Open Access Article
Open Access ArticleAssessment of a host–guest interaction in a bilayer membrane model†
Harshita Kumari *a,
Saeedeh Neginb,
Andrew Eisenhartd,
Mohit B. Patel
*a,
Saeedeh Neginb,
Andrew Eisenhartd,
Mohit B. Patel b,
Thomas L. Beckde,
Frank Heinrichfg,
Helena J. Spikesb and
George W. Gokel
b,
Thomas L. Beckde,
Frank Heinrichfg,
Helena J. Spikesb and
George W. Gokel *bc
*bc
aJames L. Winkle College of Pharmacy, University of Cincinnati, Cincinnati, Ohio, USA 45267-0514. E-mail: kumariha@ucmail.uc.edu
bChemistry & Biochemistry, University of Missouri-St. Louis, 1 University Blvd., St. Louis, MO 63121, USA. E-mail: gokelg@umsl.edu
cBiology, University of Missouri-St. Louis, 1 University Blvd., St. Louis, MO 63121, USA
dDepartment of Chemistry, University of Cincinnati, OH 45267, USA
eNational Center for Computational Sciences, Oak Ridge National Laboratory, Oak Ridge, TN 37830, USA
fDepartment of Physics, Carnegie Mellon University, Pittsburgh, PA 15213, USA
gNIST Center for Neutron Research, National Institute of Standards and Technology, Gaithersburg, MD 20899, USA
First published on 10th November 2022
Abstract
Supramolecular interactions are well recognized and many of them have been extensively studied in chemistry. The formation of supramolecular complexes that rely on weak force interactions are less well studied in bilayer membranes. Herein, a supported bilayer membrane is used to probe the penetration of a complex between tetracycline and a macrocyclic polyether. In a number of bacterial systems, the presence of the macrocycle has been found to significantly enhance the potency of the antimicrobial in vitro. The crown·tetracycline complex has been characterized in solution, neutron reflectometry has probed complex penetration, and the phenomena have been modeled by computational methods.
Introduction
The field of supramolecular chemistry has grown from ion complexation studies conducted five decades ago1 to encompass gas phase, solution, and solid-state association and complexation interactions.2 More recently, numerous studies have focused on transport of ions through bilayer membranes by using amphiphilic molecules that show ion conduction by dint of channel function or pore formation.3 In some cases, these synthetic channels are designed to function as unimolecular conductors4 while other pore-formers self-assemble. As part of our effort in this area, we found that crown ethers having alkyl side arms can form assemblies that exhibit evidence of ion conduction rather than function as simple carriers.5 The assemblies formed by these bibracchial lariat ethers show ion transport in planar bilayer voltage clamp experiments.We had previously shown that the compounds we call hydraphiles, which consist of three linked macrocycles, insert in bilayer membranes and function as ion channels.4 When hydraphiles were co-administered at low concentrations with antibiotics, the potency of the antimicrobial against bacteria was enhanced.6 Surprisingly, similar antimicrobial potency increases were apparent when dialkyl lariat ethers were the antibiotic potentiators.5 The hypothesis was that formation of a pore caused increased membrane permeability resulting from a local disordering effect within the membrane. This could enhance the entry of an antimicrobial into the bacterial cell, resulting in a higher effective concentration. Further, any transmembrane ion conduction function would disrupt ion homeostasis. This, in turn, would disrupt the function of efflux pumps or other enzymes that are regulated by ion balance.7
To be sure, bacterial membranes are more complicated than phospholipid bilayers. The lack of details concerning membrane penetration by the lariat ether and any resultant effect on antimicrobial penetration encouraged the neutron reflectometry study reported here. The present study was further encouraged by the urgent need for any means to enhance antimicrobial potency and/or reverse resistance. Both the Centers for Disease Control and Prevention (CDC)8 and the World Health Organization (WHO)9 have issued lengthy reports describing the concerning antimicrobial situation. Indeed, the CDC proclaimed that we have already entered a “post-antibiotic” era. Each year in the United States alone, 2.8 million individuals contract resistant infections, many of which prove to be mortal.
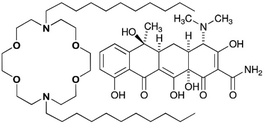
Fig. 1 shows a lariat ether that possesses a crown ether module within it. Some crown ethers have long been known to exhibit biological activity.10 As early as the 1970s, Leong et al. reported the effect of 12-crown-4 on mice11 and Takayama et al. reported toxicity to dogs12 as did crown ether inventor Pedersen.13 In the intervening years, studies have been reported of crown ether effects on mammalian tissues,14 whole animals,15 and higher plants.16 By far, however, the largest number of reports has dealt with a range of antimicrobial activities against bacteria and fungi.17
At half its minimum inhibitory concentration (MIC), N,N′-di-n-octyl-4,13-diaza-18-crown-6 (C8LE) enhanced the potency of rifampicin (RIF) and tetracycline hydrochloride (TET) against E. coli DH5α by 21-fold and 24-fold, respectively. Likewise, at 1/2 MIC, C11LE showed a 10-fold potency increase of RIF. At 1/2MIC, the enhancements of TET potency for C8LE and C11LE were 24-fold and 12-fold, respectively. It is interesting to note that the MIC for C8LE itself against this strain of E. coli was 120 μM, which indicates that it is nearly inactive as an antimicrobial against this organism.
The evidence for membrane insertion and channel formation by dialkyl lariat ethers was evident from the planar bilayer voltage clamp study noted above.5 Open-close behavior was detected from trimer, tetramer, and pentamer aggregates. Instead of being complexed within the crown's interior donor cavity, ions presumably passed through the internal channel formed by the aggregated lariat ethers, a mechanism reminiscent of that proposed for the natural channel melittin.18 We speculate that the n-alkyl groups can insert in the membrane's lipid chains in a fashion reminiscent of protein insertion in bilayers to form porins. Of course, some carrier function not detected in the planar bilayer experiment, cannot be excluded.19
Results
The LE-enhanced potency of TET against selected bacteria implies that there is an increased cytosolic concentration of the antimicrobial agent. However, the interactions of lariat ethers with a bacterial surface, perhaps altering it in some way (e.g. carpet model20) or actually within the boundary membranes remains unclear. The ability of tetracycline to penetrate bacterial membranes is, of course, well established. It was unknown, however, whether any direct interaction occurred between C10LE and TET. In order to determine this, a series of complexation experiments was conducted using NMR analysis as the primary reporter.The NMR spectra of C10LE, TET, and a mixture of the two were obtained in CD2Cl2. It was presumed that any potential interaction would be masked by the hydrogen bonding ability of bulk water, so D2O was dismissed as a potential solvent. Because increased potency of TET was observed in bacteria, it seemed reasonable to conduct the NMR experiment under conditions that mimicked a membrane. Membrane partition, as log P, estimates the water and n-octanol partition of a substance. The dielectric constant of n-octanol is reported in various sources (at 20 °C) to be 11.3, 10.34, and 8.2 (average = 9.9). We desired a solvent that is immiscible with water that had a dielectric constant in this range. Dichloromethane (as CD2Cl2 in the NMR experiment) has a dielectric constant reported to be 9.14. It was this solvent that was chosen for the complexation experiments.
The 1H-NMR spectra (Fig. 2) of C10LE and TET were both determined in CD2Cl2. The lariat ether was soluble, but only an extremely weak spectrum was observed for a saturated solution of tetracycline hydrochloride after filtration even at 600 MHz after sonication. A mixture of TET and LE was prepared by finely grinding TET and slurrying it with a solution of C10LE in CD2Cl2, followed by filtration. After filtering, no residual solid remained in the solvent–solute mixture. The 1H-NMR spectrum was recorded and the ratio of appropriately chosen integrals for TET and C10LE gave an average value (three replicates) of 1.00 ± 0.03. The results are shown in the spectra of Fig. 2.
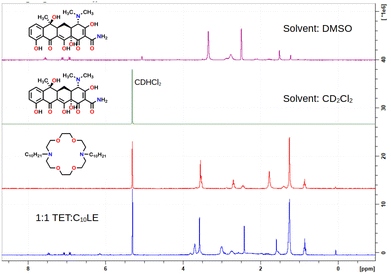 | ||
| Fig. 2 1H NMR stack plot of tetracycline (alone, top), C10LE (alone, middle), and C10LE complexed with tetracycline all in CD2Cl2 solution. | ||
Control experiments were done to confirm that 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation between C10LE and TET was not coincidental. Two other tetracycline derivatives were studied, minocycline and chlortetracycline, both also hydrochlorides. The structures of the three compounds are shown in Fig. 3, in which only the R group in the D ring varies. Like tetracycline, minocycline proved to be insoluble in both CDCl3 and CD2Cl2, but was solubilized on a 1
1 complexation between C10LE and TET was not coincidental. Two other tetracycline derivatives were studied, minocycline and chlortetracycline, both also hydrochlorides. The structures of the three compounds are shown in Fig. 3, in which only the R group in the D ring varies. Like tetracycline, minocycline proved to be insoluble in both CDCl3 and CD2Cl2, but was solubilized on a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar basis by C10LE (see Fig. S8†). Since it seemed possible that solubilization might be due to the presence of the hydrochloride salt rather than the free base, a similar study was conducted with chlortetracycline hydrochloride. No solubilization was observed in either CD2Cl2 or CDCl3 in the presence of absence of C10LE.
1 molar basis by C10LE (see Fig. S8†). Since it seemed possible that solubilization might be due to the presence of the hydrochloride salt rather than the free base, a similar study was conducted with chlortetracycline hydrochloride. No solubilization was observed in either CD2Cl2 or CDCl3 in the presence of absence of C10LE.
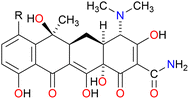 | ||
Fig. 3 Chemical structures of tetracycline (R![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) H), minocycline (R H), minocycline (R![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N(CH3)2), and chlortetracycline (R N(CH3)2), and chlortetracycline (R![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cl). Cl). | ||
Similar results were reported recently for the solubilization of tamoxifen by a pillarene. The insoluble drug was dissolved in D2O in the presence of the pillarene, but not at all in its absence.20 In that case, membrane behaviour was not probed. TET is a polyoxygenated molecule of considerable polarity. The hydrochloride salt remains protonated at the dimethylamino group at physiological pH and the cation has a calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of −4.28. The two nitrogen atoms in C10LE are also protonated at pH 7.4. We note that the pKAs of dibutyldiaza-18-crown-6 (C4LE) have been reported: pK1 = 9.40, pK2 = 7.97.21 The crown is considerably less polar in its protonated form (log
P value of −4.28. The two nitrogen atoms in C10LE are also protonated at pH 7.4. We note that the pKAs of dibutyldiaza-18-crown-6 (C4LE) have been reported: pK1 = 9.40, pK2 = 7.97.21 The crown is considerably less polar in its protonated form (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 0.83) than tetracycline, but it is still rich in ether oxygen atoms. The hydroxyl groups of tetracycline and the crown ether oxygens can interact with each other to form a complex that clearly favors the low polarity solvent and presumably the nonpolar regime of the bilayer.
P = 0.83) than tetracycline, but it is still rich in ether oxygen atoms. The hydroxyl groups of tetracycline and the crown ether oxygens can interact with each other to form a complex that clearly favors the low polarity solvent and presumably the nonpolar regime of the bilayer.
The structure of TET has a significant bend at the A:B ring junction. Even so, it presents a nearly linear line of four oxygens. The carbonyl group on the A-ring and the hydroxyl group at the A/D ring junction project nearly perpendicularly to the phenol-ketone-enol-amide carbonyl array. A solid-state structure has been reported for the hexahydrate.22 A stick representation is shown in Fig. 4 in two views that illustrate the bend in the structure, the extensive oxygenation, and a cluster of six water molecules. In principle, the heteroatoms within the macrocycle could interact as both H-bond donors (N+–H) or acceptors (>O). Likewise, TET has both H-bond donors (5OH) and acceptors (3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and 3° N), some of which are involved in intramolecular interactions.
O and 3° N), some of which are involved in intramolecular interactions.
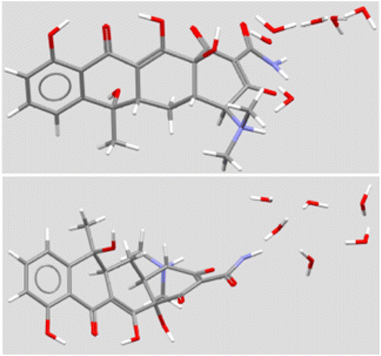 | ||
| Fig. 4 Solid state structure of tetracycline (two views) from CSD TETCYH01.28 The structure illustrated in the top panel shows the molecule with the aromatic D ring at the left. The molecule is illustrated with the long axis of the structure rotated forward by 50°. The same molecule is illustrated in the lower panel, with the upper structure rotated forward by 140°. | ||
No obvious, single 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interaction was apparent from an examination of molecular models, the lariat ether seems likely to interact with the TET structure within the hydrocarbon regime of the bilayer. The polyoxygenated array is consistent across tetracycline, minocycline, and chlortetracycline. It is unclear why chlortetracycline fails to form a complex when the two analogs do so. Chlortetracycline hydrochloride was used only as a control to assess whether HCl was playing a key role.
1 interaction was apparent from an examination of molecular models, the lariat ether seems likely to interact with the TET structure within the hydrocarbon regime of the bilayer. The polyoxygenated array is consistent across tetracycline, minocycline, and chlortetracycline. It is unclear why chlortetracycline fails to form a complex when the two analogs do so. Chlortetracycline hydrochloride was used only as a control to assess whether HCl was playing a key role.
The stack plot shown in Fig. 2, above, may not convey details of the changes observed in the chemical shifts and multiplicities. The spectra are included in the ESI along with a table (Table S-1†) that tabulates the data. We note that although the variations are modest, the most pronounced changes occur proximate to the macrocycle nitrogen atoms. This suggests at least some role for hydrochloride in the complexation. Why such an interaction would not assist in solubilizing chlortetracycline hydrochloride remains unclear. Neither it nor minocycline have been studied in the context of any membrane, which studies remain beyond the scope of the present work.
Membrane interactions
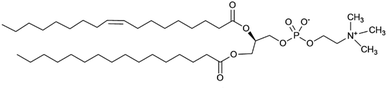
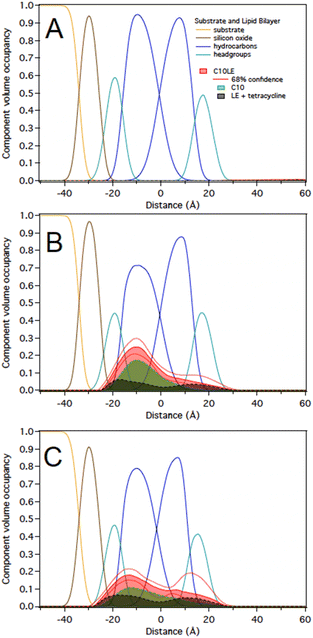
The challenge of demonstrating that the lariat ethers and antimicrobials were present in the membrane simultaneously was addressed by using neutron reflectometry (NR). This technique permits direct observation of components within an anchored bilayer membrane. The neutral charge and high penetrating property of neutrons coupled with their wavelength comparable to molecular sizes and intermolecular distances render them suitable for studying the structure and dynamics of complex materials such as bilayers and membranes.21 Moreover, the neutron's ability to detect biomolecular architectures of membrane-associated proteins and lipid membranes without destroying the samples make them uniquely suitable to study complex materials. In this case, the bilayer that was studied was formed from palmitoyloleoylphosphatidylcholine, POPC, shown in Fig. 5.An initial effort was made by using soybean asolectin membranes, in which lariat ether channels were known to form.20 This membrane proved to be unstable and therefore untenable for the study. Instead, POPC was connected to a silicon oxide matrix. The anchored POPC bilayer membrane consists of two distinct regimes that mirror each other within the structure. The arrangement shown graphically in Fig. 6 represents the anchored bilayer as substrate||head group||lipid chains:lipid chains||head group. In Fig. 6, the midplane of the bilayer, the region at which the lipid chains meet, is denoted as the zero point. In the graphs presented in the following sections, peaks correspond to each of the four regimes. The initial separation at the far left of the abscissa corresponds to the anchoring surface.
A broad range of antibacterial data were determined for C10LE and C12LE against TET with both Gram negative and Gram positive organisms.19 The data for membrane insertion and channel function of C8LE and C11LE20 made the choice of N,N′-di-n-decyl-4,13-diaza-18-crown-6 (C10LE) the most favorable candidate for study.
Neutron reflectometry
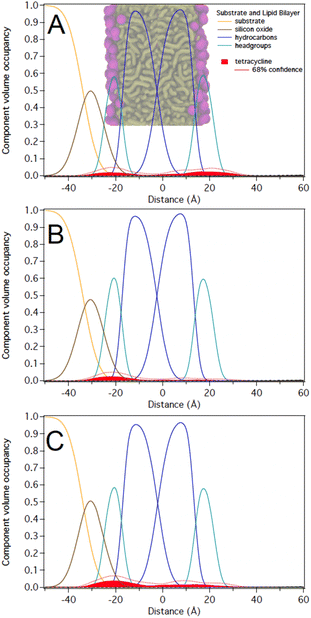
As noted above, C10LE complexed with TET in solution and the MIC of TET was reduced (potency increased) when it was present with low concentrations of C10LE.20,24 However, the interaction of C10LE, TET, or both with the membrane is unclear. To test for specific interactions with membranes, we subsequently added low and high concentrations of TET (5 μM and 36 μM), C10LE (12 μM and 58 μM), and pre-mixed C10LE and TET (12 μM C10LE/5 μM TET and 87 μM C10LE/36 μM TET) to POPC membranes. C10LE was combined with TET (C10LE:TET = 2.4) and the system/mixture was allowed to equilibrate overnight before NR experiments. As-prepared POPC membranes were structurally characterized before adding C10LE or TET, and final measurements after rinsing with pure water were performed after each concentration series. Tabulated fit results (Tables S2–S4†), NR curves (Fig. S9–S11, S15–S17, and S21–S23†), and best-fit nSLD profiles (Fig. S12–S14, S18–S20, and S24–S26†) can be found in the ESI.†Note that all reported volume occupancy profiles have some degree of asymmetry. For a symmetric membrane one can expect a symmetric distribution of C10LE/TET material. In our case, the presence of a solid substrate underneath the lipid bilayer breaks this symmetry. A lower or higher amount of C10LE and TET at this side of the lipid bilayer can either be attributed to an interaction of the small molecules with the interface or an effect related to the confined space underneath the bilayer. We are generally careful not to overinterpret such differences between the two bilayer leaflets and focus instead on the relative position of the material with respect to the hydrocarbon and headgroup regions in both leaflets. Any material found towards the substrate side of the bilayer indicates that it can easily penetrate the lipid bilayer. Studies with non-penetrating compounds have shown the inner leaflet to be void of additional material.23,25
At 12 μM, we observed that C10LE is predominantly located at the outer lipid headgroups of the bilayer with little penetration into the hydrocarbon region and the inner headgroup region adjacent to the solid substrate (Fig. 7A). At 58 μM, C10LE penetrates the hydrocarbon region and most of the membrane-associated LE headgroups associate with the inner lipid headgroups. At 12 μM and 58 μM the amount of associated C10LE is 3.0 ± 0.4 Å3 Å−2 and 7.4 ± 0.4 Å3 Å−2, and it coincides with a significant decrease in hydrocarbon thickness by 0.7 ± 0.2 Å and 1.7 ± 0.2 Å per leaflet, respectively (Fig. 7A and B). The completeness of the lipid bilayer increases slightly from on average 0.97 for the as-prepared h31-POPC and d31-POPC membranes to on average 0.99 due to observed bilayer thinning and the related increase in area per lipid (Table S3†). Upon rinsing with 10 cell volumes of water, C10LE remains associated with the membrane and within uncertainties, the CVO profiles of C10LE and the bilayer remain unchanged (Fig. 7C) The median fit parameters of fitting are reported in the ESI.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in solution at two different concentrations of 12 μM C10LE/5 μM tetracycline and 87 μM C10LE/36 μM tetracycline. The solution was allowed to equilibrate overnight to ensure complexation. We investigated the interaction of the complex with a POPC bilayer at both concentrations and after rinsing with pure water.
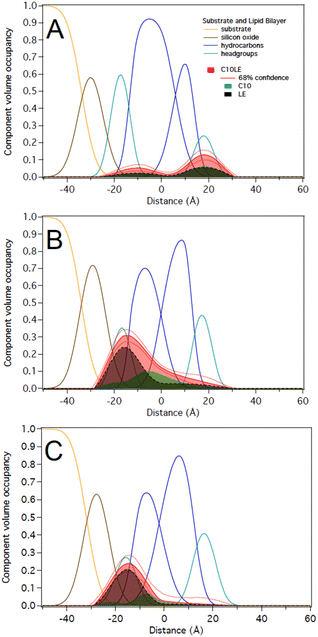
1 in solution at two different concentrations of 12 μM C10LE/5 μM tetracycline and 87 μM C10LE/36 μM tetracycline. The solution was allowed to equilibrate overnight to ensure complexation. We investigated the interaction of the complex with a POPC bilayer at both concentrations and after rinsing with pure water.We performed experiments on both protiated and deuterated POPC membranes and combined both data sets for simultaneous analysis. The mixture of C10LE and TET cannot be unambiguously resolved individually. We therefore dissected C10LE into components. The sidearms, C10, were modeled as decane and the macrocycle alone was modeled as 4,13-diaza-18-crown-6. This separated contributions to the scattering from decyl/C10 (−0.4 × 10−6 Å−2) on one side, and a mixture of LE and TET (1.32 × 10−6 and 3.0 × 10−6 Å−2, respectively) on the other side, thereby, grouping hydrophilic and hydrophobic components.
At 12 μM C10LE/5 μM TET, an insignificant amount of material was found associated with the lipid bilayer (Fig. 8A). The data fitting indicated that at low concentration (12 μM C10LE/5 μM TET), no significant association of C10LE or TET was observed with the bilayer. At 87 μM C10LE/36 μM tetracycline, C10LE is located primarily within the hydrocarbon region of the POPC membrane and C10LE/tetracycline associates with the lipid headgroups (Fig. 8B). The amount of C10LE/TET complex associated with bilayer increases from 0.1 ± 0.1 Å3 Å−2 at 12 μM C10LE/5 μM TET concentration to 5.6 ± 0.6 Å3 Å−2 at 87 μM C10LE/36 μM TET concentration, and the lipid bilayer thins by 0.1 ± 0.1 Å and 0.5 ± 0.3 Å, respectively. The bilayer completeness is not affected by incubation with the complex (Table S2†). The complex remains irreversibly bound after rinsing with pure water (Fig. 8C). As with C10LE by itself, there is a preference of the complex to associate with the inner lipid leaflet of the bilayer. The CVO profile of C10LE/tetracycline is not significantly different from that obtained from C10LE by itself. The complexation with tetracycline does not prevent C10LE from deeply penetrating the lipid membrane. The membrane thinning by the C10LE complex is less pronounced with −0.5 ± 0.3 Å per leaflet on average. (The thinning of individual leaflets cannot be resolved by the NR experiment so an average thinning is modeled)
Molecular dynamics results
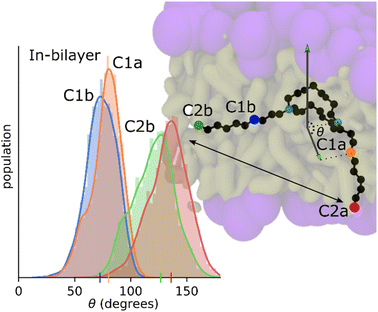
MD simulation results indicate that the tetracycline is typically parallel to the bilayer surface, but this preference is spread out due to the time it spends in the bulk solvent area where it is free to reorient. Fig. 9 shows the orientation of the C10LE molecule when embedded in the POPC bilayer. The distributions shown are the distributions of the four angles defined in the system. These four angles are each composed of two vectors, the first of these vectors is always the normal of the bilayer, and the second is drawn from a nitrogen on the crown ring to one of the carbons on the same side of the molecule. For instance, the drawn angle in Fig. 9 is showing the vector from one of the ring nitrogens to the carbon labeled C1a, this distribution is shown in the color that corresponds to the color used to highlight the carbon, in this case green. These distributions shown together find that the C10LE molecule when at equilibrium in the bilayer seems to prefer pushing one ‘arm’ further into the bilayer (C1a and C2a carbons), and one ‘arm’ prefers to lay parallel to the surface of the bilayer (C1b, C2b).Umbrella or non-equilibrium simulations
The PMFs (Fig. 10) follow the profiles of each species of interest's partitioning into the POPC bilayer. The C10LE macrocycle shows the largest favorability for the center of the POPC lipid bilayer (r = 0 nm), displaying an energy well larger than 2 kcal mol−1. Herein, the energy well is defined as the difference between the lowest points on the potential mean force inside the bilayer and maximum point at the surface of the bilayer. This energy well is larger than both the complex (1 kcal mol−1), and lone TET molecule (∼0 kcal mol−1). The other free energy value of note is the penalty for passing the hydrophilic heads (r = ∼1.8–2 nm). This is similar for each of the species (4–5 kcal mol−1) and would lead to slow penetration of molecules, even those that sit favorably inside the lipid bilayer itself. The complex being a larger structure than the other two lone species has a slightly larger barrier for entry followed by TET, and then C10LE.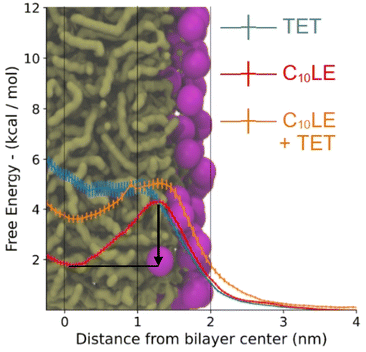 | ||
| Fig. 10 Free energy of tetracycline, C10LE, and TET·C10LE complex as a function of bilayer depth profile. The black arrow shows the energy well for C10LE molecule. | ||
These results correlate in several ways to the NR results. First, the C10LE sitting comfortably near the center of the membrane agrees with the density profiles, and secondly the profile for the TET shows little penetration of the TET molecule into the bilayer. The complex shows intermediate positioning compared to lone TET and lone C10LE molecules.
The neutron reflectance data show improved membrane penetration for TET in the presence of C10LE compared to its absence. It seems likely that some supramolecular interactions contribute to this enhancement. To the extent that the amine groups are protonated on either the lariat ether or the antibiotic, complementary interactions are possible there as well. The CPK molecular models studied showed many contacts that seemed productive, but there was no single orientation of the two that suggested a particularly favorable complex.
Experimental details
Preparation of bilayer lipid membranes for neutron reflectometry
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) dissolved in chloroform was purchased from Avanti Polar Lipids, Inc (Birmingham, AL, United States). Chloroform was removed from the lipid solution via evaporation under vacuum for 12 h. The dry lipid films were hydrated in a high-salt aqueous solution (2 M NaCl, pH 7) to a lipid concentration of 5 mg mL−1 and then sonicated until clear. Wafers (3′′ diameter, 5 mm thick n-type Si:P[100] wafers, El-Cat Inc., Ridgefield Park, NJ, United States) were cleaned with 5% Hellmanex solution (Hellma Analytics, Müllheim, Germany), sulfuric acid plus Nochromix (Godax Laboratories, St. Louis, MO, United States) followed by rinsing with ultrapure water (MilliporeSigma, Burlington, MA, United States) and pure ethanol (EtOH, Sigma-Aldrich, Shelbyville, KY) and dried in a N2 gas stream. A silicon wafer was assembled in a NCNR fluids cell and incubated with vesicle solution for ∼1 h. Thereafter, the cell was rinsed with pure water, which completes the vesicle fusion and creates a single solid-supported bilayer lipid membrane (BLM).22For the measurements of C10LE and C10LE/TET, two sets of NR experiments were conducted, one with a hydrogenated h31-POPC bilayer and another otherwise identical set of experiments with a deuterated d31-POPC bilayer. The measurements probing the membrane-interaction of TET on its own used only a h31-POPC bilayer. After measurement of the as-prepared bilayers in H2O and D2O, C10LE, TET, or C10LE/TET were added to the sample cell dissolved in H2O and D2O. NR measurements were conducted while the bilayer was in contact with either solution. Each compound was subsequently measured at two concentrations. The samples were measured again after rinsing with H2O and D2O, respectively. If measured, data for d31-POPC and h31-POPC bilayers were co-refined sharing conserved model parameters across data sets, in particular those associated with the volume profile of C10LE and TET. The co-refinement of data from two lipid bilayers with differently labelled hydrocarbon chains significantly boosts the effective resolution of the measurement.
1D-Component volume occupancy (CVO) profiles along the lipid bilayer normal were obtained as previously described.27 Bilayer fit parameters were the hydrocarbon thickness for each bilayer subsection, the bilayer completeness, and the thickness of the sub-membrane space. A single roughness parameter was applied to all distributions. The thin layer of native silicon oxide was modelled by a single distribution with individual roughness and thickness parameters. Hermite splines defined by control points that were on average 15 Å apart were used to model the CVO profiles of C10LE and TET. The number of control points was iteratively refined during model optimization for each CVO profile.
Fit parameters associated with each control point were a volume occupancy, a deviation from equidistant separation, and (for the CVO profile of the complex) nSLD (neutron scattering length density) value between those of C10LE and TET. Such a variable nSLD per control point allowed us to separate individual CVO profiles and amounts of associated material for C10 and LE/TET. The exchange of labile protons in isotopically different buffers affects the nSLD of molecular components and was taken into account during data analysis. Molecular volumes and scattering lengths are listed in Table S1.† Optimization of model parameters was performed using the ga_refl and Refl1D software packages developed at the NCNR. A Monte Carlo Markov Chain-based global optimizer was used to determine fit parameter confidence limits.
The constructed systems were equilibrated beginning with a steep energy minimization, then coupled to NVT (Nose–Hoover Thermostat: 300 K) and NPT (Parrinello–Rahman barostat: 1 atm) ensembles successively.31–33 Production simulations were collected under the NVT ensemble with a time step of 2 femtoseconds and a t-coupling value of 0.1 picoseconds. The Verlet cut-off scheme with a 0.9 nm short range cut-off was used in conjunction with the particle mesh ewald method to model the system's short and long range interactions. The LINCS constraint algorithm was also used to constrain all hydrogen bonds in the system.34 The production simulations were run for a total of 1 ms for each system, taking the final 200 ns for calculations of density and molecular orientation. All molecular dynamics and analyses were done using the GROMACS-2019.2 suite of programs.35–37
Umbrella biased simulations were run to probe the energy barrier of membrane penetration for the TET, C10LE, and TET-C10LE systems. Initial positions were chosen to be 5 nm from the bilayer center solvated in the bulk water phase, from there pulling simulations moved the species of interest at a rate of 0.0025 nm ps−1 in the negative z direction (towards the bilayer center). Three hundred symmetrically spaced windows were chosen from the pulling trajectories. These windows were then equilibrated for 1 ns, before production simulations were run for 10 ns collecting the positions and forces of the system. The GROMACS WHAM module was used to partition the data into the resulting potentials of mean force (PMF).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 solution of TET and C10LE (5 mM in each) was prepared in CDCl3. The solutions were capped and covered with parafilm to prevent evaporation and allowed to stand for 1 h at ambient temperature with occasional swirling. After 1 h, each solution was filtered through glass wool directly into an NMR tube. The NMR spectrum of each solution was obtained at 300 MHz (data shown in Fig. 2).
1 solution of TET and C10LE (5 mM in each) was prepared in CDCl3. The solutions were capped and covered with parafilm to prevent evaporation and allowed to stand for 1 h at ambient temperature with occasional swirling. After 1 h, each solution was filtered through glass wool directly into an NMR tube. The NMR spectrum of each solution was obtained at 300 MHz (data shown in Fig. 2).Conclusions and key observations
The NR studies indicate that TET (control) associates at a low level with the lipid headgroups of POPC bilayer. C10LE (control), on the other hand, associates with the lipid bilayer at the measured solution concentration of 58 μM, occupying the entire hydrocarbon region; however, the binding of C10LE with membrane is partially reversible.Interestingly, the lack of any significant C10LE/TET complex at low concentrations suggests that a host–guest complex is formed which doesn't interact with membrane at low concentrations. This means that dissolved C10LE successfully competes for binding to tetracycline and wins over the membrane. Tetracycline is known to act within the cell (at the ribosome) absent any adjuvant. C10LE acts at the membrane, potentially forming pores or short-lived defects, but it can also function as a cation carrier, at least in bulk phase.
Notably, the same complex (combining ratio of 2.4) at high concentration of 87 μM C10LE/36 μM TET binds with the POPC membrane. A plausible explanation is as follows. At higher concentrations (same ratio), and beyond a certain threshold concentration there is significant interaction with the membrane. This is detectable, but at lower concentrations the complex is more polar than C10LE alone, which leads to insufficient penetration within the membrane. The profiles of the complex binding the membrane are different from the individual components in that the molecules are mostly located in the inner lipid leaflet and no longer span the entire hydrocarbon region. Furthermore, the binding is irreversible which shows a synergistically38 improved antibacterial activity of the complex suggesting that the C10LE holds TET longer (irreversible) and in deeper layers of membrane that leads to improved efficacy.
Abbreviations
| C8LE | N,N′-Di-n-octyl-4,13-diaza-18-crown-6 |
| C10 | Decane |
| C10LE | N,N′-Di-n-decyl-4,13-diaza-18-crown-6 lariat ether |
| C11LE | N,N′-Di-n-undecyl-4,13-diaza-18-crown-6 |
| CVO | Component volume occupancy |
| HAP | Hospital acquired pneumonia |
| LE | 4,13-Diaza-18-crown-6 |
| M | Moles per liter |
| MD | Molecular dynamics |
| MIC | Minimal inhibitory concentration |
| NCNR | NIST Center for Neutron Research |
| NR | Neutron reflectance |
| nSLD | Neutron scattering length density |
| PMF | Potential mean force |
| POPC | Palmitoyloleoylphosphatidylcholine |
| TET | Tetracycline |
| VAP | Ventilator associated pneumonia |
Author contributions
The manuscript was written or edited by all authors./All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
GWG thanks the NSF for a grant (NSF: CHE 1710549) and TLB acknowledges financial support from NSF (NSF: CHE 1955161) and from the NIH MBArC program that supported portions of this work. HK thanks UC (start-up funds). FH acknowledges support from the U.S. Department of Commerce (Award 70NANB17H299). We thank Prof. Jerry L. Atwood for his initial support of this work. Research was performed in part at the National Institute of Standards and Technology (NIST) Center for Nanoscale Science and Technology. Certain commercial materials, equipment and instruments are identified in this work to describe the experimental procedure as completely as possible. In no case does such an identification imply a recommendation or endorsement by NIST, nor does it imply that the materials, equipment, or instrument identified are necessarily the best available for the purpose.References
- (a) C. J. Pedersen, Cyclic polyethers and their complexes with metal salts, J. Am. Chem. Soc., 1967, 89, 7017–7036 CrossRef CAS; (b) C. J. Pedersen and H. K. Frensdorff, Macrocyclic polyethers and their complexes, Angew. Chem., Int. Ed. Engl., 1972, 11, 16–25 CrossRef CAS PubMed.
- (a) J. M. Lehn, Supramolecular Chemistry, VCH, Weinheim, 1995, p. 271 Search PubMed; (b) J. W. Steed and J. L. Atwood, Supramolecular Chemistry, Wiley, New York, 3rd edn, 2022, p. 1216 Search PubMed.
- (a) J. D. Lear, Z. R. Wasserman and W. F. DeGrado, Science, 1988, 240, 1177 CrossRef CAS PubMed; (b) G. W. Gokel, R. Ferdani, J. Liu, R. Pajewski, H. Shabany and P. Uetrecht, Chemistry, 2001, 7, 33 CrossRef CAS PubMed; (c) S. Matile, Chem. Rec., 2001, 1, 162 CrossRef CAS PubMed; (d) P. J. Cragg, Sci Prog., 2002, 85, 219 CrossRef CAS PubMed; (e) G. W. Gokel and I. A. Carasel, Chem. Soc. Rev., 2007, 36, 378 RSC; (f) R. Wesolowski, A. Sommer, H. D. Arndt, U. Koert, P. Reiss, S. Wimmers and O. Strauss, ChemBioChem, 2007, 8, 513 CrossRef CAS PubMed; (g) F. Otis, C. Racine-Berthiaume and N. Voyer, J. Am. Chem. Soc., 2011, 133, 6481 CrossRef CAS PubMed; (h) J. K. Chui and T. M. Fyles, Chem. Soc. Rev., 2012, 41, 148 RSC; (i) F. De Riccardis, I. Izzo, D. Montesarchio and P. Tecilla, Acc. Chem. Res., 2013, 46, 2781 CrossRef CAS PubMed; (j) T. M. Fyles, Acc. Chem. Res., 2013, 46, 2847 CrossRef CAS PubMed; (k) G. W. Gokel and S. Negin, Acc. Chem. Res., 2013, 46, 2824 CrossRef CAS PubMed; (l) B. Gong and Z. Shao, Acc. Chem. Res., 2013, 46, 2856 CrossRef CAS PubMed; (m) J. Montenegro, M. R. Ghadiri and J. R. Granja, Acc. Chem. Res., 2013, 46, 2955 CrossRef CAS PubMed; (n) P. Reiss and U. Koert, Acc. Chem. Res., 2013, 46, 2773 CrossRef CAS PubMed; (o) M. Barboiu, Y. Le Duc, A. Gilles, P. A. M. CazadeMichau, Y. Marie Legrand, A. van der Lee, B. Coasne, P. Parvizi, J. Post and T. Fyles, Nat. Commun., 2014, 5, 4142 CrossRef CAS PubMed; (p) W. Si, P. Xin, Z. T. Li and J. L. Hou, Acc. Chem. Res., 2015, 48, 1612 CrossRef CAS PubMed; (q) A. V. Jentzsch and S. Matile, Top. Curr. Chem., 2015, 358, 205 CrossRef CAS PubMed; (r) C. Lang, W. Li, Z. Dong, X. Zhang, F. Yang, B. Yang, X. Deng, C. Zhang, J. Xu and J. Liu, Angew. Chem., Int. Ed., 2016, 55, 9723 CrossRef CAS PubMed; (s) C. Ren, J. Shen and H. Zeng, J. Am. Chem. Soc., 2017, 139, 12338 CrossRef CAS PubMed; (t) S. Chen, Y. Wang, T. Nie, C. Bao, C. Wang, T. Xu, Q. Lin, D. H. Qu, X. Gong, Y. Yang, L. Zhu and H. Tian, J. Am. Chem. Soc., 2018, 140, 17992 CrossRef CAS PubMed; (u) R. S. Krishnan, R. Satheesan, N. Puthumadathil, K. S. Kumar, P. Jayasree and K. R. Mahendran, J. Am. Chem. Soc., 2019, 141, 2949 CrossRef PubMed; (v) S. P. Zheng, J. J. Jiang, A. van der Lee and M. Barboiu, Angew. Chem., Int. Ed., 2020, 59, 18920 CrossRef CAS PubMed; (w) K. Sato, T. Muraoka and K. Kinbara, Acc. Chem. Res., 2021, 54, 3700 CrossRef CAS PubMed; (x) H. Yang, J. Yi, S. Pang, K. Ye, Z. Ye, Q. Duan, Z. Yan, C. Lian, Y. Yang, L. Zhu, D. H. Qu and C. A. Bao, Angew. Chem., Int. Ed., 2022, e202204605 CAS.
- G. W. Gokel, Chem. Commun., 2000, 1–9 RSC.
- S. Negin, M. B. Patel, M. R. Gokel, J. W. Meisel and G. W. Gokel, ChemBioChem, 2016, 17(22), 2153–2161 CrossRef CAS PubMed.
- J. L. Atkins, M. B. Patel, Z. Cusumano and G. W. Gokel, Chem. Commun., 2010, 46(43), 8166–8167 RSC.
- M. B. Patel, E. G. Garrad, J. W. Meisel, S. Negin, M. R. Gokel and G. W. Gokel, RSC Adv., 2018, 9, 2217–2230 RSC.
- Centers for Disease Control and Prevention, Antibiotic Resistance Threats In The United States, 2019, https://www.cdc.gov/drugresistance/biggest-threats.html Search PubMed.
- World Health Organization, Antibiotic Resistance, https://www.who.int/health-topics/antimicrobial-resistance Search PubMed.
- R. R. Hendrixson, M. P. Mack, R. A. Palmer, A. Ottolenghi and R. G. Ghirardelli, Toxicol. Appl. Pharmacol., 1978, 44(2), 263–268 CrossRef CAS PubMed.
- K. Leong, T. O. Ts'o and M. B. Chenoweth, Toxicol. Appl. Pharmacol., 1974, 27(2), 342–354 CrossRef PubMed.
- K. Takayama, S. Hasegawa, S. Sasagawa, N. Nambu and T. Nagai, Chem. Pharm. Bull., 1977, 25(11), 3125–3130 CrossRef CAS PubMed.
- (a) C. J. Pedersen, Org. Synth., 1972, 52, 66–74 CrossRef CAS; (b) C. J. Pedersen, Science, 1988, 241, 536–540 CrossRef CAS PubMed.
- (a) R. C. Kolbeck, L. B. Hendry, E. D. Bransome and W. A. Speir, Experientia, 1984, 40(7), 727–731 CrossRef CAS PubMed; (b) H. Kristbjarnarson and P. Aarhem, Acta Physiol. Scand., 1985, 123, 261–268 CrossRef CAS PubMed; (c) E. W. Bethge, K. H. Bohuslavizki, W. Hänsel, A. Kneip and E. Koppenhôfer, Gen. Physiol. Biophys., 1991, 10, 225–244 CAS.
- (a) E. K. Plotnikova, N. Y. Golovenko, V. G. Zin'kovskii, N. G. Luk'yanenko, O. V. Zhuk and S. S. Basok, Vopr. Med. Khim., 1987, 33, 62–66 CAS; (b) G. I. Van’kin, N. V. Lukoyanov, T. G. Galenko and O. A. Raevskii, Khim.-Farm. Zh., 1988, 22, 962–965 Search PubMed; (c) N. V. Lukoyanov, G. I. Van'kin, A. M. Sapegin and O. A. Raevskii, Khim.-Farm. Zh., 1990, 24, 48–51 CAS.
- (a) D. Huang, D. Wang, J. Zhang, L. Huang, T. Fu, H. Ou, R. Que and Z. Zhang, J. Nanjing Univ., Nat. Sci., 1980, 2, 33–44 Search PubMed; (b) A. E. S. Macklon, A. Sim, D. G. Parsons, M. R. Truter and J. N. Wingfield, Ann. Bot., 1983, 52, 345–356 CrossRef CAS; (c) M. A. Pemadasa, New Phytol., 1983, 93, 13–24 CrossRef; (d) D. Huang, D. Wang, T. Fu, R. Que, J. Zhang, L. Huang, H. Ou and Z. Zhang, J. Nanjing Univ., Nat. Sci., 1988, 2, 33–44 Search PubMed; (e) M. B. Patel, E. G. Garrad, S. Korb, S. Negin, M. R. Gokel, S. Sedinkin and G. W. Gokel, Chem. Sci. Int. J., 2019, 27(4), CSIJ.50645 Search PubMed; (f) G. W. Goke and M. B. Patel, US Pat. no. 10548319, issued, February 4, 2020.
- (a) N. Kato, I. Ikeda, M. Okahara and I. Shibasaki, Res. Soc. Antibac. Antifung. Agents Jpn, 1980, 8, 532–533 CAS; (b) N. Kato, I. Ikeda, M. Okahara and I. Shibasaki, Bokin Bobai, 1980, 8, 415–420 CAS; (c) N. Kato, Kenkyu Kiyo - Konan Joshi Daigaku, 1985, 585–596 Search PubMed; (d) L. A. Konup, I. P. Konup, V. E. Sklyar, K. N. Kosenko, V. P. Gorodnyuk, G. V. Fedorova, E. I. Nazarov and S. A. Kotlyar, Khim.-Farm. Zh., 1989, 23, 578–583 CAS; (e) A. Gumus, S. Karadeniz, H. I. Ugras, M. Bulut, U. Cakir and A. C. Gorend, J. Heterocycl. Chem., 2010, 47, 1127–1133 CrossRef CAS; (f) Z. Hayvali, H. Guler, H. Ogutcu and N. Sari, Med. Chem. Res., 2014, 23, 3652–3661 CrossRef CAS.
- B. Bechinger, J. Membrane Biol., 1997, 156, 197 CrossRef CAS PubMed.
- W. Carrasquel-Ursulaez, R. D. Reeves, M. Dehghany, C. Jones, J. M. Schomaker and B. Chanda, RSC Adv., 2020, 10, 40391 RSC.
- L. Shangguan, Q. Chen, B. Shi and F. Huang, Chem. Commun., 2017, 53, 9749 RSC.
- O. Murillo, S. Watanabe, A. Nakano and G. W. Gokel, J. Am. Chem. Soc., 1995, 117, 7665–7679 CrossRef CAS.
- M. R. Caira, L. R. Nassimbeni and J. C. Russell, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 2007, 33, 1171–1176, DOI:10.1107/S0567740877005603.
- A. Kuhn, Bacterial Cell Walls and Membranes, Springer, Berlin, 2019, 501 Search PubMed.
- (a) A. Dura, Rev. Sci. Instrum., 2006, 77, 74301–743011 CrossRef PubMed; (b) S. Qian, V. K. Sharma and L. A. Clifton, Langmuir, 2020, 36, 15189–15211 CrossRef CAS PubMed; (c) R. Eells, D. P. Hoogerheide, P. A. Kienzle, M. Lösche, C. F. Majkrzak and F. Heinrich, Structural Investigations of Membrane-Associated Proteins by Neutron Reflectometry, in Characterization of Biological Membranes, ed. Nieh M.-P., Heberle F. A. and Katsaras J., De Gruyter STEM, De Gruyter, Berlin, Boston, 2019, pp. 87–130, DOI:10.1515/9783110544657-003.
- A. Benedetto, F. Heinrich, M. A. Gonzalez, G. Fragneto, E. Watkins and P. Ballone, J. Phys. Chem. B118, 2014, 42, 12192–12206 CrossRef PubMed.
- O. Soubias, S. Pant, F. Heinrich, Y. Zhang, N. S. Roy, J. Li, X. Jian, M. E. Yohe, P. A. Randazzo, M. Lösche, E. Tajkhorshid and R. A. Byrd, Arf1, Sci. Adv., 2020, 6(40), eabd1882, DOI:10.1126/sciadv.abd1882.
- F. Heinrich and M. Lösche, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 2341–2349 CrossRef CAS PubMed.
- M. J. Robertson, J. Tirado-Rives, L. William and J. L. Jorgensen, J. Chem. Theory Comput., 2015, 11(7), 3499–3509 CrossRef CAS PubMed.
- L. S. Dodda, I. Cabeza de Vaca, J. Tirado-Rives and W. L. Jorgensen, Nucleic Acids Res.45, 2017, 1, W331–W336 CrossRef PubMed.
- D. J. Price and C. L. Brooks III, J. Chem. Phys., 2004, 121(20), 10096–10103 CrossRef CAS PubMed.
- S. Nosé, J. Chem. Phys., 1984, 81(1), 511–519 CrossRef.
- W. G. Hoover, Phys. Rev., A, 1985, 31(3), 1695 Search PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1982, 52(12), 7182–7190 CrossRef.
- (a) T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98(12), 10089–10092 CrossRef CAS; (b) B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, J. Comp. Chem., 1997, 18(12), 1463–1472 CrossRef CAS.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. Van Der Spoel and B. Hess, Bioinform., 2013, 29(7), 845–854 CrossRef CAS PubMed.
- S. Páll, M. J. Abraham, C. Kutzner, B. Hess and E. Lindahl, Tackling exascale software challenges in molecular dynamics simulations with GROMACS, in Solving software challenges for exascale, ed. S. Markidis and E. Laure, Springer International Publishing Switzerland, London, 2015, pp. 3–27 Search PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- (a) M. C. Berenbaum, Pharmacol. Rev., 1989, 41(2), 93–141 CAS; (b) R. J. Tallarida, J Pharmacol. Exp. Ther., 2001, 298(3), 865–872 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra03851j |
| This journal is © The Royal Society of Chemistry 2022 |