Open Access Article
Open Access ArticleIn vitro and computational investigations of novel synthetic carboxamide-linked pyridopyrrolopyrimidines with potent activity as SARS-CoV-2-MPro inhibitors†
Ateyatallah Aljuhani‡
a,
Hany E. A. Ahmed‡*bc,
Saleh K. Ihmaidbd,
Abdelsattar M. Omarefg,
Sultan S. Althagfanh,
Yaser M. Alahmadi h,
Iqrar Ahmadi,
Harun Pateli,
Sahar Ahmedbj,
Mohannad A. Almikhlafik,
Ahmed M. El-Agrodyl,
Mohamed F. Zayedgm,
Safaa Abdulrahman Turkistanim,
Shorouk H. Abulkhair
h,
Iqrar Ahmadi,
Harun Pateli,
Sahar Ahmedbj,
Mohannad A. Almikhlafik,
Ahmed M. El-Agrodyl,
Mohamed F. Zayedgm,
Safaa Abdulrahman Turkistanim,
Shorouk H. Abulkhair p,
Mohammed Almaghrabib,
Samir A. Salaman,
Ahmed A. Al-Karmalawy
p,
Mohammed Almaghrabib,
Samir A. Salaman,
Ahmed A. Al-Karmalawy o and
Hamada S. Abulkhair
o and
Hamada S. Abulkhair *co
*co
aChemistry Department, College of Sciences, Taibah University, Al-Madinah Al-Munawarah 41477, Saudi Arabia
bPharmacognosy and Pharmaceutical Chemistry Department, College of Pharmacy, Taibah University, Al-Madinah Al-Munawarah, Saudi Arabia
cPharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Al-Azhar University, Nasr City 11884, Cairo, Egypt. E-mail: hamadaorganic@azhar.edu.eg
dPharmaceutical Chemistry Department, Faculty of Pharmacy, Jadara University, Irbid, Jordan
eDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, King Abdulaziz University, Alsulaymanyah, Jeddah 21589, Saudi Arabia
fCenter for Artificial Intelligence in Precision Medicines, King Abdulaziz University, Jeddah 21589, Saudi Arabia
gDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Al-Azhar University, Nasr City 11884, Cairo, Egypt
hClinical and Hospital Pharmacy Department, College of Pharmacy, Taibah University, Al-Madinah Al-Munawarah, Saudi Arabia
iDivision of Computer Aided Drug Design, Department of Pharmaceutical Chemistry, R. C. Patel Institute of Pharmaceutical Education and Research, Shirpur, 425405, Maharashtra, India
jDepartment of Medicinal Chemistry, Faculty of Pharmacy, Assiut University, Assuit, Egypt
kPharmacology and Toxicology Department, College of Pharmacy, Taibah University, Al-Madinah Al-Munawarah, Saudi Arabia
lChemistry Department, Faculty of Science, Al-Azhar University, Nasr City, Cairo, Egypt
mPharmaceutical Sciences Department, Fakeeh College for Medical Sciences, Jeddah 21461, Saudi Arabia
nDivision of Biochemistry, Department of Pharmacology, College of Pharmacy, Taif University, P.O. Box 11099, Taif 21944, Saudi Arabia
oPharmaceutical Chemistry Department, Faculty of Pharmacy, Horus University – Egypt, International Coastal Road, New Damietta 34518, Egypt
pDepartment of Biochemistry, Faculty of Medicine, Al-Azhar University (Girls), Nasr City 11754, Cairo, Egypt
First published on 22nd September 2022
Abstract
An essential target for COVID-19 is the main protease of SARS-CoV-2 (Mpro). With the objective of targeting this receptor, a novel set of pyrido[1,2-a]pyrrolo[2,3-d]pyrimidines with terminal carboxamide fragments was designed, synthesized, and considered as an initial motif for the creation of effective pan-coronavirus inhibitors. Accordingly, nine derivatives (21–29) have been introduced for in vitro assay to evaluate their antiviral activity and cytotoxicity effect against COVID-19 virus using Vero cells. The obtained data revealed that the majority of these derivatives showed potent cellular anti-COVID-19 activity and prevent viral growth by more than 90% at two different concentrations with weak or even no detectable cytotoxic effect on Vero cells. Extensive molecular docking simulations highlighted proper non-covalent interaction of new compounds within the binding pocket of Mpro as a potential target for their antiviral activity. In vitro assay for all the synthesized derivatives against the viral Mpro target indicated that compounds 25 and 29 have promising inhibitory activity with IC50 values at low micromolar concentrations. The molecular dynamic simulation results predicted the stability of compound 29 in the binding cavity of SARS-CoV-2 Mpro and hence supported the high inhibitory activity shown by the In vitro assay. These results suggested that compounds 25 and 29 merit further investigations as promising drug candidates for the management of SARS-CoV-2.
Introduction
Coronavirus disease (COVID-19) is a life-threatening infectious disease caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which was discovered first in Wuhan city in China and subsequently spread worldwide.1 Based on the epidemiological report of the WHO on 21 June 2022 there have been more than 537![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 confirmed cases of COVID-19, including 6
000 confirmed cases of COVID-19, including 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 319
319![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 395 confirmed deaths globally.2 Until the end of 2021, there was no approved specific antiviral drug for treatment, and all options were based on symptomatic treatment and oxygen therapy to manage respiratory impairment. Recently, the Food and Drug Administration (FDA) has authorized the combination of nirmatrelvir and ritonavir for the emergency use only in mild-to-moderate cases of COVID-19. Consequently, there is still unmet needs for effective treatment options against the SARS‐CoV‐2 virus. One strategy that has been followed by scientists to combat the widespread of new viral strains is to use the already approved antiviral medications.3–5 Most of the currently used antiviral drugs are nucleoside analogs6 such as Remdesivir (1; Fig. 1), Ribavirin (2), Sofosbuvir (3), Galidesivir (4), and Tenofovir (5).
395 confirmed deaths globally.2 Until the end of 2021, there was no approved specific antiviral drug for treatment, and all options were based on symptomatic treatment and oxygen therapy to manage respiratory impairment. Recently, the Food and Drug Administration (FDA) has authorized the combination of nirmatrelvir and ritonavir for the emergency use only in mild-to-moderate cases of COVID-19. Consequently, there is still unmet needs for effective treatment options against the SARS‐CoV‐2 virus. One strategy that has been followed by scientists to combat the widespread of new viral strains is to use the already approved antiviral medications.3–5 Most of the currently used antiviral drugs are nucleoside analogs6 such as Remdesivir (1; Fig. 1), Ribavirin (2), Sofosbuvir (3), Galidesivir (4), and Tenofovir (5).
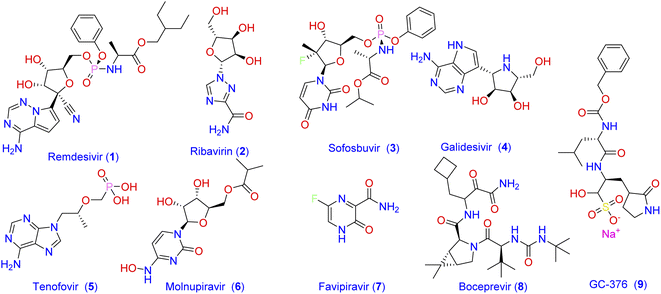 | ||
| Fig. 1 Frequently prescribed antiviral drugs and recently identified molecules with prominent activity against SARS-CoV-2. | ||
Despite the recommendation of such antiviral agents together with the antibacterial and other antiviral medications azithromycin, molnupiravir (6), and favipiravir (7) to manage the emergency cases, no effective drug has yet been introduced to treat specific structures of viral components of SARS-CoV-2.7 The main protease Mpro (also called 3CLpro) of coronavirus is a key mediator in viral replication and transcription. This functional importance, together with the absence of a closely related analogous target in humans, made the SARS-CoV-2 main protease Mpro an interesting target for the design of antiviral agents.5,8,9 Consequently, many attempts have been adopted to develop Mpro inhibitor-based antiviral medication. As a result, a number of antiviral agents with prominent activity against SARS-CoV-2 were discovered. These include boceprevir (8) and GC-376 (9) which were verified as inhibitors of Mpro through binding to its catalytic active site.10
Computational methods play an important role in the design of novel Mpro antagonistic drugs.4,11–14 In this context, a structure-based virtual screening was conducted by Jin and co-workers to evaluate a library of more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 well-known compounds as potential inhibitors of Mpro.15 Among compounds of the tested library, six inhibited Mpro with IC50 values range of 0.67–21.4 µM.16 Additionally, MUT056399 (10; Fig. 2) has been identified as a carboxamide-incorporating inhibitor of SARS-CoV-2-MPro with an EC50 value of 38.24 µM with the preferential quality of low cytotoxicity (CC50 > 100 µM). The carboxamide group of this latter forms two hydrogen bonds with the amino acid residues His163 and Phe140 in the S1 subsite of SARS-CoV-2-MPro.17,18 ZINC02123811 (11) is another carboxamide derivative derived from a natural source and has the affinity to bind with the active pocket of SARS-CoV-2 Mpro.19 The root-mean-square fluctuation (RMSF) analysis indicates that the complex of Mpro with this carboxamide derivative is pretty stable all over the simulation course.
000 well-known compounds as potential inhibitors of Mpro.15 Among compounds of the tested library, six inhibited Mpro with IC50 values range of 0.67–21.4 µM.16 Additionally, MUT056399 (10; Fig. 2) has been identified as a carboxamide-incorporating inhibitor of SARS-CoV-2-MPro with an EC50 value of 38.24 µM with the preferential quality of low cytotoxicity (CC50 > 100 µM). The carboxamide group of this latter forms two hydrogen bonds with the amino acid residues His163 and Phe140 in the S1 subsite of SARS-CoV-2-MPro.17,18 ZINC02123811 (11) is another carboxamide derivative derived from a natural source and has the affinity to bind with the active pocket of SARS-CoV-2 Mpro.19 The root-mean-square fluctuation (RMSF) analysis indicates that the complex of Mpro with this carboxamide derivative is pretty stable all over the simulation course.
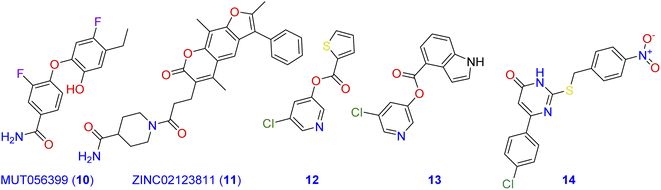 | ||
| Fig. 2 Carboxamide, pyridine, and pyrimidine incorporating molecules with potent SARS-CoV-2-MPro antagonistic activity. | ||
On the other hand, pyridine, pyrimidine, and pyrrole have long been considered privileged fragments in the construction of antiviral candidates and potential inhibitors of SARS-CoV-2-MPro in particular.20,21 Compound 12 is a pyridine derivative that has been evaluated for its activity against the Frankfurt-1 strain of SARS-CoV by MTT assay in Vero E6 cells. Results of the antiviral evaluation displayed a good potency and selectivity of 12 toward SARS-CoV-2-MPro with an IC50 value of 0.50 µM.22 In silico docking studies were also performed on compound 13 against the binding pocket of SARS-CoV-2-MPro. Results suggested that it is a promising anti-SARS-CoV agent (IC50 = 30 nM), and revealed a good fitting within the active site of the protein target through the formation of three hydrogen bonds with the amino acid residues Cys145, Ser144, and Gly143.23 Investigating the inhibitory potency of the pyrimidine-based molecule 14 against SARS-CoV-MPro demonstrated its good activity with an IC50 value of 6.10 µM.
Rationale and aim of the work
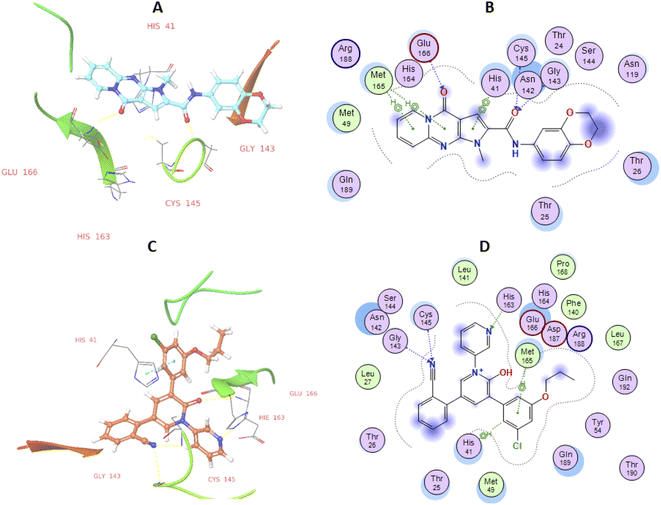
High-throughput screening of a library of small synthetic molecules against the infectious hepatitis C virus (HCV) identified the iminodipyridinopyrimidine (IDPP; Fig. 3) scaffold as a potent inhibitor in the early and the late steps of the HCV lifecycle.24,25 This scaffold showed also high levels of safety and metabolic stability. The 3D structure of HCV protease has a high level of similarity with the SARS-CoV-2-MPro (Z score = +8.40).26,27 Like Mpro, the HCV protease also has a double β-barrel fold, with relatively similar orientations to those of the SARS-CoV-2 Mpro, and a substrate-binding site located in a shallow cleft between its two six-to eight-stranded antiparallel β-barrels.28 The superimposition of these two proteases suggested the superimposition of their substrate binding pockets and their active-site catalytic residues.28,29 Accordingly, the IDPP scaffold is suggested to have a broad antiviral spectrum against both HCV and SARS-CoV-2-MPro. In continuation to our recent reports on the design of suggested bioactive molecules,30–33 the objective of the current research is to modify the structure of the reported HCV antiviral agents IDPP into pyridopyrrolopyrimidine new scaffolds and to assess the potential of new candidates to inhibit SARS-CoV-2-MPro. Hence, while keeping the carboxamide functionality linked to the new scaffold, two structural modifications of IDPP analog were conducted to enhance the ligand binding and consequently the potency: the six-membered pyridine-imine fragment was replaced with the pyrrole aromatic system; the benzyl substituent attached to the pyridine-nitrogen was replaced with a smaller hydrophobic moiety. Accordingly, the new scaffold possesses the advantages of incorporating the privileged fragments pyridine, pyrimidine, pyrrole, and carboxamide which have been reported as pharmacophoric groups in certain SARS-CoV-2-MPro inhibitors.20,21,34 A set of in vitro biological evaluation studies has been performed. The effect of new compounds to inhibit the viral replication of SARS-CoV-2 and antagonize its main protease was evaluated. As well, cell growth inhibition after treatment with new ligands has been assessed. A structure-based approach was utilized using molecular docking simulation of the reported active IDPP analog compared to the newly designed pyridopyrrolopyrimidines within the Mpro protease target of SARS-CoV; PDB 7L11. The pre-docking screening of designed ligands within the protease binding pocket (Fig. 3) showed three promising findings: (1) an edge-to-face aryl–aryl interaction with the catalytic residue His41; (2) terminal amide carbonyl is directed well at the NH of Cys145; (3) the carbonyl of the fused system appears with proper placement with the polar environment, including the side chains of Met165, His164, and Glu166. In contrast, the behavior of IDPP analog with pyridine-imine part fails to participate in the good orientation of fused system through these major and conserved interactions within the pocket due to loss of aromaticity and false ordinations of terminal amide and the carbonyl group of fused system. Moreover, there is an unfavorable benzyl substitution attached to the pyridine-N that is located at the polar binding pocket. Molecular dynamic simulations were used for confirming these findings and measuring the stability within the pocket.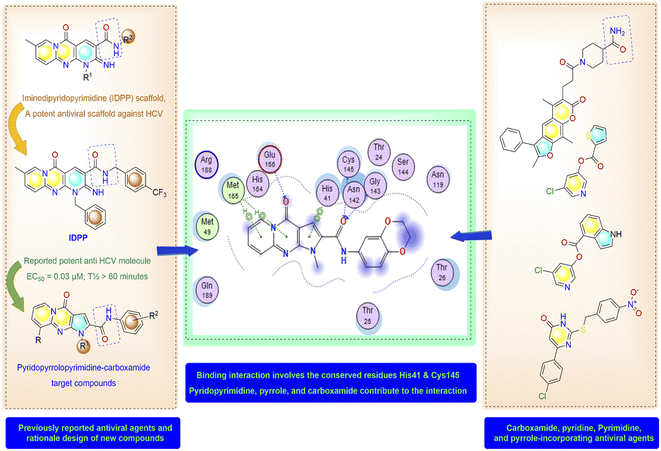 | ||
| Fig. 3 Molecular design and rationale analysis for the binding interactions of novel pyridopyrrolopyrimidines within active pocket of SARS-CoV-2-MPro. | ||
Results and discussion
Chemistry
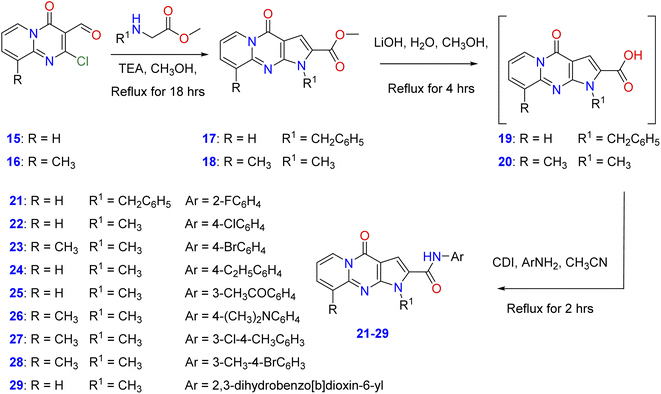
In order to elucidate the scope and features of the cyclization reaction, the starting methyl-1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-b]pyrrolo[2,3-d]pyrimidine-2-carboxylate and its 9-methyl derivatives (17, 18) were prepared in moderate to good yields by the reaction of 2-chloro-4-oxo-4H-pyrido[1,2-a]pyrimidine-3-carbaldehyde35 and its 9-methyl derivatives (15, 16) with amino acid methyl esters namely, methyl-N-methyl(N-benzyl)glycinate or methyl methyl-N-benzyl glycinate in a mixture of methanol and triethylamine under reflux as shown in Scheme 1. Hydrolysis of the ester derivatives 17, 18 by aqueous lithium hydroxide affords the corresponding intermediate, 1-benzyl/methyl-4-oxo-1,4-dihydro-pyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxylic acid and its 9-methyl derivatives (19, 20). Condensation of these carboxylic acid intermediates with appropriate aniline derivatives in the presence of 1,1′-carbonyldiimidazole (CDI) and acetonitrile gives the target N-aryl-1-benzyl/methyl-4-oxo-1,4- dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxamides (21–29) as shown in Scheme 1 with different substitutions on the original scaffold at N-1 and carboxamide functionality.Spectroscopic data
The 1H-NMR spectra of 21–29 showed singlet signals attributable to NH proton of the carboxamide functionality at the regions of 10.85–11.12 ppm. Moreover, compounds 21–29 exhibited the signals of the methylene and N-methyl group protons at δ 5.88 and δ 4.01–4.09, respectively. In addition, the elemental analysis and mass spectra of compounds 21–29 confirmed their proposed structures, see ESI†.Biological screening
| Comp. ID | Cytotoxicity IC50 (µM ± SEa) | Antiviral assay (qPCR) | SIb | |||
|---|---|---|---|---|---|---|
| Inhibition % | EC50 and EC90 (µM ± SEa) | |||||
| 1 µM | 10 µM | EC50 | EC90 | |||
| a SE represents standard error from triplicate readings.b Selectivity index (SI) was calculated by dividing of IC50 value for Vero cell cytotoxicity by the EC50 of the antiviral assay. | ||||||
| 17 | >200 | 22.25 | 42.51 | 22.1 ± 1.012 | 36.13 ± 2.31 | 9 |
| 21 | 48.8 ± 3.72 | 83.49 | 95.92 | 0.0138 ± 0.00198 | 3.60 ± 0.42 | 3544 |
| 22 | 193.0 ± 14.7 | 88.16 | 94.10 | 0.1093 ± 0.01105 | 1.738 ± 0.54 | 1766 |
| 23 | 47.6 ± 3.62 | 78.96 | 92.92 | 0.0216 ± 0.00656 | 3.11 ± 1.61 | 2201 |
| 24 | 21.6 ± 1.64 | 94.01 | 96.22 | 0.0762 ± 0.00973 | 3.437 ± 0.55 | 283.3 |
| 25 | 121.0 ± 9.21 | 92.48 | 97.08 | 0.1066 ± 0.00115 | 2.50 ± 0.841 | 1135 |
| 26 | 87.3 ± 6.65 | 87.61 | 94.05 | 0.0376 ± 0.00423 | 3.801 ± 0.041 | 2323 |
| 27 | 64.4 ± 4.91 | 94.36 | 96.52 | 0.1035 ± 0.00344 | 2.601 ± 1.02 | 622.2 |
| 28 | 139.0 ± 10.6 | 92.71 | 95.60 | 0.108 ± 0.00304 | 1.41 ± 0.91 | 1278 |
| 29 | 151.0 ± 11.5 | 94.10 | 96.39 | 0.0519 ± 0.00512 | 8.953 ± 2.01 | 2909 |
| Remdesivir | 43.1 ± 3.28 | 94.23 | 97.58 | 0.0099 ± 0.00175 | 0.0835 ± 1.88 | 7195 |
The selectivity analysis of these compounds compared to their cytotoxic effect has also been investigated. Good antiviral candidate should be highly selective with a minimum SI value of 5 or higher.36–39 Almost all the new pyridopyrrolopyrimidines showed good SI values and proved the prominent antiviral activity. Comparing the performance of novel compounds to the reference remdesivir, SI was calculated and showed 4000 to 283 ranges with promising antiviral selectivity. Some of the designed derivatives did not function positively in the antiviral activity in vitro, and their relative abilities to inhibit viral replication did not always correlate directly with in vitro inhibition parameters toward Mpro.40
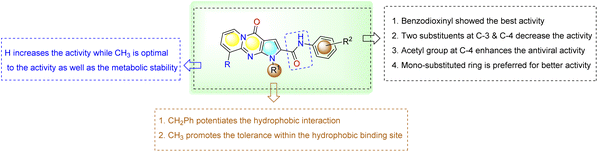
Herein, N-aryl-1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxamide derivatives were tested by in vitro assay for inhibition of SARS-CoV-2 Mpro activity and compared with the known references Lopinavir (non-specific) and GC376 (specific) Mpro inhibitors.48 As shown in Table 2, intermediate 17 seems to be functionally inactive against the Mpro enzyme as seen from its IC50 value. However, both 25 and 29exhibited high SARS-CoV-2 Mpro inhibition activity with IC50 values of 5.42 and 3.22 µM, respectively, which are more potent than the reference drugs Lopinavir and GC376 (82.17, 12.85 µM). On the other hand, introducing a halogen substituent to the terminal aryl ring, as in cases of 27, 28, 23 and 22, leads to a dramatical fall in the activity (IC50 values = 165.20 µM, 45.96 µM, 65.92 µM and 236.50 µM, respectively). As well, compound 26 with N,N-dimethylamino group at the para position of the terminal aryl showed a dramatic reduction in the inhibition activity with (IC50 values 407.50 µM). In combination with results presented in Table 1, this indicates that the 3,4-dihydrodioxane or 3-acetyl group attached to the phenyl ring is necessary for activity against Mpro of these compounds. The results all partially conveyed the proposed inhibitory effect of these derivatives on the Mpro, but more mechanistic investigations are still needed to specify the major mechanism of action whether it is competitive on non-competitive.
| Compound's ID | IC50 (µM ± SEa) | Compound's ID | IC50 (µM ± SE*) |
|---|---|---|---|
| a SE represents standard deviation from triplicate readings. | |||
| 17 | 329.5 ± 15.1 | 26 | 407.50 ± 3.44 |
| 21 | 25.36 ± 2.40 | 27 | 165.20 ± 16.70 |
| 22 | 236.50 ± 4.29 | 28 | 45.96 ± 8.62 |
| 23 | 65.92 ± 12.30 | 29 | 3.22 ± 2.83 |
| 24 | 82.17 ± 15.0 | Lopinavir | 82.17 ± 7.66 |
| 25 | 5.42 ± 21.30 | GC-376 | 12.85 ± 0.74 |
Computational investigations
| IDs | Score | Binding pocket residue interaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| His41 | Cys145 | Gly143 | His163 | Glu166 | |||||||
| VWDa | D | VWD | D | VWD | D | VWD | D | VWD | D | ||
| a VWD = van der Waals interactions (kcal mol−1) D = Distance (Å). | |||||||||||
| 29 | −7.45 | −3.86 | 2.23 | −2.81 | 3.44 | −2.48 | 2.11 | −0.44 | 4.88 | −2.57 | 2.16 |
| Ref. | −10.1 | −4.71 | 3.01 | −3.47 | 2.32 | −1.66 | 2.61 | −1.11 | 2.02 | −4.85 | 2.17 |
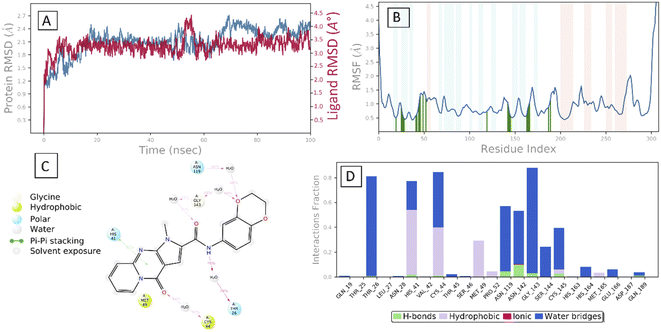
After the early fluctuation due to the equilibration, the RMSD of the compound 29 remained constant with 3.5 Å till 55 ns, after which minor RMSD fluctuation was noticed and remained stable for the rest of the simulation. The mobility of amino acid residues was assessed throughout time simulation using the protein RMSF in comparison to a reference structure. In the protein RMSF plot, α-helical and β-strand regions are represented in red and blue colors, respectively, while in the white color, the loop region is represented. Protein α-helical and β-strands regions are stiffer than the unstructured region of protein and as a result, fluctuate less than loop regions. If the active site and main chain atoms fluctuated insignificantly, it indicated that the conformational change was minimal, implying that the reported lead compound was tightly bound within the cavity of the SARS-CoV-2 Mpro protein binding pocket. The RMSF plot of 29–SARS-CoV-2 Mpro is displayed in Fig. 3B. According to the RMSF plot, it was observed that compound 29 contacted 23 amino acids of SARS-CoV-2 Mpro binding pocket, namely, Gln19, Thr25, Thr26, Leu27, Asn28, His41, Val42, Cys44, Thr45, Ser46, Met49, Pro52, Asn119, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Asp187, and Gln189. All this interacted residue is marked in green-colored vertical bars, having a RMSF value of less than 1.5 Å. Compound 29 was able to tightly bind to the active site through multiple interactions, including 42% simulation time of π–π stacking with catalytic dyed residue His 41 (Fig. 6C). Fig. 3D indicates that Compound 29 forms hydrophobic and water mediated hydrogen bonding with Tyr25, Asn28, Cys44, Met49, Asn119, Asn142, Gly143, Ser144 and Cys145. It visible that compound 29 made all three types (hydrogen, hydrophobic and water mediated hydrogen bonding) of interaction with crucial residues His 41 and Cys145. All the amino acid interactions identified during docking studies of the target molecule were also displayed during the dynamic study.
Conclusion
In conclusion, we have designed a new set of novel pyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxamides and evaluated their antiviral activity against COVID-19. Most of the designed compounds exhibited potent antiviral activity. SAR analysis of the novel scaffold was estimated based on biological and theoretical results obtained for these compounds. The benzodioxan derivative 29 showed very potent anti-SARS-CoV-2 activity at the concentrations of 1 and 10 µM by 94.10 and 96.39% inhibition of the viral growth with EC50 = 0.0519 µM. These novel derivatives were subjected to Mpro protease inhibitory assay and molecular docking study to better understand their underlying mechanism. Compound 29 strongly inhibited the activity of Mpro with an IC50 value of 3.22 µM compared to 12.85 µM for the reference control GC-376. Results of molecular docking study exhibited that compound 29 works through the non-covalent interaction with the main protease of SARS-CoV. Also, a molecular dynamic simulation study has been performed. Docking and dynamic simulations studies proved the prominent interaction behaviour and stability of 29 within the binding pocket through the conserved His41, Glu166, and Cys145 residues. More mechanistic investigations are still needed to decide whether this compound is a competitive or non-competitive inhibitor of the SARS-CoV Mpro.Materials and method
General
All chemicals were purchased from Sigma-Aldrich Chemical Co. (Sigma-Aldrich Corp., St. Louis, MO, USA). All the melting points were measured with a Stuart Scientific Co. Ltd apparatus, which are uncorrected. The 1H (500 MHz) NMR spectra were measured on a BRUKER AV 500 MHz spectrometer in DMSO-d6, a solvent, using the tetramethylsilane (TMS) as an internal standard. The mass spectra were determined on Agilent 1100 LC MSD Model G1315B Mass Spectrometer (Agilent Technologies, United States). The elemental analysis was carried out at the Regional Centre for Mycology and Biotechnology (RCMP), Al-Azhar University, Cairo, Egypt, and the results were within ± 0.25%. The reaction courses and product mixtures were routinely monitored by the thin layer chromatography (TLC) on silica gel pre-coated F254 Merck plates.General procedure for synthesis of methyl 1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxylate and 9-methyl derivative (17, 18)
A reaction mixture of 2-chloro-4-oxo-4H-pyrido[1,2-a]pyrimidine-3-carbaldehyde or its 9-methyl derivative (15, 16) (0.01 mol)55 with methyl-N-methyl(N-benzyl)glycinate or methyl methyl-N-benzyl glycinate (0.01 mol) in methanol (20 ml) and triethylamine (0.5 ml) was heated under reflux conditions for 4 h. After the reaction reached completion, the reaction mixture was cooled to room temperature, and the precipitated solid was filtered off, washed with cold methanol, and recrystallized from ethanol/benzene to give compounds 17 and 18 are as previously described.12General procedure for synthesis of 1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxylic acid (19, 20) and N-aryl-1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxamide (21–29)
A reaction mixture of methyl 1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]-pyrimidine-2-carboxylate or its 9-methyl derivatives (17, 18) (0.01 mol) in aq. lithium hydroxide (0.01 mol) in methanol (20 ml) was refluxed for 2 h. After the reaction reached completion, the reaction mixture was cooled to room temperature, andthe methanol was removed under reduced pressure till dryness, the obtained oil intermediate, 1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxylic acid or its 9-methyl derivatives (19, 20) was then washed with 10% HCl then H2O and the remaining oil was extracted by acetonitrile. The latter was reacted withvarious aromatic amines (0.01 mol) in the presence of 1,1′-carbonyldiimidazole (CDI) (0.01 mol) and acetonitrile (20 ml) under reflux for 4 h. The resultant precipitates were filtered off and the filtrate was evaporated to dryness. The residue was subjected to a column chromatography on silica gel with hexane/ethyl acetate (EtOAc) (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as an eluent to afford the target N-aryl-1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxamide (21–29). The physical and spectral data of compounds (21–29) are as follows:
1) as an eluent to afford the target N-aryl-1-benzyl/methyl-4-oxo-1,4-dihydropyrido[1,2-a]pyrrolo[2,3-d]pyrimidine-2-carboxamide (21–29). The physical and spectral data of compounds (21–29) are as follows:
Biological evaluation studies
Computational analysis methods
Author contributions
A. Aljuhani, H. E. A. Ahmed, and S. Ihmaid were responsible for the conception and rational design of the work. A. El-Agrody, A. M. Omar, M. F. Zayed, S. A. Salama, H. S. Abulkhair, and A. Aljuhani, were responsible for collection of data and synthesis of new compounds. S. Althagfan, Y. M. Alahmadi, S. Ahmed, and H. Ahmed, performed the molecular docking study. Molecular dynamic simulations have been conducted by H. E. A. Ahmed, S. Ihmaid, and A. A. Al-Karmalawy. A. El-Agrody, I. Ahma, M. A. Almikhlafi, S. H. Abulkhair, A. M. El-Agrody, S. A. Turkistani, M. Almaghrabi and, and H. S. Abulkhair were responsible for analyzing the spectral data and preparation of the ESI.† All authors read and approved the final form of the manuscript.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
We greatly thank Taif University for providing fund for this work through Taif University Researcher Supporting Project number (TURSP-2020/52), Taif University, Taif, Saudi Arabia. We would also like to thank Professor Ali H. El-Far, Department of Biochemistry, Faculty of Veterinary Medicine, Damanhour University, Egypt, for his considerable help throughout revision stages of the manuscript.References
- P. Gautret, J.-C. Lagier, P. Parola, V. T. Hoang, L. Meddeb, M. Mailhe, B. Doudier, J. Courjon, V. Giordanengo, V. E. Vieira, H. Tissot Dupont, S. Honoré, P. Colson, E. Chabrière, B. La Scola, J.-M. Rolain, P. Brouqui and D. Raoult, Int. J. Antimicrob. Agents, 2020, 56, 105949 CrossRef CAS PubMed.
- World Health Organization, WHO Coronavirus (COVID-19) Dashboard, https://covid19.who.int/, (accessed 7 July 2022) Search PubMed.
- A. R. Maher, M. Maglione, S. Bagley, M. Suttorp, J.-H. Hu, B. Ewing, Z. Wang, M. Timmer, D. Sultzer and P. G. Shekelle, JAMA, 2011, 306, 1359 CrossRef CAS PubMed.
- A. Mahmoud, A. Mostafa, A. A. Al-Karmalawy, A. Zidan, H. S. Abulkhair, S. H. Mahmoud, M. Shehata, M. M. Elhefnawi and M. A. Ali, Heliyon, 2021, 7, e07962 CrossRef PubMed.
- M. I. A. Hamed, K. M. Darwish, R. Soltane, A. Chrouda, A. Mostafa, N. M. Abo Shama, S. S. Elhady, H. S. Abulkhair, A. E. Khodir, A. A. Elmaaty and A. A. Al-karmalawy, RSC Adv., 2021, 11, 35536–35558 RSC.
- A. J. Pruijssers and M. R. Denison, Curr. Opin. Virol., 2019, 35, 57–62 CrossRef CAS PubMed.
- G. G. Plata, The BMJ, 2022, o1282 CrossRef PubMed.
- A. Mahmoud, A. Mostafa, A. A. Al-Karmalawy, A. Zidan, H. S. Abulkhair, S. H. Mahmoud, M. Shehata, M. M. Elhefnawi and M. A. Ali, Heliyon, 2021, 7, e07962 CrossRef PubMed.
- O. Kutkat, Y. Moatasim, A. A. Al-Karmalawy, H. S. Abulkhair, M. R. Gomaa, A. N. El-Taweel, N. M. Abo Shama, M. GabAllah, D. B. Mahmoud, G. Kayali, M. A. Ali, A. Kandeil and A. Mostafa, Sci. Rep., 2022, 12, 12920 CrossRef CAS PubMed.
- L. Fu, F. Ye, Y. Feng, F. Yu, Q. Wang, Y. Wu, C. Zhao, H. Sun, B. Huang, P. Niu, H. Song, Y. Shi, X. Li, W. Tan, J. Qi and G. F. Gao, Nat. Commun., 2020, 11, 4417 CrossRef CAS.
- M. M. Hammoud, M. Khattab, M. Abdel-Motaal, J. Van der Eycken, R. Alnajjar, H. S. Abulkhair and A. A. Al-Karmalawy, J. Biomol. Struct. Dyn., 2022 DOI:10.1080/07391102.2022.2082533 , https://www.tandfonline.com/doi/abs/10.1080/07391102.2022.2082533?journalCode=tbsd20.
- A. A. Gaber, A. M. El-Morsy, F. F. Sherbiny, A. H. Bayoumi, K. M. El-Gamal, K. El-Adl, A. A. Al-Karmalawy, R. R. Ezz Eldin, M. A. Saleh and H. S. Abulkhair, Arch. Pharm., 2021 DOI:10.1002/ardp.202100258 , https://onlinelibrary.wiley.com/doi/10.1002/ardp.202100258.
- C. Choudhury, J. Biomol. Struct. Dyn., 2021, 39, 3733–3746 CrossRef CAS PubMed.
- I. Antonopoulou, E. Sapountzaki, U. Rova and P. Christakopoulos, Comput. Struct. Biotechnol. J., 2022, 20, 1306–1344 CrossRef CAS PubMed.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, J. Yu, L. Wang, K. Yang, F. Liu, R. Jiang, X. Yang, T. You, X. Liu, X. Yang, F. Bai, H. Liu, X. Liu, L. W. Guddat, W. Xu, G. Xiao, C. Qin, Z. Shi, H. Jiang, Z. Rao and H. Yang, Nature, 2020, 582, 289–293 CrossRef CAS PubMed.
- K. Anand, J. Ziebuhr, P. Wadhwani, J. R. Mesters and R. Hilgenfeld, Science, 2003, 300, 1763–1767 CrossRef CAS PubMed.
- S. Günther, P. Y. A. Reinke, Y. Fernández-García, J. Lieske, T. J. Lane, H. M. Ginn, F. H. M. Koua, C. Ehrt, W. Ewert, D. Oberthuer, O. Yefanov, S. Meier, K. Lorenzen, B. Krichel, J.-D. Kopicki, L. Gelisio, W. Brehm, I. Dunkel, B. Seychell, H. Gieseler, B. Norton-Baker, B. Escudero-Pérez, M. Domaracky, S. Saouane, A. Tolstikova, T. A. White, A. Hänle, M. Groessler, H. Fleckenstein, F. Trost, M. Galchenkova, Y. Gevorkov, C. Li, S. Awel, A. Peck, M. Barthelmess, F. Schlünzen, P. Lourdu Xavier, N. Werner, H. Andaleeb, N. Ullah, S. Falke, V. Srinivasan, B. A. França, M. Schwinzer, H. Brognaro, C. Rogers, D. Melo, J. J. Zaitseva-Doyle, J. Knoska, G. E. Peña-Murillo, A. R. Mashhour, V. Hennicke, P. Fischer, J. Hakanpää, J. Meyer, P. Gribbon, B. Ellinger, M. Kuzikov, M. Wolf, A. R. Beccari, G. Bourenkov, D. von Stetten, G. Pompidor, I. Bento, S. Panneerselvam, I. Karpics, T. R. Schneider, M. M. Garcia-Alai, S. Niebling, C. Günther, C. Schmidt, R. Schubert, H. Han, J. Boger, D. C. F. Monteiro, L. Zhang, X. Sun, J. Pletzer-Zelgert, J. Wollenhaupt, C. G. Feiler, M. S. Weiss, E.-C. Schulz, P. Mehrabi, K. Karničar, A. Usenik, J. Loboda, H. Tidow, A. Chari, R. Hilgenfeld, C. Uetrecht, R. Cox, A. Zaliani, T. Beck, M. Rarey, S. Günther, D. Turk, W. Hinrichs, H. N. Chapman, A. R. Pearson, C. Betzel and A. Meents, Science, 2021, 372, 642–646 CrossRef PubMed.
- I. Antonopoulou, E. Sapountzaki, U. Rova and P. Christakopoulos, Comput. Struct. Biotechnol. J., 2022, 20, 1306–1344 CrossRef CAS PubMed.
- D. S. Jairajpuri, A. Hussain, K. Nasreen, T. Mohammad, F. Anjum, M. Tabish Rehman, G. Mustafa Hasan, M. F. Alajmi and M. Imtaiyaz Hassan, Saudi J. Biol. Sci., 2021, 28, 2423–2431 CrossRef CAS PubMed.
- A. K. Ghosh, G. Gong, V. Grum-Tokars, D. C. Mulhearn, S. C. Baker, M. Coughlin, B. S. Prabhakar, K. Sleeman, M. E. Johnson and A. D. Mesecar, Bioorg. Med. Chem. Lett., 2008, 18, 5684–5688 CrossRef CAS PubMed.
- Z. Jin, Y. Zhao, Y. Sun, B. Zhang, H. Wang, Y. Wu, Y. Zhu, C. Zhu, T. Hu, X. Du, Y. Duan, J. Yu, X. Yang, X. Yang, K. Yang, X. Liu, L. W. Guddat, G. Xiao, L. Zhang, H. Yang and Z. Rao, Nat. Struct. Mol. Biol., 2020, 27, 529–532 CrossRef CAS.
- M. Negi, P. A. Chawla, A. Faruk and V. Chawla, Bioorg. Chem., 2020, 104, 104315 CrossRef CAS PubMed.
- A. K. Ghosh, G. Gong, V. Grum-Tokars, D. C. Mulhearn, S. C. Baker, M. Coughlin, B. S. Prabhakar, K. Sleeman, M. E. Johnson and A. D. Mesecar, Bioorg. Med. Chem. Lett., 2008, 18, 5684–5688 CrossRef CAS PubMed.
- D.-S. Park, E. Jo, J. Choi, M. Lee, S. Kim, H.-Y. Kim, J. Nam, S. Ahn, J. Y. Hwang and M. P. Windisch, Eur. J. Med. Chem., 2017, 140, 65–73 CrossRef CAS PubMed.
- M. Lee, J. Yang, E. Jo, J.-Y. Lee, H.-Y. Kim, R. Bartenschlager, E.-C. Shin, Y.-S. Bae and M. P. Windisch, Sci. Rep., 2017, 7, 44676 CrossRef.
- K. Bafna, R. M. Krug and G. T. Montelione, ChemRxiv, 2020 DOI:10.26434/chemrxiv.12153615.
- K. Bafna, K. White, B. Harish, R. Rosales, T. A. Ramelot, T. B. Acton, E. Moreno, T. Kehrer, L. Miorin, C. A. Royer, A. García-Sastre, R. M. Krug and G. T. Montelione, Cell Rep., 2021, 35, 109133 CrossRef CAS PubMed.
- K. Bafna, K. White, B. Harish, R. Rosales, T. A. Ramelot, T. B. Acton, E. Moreno, T. Kehrer, L. Miorin, C. A. Royer, A. García-Sastre, R. M. Krug and G. T. Montelione, Cell Rep., 2021, 35, 109133 CrossRef CAS.
- R. Oerlemans, A. J. Ruiz-Moreno, Y. Cong, N. Dinesh Kumar, M. A. Velasco-Velazquez, C. G. Neochoritis, J. Smith, F. Reggiori, M. R. Groves and A. Dömling, RSC Med. Chem., 2021, 12, 370–379 RSC.
- H. S. Abulkhair, A. Turky, A. Ghiaty, H. E. A. Ahmed and A. H. Bayoumi, Bioorg. Chem., 2020, 100, 103899 CrossRef CAS.
- M. H. El-Shershaby, K. M. El-Gamal, A. H. Bayoumi, K. El-Adl, H. E. A. Ahmed and H. S. Abulkhair, Arch. Pharm., 2021, 354, e2000277 CrossRef PubMed.
- A. M. Omar, S. Ihmaid, E. S. E. Habib, S. S. Althagfan, S. Ahmed, H. S. Abulkhair and H. E. A. Ahmed, Bioorg. Chem., 2020, 99, 103781 CrossRef CAS PubMed.
- E. M. Othman, E. A. Fayed, E. M. Husseiny and H. S. Abulkhair, New J. Chem., 2022, 46, 12206–12216 RSC.
- D. S. Jairajpuri, A. Hussain, K. Nasreen, T. Mohammad, F. Anjum, M. Tabish Rehman, G. Mustafa Hasan, M. F. Alajmi and M. Imtaiyaz Hassan, Saudi J. Biol. Sci., 2021, 28, 2423–2431 CrossRef CAS.
- M. Abass, M. Ismail, W. Abdel-Monem and A. Mayas, Chem. Pap., 2010, 64 DOI:10.2478/s11696-009-0100-0 , https://link.springer.com/article/10.2478/s11696-009-0100-0.
- M. F. Maioral, C. do N. Bodack, N. M. Stefanes, Á. Bigolin, A. Mascarello, L. D. Chiaradia-Delatorre, R. A. Yunes, R. J. Nunes and M. C. Santos-Silva, Biochimie, 2017, 140, 48–57 CrossRef CAS.
- M. H. El-Shershaby, A. Ghiaty, A. H. Bayoumi, A. A. Al-Karmalawy, E. M. Husseiny, M. S. El-Zoghbi and H. S. Abulkhair, Bioorg. Med. Chem., 2021, 42, 116266 CrossRef CAS PubMed.
- E. M. Othman, E. A. Fayed, E. M. Husseiny and H. S. Abulkhair, Bioorg. Chem., 2022, 123, 105762 CrossRef CAS PubMed.
- E. M. Othman, E. A. Fayed, E. M. Husseiny and H. S. Abulkhair, Bioorg. Chem., 2022, 127, 105968 CrossRef CAS PubMed.
- J. Osipiuk, S.-A. Azizi, S. Dvorkin, M. Endres, R. Jedrzejczak, K. A. Jones, S. Kang, R. S. Kathayat, Y. Kim, V. G. Lisnyak, S. L. Maki, V. Nicolaescu, C. A. Taylor, C. Tesar, Y.-A. Zhang, Z. Zhou, G. Randall, K. Michalska, S. A. Snyder, B. C. Dickinson and A. Joachimiak, Nat. Commun., 2021, 12, 743 CrossRef CAS PubMed.
- A. Abo Elmaaty, M. I. A. Hamed, M. I. Ismail, E. B. Elkaeed, H. S. Abulkhair, M. Khattab and A. A. Al-Karmalawy, Molecules, 2021, 26, 3772 CrossRef.
- T. Pillaiyar, M. Manickam, V. Namasivayam, Y. Hayashi and S.-H. Jung, J. Med. Chem., 2016, 59, 6595–6628 CrossRef CAS PubMed.
- R. L. Hoffman, R. S. Kania, M. A. Brothers, J. F. Davies, R. A. Ferre, K. S. Gajiwala, M. He, R. J. Hogan, K. Kozminski, L. Y. Li, J. W. Lockner, J. Lou, M. T. Marra, L. J. Mitchell, B. W. Murray, J. A. Nieman, S. Noell, S. P. Planken, T. Rowe, K. Ryan, G. J. Smith, J. E. Solowiej, C. M. Steppan and B. Taggart, J. Med. Chem., 2020, 63, 12725–12747 CrossRef CAS PubMed.
- C. Ma, Y. Hu, J. A. Townsend, P. I. Lagarias, M. T. Marty, A. Kolocouris and J. Wang, ACS Pharmacol. Transl. Sci., 2020, 3, 1265–1277 CrossRef CAS PubMed.
- W. Vuong, M. B. Khan, C. Fischer, E. Arutyunova, T. Lamer, J. Shields, H. A. Saffran, R. T. McKay, M. J. van Belkum, M. A. Joyce, H. S. Young, D. L. Tyrrell, J. C. Vederas and M. J. Lemieux, Nat. Commun., 2020, 11, 4282 CrossRef CAS PubMed.
- H. Abul-Khair, S. Elmeligie, A. Bayoumi, A. Ghiaty, A. El-Morsy and M. H. Hassan, J. Heterocycl. Chem., 2013, 50, 1202–1208 CAS.
- A. K. Ghosh, M. Brindisi, D. Shahabi, M. E. Chapman and A. D. Mesecar, ChemMedChem, 2020, 15, 907–932 CrossRef CAS PubMed.
- K. Sharun, R. Tiwari and K. Dhama, Ann. Med. Surg., 2021, 61, 122–125 CrossRef.
- C.-H. Zhang, E. A. Stone, M. Deshmukh, J. A. Ippolito, M. M. Ghahremanpour, J. Tirado-Rives, K. A. Spasov, S. Zhang, Y. Takeo, S. N. Kudalkar, Z. Liang, F. Isaacs, B. Lindenbach, S. J. Miller, K. S. Anderson and W. L. Jorgensen, ACS Cent. Sci., 2021, 7, 467–475 CrossRef CAS.
- T. A. Halgren, R. B. Murphy, R. A. Friesner, H. S. Beard, L. L. Frye, W. T. Pollard and J. L. Banks, J. Med. Chem., 2004, 47, 1750–1759 CrossRef CAS PubMed.
- A. A. Zaki, M. M. Y. Kaddah, H. S. Abulkhair and A. Ashour, RSC Adv., 2022, 12, 2980–2991 RSC.
- H. S. Abulkhair, S. Elmeligie, A. Ghiaty, A. El-Morsy, A. H. Bayoumi, H. E. A. Ahmed, K. El-Adl, M. F. Zayed, M. H. Hassan, E. N. Akl and M. S. El-Zoghbi, Arch. Pharm., 2021, 354, 2000449 CrossRef CAS PubMed.
- S. K. Ihmaid, A. Aljuhani, M. Alsehli, N. Rezki, A. Alawi, A. J. Aldhafiri, S. A. Salama, H. E. A. Ahmed and M. R. Aouad, J. Mol. Struct., 2022, 1249, 131568 CrossRef CAS.
- M. H. El-Shershaby, A. Ghiaty, A. H. Bayoumi, H. E. A. Ahmed, M. S. El-Zoghbi, K. El-Adl and H. S. Abulkhair, New J. Chem., 2021, 45, 11136–11152 RSC.
- A. De, S. Sarkar and A. Majee, Chem. Heterocycl. Compd., 2021, 57, 410–416 CrossRef CAS PubMed.
- Y.-S. Chung, N.-J. Lee, S. H. Woo, J.-M. Kim, H. M. Kim, H. J. Jo, Y. E. Park and M.-G. Han, Sci. Rep., 2021, 11, 14817 CrossRef CAS.
- Genesig, Coronavirus COVID-19 genesig® Real-Time PCR assay, https://www.genesig.com/assets/files/Path_COVID_19_CE_STED_IFU_Issue_500.pdf, (accessed 12 March 2022) Search PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- K. El-Adl, H. M. Sakr, R. G. Yousef, A. B. M. Mehany, H. S. Abulkhair and I. H. Eissa, Arch. Pharm., 2022, 355(7), 2200048 CrossRef CAS PubMed , https://onlinelibrary.wiley.com/doi/10.1002/ardp.202200048.
- K. El-Adl, M. K. Ibrahim, F. Khedr, H. S. Abulkhair and I. H. Eissa, Arch. Pharm., 2022, 355, 2100278 CrossRef CAS PubMed.
- F. Khedr, M. K. Ibrahim, I. H. Eissa, H. S. Abulkhair and K. El-Adl, Arch. Pharm., 2021, 354, e2100201 CrossRef PubMed.
- K. El-Adl, H. Sakr, S. S. A. El-Hddad, A. G. A. El-Helby, M. Nasser and H. S. Abulkhair, Arch. Pharm., 2021, 354(7) DOI:10.1002/ardp.202000491 , https://onlinelibrary.wiley.com/doi/10.1002/ardp.202000491.
- W. Zhu, M. Xu, C. Z. Chen, H. Guo, M. Shen, X. Hu, P. Shinn, C. Klumpp-Thomas, S. G. Michael and W. Zheng, Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-throughput Screening, 2020 Search PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- J. S. Morse, T. Lalonde, S. Xu and W. R. Liu, ChemBioChem, 2020, 21, 730–738 CrossRef CAS PubMed.
- R. A. Friesner, R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood, T. A. Halgren, P. C. Sanschagrin and D. T. Mainz, J. Med. Chem., 2006, 49, 6177–6196 CrossRef CAS PubMed.
- K. El-Adl, A. G. A. El-Helby, H. Sakr, R. R. Ayyad, H. A. Mahdy, M. Nasser, H. S. Abulkhair and S. S. A. El-Hddad, Arch. Pharm., 2021, 354, e202000279 Search PubMed.
- H. G. Ezzat, A. H. Bayoumi, F. F. Sherbiny, A. M. El-Morsy, A. Ghiaty, M. Alswah and H. S. Abulkhair, Mol. Diversity, 2021, 25, 291–306 CrossRef CAS PubMed.
- R. G. Yousef, H. M. Sakr, I. H. Eissa, A. B. M. Mehany, A. M. Metwaly, M. A. Elhendawy, M. M. Radwan, M. A. ElSohly, H. S. Abulkhair and K. El-Adl, New J. Chem., 2021, 45, 16949–16964 RSC.
- I. Ahmad, M. Shaikh, S. Surana, A. Ghosh and H. Patel, J. Biomol. Struct. Dyn., 2022, 40, 3046–3059 CrossRef CAS PubMed.
- D. E. Shaw Research, 2021, https://www.schrodinger.com/products/desmond.
- W. L. Jorgensen, D. S. Maxwell and J. Tirado-Rives, J. Am. Chem. Soc., 1996, 118, 11225–11236 CrossRef CAS.
- I. Ahmad, H. Jadhav, Y. Shinde, V. Jagtap, R. Girase and H. Patel, In Silico Pharmacol., 2021, 9, 23 CrossRef PubMed.
- G. Kalibaeva, M. Ferrario and G. Ciccotti, Mol. Phys., 2003, 101, 765–778 CrossRef CAS.
- G. J. Martyna, Phys. Rev. C: Nucl. Phys., 1994, 50, 3234–3236 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04015h |
| ‡ These authors contributed equally to this work and share first authorship. |
| This journal is © The Royal Society of Chemistry 2022 |