Open Access Article
Open Access ArticleThe role of electrochemical biosensors in SARS-CoV-2 detection: a bibliometrics-based analysis and review
Shudan Mao*a,
Li Fu *b,
Chengliang Yincd,
Xiaozhu Liue and
Hassan Karimi-Maleh
*b,
Chengliang Yincd,
Xiaozhu Liue and
Hassan Karimi-Maleh fgh
fgh
aKey Laboratory of Pollution Exposure and Health Intervention of Zhejiang Province, Interdisciplinary Research Academy, Zhejiang Shuren University, Hangzhou 310021, PR China
bKey Laboratory of Novel Materials for Sensor of Zhejiang Province, College of Materials and Environmental Engineering, Hangzhou Dianzi University, Hangzhou 310018, China. E-mail: fuli@hdu.edu.cn
cNational Engineering Laboratory for Medical Big Data Application Technology, Chinese PLA General Hospital, Beijing, China
dMedical Big Data Research Center, Medical Innovation Research Division of PLA General Hospital, Beijing, China
eDepartment of Cardiology, The Second Affiliated Hospital of Chongqing Medical University, Chongqing 400010, China
fSchool of Resources and Environment, University of Electronic Science and Technology of China, Xiyuan Ave, 611731, Chengdu, China
gDepartment of Chemical Engineering, Quchan University of Technology, Quchan 9477177870, Iran
hDepartment of Chemical Sciences, University of Johannesburg, Doornfontein Campus, 2028, Johannesburg 17011, South Africa
First published on 12th August 2022
Abstract
The global pandemic of COVID-19, which began in late 2019, has resulted in extremely high morbidity and severe mortality worldwide, with important implications for human health, international trade, and national politics. Severe acute respiratory syndrome coronavirus (SARS-CoV-2) is the primary pathogen causing COVID-19. Analytical chemistry played an important role in this global epidemic event, and detection of SARS-CoV-2 even became a part of daily life. Analytical chemists have devoted much effort and enthusiasm to this event, and different analytical techniques have shown very rapid development. Electrochemical biosensors are highly efficient, sensitive, and cost-effective and have been used to detect many highly pathogenic viruses long before this event. However, another fact is that electrochemical biosensors are not the technology of choice for most detection applications. This review describes for the first time the role played by electrochemical biosensors in SARS-CoV-2 detection from a bibliometric perspective. This paper analyzed 254 relevant research papers up to June 2022. The contributions of different countries and institutions to this topic were analyzed. Keyword analysis was used to explore different methodological attempts of electrochemical detection techniques. More importantly, we are trying to find an answer to the question: do electrochemical biosensors have the potential to become a genuinely employable detection technology in an outbreak of infectious disease?
Introduction
SARS-CoV-2 was first identified in December 2019 by the International Committee on Taxonomy of Viruses (ICTV).1 The virus causes fatal acute respiratory distress syndrome, heart disease, multi-organ dysfunction, and death when it infects humans. The composition of SARS-CoV-2 mainly includes structural proteins, genetic material single-stranded RNA, non-structural proteins, and accessory proteins.2 The analytes currently tested fall into four categories: nucleic acids,3 antigens,4 antibodies,5 and viral particles.6 To date, RT-PCR-based nucleic acid testing is still the gold standard for diagnosing COVID-19.7 Developing an accurate and efficient SARS-CoV-2 detection method is essential to reducing the rate of COVID-19 transmission.8–10 In the past two years, many papers have proposed new methods to detect SARS-CoV-2.Biosensor technology is one of the most promising approaches to solving the problems of low sensitivity and high cost in clinical diagnosis.11 It consists of two most important components: the biological part reacting with the target material and the detection signal generation. The choice of these two components is crucial for the sensitivity and specificity of the biosensor. The principle classification of biosensor devices can be divided into thermal biosensors,12 field-effect transistor biosensors,13 piezoelectric biosensors,14 electrochemical biosensors,15 optical biosensors,16 acoustic wave biosensors,17 etc. Electrochemical biosensors are based on electrochemical analysis (including potentiometric analysis,18 conductivity analysis,19 voltammetry,20 capacitance analysis,21 etc.) to convert the chemical quantity of the analyte to be measured into an electrical quantity to achieve the monitoring of the target analyte, which has the potential for real-time monitoring and rapid detection in the field for virus detection. According to the different biomaterials used for sensitive elements, electrochemical biosensors can be divided into enzyme electrode biosensors,22 microbial electrode biosensors,23 electrochemical immunosensors,24 and electrochemical nucleic acid biosensors.25 Among them, the primary biosensors commonly used to detect viruses are immunosensors and nucleic acid biosensors. They have been successfully used to detect hepatitis virus,26 influenza virus,27 human papilloma virus,28 Zika virus,29 etc.
Although there have been tremendous methodological breakthroughs in detecting viruses by electrochemical biosensors, it cannot be denied that these efforts have been mainly investigated and discussed at the scientific level and have not presented an undeniable commercial value. This is because there are still many challenges facing the current electrochemical biosensors in virus detection.30–32 Virus field diagnosis requires simple and efficient detection methods with accurate results and universal applicability. Current biosensors of this type still require demanding operating conditions. In particular, the activity of biosensitive materials can directly affect the accuracy of results, and most materials are only maximally active at optimal pH, ionic strength, temperature, etc. Moreover, many materials remain active for a short period at room temperature and are only suitable for testing under ideal laboratory conditions. In addition, the complex composition of clinical samples may result in false positive or false negative test results, affecting the accuracy of the test results.
Electrochemical biosensors can be used to detect and quantify SARS-CoV-2 through several strategies. SARS-CoV-2 related antigens and antibodies were the most selected analytes. Secondly, markers of inflammation, heart biomarkers, liver biomarkers, kidney biomarkers, blood biomarkers, and biochemical biomarkers altered during disease progression can also be targeted for viral detection.33,34 Because electrochemical biosensors are a highly sensitive analytical method and a candidate technology in the COVID-19 global pandemic, related publications are not only cited and discussed in academic journals but are even competitively covered by social media.35 Some of these reports are considered to have the potential for practical application. So far, electrochemical biosensor-based detection in COVID-19 has been reviewed by several papers.36–38 These reviews introduce the different methodologies and interpret the highlighted work. In this review, we attempt to analyse and review this topic statistically using a bibliometric approach. Bibliometric analysis is a literature and information mining method based on mathematical statistics. It can reflect research trends and hotspots through clustering relationships of keywords in the literature and has become an important tool for global analysis in various scientific fields.39–47 This article hopes to analyse the collaborative networks and directions of investigation on this topic. We try to be able to give answers to the following questions:
(1) Which detection methodologies in electrochemical biosensors have been applied to SARS-CoV-2 detection?
(2) Which countries and institutions have majorly contributed to this topic? Do they form an international collaboration?
(3) Do electrochemical biosensors already meet the sensitivity requirement for SARS-CoV-2 detection? Do scientists need to explore further ways to lower the limit of detection?
(4) Is the current stage of miniaturization efforts of electrochemical biosensors sufficient for field detection?
(5) What are the most significant challenges facing electrochemical biosensors for SARS-CoV-2 detection?
Data and analysis method
Two bibliometrics software have been used in this systematic literature review. The first is CiteSpace, developed by Dr Chaomei Chen, a professor at the Drexel University School of Information Science and Technology.48–51 CiteSpace 6.1R2 was used to calculate and analyze all documents. COOC is another emerging bibliometrics software.52 COOC12.6 was used to analysis of country contribution and keywords co-occurrence. We used the core collection on Web of Science as a database to assure the integrity and academic quality of the studied material. “electrochemical COVID-19” or “electrochemical ‘SARS CoV 2’” has been used as a “Topic”. The retrieval period was indefinite, and the date of retrieval was June, 2022. 254 articles (including 254 articles and 9 early access) were retrieved (review article was not included in this survey).Developments in the research field
Literature development trends
Since the COVID-19 pandemic started in 2019, the annual publication number is not of significant value on this topic. Electrochemical biosensors in the detection of COVID-19 did not show a spurt in 2020, with only 18 papers published. However, this number reached 150 articles in 2021. The growth of this data is influenced by many factors, such as our understanding of COVID-19, experiments and the publication cycle of the paper. By June 2022, 86 papers on this topic had been indexed by WOS. It can be predicted that the annual publication of this topic in 2022 should be no less than that in 2021, but it will not show very rapid growth. Academic attention to a topic during an outburst public health event is not led by free inquiry but can be heavily influenced by politics and policy. 22 papers out of 254 are now marked as highly cited papers in WOS, accounting for 8.66%. In contrast, only about 6% of COVID-19-related articles are highly cited (“COVID-19” or “SARS CoV 2” has been used as a “Topic”). Therefore, applying electrochemical biosensors in COVID-19 detection is a hot topic. This may be because electrochemical biosensors have been considered throughout their development history as an analytical technique with the potential for rapid detection. The methodology it has been developed possesses excellent specificity and sensitivity.Journals, cited journals and research subjects
Fig. 1 shows the 9 journals that published the highest number of electrochemical biosensors in SARS-CoV-2 detection. As seen from the figure, Biosensors and Bioelectronics presents an absolute dominance with a total of 44 publications, accounting for approximately 17% of the total number of articles on this topic. Biosensors and Bioelectronics is a top journal in the field of biosensing, and electrochemical biosensors are a methodology of great interest to this journal. All the remaining journals in Fig. 1, except Scientific Reports, are in the field of analytical chemistry. Therefore, the establishment and innovation of detection technology have been the most important focus of this topic during these three years.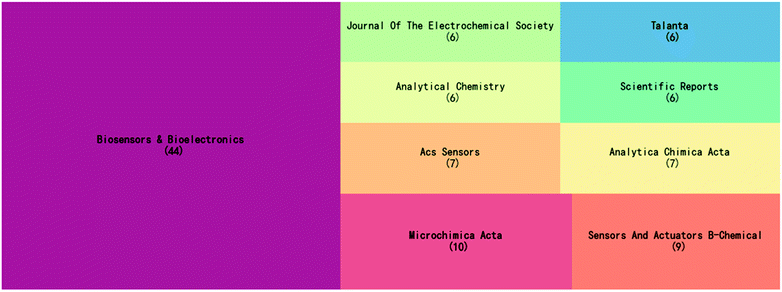 | ||
| Fig. 1 Top 9 journals that published articles on the electrochemical biosensors in SARS-CoV-2 detection. | ||
In addition to the number of papers published by the journal on the topic, the frequency with which the journal is cited by papers related to the topic is also an important indicator. Table 1 shows the top 15 cited journals on the electrochemical biosensors in SARS-CoV-2 detection.
| No. | Count | Cited journal |
|---|---|---|
| 1 | 180 | Biosensors and Bioelectronics |
| 2 | 146 | Sensors and Actuators B: Chemical |
| 3 | 141 | Analytical Chemistry |
| 4 | 133 | ACS Nano |
| 5 | 87 | Sensors |
| 6 | 85 | Nature |
| 7 | 85 | Microchimica Acta |
| 8 | 84 | Talanta |
| 9 | 83 | Scientific Reports |
| 10 | 79 | ACS Sensors |
| 11 | 78 | Analytica Chimica Acta |
| 12 | 78 | Electrochimica Acta |
| 13 | 75 | ACS Applied Materials & Interfaces |
| 14 | 74 | Nature Communications |
| 15 | 71 | Science |
The journals in Fig. 1 can be found in Table 1, except for the Journal of the Electrochemical Society. Biosensors and Bioelectronics is also ranked first in Table 1, further indicating that it plays a very important role in this topic in terms of publications and citations. Also appearing in Table 1 is a series of comprehensive journals, including Science, Nature, and Nature Communication, representing papers published in these journals (not necessarily relevant to electrochemical biosensors, but at least relevant to the COVID-19 pandemic) that have also influenced the development of the topic of electrochemical biosensors in SARS-CoV-2 detection. Sensors, ranked fifth, deserve special attention. It does not publish many papers related to electrochemical biosensors in SARS-CoV-2 detection, but the papers published in this journal on other types of biosensors clearly impact this topic. The development of electrochemical techniques and the investigation of electrochemical behavior is also directly related to this topic, hence the inclusion of Electrochimica Acta in Table 1. On the other hand, the development of materials science has been closely related to electrochemical biosensors. Superior materials can improve electrode performance and increase the efficiency of the recognition element immobilization. In addition, electrochemical biosensors rely on interfacial reactions. Therefore, ACS Applied Materials & Interfaces is also included in Table 2.
| Year | Category |
|---|---|
| 2020 | Chemistry, Analytical; Geosciences, Multidisciplinary; Geology; Energy & Fuels; Pharmacology & Pharmacy; Engineering, Electrical & Electronic; Materials Science, Coatings & Films; Chemistry, Physical; Physics, Applied; Instruments & Instrumentation; Materials Science, Multidisciplinary; Biotechnology & Applied Microbiology; Chemistry, Multidisciplinary; Biophysics; Electrochemistry; Nanoscience & Nanotechnology |
| 2021 | Clinical Neurology; Medicine, General & Internal; Computer Science, Theory & Methods; Infectious Diseases; Engineering; Biomedical; Computer Science, Artificial Intelligence; Physiology; Acoustics; Computer Science, Hardware & Architecture; Neurosciences; Immunology; Chemistry, Inorganic & Nuclear; Education, Scientific Disciplines; Physics, Atomic, Molecular & Chemical; Computer Science, Information Systems; Telecommunications; Public, Environmental & Occupational Health; Virology; Spectroscopy; Metallurgy & Metallurgical Engineering; Engineering, Environmental; Food Science & Technology; Biochemical Research Methods; Engineering, Chemical; Environmental Sciences; Biochemistry & Molecular Biology; Physics, Condensed Matter; Multidisciplinary Sciences |
| 2022 | Nutrition & Dietetics; Chemistry, Applied; Medical Laboratory Technology; Engineering, Multidisciplinary; Biology |
The above analysis can be used to understand the most important and popular journals on this topic, but these data cannot present the latest developments. Journals publishing papers on this topic for the first time in 2022 can be used to understand the most cutting-edge research trends. Although journals in materials science are already included in Table 1, this topic was first published in 2D Materials and Flatchem in 2022. This means that two-dimensional materials have been used for the assembly of electrochemical biosensors. Pola et al.53 proposed an electrochemical immunosensor using graphene ink and aerosol jet printing technology that enables the detection of SARS-CoV-2 in saliva samples. Ang et al.54 used graphene oxide nanocolloids as a transduction platform and electroactive tag to enable the detection of SARS-CoV-2 genomic sequences. A paper on this topic was also first published in Dalton Transactions, which mainly publishes papers related to organometallic chemistry. Cui et al.55 constructed a novel porphyrin-based porous organic polymer by a solvothermal method that can strongly bind to SARS-CoV-2 N-gene targeting antibodies. This new material can be used for the construction of electrochemical aptamersensor and immunosensor. Since SARS-CoV-2 can survive in the environment, detecting SARS-CoV-2 is not limited to body fluid samples. Ramírez-Chavarría et al.56 reported an electrochemical biosensor for the detection of the N and ORF1ab genes of the SARS-CoV-2 genome. This biosensor enabled the detection of SARS-CoV-2 in wastewater. This work was published in the Journal of Environmental Chemical Engineering, a journal related to environmental science.
The category analysis provides further insight into how this topic changes in different areas. Table 2 summarizes the evolution of this topic in the category from 2010 to the present. Electrochemical biosensors for detecting SARS-CoV-2 did not enter the clinical and pharmaceutical-related category in 2020. On the other hand, computer science-related categories such as Information Systems and Artificial Intelligence appear in this theme in 2021, representing the data processing of electrochemical biosensors that have begun to be coupled with intelligent systems. At the same time, the topic also began to cover Engineering, Environmental; and Food Science & Technology. In 2022, some of the more secondary areas, such as Nutrition & Dietetics and Medical Laboratory Technology, begin to be included.
Geographic distribution
Fig. 2 shows a Rose plot of the publication number by different countries. China was involved in 50 of these papers (>16%). The USA is similar, participating in >15% of the papers. India, Turkey, Brazil, and South Korea were all involved in >5% of the papers. According to the distribution of countries, it is known that this topic attracts the attention of scientists and doctors from all regions of the world. However, as this topic is in its explosive phase, it will continue attracting new academic groups to the survey. For example, in the first six months of 2022, German, Lithuania, United Arab Emirates, Israel, Romania, Latvia, and Netherlands all published papers for the first time on this topic.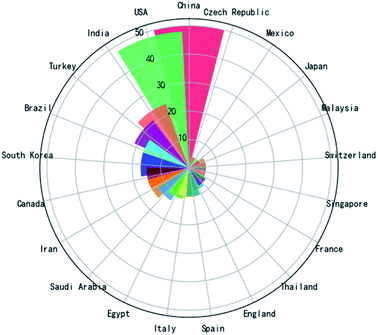 | ||
| Fig. 2 Rose plot of papers related to the electrochemical biosensors in SARS-CoV-2 detection contributed by different countries (countries with fewer than 4 papers are not included). | ||
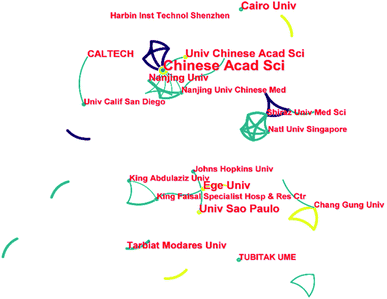
Fig. 3 illustrates the cooperation network between the different institutions on this topic. Although this topic has only been going on for less than three years, three incipient cooperation networks have already been formed. The Chinese Academy of Sciences, Nanjing University, and the Nanjing University of Chinese Medicine led the largest collaborative network. This collaborative network includes universities and research institutions from China, universities in the United States (Massachusetts Institute of Technology and University of California, Los Angeles), and the United Arab Emirates (Imam Abdulrahman Bin Faisal University). The second cooperation network is mainly led by Ege Üniversitesi and the University of São Paulo. This collaborative network presents a wide range of international collaborations, including universities and research institutions in Turkey, Brazil, the United Arab Emirates and the USA. The third collaborative network comprises universities and research institutes in Singapore and Iran, led primarily by Shiraz University of Medical Sciences and the National University of Singapore. Although this academic topic is only three years old, it already presents a wide range of international collaborations. This is not to be taken as the usual pattern for developing academic topics. As COVID-19 was an outburst global public health event, breakthroughs in the medical field had a decisive impact on the resolution of the event. As a result, a great deal of attention and resources were devoted to this topic, which in turn contributed to effective international cooperation's success in a short time.
Keyword analysis and evolution of the field
The most effective way to understand the direction of investigating concerns in a topic is the analysis of keywords. Table 3 lists the top 15 keywords in this topic. Most of these keywords are directly related to the topic and do not require further explanation, such as Biosensor, Sensor, COVID 19, etc. However, some of these keywords can be used for further interpretation. For example, DNA was ranked 7th as a keyword appearing 14 times. Based on the published RNA sequence of SARS-CoV-2, the synthesis of single-stranded DNA matching it is already commercially customizable. Zhao et al.57 prepared a supersandwich-type electrochemical biosensor, which can read the electrochemical signal by smartphone and then realize the qualitative detection of SARS-CoV-2. On the other hand, the RNA of SARS-CoV-2 can trigger the catalytic hairpin assembly circuit. This can initiate terminal deoxynucleotidyl transferase-mediated DNA polymerization. A large amount of long single-stranded DNA product is produced and bound to Ru(NH3)63+ to generate a current signal. Peng et al.58 used this hairpin structure to design an ultrasensitive electrochemical biosensor to achieve linear detection of SARS-CoV-2 RNA in the range of 0.1–1000 pM. The response part of an electrochemical sensor is on the electrode surface, so the electrode characteristics are critical to the biosensor's performance. Electrode is ranked 13th in Table 3, implying that the selection and modification of electrodes is an important direction in the assembly of biosensors. Witt et al.59 investigated the effectiveness of boron-doped diamond electrodes in detecting SARS-CoV-2 S1 protein. Graphitic carbon foil was investigated by Adeel et al.60 for the response performance of SARS-CoV-2 S1 protein. Butler and Ebrahimi61 investigated the performance of interdigitated electrodes in detecting SARS-CoV-2 virus particles. Nanoparticles are ranked 12th in Table 3. Nanoparticles have been a frequently chosen material for electrode surface modification for electrochemical biosensors. They can not only increase the specific surface area of the electrode and facilitate the adsorption of the analytes but also trigger some electrocatalytic responses. Also, some nanomaterials useful for redox signals can be used as probes for quantitative identification of the concentration of analytes. For example, Aydın et al.62 coated 11-mercaptoundecanoic acid with gold nanoparticles to detect COVID 19 specific spike receptor-binding domain protein. Feng et al.63 developed an electrochemical and colorimetric biosensor based on peptide-triggered gold nanoparticle assembly to detect SARS-CoV-2 major protease.| No. | Freq | Centrality | Keywords |
|---|---|---|---|
| 1 | 31 | 0.18 | Biosensor |
| 2 | 24 | 0.2 | Sensor |
| 3 | 22 | 0.12 | Electrochemical Sensor |
| 4 | 20 | 0.1 | COVID 19 |
| 5 | 19 | 0.08 | Electrochemical Biosensor |
| 6 | 16 | 0.06 | Assay |
| 7 | 14 | 0.05 | DNA |
| 8 | 13 | 0.08 | Virus |
| 9 | 13 | 0.07 | Electrochemical Detection |
| 10 | 12 | 0.17 | Gold Nanoparticle |
| 11 | 12 | 0.05 | Rapid Detection |
| 12 | 11 | 0.06 | Nanoparticle |
| 13 | 11 | 0.06 | Electrode |
| 14 | 11 | 0.02 | SARS CoV 2 |
| 15 | 11 | 0.02 | Coronavirus |
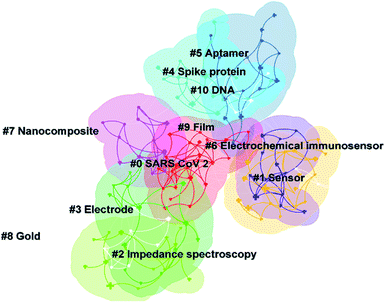
Cluster analysis can further understand the different directions of investigation in this topic. Fig. 4 shows that 11 clusters were formed after clustering the keywords. Some of these clusters overlap with each other, representing that these clusters share some similar content. Table 4 describes the clusters and their ID, size, silhouette, and respective keywords. The silhouette values of most of the clusters in Table 4 are below 0.9, representing that the clustering results are not particularly satisfactory. This may be because there is no particular uniform direction of investigation on this topic. Different works have tried to build electrochemical biosensors for SARS-CoV-2 detection with different sensing strategies. Therefore, different sensing strategies and detection targets in electrochemical biosensors are included in Table 4. By reading the specific literature, the strategies for SARS-CoV-2 detection using electrochemical biosensors at this stage can be summarized as follows:
| Cluster ID | Size | Silhouette | Keywords | Ref. |
|---|---|---|---|---|
| 0 | 23 | 0.818 | SARS CoV 2; platform; amplification | 31, 38, 62, 64–71 |
| 1 | 18 | 0.867 | Sensor; electrochemical biosensor; rapid detection; binding | 31, 64, 72–84 |
| 2 | 16 | 0.882 | Biosensor; Impedance spectroscopy; Point of care; Electrochemical impedance spectroscopy; Protein | 53, 76, 85–96 |
| 3 | 16 | 0.851 | Electrochemical; Detection; Electrode; Antibody; Immunosensor | 38, 77, 87, 97–105 |
| 4 | 16 | 0.868 | Electrochemical sensor; COVID 19; Spike protein; Performance; Nanosheet | 56, 72, 85, 106–117 |
| 5 | 15 | 0.918 | Virus; Coronavirus; Point; Aptamer; Discovery | 118–128 |
| 6 | 13 | 0.858 | Assay; Electrochemical immunosensor; Nucleocapsid protein; Surface | 63, 85, 90, 129–133 |
| 7 | 12 | 0.843 | Nanoparticle; Graphene; Nanocomposite; Oxide; | 55, 85, 86, 98, 106, 134–136 |
| 8 | 7 | 0.923 | Gold; Nanoparticle; Polymer; Impedimetric biosensor; Acid | 86, 89, 137, 138 |
| 9 | 6 | 0.838 | Film; Ion; Electrochemical sensing; Plasma; System; C-reactive protein | 99, 139 |
| 10 | 6 | 0.950 | DNA; Biomarker; Multiplexed detection; PCR; Electrochemical immunosensing | 85, 101, 140–143 |
SARS-CoV-2 genome consists of genes encoding structural proteins, including spike (S), envelope (E), membrane (M), and core–shell (N) proteins. Among them, the S-protein protrudes from the lipid bilayer surrounding SARS-COV-2. S-protein can be used as a biometric element for electrochemical immunosensor because SARS-COV-2 infection induces an immune response and releases specific antibodies. Liustrovaite et al.130 recombinant SARS-CoV-2 spike protein (SCoV2-rS) and used covalent immobilization on the surface of a pre-modified gold electrode. SCoV2-rS can interact affinely with SARS-COV-2 infection-induced anti-S-protein and alter the electrical signal. This biosensor can confirm COVID-19 infection or immune response in humans after vaccination. Karaman et al.112 chose nucleocapsid protein as the analyte. They modified the electrode surface with Bi2WO6/Bi2S3 to efficiently immobilize the capture antibody. Under optimal conditions, the detection limit of this biosensor can reach 3 fg mL−1. Organic electrochemical transistors (OECTs) are often used to detect nucleic acids, proteins, and metabolites. It has also been reported for the detection of SARS-CoV-2. Liu et al.144 prepared a simple OECT to detect SARS-CoV-2 IgG. SARS-CoV-2 S-protein was immobilized on the gate electrode by covalent binding, and SARS-CoV-2 IgG was bound to S-protein by antibody–antigen reaction during incubation leading to the OECT response. Field-effect transistors (FETs) can also be used in combination with immunoassays for S-protein detection. Cui et al.67 prepared FET using laser-induced graphene and immobilized specific antibodies. The FET can achieve detection limits of 1 pg mL−1 and 1 ng mL−1 in PBS and human serum, respectively.
Enzymatic electrochemistry is a sensing strategy based on the specific binding of enzymes and substrates. The receptor-binding domain of the S protein of SARS-CoV-2 can bind to the human angiotensin-converting enzyme 2 (ACE2) receptor and mediate coronavirus entry into host cells. Therefore, Nascimento et al.145 developed an ultrasensitive electrochemical biosensor of magnetic beads and gold nanoparticles bound to ACE2 peptide for highly sensitive detection of S protein (Fig. 5). Similar work has been reported by Liu et al.146 They proposed peptides bind to ACE2 in serum and then form a gel-like 2D protein network with serum proteins. The detection limit of this strategy can reach 1 pM.
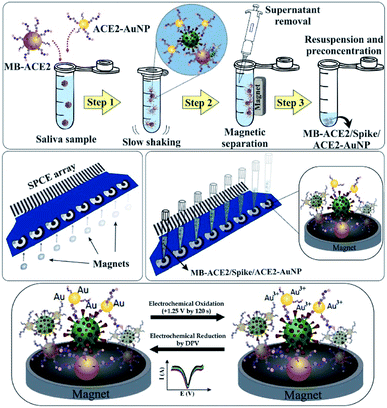 | ||
| Fig. 5 Scheme of MIP-based electrochemical biosensor for N-protein detection. Reprinted with permission.145 Copyright 2022, Elsevier. | ||
Molecular imprinting (MIP) is often used in electrochemical sensing to identify substances that are not significantly electrochemically active. It synthesizes MIP by mixing functional monomers, crosslinkers, and initiators with appropriate templating agents. After synthesis, removal of the template from the MIP leaves a template-specific binding site that allows the analyte to bind selectively. This detection strategy has been explored for various viral assays such as HIV-1,147 HIV-p24,148 Zika,149 and Dengue.150 For SARS-COV-2, detection with MIP was also attempted by Raziq et al.151 (Fig. 6A). SARS-CoV-2 N-protein and phenylenediamine were chosen as the template molecule and functional monomer, respectively. This MIP electrochemical biosensor can detect N-protein in the range of 2.22–111 fM. The detection limit can reach 15 fM. Sukjee et al.132 used the MIP strategy to detect viral particles, and the detection limit could reach 0.1 fM. Similar work has been reported by Sharif et al.90 for whole virus detection via MIP. Other functional monomers have been chosen to construct MIP electrochemical biosensors, such as S spike glycoprotein.152
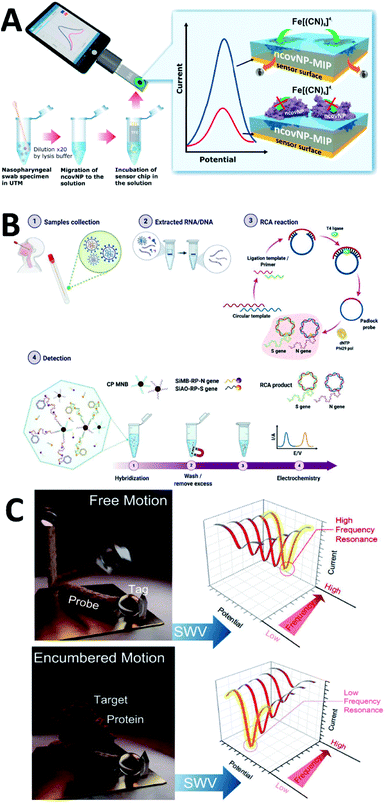 | ||
| Fig. 6 (A) Scheme of MIP-based electrochemical biosensor for N-protein detection. (B) Detection workflow of SARS-CoV-2 using the electrochemical biosensor with RCA of the N and S genes. (C) Using sampling frequency to“resonance” with the molecular motion for SARS-CoV-2 labelled protein detection. Reprinted with permission.151,157,162 Copyright 2022, Elsevier, Springer Nature, American Chemical Society. | ||
The electrochemical genosensor mainly detects hybridization reactions of nucleic acids. It consists of an electrode as a transducer, a single-stranded or gene probe as a sensitive element, and an electroactive indicator that recognizes the hybridization information. Tailoring single-stranded DNA for specific identification based on the reported genetic information of SARS-CoV-2 has been reported in many papers.58,96,153–156 Isothermal rolling circle amplification (RCA) can be used to assist in electrochemical genosensor. RCA amplicons can hybridize with probes that have redox signals for the identification of N and S genes in SARS-CoV-2 (ref. 157) (Fig. 6B). Gene sensing strategy can also be combined with immunosensing to detect different analytes simultaneously. Jiang et al.158 proposed a biochip for simultaneous detection of S-protein, N-protein, and anti-IgG by two strategies. FET can also assist genosensor in recognition of nucleic acids. Kong et al.159 constructed a graphene-based FET containing y-shaped DNA dual probes for highly sensitive recognition of ORF1ab and N genes of SARS-CoV-2 nucleic acids. In addition to conventional electrochemical signals, photoelectrochemical signals can also be used for detection. Tabrizi et al.160 assembled an aptamer-based photoelectrochemical biosensor to detect S-protein RBD. The photogenerated current was used as the detection signal and could be detected linearly in the range of 0.5–32.0 nM, with a detection limit of 0.12 nM. Information visualization and machine learning have been explored for data analysis of genosensor.161
Potential scanning can be used to sense intermolecular resonances. Zhou et al.162 used this methodology to capture the motion of SARS-CoV-2 labeled proteins during their interaction with peptide probes. After a single sampling, a sudden change in potential causes a current to be generated between the electroactive molecule and the electrode surface. The currents of molecules that move too fast are not sampled in real-time, but the current values of molecules that move too slowly cannot be captured before the data are sampled (Fig. 6C). By this property, they can identify the movement before and after covalent binding to SARS-CoV-2 tagged proteins with peptide probes.
Table 5 summarizes the published electrochemical biosensors for SARS-CoV-2 detection. The following information can be obtained by combining the above discussion:
| Methodology | Electrochemical technique | Analytes | LOD | Ref. |
|---|---|---|---|---|
| Electrochemical immunoassay | CV, EIS | Anti-S-protein | 2.53 nM (CV); 1.99 nM (EIS) | 130 |
| DPV | N-protein | 3 fg mL−1 | 112 | |
| SWV | IgG and IgM | 20 nM | 163 | |
| EIS | Anti-N-protein | 0.4 ng mL−1 | 89 | |
| SWV | Anti-S-protein | 0.01 ag/mL | 95 | |
| EIS | S-protein RBD | 150 ng mL−1 | 114 | |
| EIS | S-protein | 100.0 fg mL−1 | 164 | |
| DPV | S-protein | 25 pg mL−1 | 60 | |
| SWV | Anti-S-protein | 1 pg mL−1 | 165 | |
| EIS | Anti-S-protein | — | 166 | |
| EIS | Anti-S-IgG | 10 ng mL−1 | 167 | |
| SWV | Anti-S-protein | 9.3 ag/mL | 168 | |
| EIS | Anti-N-protein | 21 fg mL−1 | 85 | |
| EIS | Anti-N-protein | 13 fM | 169 | |
| EIS | Anti-S-protein, IgG | 0.25 fg mL−1 | 117 | |
| EIS | S-protein | 15 ng mL−1 | 92 | |
| EIS | S-protein | 0.04 fg mL−1 | 108 | |
| EIS | S-protein | 3.16 pM | 170 | |
| EIS | S-protein | 189 fM | 111 | |
| EIS | S-protein | 1 fg mL−1 | 59 | |
| EIS | S-protein | 0.54 ng mL−1 | 133 | |
| EIS | S-protein RBD | 0.577 pg mL−1 | 62 | |
| DPV | S-protein, N-protein | 19 ng mL−1, 8 ng mL−1 | 171 | |
| DPV | S-protein | 0.3 fg mL−1 | 172 | |
| DPV | S-protein | 0.1 ng mL−1 | 173 | |
| DPV | S-protein 1, S-protein 2 | 0.53 ng mL−1, 0.75 ng mL−1 | 174 | |
| SWV | N-protein | 0.4 pg mL−1 | 175 | |
| SWV | S-protein | 276 fM | 76 | |
| Chronoamperogram | N-protein | 50 PFU/Ml | 176 | |
| EIS | N-protein | 0.227 ng mL−1 | 177 | |
| EIS | N-protein | 0.48 fg mL−1 | 138 | |
| I-T | S-protein | — | 178 | |
| DPV | Anti-S-protein | 0.0072 ng mL−1 | 179 | |
| DPV | Anti-S-protein | 0.001 fg mL−1 | 180 | |
| Chronoamperogram | N-protein | 10 pg mL−1 | 181 | |
| DPV | S-protein | 0.73 fM | 182 | |
| SWV | Anti-S-protein | 0.8 pg mL−1 | 183 | |
| SWV | S-protein | 1 ag/mL | 184 | |
| SWV | Anti-S-protein | 0.03 fg mL−1 | 185 | |
| DPV | S-protein | 2.9 ng mL−1 | 186 | |
| SWV | S-protein | 20 μg mL−1 | 187 | |
| EIS | N-protein | 6 pg mL−1 | 129 | |
| MIP | DPV | N-protein | 15 fM | 151 |
| CV | Virus particle | 0.1 fM | 132 | |
| Pulsed amperometric detection | S-glycoprotein | — | 152 | |
| DPV | S-protein | 64 fM | 188 | |
| EIS | S-protein RBD | 0.7 pg mL−1 | 189 | |
| EIS | Virus particle | 57 pg mL−1 | 70 | |
| EIS | Virus particle | 4.8 pg mL−1 | 90 | |
| Electrochemical genosensor | DPV | RNA | 200 copy/mL | 57 |
| CV | RNA | 1 fM | 143 | |
| I-T | RNA | 807 fM | 153 | |
| I-V | RNA | 5.0 and 6.9 copy/μL | 154 | |
| SWV | S and Orf1ab genes | 6.8 ag/μL | 155 | |
| DPV | RNA | 26 fM | 58 | |
| DPV | RNA | 45 fM | 156 | |
| Capacitive measurement | RdRP gene | 0.843 nF nM−1 | 190 | |
| EIS | S-protein RBD | 7.0 pM | 191 | |
| CV, EIS | D-protein | 258.01 copies/μL | 96 | |
| DPV | RdRP gene, N gene | 0.972 fg μL−1, 3.925 fg μL−1 | 192 | |
| DPV | N gene | 15 nM | 127 | |
| EIS, DPV | RdRP gene | 0.186 μM | 54 | |
| DPV | RNA | 0.48 aM | 193 | |
| DPV | RdRP gene | 0.15 pM | 194 | |
| DPV | S-glycoprotein | 0.07 fM | 121 | |
| SWV | S-glycoprotein | 0.03 fg mL−1 | 124 | |
| DPV | RNA | 22.2 fM | 195 | |
| Chronoamperogram | RNA | 1 pM | 196 | |
| DPV | MicroRNA of N-gene | — | 118 | |
| DPV | ORF, S genes | 0.044 fg mL−1, 0.081 fg mL−1 | 197 | |
| DPV | ORF1ab | 100 fg mL−1 | 198 | |
| EIS | S-protein | 1000 copies per mL | 72 | |
| EIS | D-protein | 0.389 fM | 199 | |
| EIS | S-protein | 1.30 pM | 200 | |
| DPV | N-protein | 1.7 fM | 65 | |
| DPV | N-protein | 16.5 pg mL−1 | 201 | |
| DPV | N-protein | 8.33 pg mL−1 | 202 | |
| SWV | S-protein | — | 203 | |
| DPV | N and S genes target sequences | 1 copy per μL | 157 | |
| EIS | N-protein | 0.16 ng mL−1 | 204 | |
| Enzymatic electrochemical sensor | DPV | S-protein | 0.35 ag mL−1 | 145 |
| DPV | ACE2 | 1 pM | 146 | |
| SWV | ACE2 | 229 fg mL−1 | 205 | |
| EIS | S-protein | 0.1 μg mL−1 | 88 | |
| EIS | S-protein | 299.30 ng mL−1 | 206 |
(1) Electrochemical immunosensors and electrochemical genosensor (aptamer biosensors) are the most investigated methodologies. Several other technologies are also used to assist in efficiently operating these biosensors.
(2) S-protein and N-protein are the most commonly used fractions for recognizing SARS-CoV-2. They can be specifically recognized by antigen–antibody binding.
(3) By customizing the sequence of ssDNA, the genosensor can recognize the RNA of SARS-CoV-2. Specific sequence segments were also used for efficient identification.
(4) Enzyme-based electrochemical biosensors rely mainly on the specific reaction between S-protein and ACE2.
(5) MIP can recognize not only specific proteins but also whole viral particles.
(6) Regardless of which fraction of SARS-CoV-2 is used as the analyte, they do not possess their electrochemical activity. Therefore, voltammetry-based detection requires the inclusion of a probe in the biosensor's design. Another approach is to determine the change of the electrode surface by EIS.
(7) Due to methodological differences, different works are difficult to compare directly in terms of assay performance. This difficulty is simultaneously amplified by the different concentration units used for the papers.
Key publications analysis
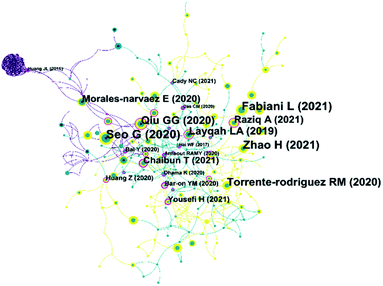
Through reference analysis, bibliometrics can analyse papers that have significantly impacted a topic. Fig. 7 shows the reference network diagram of literature on electrochemical biosensors in SARS-CoV-2 detection. As seen in the figure, some of these important co-cited papers are earlier than 2020. These papers are not related to COVID-19, but their contents have significantly impacted the development of these electrochemical biosensors in SARS-CoV-2 detection. For example, the paper published by Hai et al.207 in 2017 is at the centre of the network. This work achieved label-free detection of human influenza virus by quartz crystal microbalance and potentiometric detection method. This methodology provides ideas for subsequent virus detection. On the far left side of the co-citation network, an electrochemical detection technique for avian influenza virus H7 (AIV H7) was proposed by Huang et al.208 in 2016. Biosensors modified with graphene and silver nanoparticles were used to efficiently immobilize antibodies and subsequently for specific recognition of AIV H7. The methodology of this electrochemical biosensor is very similar to most electrochemical immunoassays mentioned in Table 4. Another similar electrochemical immunosensor was published by Layqah and Eissa209 in 2019. The biosensor proposed in this work is for detecting Middle East respiratory syndrome coronavirus (MERS-CoV). The S-protein of MERS-CoV is the analyte used for quantitative analysis. Although the coronavirus species are different, the detection strategy and the target of detection in it have been very similar to the most commonly used strategies in the electrochemical biosensors in SARS-CoV-2 detection.Some of these papers have high citation rates, representing that they establish excellent sensing strategies or are often used as comparisons. The FET-based biosensing device reported by Seo et al.210 allows highly sensitive detection of the S-protein of SARS-CoV-2. Fabiani et al.171 used magnetic beads to support the immune chain in preparing the biosensor. This biosensor can be used to detect S-protein and N-protein in SARS-CoV-2. This work was cited 41 times in all the articles analysed. Qiu et al.211 developed a genosensor by combining the ionophore photothermal effect with localized surface plasmon resonance. This work was cited 33 times in all the articles analysed. Electrochemical biosensors also play an important role in this network. The electrochemical immunosensor prepared by Zhao et al.57 was widely recognized and was cited 38 times in these papers. The MIP biosensor proposed by Raziq et al.151 has also received attention and has been cited 23 times. The electrochemical biosensor (Fig. 6B) constructed by Chaibun et al.157 mentioned in the previous section has also received a series of citations.
Other biosensors with various technologies are often applied for comparison in papers on this topic, such as plasmonic biosensor,212 fluorescent plasmonics biosensor,213,214 and CRISPR-based assay.215,216 In addition, some reviews and perspectives on COVID217–221 are often cited in papers on this topic.
Product analysis
In addition to the establishment and innovation of the methodology, some of these works have gone a step further by proposing ideas for products and even producing model products. The following is a brief description and summary of the products that have been proposed.COVID-19 ePAD222 (Fig. 8A): It can be printed by a wax printer and consists of 3 folded layers. The wax barrier is used to control the reaction of the liquid sample in the test area. The sample can be encapsulated by folding after sample collection, ensuring that the sample is not contaminated before testing. The ePAD works by immobilizing the spike protein receptor-binding domain (SP RBD) of SARS-CoV-2 on a working electrode and is intended to detect SARS-CoV-2 antibodies.
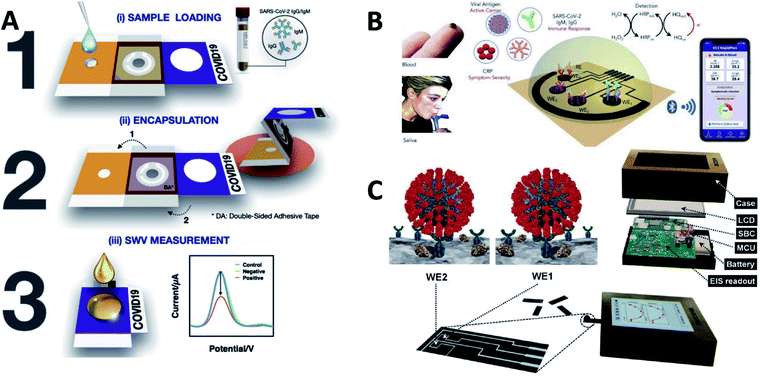 | ||
| Fig. 8 (A) Scheme of detection principle of the COVID-19 ePAD. (B) Scheme of SARS-CoV-2 RapidPlex for rapid and multiplex electrochemical detection of SARS-CoV-2 in blood and saliva. (C) Scheme of ECDAQ platform. Reprinted with permission.140,222,223 Copyright 2022, Elsevier. | ||
SARS-CoV-2 RapidPlex223 (Fig. 8B): Four laser engraved graphene electrodes are used as working electrodes to detect viral antigenic nucleocapsid proteins, IgM antibodies, IgG antibodies, and C-reactive proteins. This biosensor can detect blood and saliva. The cost of each biosensor and reagent can be kept below $1.
SPEEDS:224 This is a portable, low-cost electrochemical immunosensor. It allows for the rapid, quantitative determination of IgG and IgM antibodies against SARS-CoV-2 S-protein in human serum. This biosensor also comes with a device stabilization and storage strategy that allows for stable performance at room temperature for up to 24 weeks.
RAPID:225 Real-time Accurate Portable Impedance Detection Prototype 1.0 (RAPID) is based on EIS. The product was tested on 321 anterior nasal swab samples with sensitivity, specificity, and accuracy of 80.6%, 89.0%, and 88.2%, respectively.
MARK-B:226 This is a magnetically assisted electrochemical immunosensor. It was tested on 170 nasopharyngeal swab specimens with an overall sensitivity and specificity of 90.0% and 99.0%, respectively. The semi-quantitative determination of the results can be achieved in conjunction with a fully automated portable device.
SenSARS:91 This is also a portable biosensor that detects S-protein with EIS. The cost per test is about $5, and the test time is less than 10 minutes. The paper on SenSARS is different from other papers in that the authors provide its hardware architecture, power supply circuit, signal generation module, and signal conditioning module.
Bi-ECDAQ140 (Fig. 8C): It is a biosensor with a potentiostat and contains a two-electrode design. The EIS response was used to determine the specific binding of S-protein to the antigen on the electrode surface. Embedded microcontrollers and single-board computers will process the measured data and display it to the user through an LCD.
Conclusion and future perspectives
COVID-19 has changed our lives while also having a profound impact on academic research. How to rapidly detect SARS-CoV-2 is a very important topic in the COVID-19 pandemic. In this bibliometric-based review, the development history and the current status of electrochemical biosensors in SARS-CoV-2 detection are summarized and expressed. We present a statistical analysis and data visualization of this topic through contributions from different countries, collaborative relationships between different institutions, keyword analysis/clustering and citation analysis. Based on the results of the analysis, the following tentative conclusions can be drawn:(1) Electrochemical biosensors are a technology with applications worthy of being explored, as many of these methodologies can genuinely meet the needs of field detection. This challenge has been accomplished in some existing initial products.
(2) Electrochemical immunosensors and genosensor are the most investigated methodologies at this stage. The most commonly used detection targets are the S-protein and N-protein of SARS-CoV-2.
(3) The rapid achievement of electrochemical detection for SARS-CoV-2 has been made possible by the methodology established by developing the field of electrochemical sensing over the years. Although SARS-CoV-2 is a new virus, the techniques used in the detection have been successfully validated in other viruses, such as influenza virus and MERS-CoV.
(4) China and USA are the countries contributing the highest number of papers on this topic but do not present an absolute dominance. In general, the participants in this topic do not present local geographical limitations because the COVID-19 pandemic attracts worldwide attention.
(5) This topic has only been developing for less than 3 years but has already formed three initial international cooperation networks. This rapid international cooperation has benefited from the global impact caused by this public health event. At the same time, political intervention should have played an important role in this cooperation.
(6) Some initial electrochemical biosensors have been assembled, and efficient field detection has been achieved. However, it is undeniable that electrochemical biosensors are still not particularly competitive in terms of cost. The lowest cost price (electrode + reagent) reported is $1.
On the other hand, based on the clues given by bibliometrics and the changes in national policies for the COVID-19 pandemic, we can give the following perspectives:
(1) The sensitivity and detection limits of electrochemical biosensors are already sufficient for confirming COVID-19, so further developing more sensitive electrochemical biosensors is not the most important challenge. Instead, reducing the cost of detection is a significant challenge for this detection technology to be practically applied.
(2) Ensuring the stability of electrochemical biosensors is also a significant challenge in practical applications. Since this technology is designed for field detection, how to avoid the deactivation of biological reagents and interference by environmental factors is a direction that deserves further exploration.
(3) Variants of SARS-CoV-2 have been an important factor contributing to the spread of COVID-19. The direct determination of SARS-CoV-2 variants by electrochemistry is a direction worth exploring.
(4) Based on the data for the first 6 months of 2022, the number of papers published on this topic this year should not be less than the annual publication last year. However, shifts in attitudes toward COVID-19 in different countries can significantly affect scholars' enthusiasm for the topic. The commercialization of electrochemical biosensors must be supported by very large and sustained market behaviour. It is doubtful whether this topic has any chance to continue to attract researchers. Therefore, whether electrochemical biosensors can have a broad market does not possess a bright future.
(5) COVID-19 pandemic is a special event. A retrospective analysis of the development of different academic fields and topics through this time is worth discussing.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author would like to acknowledge Figdraw for providing the drawing materials and platform for Graphic Abstract design.References
- Y. Wu, W. Ho, Y. Huang, D.-Y. Jin, S. Li, S.-L. Liu, X. Liu, J. Qiu, Y. Sang and Q. Wang, Lancet, 2020, 395, 949–950 CrossRef CAS.
- M.-Y. Wang, R. Zhao, L.-J. Gao, X.-F. Gao, D.-P. Wang and J.-M. Cao, Front. Cell. Infect. Microbiol., 2020, 10, 587269 CrossRef CAS PubMed.
- S. Cao, Y. Gan, C. Wang, M. Bachmann, S. Wei, J. Gong, Y. Huang, T. Wang, L. Li, K. Lu, H. Jiang, Y. Gong, H. Xu, X. Shen, Q. Tian, C. Lv, F. Song, X. Yin and Z. Lu, Nat. Commun., 2020, 11, 5917 CrossRef CAS PubMed.
- G. C. Mak, P. K. Cheng, S. S. Lau, K. K. Wong, C. Lau, E. T. Lam, R. C. Chan and D. N. Tsang, J. Clin. Virol., 2020, 129, 104500 CrossRef CAS PubMed.
- M. K. Özçürümez, A. Ambrosch, O. Frey, V. Haselmann, S. Holdenrieder, M. Kiehntopf, M. Neumaier, M. Walter, F. Wenzel, R. Wölfel and H. Renz, J. Allergy Clin. Immunol., 2020, 146, 35–43 CrossRef.
- H. Yousefi, A. Mahmud, D. Chang, J. Das, S. Gomis, J. B. Chen, H. Wang, T. Been, L. Yip, E. Coomes, Z. Li, S. Mubareka, A. McGeer, N. Christie, S. Gray-Owen, A. Cochrane, J. M. Rini, E. H. Sargent and S. O. Kelley, J. Am. Chem. Soc., 2021, 143, 1722–1727 CrossRef CAS.
- N. N. Y. Tsang, H. C. So, K. Y. Ng, B. J. Cowling, G. M. Leung and D. K. M. Ip, Lancet Infect. Dis., 2021, 21, 1233–1245 CrossRef CAS PubMed.
- G. Das, S. Ghosh, S. Garg, S. Ghosh, A. Jana, R. Samat, N. Mukherjee, R. Roy and S. Ghosh, RSC Adv., 2020, 10, 28243–28266 RSC.
- R. Chakravarti, R. Singh, A. Ghosh, D. Dey, P. Sharma, R. Velayutham, S. Roy and D. Ghosh, RSC Adv., 2021, 11, 16711–16735 RSC.
- U. Amara, S. Rashid, K. Mahmood, M. H. Nawaz, A. Hayat and M. Hassan, RSC Adv., 2022, 12, 8059–8094 RSC.
- J. A. Goode, J. V. H. Rushworth and P. A. Millner, Langmuir, 2015, 31, 6267–6276 CrossRef CAS PubMed.
- K. Ramanathan and B. Danielsson, Biosens. Bioelectron., 2001, 16, 417–423 CrossRef CAS.
- C.-A. Vu and W.-Y. Chen, Sensors, 2019, 19, 4214 CrossRef CAS.
- P. Skládal, Past, Present and Future Challenges of Biosensors and Bioanalytical Tools in Analytical Chemistry: A Tribute to Professor Marco Mascini, 2016, vol. 79, pp. 127–133 Search PubMed.
- N. J. Ronkainen, H. B. Halsall and W. R. Heineman, Chem. Soc. Rev., 2010, 39, 1747–1763 RSC.
- S. M. Borisov and O. S. Wolfbeis, Chem. Rev., 2008, 108, 423–461 CrossRef CAS PubMed.
- M.-I. Rocha-Gaso, C. March-Iborra, Á. Montoya-Baides and A. Arnau-Vives, Sensors, 2009, 9, 5740–5769 CrossRef CAS.
- J. Ding and W. Qin, TrAC, Trends Anal. Chem., 2020, 124, 115803 CrossRef CAS.
- M. Nasir, D. A. Ateya, D. Burk, J. P. Golden and F. S. Ligler, Biosens. Bioelectron., 2010, 25, 1363–1369 CrossRef CAS PubMed.
- M. Trojanowicz, Electrochem. Commun., 2014, 38, 47–52 CrossRef CAS.
- C. Berggren, B. Bjarnason and G. Johansson, Electroanalysis, 2001, 13, 173–180 CrossRef CAS.
- P. Das, M. Das, S. R. Chinnadayyala, I. M. Singha and P. Goswami, Biosens. Bioelectron., 2016, 79, 386–397 CrossRef CAS.
- X. Xu and Y. Ying, Food Rev. Int., 2011, 27, 300–329 CrossRef.
- F. Mollarasouli, S. Kurbanoglu and S. A. Ozkan, Biosensors, 2019, 9, 86 CrossRef CAS PubMed.
- D. Zhang, Y. Yan, H. Que, T. Yang, X. Cheng, S. Ding, X. Zhang and W. Cheng, ACS Sens., 2020, 5, 557–562 CrossRef CAS.
- S. Hassanpour, B. Baradaran, M. de la Guardia, A. Baghbanzadeh, J. Mosafer, M. Hejazi, A. Mokhtarzadeh and M. Hasanzadeh, Microchim. Acta, 2018, 185, 568 CrossRef.
- Y. Tepeli and A. Ülkü, Sens. Actuators, B, 2018, 254, 377–384 CrossRef CAS.
- E. Rasouli, Z. Shahnavaz, W. J. Basirun, M. Rezayi, A. Avan, M. Ghayour-Mobarhan, R. Khandanlou and M. R. Johan, Anal. Biochem., 2018, 556, 136–144 CrossRef CAS PubMed.
- A. Kaushik, S. Tiwari, R. D. Jayant, A. Vashist, R. Nikkhah-Moshaie, N. El-Hage and M. Nair, Trends Biotechnol., 2017, 35, 308–317 CrossRef CAS.
- Z. Zhao, C. Huang, Z. Huang, F. Lin, Q. He, D. Tao, N. Jaffrezic-Renault and Z. Guo, TrAC, Trends Anal. Chem., 2021, 139, 116253 CrossRef CAS PubMed.
- K. Y. Goud, K. K. Reddy, A. Khorshed, V. S. Kumar, R. K. Mishra, M. Oraby, A. H. Ibrahim, H. Kim and K. V. Gobi, Biosens. Bioelectron., 2021, 180, 113112 CrossRef CAS PubMed.
- D. Lu, D. Z. Zhu, H. Gan, Z. Yao, Q. Fu and X. (Jackie) Zhang, Sci. Total Environ., 2021, 777, 146239 CrossRef CAS.
- S. Madhurantakam, S. Muthukumar and S. Prasad, ACS Omega, 2022, 7, 12467–12473 CrossRef CAS.
- B. E. García-Pérez, J. A. González-Rojas, M. I. Salazar, C. Torres-Torres and N. S. Castrejón-Jiménez, Cells, 2020, 9, 2679 CrossRef.
- B. Amen, S. Faiz and T.-T. Do, Pattern Recognit., 2022, 123, 108404 CrossRef PubMed.
- S. Kotru, M. Klimuntowski, H. Ridha, Z. Uddin, A. A. Askhar, G. Singh and M. M. R. Howlader, TrAC, Trends Anal. Chem., 2021, 136, 116198 CrossRef CAS.
- J. Kudr, P. Michalek, L. Ilieva, V. Adam and O. Zitka, TrAC, Trends Anal. Chem., 2021, 136, 116192 CrossRef CAS.
- S. S. Mahshid, S. E. Flynn and S. Mahshid, Biosens. Bioelectron., 2021, 176, 112905 CrossRef CAS.
- Y. Zheng, H. Karimi-Maleh and L. Fu, Chemosensors, 2022, 10, 194 CrossRef CAS.
- Y. Zheng, S. Mao, J. Zhu, L. Fu, N. Zare and F. Karimi, Food Chem. Toxicol., 2022, 164, 113019 CrossRef CAS.
- Y. Shen, S. Mao, F. Chen, S. Zhao, W. Su, L. Fu, N. Zare and F. Karimi, Food Chem. Toxicol., 2022, 163, 112960 CrossRef CAS PubMed.
- Y. Zheng, H. Karimi-Maleh and L. Fu, Sensors, 2022, 22, 3238 CrossRef CAS.
- L. Fu, S. Mao, F. Chen, S. Zhao, W. Su, G. Lai, A. Yu and C.-T. Lin, Chemosphere, 2022, 297, 134127 CrossRef CAS PubMed.
- M. Jin, J. Liu, W. Wu, Q. Zhou, L. Fu, N. Zare, F. Karimi, J. Yu and C.-T. Lin, Chemosphere, 2022, 300, 134599 CrossRef CAS PubMed.
- S. Xiang, S. Mao, F. Chen, S. Zhao, W. Su, L. Fu, N. Zare and F. Karimi, Chemosphere, 2022, 306, 135517 CrossRef CAS PubMed.
- Y. Pan, C. Yin, C. Fernandez, L. Fu and C.-T. Lin, Polymers, 2022, 14, 3011 CrossRef PubMed.
- X. Li, Y. Zheng, W. Wu, M. Jin, Q. Zhou, L. Fu, N. Zare, F. Karimi and M. Moghadam, Chemosphere, 2022, 307, 135720 CrossRef CAS PubMed.
- K. Börner, C. Chen and K. W. Boyack, Annu. Rev. Inf. Sci. Technol., 2003, 37, 179–255 CrossRef.
- C. Chen, J. Am. Soc. Inf. Sci. Technol., 2006, 57, 359–377 CrossRef.
- C. Chen, Proc. Natl. Acad. Sci., 2004, 101, 5303–5310 CrossRef CAS.
- C. Chen, F. Ibekwe-SanJuan and J. Hou, J. Am. Soc. Inf. Sci. Technol., 2010, 61, 1386–1409 CrossRef.
- D. Xueshu and J. Wenxian, COOC is a software for bibliometrics and knowledge mapping[CP/OL], (accessed February 15, 2022), https://github.com/2088904822 Search PubMed.
- C. C. Pola, S. V. Rangnekar, R. Sheets, B. M. Szydłowska, J. R. Downing, K. W. Parate, S. G. Wallace, D. Tsai, M. C. Hersam, C. L. Gomes and J. C. Claussen, 2D Mater., 2022, 9, 035016 CrossRef CAS PubMed.
- W. L. Ang, R. R. X. Lim, A. Ambrosi and A. Bonanni, FlatChem, 2022, 32, 100336 CrossRef CAS.
- J. Cui, L. Kan, F. Cheng, J. Liu, L. He, Y. Xue, S. Fang and Z. Zhang, Dalton Trans., 2022, 51, 2094–2104 RSC.
- R. G. Ramírez-Chavarría, E. Castillo-Villanueva, B. E. Alvarez-Serna, J. Carrillo-Reyes, R. M. Ramírez-Zamora, G. Buitrón and L. Alvarez-Icaza, J. Environ. Chem. Eng., 2022, 10, 107488 CrossRef.
- H. Zhao, F. Liu, W. Xie, T.-C. Zhou, J. OuYang, L. Jin, H. Li, C.-Y. Zhao, L. Zhang, J. Wei, Y.-P. Zhang and C.-P. Li, Sens. Actuators, B, 2021, 327, 128899 CrossRef CAS PubMed.
- Y. Peng, Y. Pan, Z. Sun, J. Li, Y. Yi, J. Yang and G. Li, Biosens. Bioelectron., 2021, 186, 113309 CrossRef CAS PubMed.
- S. Witt, A. Rogien, D. Werner, J. Siegenthaler, R. Lesiyon, N. Kurien, R. Rechenberg, N. Baule, A. Hardy and M. Becker, Diamond Relat. Mater., 2021, 118, 108542 CrossRef CAS.
- M. Adeel, K. Asif, V. Canzonieri, H. R. Barai, Md. M. Rahman, S. Daniele and F. Rizzolio, Sens. Actuators, B, 2022, 359, 131591 CrossRef CAS.
- D. Butler and A. Ebrahimi, Biosens. Bioelectron., 2022, 208, 114198 CrossRef CAS.
- E. B. Aydın, M. Aydın and M. K. Sezgintürk, Sens. Actuators, B, 2021, 345, 130355 CrossRef.
- Y. Feng, G. Liu, M. La and L. Liu, Molecules, 2022, 27, 615 CrossRef CAS.
- S. Ahuja, M. S. Kumar, R. Nandeshwar, K. Kondabagil and S. Tallur, Sci. Rep., 2022, 12, 8814 CrossRef CAS PubMed.
- M. S. Kumar, R. Nandeshwar, S. B. Lad, K. Megha, M. Mangat, A. Butterworth, C. W. Knapp, M. Knapp, P. A. Hoskisson, D. K. Corrigan, A. C. Ward, K. Kondabagil and S. Tallur, Sens. Actuators, B, 2021, 343, 130169 CrossRef CAS.
- A. V. Ramya, R. Thomas and M. Balachandran, Ultrason. Sonochem., 2021, 79, 105767 CrossRef CAS.
- T.-R. Cui, Y.-C. Qiao, J.-W. Gao, C.-H. Wang, Y. Zhang, L. Han, Y. Yang and T.-L. Ren, Molecules, 2021, 26, 6947 CrossRef CAS PubMed.
- L. Fiore, V. Mazzaracchio, P. Galloni, F. Sabuzi, S. Pezzola, G. Matteucci, D. Moscone and F. Arduini, Microchem. J., 2021, 166, 106249 CrossRef CAS.
- H. Xi, H. Jiang, M. Juhas and Y. Zhang, ACS Omega, 2021, 6, 25846–25859 CrossRef CAS.
- H. A. Hussein, A. Kandeil, M. Gomaa, R. Mohamed El Nashar, I. M. El-Sherbiny and R. Y. A. Hassan, ACS Sens., 2021, 6, 4098–4107 CrossRef CAS.
- J. Muñoz and M. Pumera, Chem. Eng. J., 2021, 425, 131433 CrossRef.
- Z. Zhang, R. Pandey, J. Li, J. Gu, D. White, H. D. Stacey, J. C. Ang, C.-J. Steinberg, A. Capretta, C. D. M. Filipe, K. Mossman, C. Balion, M. S. Miller, B. J. Salena, D. Yamamura, L. Soleymani, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2021, 60, 24266–24274 CrossRef CAS PubMed.
- M. Z. H. Khan, M. R. Hasan, S. I. Hossain, M. S. Ahommed and M. Daizy, Biosens. Bioelectron., 2020, 166, 112431 CrossRef CAS.
- G. Martins, J. L. Gogola, L. H. Budni, B. C. Janegitz, L. H. Marcolino-Junior and M. F. Bergamini, Anal. Chim. Acta, 2021, 1147, 30–37 CrossRef CAS.
- E. Vermisoglou, D. Panáček, K. Jayaramulu, M. Pykal, I. Frébort, M. Kolář, M. Hajdúch, R. Zbořil and M. Otyepka, Biosens. Bioelectron., 2020, 166, 112436 CrossRef CAS PubMed.
- W. A. El-Said, A. S. Al-Bogami, W. Alshitari, D. A. El-Hady, T. S. Saleh, M. A. El-Mokhtar and J.-W. Choi, BioChip J., 2021, 15, 287–295 CrossRef CAS PubMed.
- R. Antiochia, Biosens. Bioelectron., 2021, 173, 112777 CrossRef CAS PubMed.
- H. Xi, M. Juhas and Y. Zhang, Biosens. Bioelectron., 2020, 167, 112494 CrossRef CAS.
- Z. Jiang, B. Feng, J. Xu, T. Qing, P. Zhang and Z. Qing, Biosens. Bioelectron., 2020, 166, 112471 CrossRef CAS.
- A. K. Yadav, D. Verma, A. Kumar, P. Kumar and P. R. Solanki, Mater. Today Chem., 2021, 20, 100443 CrossRef CAS.
- L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Biosens. Bioelectron., 2021, 186, 113279 CrossRef CAS.
- M. Abubakar Sadique, S. Yadav, P. Ranjan, M. A. Khan, A. Kumar and R. Khan, Mater. Lett., 2021, 305, 130824 CrossRef CAS.
- T. S. K. Sharma and K.-Y. Hwa, Inorg. Chem., 2021, 60, 6585–6599 CrossRef CAS PubMed.
- A. Galal, Y. M. Ahmed, M. H. M. Ahmed and N. F. Atta, J. Electrochem. Soc., 2021, 168, 116512 CrossRef CAS.
- F. Haghayegh, R. Salahandish, M. Hassani and A. Sanati-Nezhad, ACS Appl. Mater. Interfaces, 2022, 14, 10844–10855 CrossRef CAS PubMed.
- A. L. Lorenzen, A. M. dos Santos, L. P. dos Santos, L. da Silva Pinto, F. R. Conceição and F. Wolfart, Electrochim. Acta, 2022, 404, 139757 CrossRef CAS PubMed.
- C. Y. N. Nicoliche, A. M. Pascon, Í. R. S. Bezerra, A. C. H. de Castro, G. R. Martos, J. Bettini, W. A. Alves, M. Santhiago and R. S. Lima, ACS Appl. Mater. Interfaces, 2022, 14, 2522–2533 CrossRef CAS.
- L.-V. Kiew, C.-Y. Chang, S.-Y. Huang, P.-W. Wang, C.-H. Heh, C.-T. Liu, C.-H. Cheng, Y.-X. Lu, Y.-C. Chen, Y.-X. Huang, S.-Y. Chang, H.-Y. Tsai, Y.-A. Kung, P.-N. Huang, M.-H. Hsu, B.-F. Leo, Y.-Y. Foo, C.-H. Su, K.-C. Hsu, P.-H. Huang, C.-J. Ng, A. Kamarulzaman, C.-J. Yuan, D.-B. Shieh, S.-R. Shih, L.-Y. Chung and C.-C. Chang, Biosens. Bioelectron., 2021, 183, 113213 CrossRef CAS PubMed.
- B. M. Hryniewicz, J. Volpe, L. Bach-Toledo, K. C. Kurpel, A. E. Deller, A. L. Soares, J. M. Nardin, L. F. Marchesi, F. F. Simas, C. C. Oliveira, L. Huergo, D. E. P. Souto and M. Vidotti, Mater. Today Chem., 2022, 24, 100817 CrossRef CAS.
- H. F. EL Sharif, S. R. Dennison, M. Tully, S. Crossley, W. Mwangi, D. Bailey, S. P. Graham and S. M. Reddy, Anal. Chim. Acta, 2022, 1206, 339777 CrossRef CAS PubMed.
- S. A. Perdomo, V. Ortega, A. Jaramillo-Botero, N. Mancilla, J. H. Mosquera-DeLaCruz, D. P. Valencia, M. Quimbaya, J. D. Contreras, G. E. Velez, O. A. Loaiza, A. Gómez and J. de la Roche, IEEE Trans. Instrum. Meas., 2021, 70, 1–10 Search PubMed.
- I. Ashur, J. Alter, M. Werbner, A. Ogungbile, M. Dessau, M. Gal-Tanamy and S. Vernick, Talanta, 2022, 239, 123147 CrossRef CAS PubMed.
- T. Nakayama and R. Honda, ACS Omega, 2021, 6, 21730–21740 CrossRef CAS PubMed.
- Z. Song, Y. Ma, M. Chen, A. Ambrosi, C. Ding and X. Luo, Anal. Chem., 2021, 93, 5963–5971 CrossRef CAS.
- L. Liv, Microchem. J., 2021, 168, 106445 CrossRef CAS PubMed.
- K. Y. P. S. Avelino, G. S. dos Santos, I. A. M. Frías, A. G. Silva-Junior, M. C. Pereira, M. G. R. Pitta, B. C. de Araújo, A. Errachid, M. D. L. Oliveira and C. A. S. Andrade, J. Pharm. Biomed. Anal., 2021, 206, 114392 CrossRef CAS PubMed.
- R. Rahimi and M. Solimannejad, J. Mol. Liq., 2022, 354, 118855 CrossRef CAS.
- C. Varodi, M. Coros, F. Pogăcean, A. Ciorîţă, A. Turza and S. Pruneanu, Chemosensors, 2022, 10, 47 CrossRef CAS.
- A. Ganguly and K.-Y. Hwa, Mater. Today Chem., 2022, 24, 100862 CrossRef CAS.
- X. Mi, H. Li, R. Tan, B. Feng and Y. Tu, Biosens. Bioelectron., 2021, 192, 113482 CrossRef CAS PubMed.
- D. McGonagle, G. De Marco and C. Bridgewood, J. Autoimmun., 2021, 121, 102662 CrossRef CAS PubMed.
- H. Ibrahim, A. M. Hamdy, H. A. Merey and A. S. Saad, J. AOAC Int., 2021, 104, 975–982 CrossRef.
- I. Ciglenečki, P. Orlović-Leko, K. Vidović and V. Tasić, Environ. Res., 2021, 198, 111215 CrossRef.
- M. Garg, A. L. Sharma and S. Singh, Biosens. Bioelectron., 2021, 171, 112703 CrossRef CAS PubMed.
- A. Pradhan, P. Lahare, P. Sinha, N. Singh, B. Gupta, K. Kuca, K. K. Ghosh and O. Krejcar, Sensors, 2021, 21, 7823 CrossRef CAS.
- A. K. Mohiuddin and S. Jeon, Appl. Surf. Sci., 2022, 592, 153162 CrossRef CAS PubMed.
- Ç. Kanbeş Dindar, B. Bozal-Palabiyik and B. Uslu, Electroanalysis, 2022, 34(7), 1174–1186 CrossRef.
- Z. Rahmati, M. Roushani, H. Hosseini and H. Choobin, Microchim. Acta, 2021, 188, 105 CrossRef CAS PubMed.
- M. A. Mohamed, G. M. G. Eldin, S. M. Ismail, N. Zine, A. Elaissari, N. Jaffrezic-Renault and A. Errachid, J. Electroanal. Chem., 2021, 895, 115422 CrossRef CAS.
- B. Koçak and Y. İpek, Electrocatalysis, 2022, 13, 108–115 CrossRef PubMed.
- S. Ramanathan, S. C. B. Gopinath, Z. Hilmi Ismail and S. Subramaniam, Microchim. Acta, 2022, 189, 226 CrossRef CAS PubMed.
- C. Karaman, B. B. Yola, O. Karaman, N. Atar, İ. Polat and M. L. Yola, Microchim. Acta, 2021, 188, 425 CrossRef CAS.
- Z. S. Miripour, R. Sarrami-Forooshani, H. Sanati, J. Makarem, M. S. Taheri, F. Shojaeian, A. H. Eskafi, F. Abbasvandi, N. Namdar, H. Ghafari, P. Aghaee, A. Zandi, M. Faramarzpour, M. Hoseinyazdi, M. Tayebi and M. Abdolahad, Biosens. Bioelectron., 2020, 165, 112435 CrossRef CAS PubMed.
- G. C. Zaccariotto, M. K. Silva, G. S. Rocha and I. Cesarino, Materials, 2021, 14, 4230 CrossRef CAS.
- M. Mehmandoust, Y. Khoshnavaz, M. Tuzen and N. Erk, Microchim. Acta, 2021, 188, 434 CrossRef CAS PubMed.
- G. Veerapandi, S. Meenakshi, S. Anitta, C. Arul, P. Ashokkumar and C. Sekar, Food Chem., 2022, 382, 132251 CrossRef CAS PubMed.
- M. A. Ehsan, S. A. Khan and A. Rehman, Diagnostics, 2021, 11, 1030 CrossRef CAS PubMed.
- F. S. Halim, N. A. Parmin, U. Hashim, S. C. B. Gopinath, F. A. Dahalan, I. I. Zakaria, W. C. Ang and N. F. Jaapar, Biotechnol. Appl. Biochem., 2022 DOI:10.1002/bab.2239.
- J. A. Zakashansky, A. H. Imamura, D. F. Salgado, H. C. Romero Mercieca, R. F. L. Aguas, A. M. Lao, J. Pariser, N. Arroyo-Currás and M. Khine, Anal. Methods, 2021, 13, 874–883 RSC.
- W. A. Ameku, D. W. Provance, C. M. Morel and S. G. De-Simone, Biosensors, 2022, 12, 272 CrossRef CAS PubMed.
- M. A. Martínez-Roque, P. A. Franco-Urquijo, V. M. García-Velásquez, M. Choukeife, G. Mayer, S. R. Molina-Ramírez, G. Figueroa-Miranda, D. Mayer and L. M. Alvarez-Salas, Anal. Biochem., 2022, 645, 114633 CrossRef.
- A. Ramanujam, S. Almodovar and G. G. Botte, Processes, 2021, 9, 1236 CrossRef CAS.
- H. A. Barton, Int. J. Speleol., 2020, 49, 6 Search PubMed.
- Z. Rahmati, M. Roushani, H. Hosseini and H. Choobin, Bioelectrochemistry, 2022, 146, 108106 CrossRef CAS PubMed.
- M. C. Machado, M. Zamani, S. Daniel and A. L. Furst, MRS Bull., 2021, 46, 840–846 CrossRef CAS PubMed.
- J. Daniels, S. Wadekar, K. DeCubellis, G. W. Jackson, A. S. Chiu, Q. Pagneux, H. Saada, I. Engelmann, J. Ogiez, D. Loze-Warot, R. Boukherroub and S. Szunerits, Biosens. Bioelectron., 2021, 192, 113486 CrossRef CAS PubMed.
- A. G. Crevillen, C. C. Mayorga-Martinez, J. V. Vaghasiya and M. Pumera, Adv. Mater. Technol., 2022, 7, 2101121 CrossRef CAS.
- P. Miao, H. Chai and Y. Tang, ACS Nano, 2022, 16, 4726–4733 CrossRef CAS PubMed.
- C.-C. Wu, Y.-H. Chiang and H.-Y. Chiang, Biosensors, 2022, 12, 265 CrossRef CAS.
- V. Liustrovaite, M. Drobysh, A. Rucinskiene, A. Baradoke, A. Ramanaviciene, I. Plikusiene, U. Samukaite-Bubniene, R. Viter, C.-F. Chen and A. Ramanavicius, J. Electrochem. Soc., 2022, 169, 037523 CrossRef CAS.
- J. Liu, C. Ma, S. Shi, H. Liu, W. Wen, X. Zhang, Z. Wu and S. Wang, Biosens. Bioelectron., 2022, 207, 114182 CrossRef CAS PubMed.
- W. Sukjee, A. Thitithanyanont, S. Manopwisedjaroen, S. Seetaha, C. Thepparit and C. Sangma, Mater. Lett., 2022, 315, 131973 CrossRef CAS PubMed.
- P. Malla, H.-P. Liao, C.-H. Liu, W.-C. Wu and P. Sreearunothai, Microchim. Acta, 2022, 189, 168 CrossRef CAS.
- V. V. Tran, N. H. T. Tran, H. S. Hwang and M. Chang, Biosens. Bioelectron., 2021, 182, 113192 CrossRef CAS PubMed.
- Z. Li, G. Jiang, Y. Wang, M. Tan, Y. Cao, E. Tian, L. Zhang, X. Chen, M. Zhao, Y. Jiang, Y. Luo, Y. Zheng, Z. Ma, D. Wang, W. Fu, K. Liu, C. Tang and J. Jiang, Environ. Sci. Nano, 2022, 9, 1759–1769 RSC.
- J. Vigneshwaran, S. Abraham, B. Muniyandi, T. Prasankumar, J.-T. Li and S. Jose, Surf. Interfaces, 2021, 27, 101572 CrossRef CAS.
- F. Cui and H. S. Zhou, Biosens. Bioelectron., 2020, 165, 112349 CrossRef CAS PubMed.
- E. B. Aydın, M. Aydın and M. K. Sezgintürk, New J. Chem., 2022, 46, 9172–9183 RSC.
- K.-H. Liu, H.-Y. Lin, J. L. Thomas, C.-Y. Chen, Y.-T. Chen, C.-Y. Chen, C.-H. Yang and M.-H. Lee, Biosensors, 2022, 12, 31 CrossRef CAS PubMed.
- R. Salahandish, F. Haghayegh, G. Ayala-Charca, J. E. Hyun, M. Khalghollah, A. Zare, B. Far, B. M. Berenger, Y. D. Niu, E. Ghafar-Zadeh and A. Sanati-Nezhad, Biosens. Bioelectron., 2022, 203, 114018 CrossRef CAS PubMed.
- X. Lu, H. Lee Yu, H. Lin, Y. Cao and I.-M. Hsing, Sens. Actuators, B, 2022, 357, 131385 CrossRef CAS.
- Y.-T. Lin, A. Preet, Y.-P. Chiu, B.-S. Yip, H. H. Girault, S. Darvishi, L. Wang and T.-E. Lin, ECS J. Solid State Sci. Technol., 2020, 9, 115028 CrossRef CAS.
- J. Yoon, B. M. Conley, M. Shin, J.-H. Choi, C. K. Bektas, J.-W. Choi and K.-B. Lee, ACS Nano, 2022, 16, 5764–5777 CrossRef CAS.
- H. Liu, A. Yang, J. Song, N. Wang, P. Lam, Y. Li, H. K.-w. Law and F. Yan, Sci. Adv., 2021, 7, eabg8387 CrossRef CAS PubMed.
- E. D. Nascimento, W. T. Fonseca, T. R. de Oliveira, C. R. S. T. B. de Correia, V. M. Faça, B. P. de Morais, V. C. Silvestrini, H. Pott-Junior, F. R. Teixeira and R. C. Faria, Sens. Actuators, B, 2022, 353, 131128 CrossRef CAS PubMed.
- S. Liu, L. Han, J. Li and H. Li, Biosens. Bioelectron., 2022, 198, 113788 CrossRef CAS PubMed.
- Y. Ma, C. Liu, M. Wang and L.-S. Wang, Talanta, 2019, 196, 486–492 CrossRef CAS PubMed.
- Y. Ma, X.-L. Shen, Q. Zeng, H.-S. Wang and L.-S. Wang, Talanta, 2017, 164, 121–127 CrossRef CAS.
- C. Tancharoen, W. Sukjee, C. Thepparit, T. Jaimipuk, P. Auewarakul, A. Thitithanyanont and C. Sangma, ACS Sens., 2019, 4, 69–75 CrossRef CAS.
- R. Arshad, A. Rhouati, A. Hayat, M. H. Nawaz, M. A. Yameen, A. Mujahid and U. Latif, Appl. Biochem. Biotechnol., 2020, 191, 1384–1394 CrossRef CAS.
- A. Raziq, A. Kidakova, R. Boroznjak, J. Reut, A. Öpik and V. Syritski, Biosens. Bioelectron., 2021, 178, 113029 CrossRef CAS.
- V. Ratautaite, R. Boguzaite, E. Brazys, A. Ramanaviciene, E. Ciplys, M. Juozapaitis, R. Slibinskas, M. Bechelany and A. Ramanavicius, Electrochim. Acta, 2022, 403, 139581 CrossRef CAS PubMed.
- S. Cajigas, D. Alzate, M. Fernández, C. Muskus and J. Orozco, Talanta, 2022, 245, 123482 CrossRef CAS PubMed.
- M. Alafeef, K. Dighe, P. Moitra and D. Pan, ACS Nano, 2020, 14, 17028–17045 CrossRef CAS PubMed.
- L. Kashefi-Kheyrabadi, H. V. Nguyen, A. Go, C. Baek, N. Jang, J. M. Lee, N.-H. Cho, J. Min and M.-H. Lee, Biosens. Bioelectron., 2022, 195, 113649 CrossRef CAS PubMed.
- Y. Deng, Y. Peng, L. Wang, M. Wang, T. Zhou, L. Xiang, J. Li, J. Yang and G. Li, Anal. Chim. Acta, 2022, 1208, 339846 CrossRef CAS PubMed.
- T. Chaibun, J. Puenpa, T. Ngamdee, N. Boonapatcharoen, P. Athamanolap, A. P. O'Mullane, S. Vongpunsawad, Y. Poovorawan, S. Y. Lee and B. Lertanantawong, Nat. Commun., 2021, 12, 802 CrossRef CAS PubMed.
- F. Jiang, Z. Xiao, T. Wang, J. Wang, L. Bie, L. Saleh, K. Frey, L. Zhang and J. Wang, Chem. Commun., 2022, 58, 7285–7288 RSC.
- D. Kong, X. Wang, C. Gu, M. Guo, Y. Wang, Z. Ai, S. Zhang, Y. Chen, W. Liu, Y. Wu, C. Dai, Q. Guo, D. Qu, Z. Zhu, Y. Xie, Y. Liu and D. Wei, J. Am. Chem. Soc., 2021, 143, 17004–17014 CrossRef CAS PubMed.
- M. Amouzadeh Tabrizi, L. Nazari and P. Acedo, Sens. Actuators, B, 2021, 345, 130377 CrossRef CAS PubMed.
- J. C. Soares, A. C. Soares, V. C. Rodrigues, P. R. A. Oiticica, P. A. Raymundo-Pereira, J. L. Bott-Neto, L. A. Buscaglia, L. D. C. de Castro, L. C. Ribas, L. Scabini, L. C. Brazaca, D. S. Correa, L. H. C. Mattoso, M. C. F. de Oliveira, A. C. P. L. F. de Carvalho, E. Carrilho, O. M. Bruno, M. E. Melendez and O. N. Oliveira, Mater. Chem. Front., 2021, 5, 5658–5670 RSC.
- L. Zhou, P. Hao, H. Li and Z. Zhang, ACS Sens., 2021, 6, 1613–1620 CrossRef CAS.
- V. Kumar, K. Ghosh, A. Chandran, S. Panwar, A. Bhat, S. Konaje, S. Das, S. Srikanta, L. Jaganathan, S. Prasad, D. B. Venkatesh, C. Shivaram, P. R. Krishnaswamy and N. Bhat, J. Virol. Methods, 2021, 298, 114275 CrossRef CAS.
- M. Mehmandoust, Z. P. Gumus, M. Soylak and N. Erk, Talanta, 2022, 240, 123211 CrossRef CAS PubMed.
- E. Karakuş, E. Erdemir, N. Demirbilek and L. Liv, Anal. Chim. Acta, 2021, 1182, 338939 CrossRef PubMed.
- M. Z. Rashed, J. A. Kopechek, M. C. Priddy, K. T. Hamorsky, K. E. Palmer, N. Mittal, J. Valdez, J. Flynn and S. J. Williams, Biosens. Bioelectron., 2021, 171, 112709 CrossRef CAS PubMed.
- X. Li, Z. Qin, H. Fu, T. Li, R. Peng, Z. Li, J. M. Rini and X. Liu, Biosens. Bioelectron., 2021, 177, 112672 CrossRef CAS PubMed.
- L. Liv and H. Kayabay, ChemistrySelect, 2022, 7, e202200256 CrossRef CAS.
- Md. A. Ali, C. Hu, F. Zhang, S. Jahan, B. Yuan, M. S. Saleh, S.-J. Gao and R. Panat, J. Med. Virol., 2022, 94, 2067–2078 CrossRef CAS PubMed.
- L. C. Brazaca, A. H. Imamura, N. O. Gomes, M. B. Almeida, D. T. Scheidt, P. A. Raymundo-Pereira, O. N. Oliveira, B. C. Janegitz, S. A. S. Machado and E. Carrilho, Anal. Bioanal. Chem., 2022, 414, 5507–5517 CrossRef CAS PubMed.
- L. Fabiani, M. Saroglia, G. Galatà, R. De Santis, S. Fillo, V. Luca, G. Faggioni, N. D'Amore, E. Regalbuto, P. Salvatori, G. Terova, D. Moscone, F. Lista and F. Arduini, Biosens. Bioelectron., 2021, 171, 112686 CrossRef CAS PubMed.
- Z. Rahmati, M. Roushani, H. Hosseini and H. Choobin, Microchem. J., 2021, 170, 106718 CrossRef CAS PubMed.
- R. Nandeshwar, M. S. Kumar, K. Kondabagil and S. Tallur, IEEE Access, 2021, 9, 154368–154377 Search PubMed.
- C. Durmus, S. Balaban Hanoglu, D. Harmanci, H. Moulahoum, K. Tok, F. Ghorbanizamani, S. Sanli, F. Zihnioglu, S. Evran, C. Cicek, R. Sertoz, B. Arda, T. Goksel, K. Turhan and S. Timur, Talanta, 2022, 243, 123356 CrossRef CAS PubMed.
- S. Eissa, H. A. Alhadrami, M. Al-Mozaini, A. M. Hassan and M. Zourob, Microchim. Acta, 2021, 188, 199 CrossRef CAS.
- I. C. Samper, C. J. McMahon, M. S. Schenkel, K. M. Clark, W. Khamcharoen, L. B. R. Anderson, J. S. Terry, E. N. Gallichotte, G. D. Ebel, B. J. Geiss, D. S. Dandy and C. S. Henry, Anal. Chem., 2022, 94, 4712–4719 CrossRef CAS PubMed.
- W. Białobrzeska, M. Ficek, B. Dec, S. Osella, B. Trzaskowski, A. Jaramillo-Botero, M. Pierpaoli, M. Rycewicz, Y. Dashkevich, T. Łęga, N. Malinowska, Z. Cebula, D. Bigus, D. Firganek, E. Bięga, K. Dziąbowska, M. Brodowski, M. Kowalski, M. Panasiuk, B. Gromadzka, S. Żołędowska, D. Nidzworski, K. Pyrć, W. A. Goddard and R. Bogdanowicz, Biosens. Bioelectron., 2022, 209, 114222 CrossRef.
- J. Li, R. Lin, Y. Yang, R. Zhao, S. Song, Y. Zhou, J. Shi, L. Wang, H. Song and R. Hao, ACS Appl. Mater. Interfaces, 2021, 13, 22262–22270 CrossRef CAS PubMed.
- J. Zhao, Z. Fu, H. Li, Y. Xiong, S. Cai, C. Wang, Y. Chen, N. Han and R. Yang, Electrochim. Acta, 2022, 404, 139766 CrossRef CAS PubMed.
- S. A. Hashemi, S. Bahrani, S. M. Mousavi, N. Omidifar, N. G. G. Behbahan, M. Arjmand, S. Ramakrishna, A. M. Dimiev, K. B. Lankarani, M. Moghadami and M. Firoozsani, Talanta, 2022, 239, 123113 CrossRef CAS PubMed.
- J. Li and P. B. Lillehoj, ACS Sens., 2021, 6, 1270–1278 CrossRef CAS PubMed.
- D. Shahdeo, A. Roberts, G. J. Archana, N. S. Shrikrishna, S. Mahari, K. Nagamani and S. Gandhi, Biosens. Bioelectron., 2022, 212, 114406 CrossRef CAS.
- S. Eissa and M. Zourob, Anal. Chem., 2021, 93, 1826–1833 CrossRef CAS PubMed.
- L. Liv, G. Çoban, N. Nakiboğlu and T. Kocagöz, Biosens. Bioelectron., 2021, 192, 113497 CrossRef CAS PubMed.
- L. Liv, M. Yener, G. Çoban and Ş. A. Can, Anal. Bioanal. Chem., 2022, 414, 1313–1322 CrossRef CAS PubMed.
- T. Beduk, D. Beduk, J. I. de Oliveira Filho, F. Zihnioglu, C. Cicek, R. Sertoz, B. Arda, T. Goksel, K. Turhan, K. N. Salama and S. Timur, Anal. Chem., 2021, 93, 8585–8594 CrossRef CAS PubMed.
- B. Mojsoska, S. Larsen, D. A. Olsen, J. S. Madsen, I. Brandslund and F. A. Alatraktchi, Sensors, 2021, 21, 390 CrossRef CAS.
- A. G. Ayankojo, R. Boroznjak, J. Reut, A. Öpik and V. Syritski, Sens. Actuators, B, 2022, 353, 131160 CrossRef CAS.
- M. Amouzadeh Tabrizi, J. P. Fernández-Blázquez, D. M. Medina and P. Acedo, Biosens. Bioelectron., 2022, 196, 113729 CrossRef CAS PubMed.
- C. Hwang, N. Park, E. S. Kim, M. Kim, S. D. Kim, S. Park, N. Y. Kim and J. H. Kim, Biosens. Bioelectron., 2021, 185, 113177 CrossRef CAS PubMed.
- M. Amouzadeh Tabrizi and P. Acedo, Biosensors, 2022, 12, 142 CrossRef CAS PubMed.
- H. E. Kim, A. Schuck, S. H. Lee, Y. Lee, M. Kang and Y.-S. Kim, Biosens. Bioelectron., 2021, 182, 113168 CrossRef CAS PubMed.
- B. Hatamluyi, M. Rezayi, S. Amel Jamehdar, K. S. Rizi, M. Mojarrad, Z. Meshkat, H. Choobin, S. Soleimanpour and M. T. Boroushaki, Biosens. Bioelectron., 2022, 207, 114209 CrossRef CAS PubMed.
- M. Ali Farzin, H. Abdoos and R. Saber, Microchem. J., 2022, 179, 107585 CrossRef CAS PubMed.
- R. del Caño, T. García-Mendiola, D. García-Nieto, R. Álvaro, M. Luna, H. A. Iniesta, R. Coloma, C. R. Diaz, P. Milán-Rois, M. Castellanos, M. Abreu, R. Cantón, J. C. Galán, T. Pineda, F. Pariente, R. Miranda, Á. Somoza and E. Lorenzo, Microchim. Acta, 2022, 189, 171 CrossRef PubMed.
- S.-N. Pang, Y.-L. Lin, K.-J. Yu, Y.-E. Chiou, W.-H. Leung and W.-H. Weng, Micromachines, 2021, 12, 1171 CrossRef PubMed.
- W. Heo, K. Lee, S. Park, K.-A. Hyun and H.-I. Jung, Biosens. Bioelectron., 2022, 201, 113960 CrossRef CAS PubMed.
- S. Damiati, S. Søpstad, M. Peacock, A. S. Akhtar, I. Pinto, R. R. Soares and A. Russom, IEEE Sens. J., 2021, 21, 13060–13067 CAS.
- S. Ramanathan, S. C. B. Gopinath, Z. H. Ismail, M. K. Md Arshad and P. Poopalan, Biosens. Bioelectron., 2022, 197, 113735 CrossRef CAS PubMed.
- J. C. Abrego-Martinez, M. Jafari, S. Chergui, C. Pavel, D. Che and M. Siaj, Biosens. Bioelectron., 2022, 195, 113595 CrossRef CAS PubMed.
- C. Han, W. Li, Q. Li, W. Xing, H. Luo, H. Ji, X. Fang, Z. Luo and L. Zhang, Biosens. Bioelectron., 2022, 200, 113922 CrossRef CAS PubMed.
- J. Tian, Z. Liang, O. Hu, Q. He, D. Sun and Z. Chen, Electrochim. Acta, 2021, 387, 138553 CrossRef CAS.
- A. Idili, C. Parolo, R. Alvarez-Diduk and A. Merkoçi, ACS Sens., 2021, 6, 3093–3101 CrossRef CAS PubMed.
- N. Liu, R. Liu and J. Zhang, Bioelectrochemistry, 2022, 146, 108105 CrossRef CAS PubMed.
- L. F. de Lima, A. L. Ferreira, M. D. Torres, W. R. de Araujo and C. de la Fuente-Nunez, Proc. Natl. Acad. Sci., 2021, 118, e2106724118 CrossRef CAS PubMed.
- Y. T. Büyüksünetçi, B. E. Çitil and Ü. Anık, Analyst, 2022, 147, 130–138 RSC.
- W. Hai, T. Goda, H. Takeuchi, S. Yamaoka, Y. Horiguchi, A. Matsumoto and Y. Miyahara, ACS Appl. Mater. Interfaces, 2017, 9, 14162–14170 CrossRef CAS PubMed.
- J. Huang, Z. Xie, Z. Xie, S. Luo, L. Xie, L. Huang, Q. Fan, Y. Zhang, S. Wang and T. Zeng, Anal. Chim. Acta, 2016, 913, 121–127 CrossRef CAS PubMed.
- L. A. Layqah and S. Eissa, Microchim. Acta, 2019, 186, 224 CrossRef PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- C. M. Das, Y. Guo, G. Yang, L. Kang, G. Xu, H.-P. Ho and K.-T. Yong, Adv. Theory Simul., 2020, 3, 2000185 CrossRef CAS PubMed.
- N. C. Cady, N. Tokranova, A. Minor, N. Nikvand, K. Strle, W. T. Lee, W. Page, E. Guignon, A. Pilar and G. N. Gibson, Biosens. Bioelectron., 2021, 171, 112679 CrossRef CAS PubMed.
- P. Moitra, M. Alafeef, K. Dighe, M. B. Frieman and D. Pan, ACS Nano, 2020, 14, 7617–7627 CrossRef CAS PubMed.
- Z. Huang, D. Tian, Y. Liu, Z. Lin, C. J. Lyon, W. Lai, D. Fusco, A. Drouin, X. Yin, T. Hu and B. Ning, Biosens. Bioelectron., 2020, 164, 112316 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C.-Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- E. Morales-Narváez and C. Dincer, Biosens. Bioelectron., 2020, 163, 112274 CrossRef PubMed.
- Y. Bai, L. Yao, T. Wei, F. Tian, D.-Y. Jin, L. Chen and M. Wang, JAMA, J. Am. Med. Assoc., 2020, 323, 1406–1407 CrossRef CAS PubMed.
- Y. M. Bar-On, A. Flamholz, R. Phillips and R. Milo, eLife, 2020, 9, e57309 CrossRef PubMed.
- J. Abbasi, JAMA, J. Am. Med. Assoc., 2020, 323, 1881–1883 CrossRef PubMed.
- M. Kermali, R. K. Khalsa, K. Pillai, Z. Ismail and A. Harky, Life Sci., 2020, 254, 117788 CrossRef CAS PubMed.
- A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul and S. Chaiyo, Biosens. Bioelectron., 2021, 176, 112912 CrossRef CAS PubMed.
- R. M. Torrente-Rodríguez, H. Lukas, J. Tu, J. Min, Y. Yang, C. Xu, H. B. Rossiter and W. Gao, Matter, 2020, 3, 1981–1998 CrossRef PubMed.
- R. Peng, Y. Pan, Z. Li, Z. Qin, J. M. Rini and X. Liu, Biosens. Bioelectron., 2022, 197, 113762 CrossRef CAS PubMed.
- M. D. T. Torres, L. F. de Lima, A. L. Ferreira, W. R. de Araujo, P. Callahan, A. Dávila, B. S. Abella and C. de la Fuente-Nunez, iScience, 2022, 25, 104055 CrossRef CAS PubMed.
- S. J. Jo, S. Shin, J. Kim, S. Lee and J. Lee, Plos One, 2021, 16, e0258394 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |