·+). Therefore, the detection of BLM can be achieved by observing the color transition with the naked eye or measuring the absorbance at a wavelength of 420 nm using a UV-Vis spectrophotometer. Attributing to the specific BLM-induced DNA strand scission and the effective locking of G-tetrads in the stem of the G4HP, the colorimetric sensing strategy exhibits high sensitivity and selectivity for detection of BLM in human serum samples, which might hold great promise for BLM assay in biomedical and clinical research.
Introduction
As a class of glycopeptide-derived antibiotics isolated from Streptomyces species, bleomycins (BLMs) have been widely used clinically in combination with chemotherapy for the treatment of various tumorous diseases due to their distinct advantages of low myelosuppression and low immunosuppression.1–4 The antitumor activity of BLMs is generally attributed to the selective cleavage of single-stranded or double-stranded DNAs and possibly RNAs in the presence of oxygen and metal ions as cofactors.5–7 However, an unsuitable dose of BLMs in clinical treatment also exhibits some serious dose-limiting side effects, such as renal and lung toxicity, and bad pulmonary fibrosis.8,9 In order to achieve the desired therapeutic effect with a lower dose and toxicity, there is great anxiety to develop a reliable and sensitive method for the determination of trace amounts of BLM in both pharmaceutical analysis and clinical samples. Up to date, various methods for BLM detection have been developed, such as high-performance liquid chromatography (HPLC),10 radioimmunoassay (RIA),11 enzyme immunoassay (EIA),12 microbiological assay,13 colorimetric assay,14 fluorescent assay15–17 and electrochemical assay.18,19 Among them, colorimetric methods have attracted significant attention due to the remarkable features of low cost, convenient operation, instrument simplicity and fast response.20Recently, some colorimetric strategies have been reported for BLM assay based on the aggregation of gold nanoparticles (AuNPs),14,21 but these methods suffer from poor specificity. An alternative approach has been developed for colorimetric detection of BLM based on BLM–Fe(II) complex catalyzed oxidation of 2,2′-azino-bis(3-ethylbenzothiozoline-6-sulfonic acid) (ABTS).22 However, Fe(II)-based Fenton reaction is optimal at low pH,23 which limits its scope of application. As reported, the BLM–Fe(II) complex can active molecular oxygen to generate a ternary complex BLM–Fe(III)OOH, which is able to selectively degrade DNA at a recognition site.24,25 Various sensing strategies have been developed for the sensitive determination of BLM based on BLM–Fe(II) induced DNA strand scission.26–29 For example, Ma et al. applied the specific scission of BLM to release G-tracts DNA fragment locked in the molecular beacon (MB), which self-assembles into a G-triplex-thioflavin T (ThT) complex with strong fluorescence emission.30 He et al. reported a sensitive electrochemical assay for BLM detection based on BLM-mediated activation of Zn2+-dependent DNAzyme and a MOF-modified electrode.18 To the best of our knowledge, the application of BLM-induced DNA scission for colorimetric assay of BLM has not been reported yet. Therefore, it is still highly desirable to develop sensitive and label-free colorimetric sensing strategies for BLM detection.
Over the past decades, G-quadruplex DNAzyme has been widely applied in colorimetric, chemiluminescent and electrochemical assays because of its good stability, low cost and ease of synthesis.31–36 More importantly, the peroxidase-like activity of G-quadruplex DNAzyme can be applied to efficiently catalyze the H2O2-mediated oxidation of ABTS to free-radical cation (ABTS·+) with an obvious color change from colorless to green. Furthermore, G-quadruplex DNAzyme can be designed ingeniously, such as G-quadruplex based label-free molecular beacon, to improve the selectivity and enhance the signal response. Inspired by the BLM–Fe(II) mediated DNA scission and the catalytic activity of G-quadruplex DNAzyme, we herein propose a label-free and sensitive colorimetric method for determination of BLM based on BLM-mediated activation of G-quadruplex DNAzyme via DNA strand scission. We design a G-quadruplex based hairpin probe (G4HP) containing the scission site (5′-GT-3′) of BLM at the loop region and guanine (G)-rich sequences at its 5′-end. The G4HP undergoes an irreversible cleavage event by BLM–Fe(II) complex at 5′-GT-3′ scission site. The released G-tetrads DNA fragment further binds hemin to form a G-quadruplex-hemin DNAzyme, which can effectively catalyze the oxidation of ABTS by H2O2 to produce a green color. The determination of BLM is thus realized by measuring the absorbance at a wavelength of 420 nm using a UV-Vis spectrophotometer. The colorimetric sensing strategy shows high sensitivity and good selectivity for BLM detection, and it exhibits good assay performance in human serum samples.
Experimental section
Materials and reagents
Bleomycin (BLM) was purchased from Melone Pharmaceutical Co., Ltd. (Dalian, China). Streptomycin, kanamycin, oxytetracycline, chloramphenicol, ampicillin, tobramycin, cisplatin and 5-fluorouracil were obtained from Aladdin Industrial Corporation (Shanghai, China). Glucose, arginine, urea, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS), hemin, and doxorubicin were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). H2O2, FeCl2 and other analytical reagents were obtained from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China) and used as received without further purification. A 10 mM sodium phosphate buffer containing 100 mM NaCl, and 5 mM KCl (PBS, pH 7.5) was used in the experiments. Ultrapure water with a resistivity of 18.2 MΩ cm obtained from a Millipore Milli-Q water purification system was used throughout the experiments. Oligonucleotide (5′-TGGGTAGGGCGGGTTGGGATTAATTTGTTTTTAATCCC-3′, G4HP) was synthesized and purified by Sangon Biotech Co., Ltd. (Shanghai, China).Colorimetric detection of BLM
The oligomucleotide G4HP was used as received and diluted in PBS buffer. The BLM–Fe(II) complex was prepared by mixing BLM with FeCl2 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. Subsequently, 40 μL of G4HP (1.5 μM) and 50 μL of hemin (3 μM) were added to the as-prepared BLM–Fe(II) complex, and then incubated at 37 °C for 30 min for the oxidative cleavage of G4HP and the formation of G-quadruplex-hemin DNAzyme. Colorimetric measurements were performed by mixing the above solution with 10 μL of ABTS (4.5 μM), and 10 μL of H2O2 (30 mM). The obtained mixture was incubated at room temperature for about 10 min. Finally, the UV-Vis absorbance spectra of the resulting solution were recorded using a UV-6100 spectrophotometer (Metash, Shanghai), and the absorbance at 420 nm was measured to determine the concentration of BLM.
1 molar ratio. Subsequently, 40 μL of G4HP (1.5 μM) and 50 μL of hemin (3 μM) were added to the as-prepared BLM–Fe(II) complex, and then incubated at 37 °C for 30 min for the oxidative cleavage of G4HP and the formation of G-quadruplex-hemin DNAzyme. Colorimetric measurements were performed by mixing the above solution with 10 μL of ABTS (4.5 μM), and 10 μL of H2O2 (30 mM). The obtained mixture was incubated at room temperature for about 10 min. Finally, the UV-Vis absorbance spectra of the resulting solution were recorded using a UV-6100 spectrophotometer (Metash, Shanghai), and the absorbance at 420 nm was measured to determine the concentration of BLM.
Real samples analysis
For the determination of BLM in real complex samples, recovery experiment by using the standard addition method was carried out by adding four different concentrations of BLM (50 nM, 300 nM, 500 nM, and 800 nM) into the diluted serum sample, followed by colorimetric measurements. Human serum samples of healthy persons were kindly supplied by Sun Yat-Sen University Cancer Center (Guangzhou, China) and diluted 200 times with PBS buffer (10 mM, pH 7.5).Results and discussion
Principle of the colorimetric sensing strategy
A label-free and sensitive colorimetric sensing strategy for BLM detection was developed on the basis of BLM-mediated activation of G-quadruplex DNAzyme via DNA strand scission, which is illustrated in Scheme 1. The G-quadruplex based hairpin probe (G4HP) contains two function domains: a 5′-GT-3′ scission site of BLM at the loop region; the four G-tetrads with the first one of which is locked in the stem of the G4HP, thereby inhibiting the formation of G-quadruplex-hemin DNAzyme. In the presence of BLM, the G4HP undergoes an irreversible cleavage event at the 5′-GT-3′ site via the oxidative effect of BLM with Fe(II) as a cofactor, releasing the G-tetrads DNA fragment, which further binds hemin to form a G-quadruplex-hemin DNAzyme. The resultant G-quadruplex DNAzyme can effectively catalyze the oxidation of ABTS by H2O2 to induce a distinct color change from colorless to green. Therefore, the colorimetric detection of BLM can be achieved by observing the color transition with the naked eye or measuring the absorbance at a wavelength of 420 nm using a UV-Vis spectrophotometer.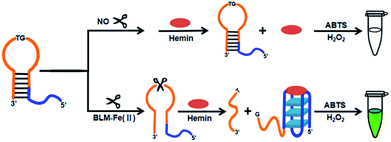 | ||
| Scheme 1 Schematic illustration of the G-quadruplex based hairpin probe (G4HP) sensing system for BLM detection. | ||
Feasibility of the colorimetric sensing strategy
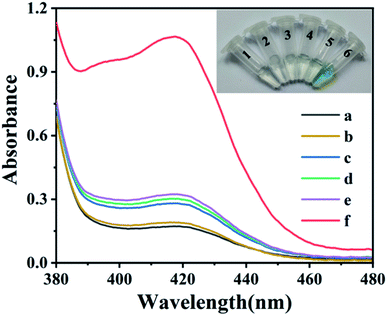
To demonstrate the feasibility of the G-quadruplex based hairpin probe (G4HP) sensing system for the colorimetric detection of BLM, the color changes and UV-Vis spectra of the H2O2–ABTS mixture under different conditions were investigated. As shown in Fig. 1, the peak absorbance at 420 nm was not obvious when adding BLM to H2O2-ABTS (curve a) or Fe(II)–H2O2–ABTS (curve b) mixtures, and the solution was colorless, which indicated that the BLM enhanced Fe(II)–H2O2–ABTS reaction displayed poor efficiency when the concentration of Fe2+ was below 5 μM.22 In addition, adding G4HP to H2O2-ABTS (vial 3, inset) or Fe(II)–H2O2–ABTS (vial 4, inset) mixtures did not induce a color change and a remarkable increase in the absorbance at 420 nm, indicating that the four G-tetrads was effectively locked in the stem of the G4HP and thus no G-quadruplex-hemin DNAzyme was generated. However, the peak absorbance at 420 nm sharply increased upon incubation with BLM–Fe(II) complex (curve f), and the color of the solution changed from colorless to green (vial 6, inset), indicating the occurrence of BLM–Fe(II) induced DNA strand scission at the loop region of G4HP to release the G-tetrads DNA fragment which can self-assemble into a G-quadruplex-hemin DNAzyme with notable peroxidase-like catalytic activity. Therefore, the G4HP based sensing system can be applied for label-free colorimetric detection of BLM.Optimization of assay conditions
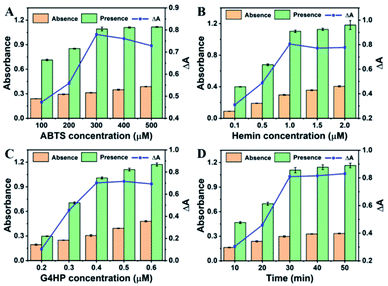
To achieve the best assay performance for BLM detection, some important experimental conditions were systematically investigated. First, the effect of ABTS concentration on the colorimetric signal was investigated. As shown in Fig. 2A, the absorbance change (ΔA = A − A0) was enhanced significantly with the increase of ABTS concentration from 100 to 300 μM, followed by the decrease beyond 300 μM (where A0 and A were the peak absorbance at 420 nm of the sensing system with or without BLM, respectively). Hence, 300 μM was chosen as the optimum ABTS concentration for the subsequent experiments. Meanwhile, the effect of hemin concentration on the sensing performance was investigated. It could be observed that the value of ΔA increased as the hemin concentration increased from 0.1 to 1.0 μM and tended to be constant when the concentration was higher than 1.0 μM (Fig. 2B). Thus, the hemin concentration of 1.0 μM was adopted in the following experiments. In addition, the effect of G4HP concentration and the reaction time on the peak absorbance of the sensing system were also investigated. As shown in Fig. 2C, with the concentration of G4HP increased from 0.2 to 0.4 μM, the value of ΔA significantly increased and leveled off when the G4HP concentration was above 0.4 μM. As shown in Fig. 2D, the ΔA increased rapidly with the reaction time until 30 min, and then tended to be constant. As a result, the G4HP concentration of 0.4 μM and reaction time of 30 min were selected in the detection of BLM.Analytical performance for BLM detection
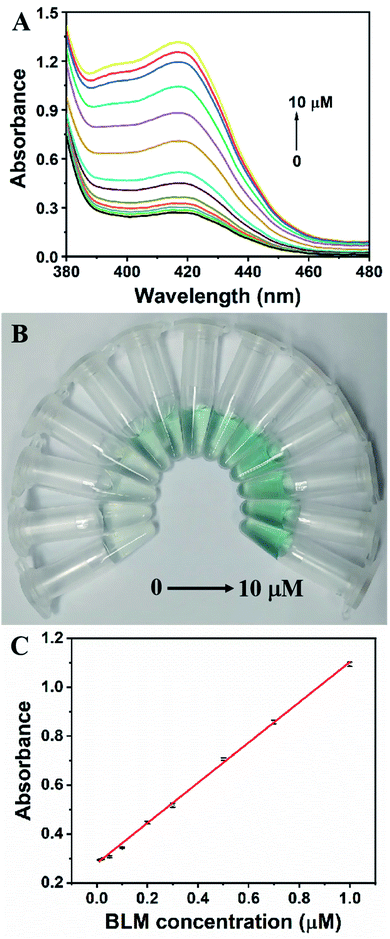
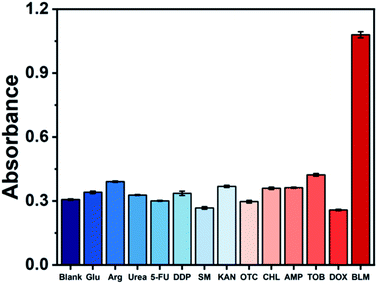
Under optimal conditions, the analytical performance of the G4HP based sensing system was investigated. Fig. 3A displayed the UV-Vis spectra corresponding to different concentrations of BLM. The absorbance at 420 nm of the sensing system increased significantly with the increase of BLM concentration ranging from 0 to 10 μM, and the color of solution gradually changed from colorless to green (Fig. 3B). A linear calibration curve was obtained by plotting the absorption intensity at 420 nm against the BLM concentration over the range from 10 nM to 1 μM (Fig. 3C). The regression equation is A = 0.28014 + 0.82307C (R2 = 0.9967), where A and C represent the absorption intensity at 420 nm and the BLM concentration, respectively. The detection limit was estimated to be as low as 5 nM with a signal-to-noise ratio of 3. Meanwhile, the assay volume of 0.15 mL and the assay time of 40 min needed to complete the detection of BLM, which are comparable to those of the existing sensing strategies for BLM detection (Table S1†). To evaluate the selectivity of the colorimetric sensing strategy, various other potential interferences including glucose (Glu), arginine (Arg), urea, 5-fluorouracil (5-FU), cisplatin (DDP), streptomycin (SM), kanamycin (KAN), oxytetracycline (OTC), chloramphenicol (CHL), ampicillin (AMP), tobramycin (TOB) and doxorubicin (DOX) were added into the sensing system, respectively. As shown in Fig. 4, only upon addition of BLM, a remarkable change in the absorbance at 420 nm could be induced due to the specific DNA scission, whereas the absorbance of one of the twelve interfering substances were fairly low and exhibited no conspicuous difference compared with that of blank sample, indicating that the G4HP based sensing system exhibited high specificity for discriminating BLM from other interfering substances.Human serum samples analysis
In order to evaluate the practical applicability of the colorimetric sensing strategy in complex biological samples, recovery experiments by applying the standard addition method in human serum samples were carried out. Four different concentrations of BLM were separately spiked in the diluted serum sample and detected by the proposed method. As shown in Table 1, the recoveries of the four samples were in the range of 95.2–103.8% with the relative standard deviations (RSDs) less than 3.11%, indicating that the developed method has potential application for BLM assay in complex real samples.| Sample | Added (nM) | Found (nM) | Recovery (%) | RSD (n = 3, %) |
|---|---|---|---|---|
| 1 | 50 | 47.6 | 95.2 | 2.76 |
| 2 | 300 | 290.0 | 96.6 | 1.66 |
| 3 | 500 | 506.8 | 101.4 | 3.11 |
| 4 | 800 | 830.0 | 103.8 | 1.34 |
Conclusions
In summary, we have developed a label-free and sensitive G-quadruplex based hairpin probe (G4HP) sensing system for colorimetric detection of BLM based on BLM-mediated activation of G-quadruplex DNAzyme via DNA strand scission. Under the oxidative effect of BLM with Fe(II) as a cofactor, the G4HP undergoes an irreversible cleavage event at the loop region that contains 5′-GT-3′ recognition site for BLM. The released G-tetrads DNA fragment further binds hemin to form a G-quadruplex-hemin DNAzyme, which can effectively catalyze the H2O2-meiated oxidation of ABTS to induce a distinct color change from colorless to green. Therefore, the detection of BLM can be achieved by observing the color transition or measuring the absorbance at a wavelength of 420 nm. Due to the high efficiency of BLM-activated G-quadruplex DNAzyme catalysis, the colorimetric sensing strategy exhibited a high sensitivity with a detection limit of 5 nM in a linear range of 10–1000 nM. Moreover, the application of the proposed method for BLM detection in human serum samples was also implemented with satisfactory results due to its excellent selectivity. Considering these merits, the colorimetric sensing strategy may provide with great potential for BLM determination in clinical samples.Author contributions
Xiaofei Liao: data curation, formal analysis, investigation, methodology. Mengyan Li: investigation, validation, visualization. Li Zou: conceptualization, funding acquisition, project administration, supervision, writing – original draft, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81903570) and the Natural Science Foundation of Guangdong Province (No. 2021A1515011519).References
- H. Umezawa, K. Maeda, T. Takeuchi and Y. Okami, J. Antibiot., 1966, 19, 200–209 CAS.
- U. Galm, M. H. Hager, S. G. Van Lanen, J. Ju, J. S. Thorson and B. Shen, Chem. Rev., 2005, 105, 739–758 CrossRef CAS PubMed.
- T. C. Bozeman, R. Nanjunda, C. Tang, Y. Liu, Z. J. Segerman, P. A. Zaleski, W. D. Wilson and S. M. Hecht, J. Am. Chem. Soc., 2012, 134, 17842–17845 CrossRef CAS PubMed.
- G. Tisman, V. Herbert, L. T. Go and L. Brenner, Blood, 1973, 41, 721–726 CrossRef CAS PubMed.
- J. Stubbe and J. W. Kozarich, Chem. Rev., 1987, 87, 1107–1136 CrossRef CAS.
- C. A. Claussen and E. C. Long, Chem. Rev., 1999, 99, 2797–2816 CrossRef CAS PubMed.
- R. A. Giroux and S. M. Hecht, J. Am. Chem. Soc., 2010, 132, 16987–16996 CrossRef CAS PubMed.
- J. Hay, S. Shahzeidi and G. Laurent, Arch. Toxicol., 1991, 65, 81–94 CrossRef CAS PubMed.
- E. Azambuja, J. F. Fleck, R. G. Batista and S. S. Menna Barreto, Pulm. Pharmacol. Ther., 2005, 18, 363–366 CrossRef CAS PubMed.
- R. P. Klett and J. P. Chovan, J. Chromatogr., Biomed. Appl., 1985, 337, 182–186 CrossRef CAS.
- J. D. Teale, J. M. Clough and V. Marks, Br. J. Cancer, 1977, 35, 822–827 CrossRef CAS PubMed.
- K. Fujiwara, M. Isobe, H. Saikusa, H. Nakamura, T. Kitagawa and S. Takahashi, Cancer Treat. Rep., 1983, 67, 363–369 CAS.
- T. Onuma, J. F. Holland, H. Masuda, J. A. Waligunda and G. A. Goldberg, Cancer, 1974, 33, 1230–1238 CrossRef CAS PubMed.
- F. Li, Y. Feng, C. Zhao and B. Tang, Biosens. Bioelectron., 2011, 26, 4628–4631 CrossRef CAS PubMed.
- X. Liu, W. Na, Q. Liu and X. Su, Anal. Chim. Acta, 2018, 1028, 45–49 CrossRef CAS PubMed.
- S. Zhu, L. Liu, J. Sun, F. Shi and X. Zhao, Spectrochim. Acta, Part A, 2021, 252, 119521 CrossRef CAS PubMed.
- D. Zhang, J. Hu, X. Yang, Y. Wu, W. Su and C. Zhang, Nanoscale, 2018, 10, 11134–11142 RSC.
- Y. He, Y. Gao, H. Gu, X. Meng, H. Yi, Y. Chen and W. Sun, Biosens. Bioelectron., 2021, 178, 113034 CrossRef CAS PubMed.
- J. Chang, P. Gai, H. Li and F. Li, Talanta, 2018, 190, 492–497 CrossRef CAS PubMed.
- Y. Song, W. Wei and X. Qu, Adv. Mater., 2011, 23, 4215–4236 CrossRef CAS PubMed.
- X. Zhang, Y. Zhang, H. Zhao, Y. He, X. Li and Z. Yuan, Anal. Chim. Acta, 2013, 778, 63–69 CrossRef CAS PubMed.
- Y. Qin, L. Zhang, G. Ye and S. Zhao, Anal. Methods, 2014, 6, 7973–7977 RSC.
- G. Xu and M. R. Chance, Chem. Rev., 2007, 107, 3514–3543 CrossRef CAS PubMed.
- R. A. Giroux and S. M. Hecht, J. Am. Chem. Soc., 2010, 132, 16987–16996 CrossRef CAS PubMed.
- B. Yin, D. Wu and B. Ye, Anal. Chem., 2010, 82, 8272–8277 CrossRef CAS PubMed.
- F. Li, Y. Feng, C. Zhao, P. Li and B. Tang, Chem. Commun., 2012, 48, 127–129 RSC.
- Y. Qin, Y. Ma, X. Jin, L. Zhang, G. Ye and S. Zhao, Anal. Chim. Acta, 2015, 866, 84–89 CrossRef CAS PubMed.
- H. Li, J. Chang, T. Hou and F. Li, Anal. Chem., 2017, 89, 673–680 CrossRef CAS PubMed.
- S. Cheng, M. Khan, L. Luo, L. Wang, S. Liu, J. Ping, J. Lin and Q. Hu, Anal. Chim. Acta, 2021, 1150, 338247 CrossRef CAS PubMed.
- L. Ma, X. Han, L. Xia, R. Kong and F. Qu, Analyst, 2018, 143, 5474–5480 RSC.
- H. Peng, A. M. Newbigging, Z. Wang, J. Tao, W. Deng, X. C. Le and H. Zhang, Anal. Chem., 2018, 90, 190–207 CrossRef CAS PubMed.
- R. Freeman, E. Sharon, C. Teller, A. Henning, Y. Tzfati and I. Willner, ChemBioChem, 2010, 11, 2362–2367 CrossRef CAS PubMed.
- Y. Lai, M. Li, X. Liao and L. Zou, Molecules, 2021, 26, 5016 CrossRef CAS PubMed.
- Y. M. Xie, F. N. Niu, A. M. Yu and G. S. Lai, Anal. Chem., 2020, 92, 593–598 CrossRef CAS PubMed.
- Y. Gao and B. X. Li, Anal. Chem., 2013, 85, 11494–11500 CrossRef CAS PubMed.
- P. Zhang, X. Y. Wu, R. Yuan and Y. Q. Chai, Anal. Chem., 2015, 87, 3202–3207 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02816f |
| This journal is © The Royal Society of Chemistry 2022 |