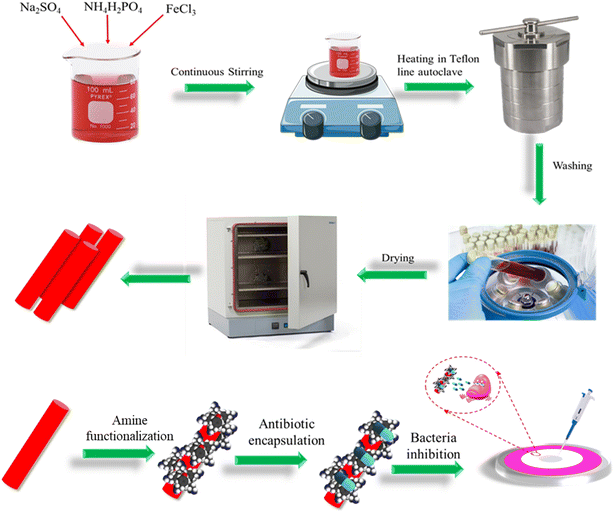
Open Access Article
Open Access ArticleDevelopment of a hematite nanotube and tyramine-based drug carrier against drug-resistant bacteria Klebsiella pneumoniae†
M. R. Aliab,
M. S. Bacchuab,
D. D. Ridoyab,
P. L. Mozumderab,
M. N. Hasanab,
S. Dasc,
M. F. H. Palashc,
S. Akterc,
N. Sakibc,
A. Khalequeab,
D. Chakroborttyd and
M. Z. H. Khan *ab
*ab
aDept. of Chemical Engineering, Jashore University of Science and Technology, Jashore 7408, Bangladesh. E-mail: zaved.khan@yahoo.com
bLaboratory of Nano-bio and Advanced Materials Engineering (NAME), Jashore University of Science and Technology, Jashore 7408, Bangladesh
cDept. of Microbiology, Jashore University of Science and Technology, Jashore 7408, Bangladesh
dGenome Centre, Jashore University of Science and Technology, Jashore 7408, Bangladesh
First published on 3rd November 2022
Abstract
In this study, hematite nanotube (HNT) and tyramine-based advanced nano-drug carriers were developed for inhibiting the growth of Klebsiella pneumoniae (K. pneumoniae). The HNT was synthesized by following the Teflon line autoclaved assisted hydrothermal process and tyramine was incorporated on the surface of the HNT to fabricate the formulated nano-drug. The nano-drug was prepared by conjugating meropenem (MP) on the surface of Tyramine-HNT and characterized using different techniques, such as scanning electron microscopy (SEM), attenuated total reflection Fourier transform infrared (ATR-FTIR), etc. Furthermore, the drug-loading efficiency and loading capacity were measured using a UV-vis spectrometer. The pH, amount of Tyr, and HNT required for drug loading were optimized. A controlled and gradual manner of pH-sensitive release profiles was found after investigating the release profile of MP from the carrier drug. The antibacterial activity of MP@Tyramine-HNT and MP was compared through the agar disc diffusion method which indicates that antibacterial properties of antibiotics are enhanced after conjugating. Surprisingly, the MP@Tyramine-HNT exhibits a minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of K. pneumoniae lower than MP itself. These results indicate the nanocarrier can reduce the amount of MP dosed to eradicate K. pneumoniae.
1. Introduction
Klebsiella pneumoniae (K. pneumoniae) is a Gram-negative and non-motile bacterium which belongs to the family of Enterobacteriaceae. Generally, K. pneumoniae is found in different parts of the human body, such as the skin, mouth, intestines, and respiratory tracts.1 But, it becomes dangerous when it moves into other parts of body. It accounts for a significant proportion of hospital-acquired urinary tract infections, cystitis, pneumonia, surgical wound from infections, and life-threatening infections, such as endocarditis and septicemia. It is also an important cause of serious community-onset infections, such as necrotizing pneumonia, pyogenic liver abscesses, endogenous endophthalmitis, septicemias, and soft tissue infections. It has a high mortality rate of approximately 50% even with antimicrobial therapy.2,3 Pneumonia is a major human health threat with a mortality rate approximately 50% under the current antibacterial treatments.4 In Bangladesh, 1.4 million children under 5 years fight against pneumonia, and, more than one child dies every hour. In the previous era, antibiotics specially carbapenem class beta-lactam antibiotic treatment is the best way for pneumonia treatment but K. pneumoniae is getting resistant to multiple antibiotics (third and fourth-generation antibiotics) day by day.5 K. pneumoniae becomes resistant to fourth-generation antibiotic carbapenem by producing carbapenemase enzyme.6 So, it is important to search for a new solution for the treatment of infectious diseases which is caused by antibiotic-resistant bacteria. For this purpose, researchers follow several strategies to solve the problem, such as chemical modification of antibiotics by cationic polymers, photothermal agents, proteins, antimicrobial peptides, and nanoparticles.7,8 Nowadays, nanomedicine is becoming a promising strategy for the treatment of antibiotic-resistant bacterial infections where nanoparticles or nanostructure materials are working as antibacterial carriers or carrier of antibacterial agents to enhance the effectiveness and targeted delivery of antibiotics.9–12 Nanomaterials as an antibacterial agent can bind the negatively charged membrane surface of bacteria and promotes the damage of cell membrane.13 Nanomaterials as a carrier of antibiotic drugs or drugs encapsulation with antibiotics are the most effective strategy to enhance the eradication of bacteria and bioavailability. The antibacterial activity and affinity to bacteria of drugs enhances after nanoparticle conjugation due to the higher surface-to-volume ratio, effective surface modification, and loading of drugs.14 The antibiotic resistance of bacteria to antibiotic reduces after conjugation of drugs with nanomaterials by projecting the drugs from different enzymatic attacks. The release properties and bioavailability of drugs become sustainable by enhancing the half-life of antibiotics after nanomaterial conjugation.15,16Different nanoparticles, such as gold, silver, iron oxide, silica, and zinc oxide nanoparticles, etc. are of growing interest in application of bacterial infection treatment.17 Among them, iron oxide is most widely used for the targeted delivery of anticancer drugs and antibiotic.18,19 Hematite (α-Fe2O3) and magnetite (Fe3O4) are the mostly available and inexpensive iron oxide found naturally in iron ore.20 For this study, we choose tube size hematite (HNT) particles delivery of meropenem to the K. pneumoniae. The iron nano particle is the most preferable nanoparticles used in nanomedicine due to its low toxicity and high stability in aqueous solution.21,22 It is proposed that different amino acid conjugated on the surface of α-Fe2O3 reduces toxicity of HNT and makes it's as more efficient for targeted antibiotic delivery.23 For this reason, naturally occurring trace amino acid tyramine was conjugated HNT to with reduce the toxicity of HNT and effective encapsulation of MP. The amine group of tyramine is easily conjugated with the carboxylic group of antibiotics.
In this work, we produce MP conjugated nanodrug carrier named MP@Tyramine-HNT for targeted antibiotic delivery to eradicate K. pnumoniae. To prepare the carrier matrix, HNT was fabricated from FeCl3 by using Teflon-line autoclave assisted hydrothermal process and conjugated with tyramine to coat the antibiotic MP. The conjugation and fabrication of HNT were confirmed by field emission scanning electron microscope (FESEM) and annulated total reflectance Fourier transforms infrared (ATR-FTIR). Finally, the antimicrobial properties of the conjugated drug were evaluated through performing minimum inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) against K. pneumoniae.
2. Materials and methods
2.1 Chemicals and reagents
In this work, ferric chloride (FeCl3), ammonium dihydrogen phosphate (NH₄H₂PO₄), sodium sulphate (Na2SO4), and sodium Chloride (NaCl) were purchased from Merck (India). Tyramine was bought from Aladdin Reagent Ltd. (China). The antibiotic Meropenem (MP) was supplied from local pharmaceutical company (ACI Ltd.). Ultrapure water (prepared by using Evoqua (Germany)) was utilized for preparing all kinds of solutions. McConkey Agar and Mueller Hinton Agar were used in this work were purchased respectively from Oxoid (UK). Mueller Hinton broth and Tryptiocase-Soy broth were bought from Himedia (India).2.2 Instruments
The surface morphology of nanoparticles and drug carriers was investigated by using a ZEISS GeminiSEM 500 field emission scanning electron microscope (FE-SEM). The interaction between nano-carrier and antibiotic was confirmed by performing a Fourier transforms infrared (FTIR) spectroscopy analysis by using NICOLERT iS20 ATR-FTIR. Shimadzu lab solution UV-VIS (UV-1900i) were utilized for investigating drug loading efficiency, Antibiotic loading capability and release profile of MP.2.3 Synthesis of hematite nano-tube structures
Hematite nano-tube (HNT) structures were synthesized by following the similar technique as reported previously.24 In short, aqueous solutions of 3.20 mL FeCl3 (0.5 mol L−1), 0.90 mL NH4H2PO4 (0.04 mol L−1), and 1.10 mL Na2SO4 (0.04 mol L−1) were mixed vigorously. To maintain the total volume to 80 mL, required amount of ultra-pure water was added. Then the mixture was placed into a Teflon-lined stainless-steel autoclave with a capacity of 100 mL for hydrothermal treatment at 200 °C for 24 hours after being stirred for 20 minutes. The autoclave spontaneously cooled at room temperature. The precipitates were separated by centrifuging and washed the precipitates with distilled water and absolute ethanol to remove unreacted substance and dried at a temperature of 80 °C.2.4 Preparation of HNT-Tyramine and MP@Tyramine-HNT
At first, 15 mg of HNT and 7.5 mg of Try were dispersed individually in 5 ml normal saline (0.85 percent NaCl) to make 10 ml Tyramine-HNT solution. Each solution was ultrasonicated for 30 minutes and then magnetically stirred for 1 hour. The solution was mixed and continuously stirred at room temperature for 6 hours with a magnetic stirrer to produce Tyramine-HNT.To fabricate MP@Tyramine-HNT, 500 mg of MP was dissolved in 10 mL normal saline to make a 50 mg mL−1 MP stock solution, which was magnetically stirred to ensure homogeneity. An aliquot of the needed volume MP was added to the Tyramine-HNT solution and stirred for 1 hour in a magnetic stirrer to make MP@Tyramine-HNT where MP concentration reached 10 μg ml−1. The step-by-step procedure for fabrication of MP@Tyramine-HNT is shown in Scheme 1.
2.5 Antibiotic loading efficiency and encapsulation capacity analysis
The MP loading efficiency of the nano-carrier and MP encapsulation efficiency was measured by following the similar methods as reported previously.17 To access the loading efficiency 10 μg of MP was loaded on the solution of nano-carrier at different pH ranging 1.5 to 7.5. The drug loading efficiency was determined by performing UV-Vis spectroscopy at the wavelength of 340 nm and amount of drug loaded was calculated by using the eqn (1) and the calibration curve in Fig. S1.† Then, the drug loading efficiency was calculated by following the eqn (2). The antibiotic encapsulation capacity analysis was performed by loading MP of different concentration of 10 μg to 80 μg at a pH of 6.5 and amount of antibiotic encapsulated was calculated by following similar method to amount of drug loading analysis.| Amount of MP loaded (μg) = μg of MP added – μg of MP found in supernatant | (1) |
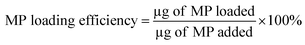 | (2) |
2.6 Antibiotic release profile analysis
The antibiotic release profile analysis was carried at different pH (2.5, 7.4 & 9.0) by using UV-Vis spectrophotometer. For this measurement, MP@Tyramine-HNT was prepared in PBS at different pH as stated above. Then, the solution was placed in water bath at 37 °C and 5 ml of supernatant of the solution was withdraw from the mother solution. The amount of antibiotic released was determined by taking UV-Vis spectroscopy at a wavelength of 340 nm. Total amount of MP released from the MP@Tyramine-HNT was calculated by subtracting amount of MP in supernatant from amount of MP loaded with nanocarrier.2.7 Bacterial stain and cultivation
In this study, we have used meropenem sensitive Klebsiella pneumoniae (ATCC 700603) strain. The ‘reference culture collection’ of Department of Microbiology, Jashore University of Science and Technology provided the stain. We have resuscitated it through inoculating 100 μl of glycerol broth stock culture into 5 ml of trypticase soy broth. After incubation at 37 °C for 24 hours it was cultured onto MacConkey agar media for isolation. Pure discrete colonies of K. pneumoniae were screened with microscopy and some biochemical tests to confirm the identity and used for further work.We have also used a meropenem resistance K. pneumoniae strain isolated from water. For isolation, ten water samples was collected from a river which flows across the city of Jashore district. Sampling sites were selected from regions where hospital effluents drain into the river. Water samples were collected into equal volume of 2× concentrated MacConkey Broth (OXOID, UK) incorporated with Meropenem antibiotic (25 μg L−1) for selective enrichment of resistant enterobacteriaceae. After incubation at 37 °C for 24 hours, the enrichment cultures were streaked onto MacConkey agar isolation and subjected to different biochemical test, such as oxidase, catalase, glucose and lactose fermentation by triple sugar iron agar test and citrate utilization test for screening. Preliminarily selected isolates were tested with API20E kit (Biomerieux, France) and analyzed by epiweb database. From the isolates, a meropenem resistant isolate was selected to include in this study.
2.8 Antibiotic susceptibility assay
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and suspended into normal physiological saline (0.85% NaCl). Turbidity of the bacterial suspensions were adjusted with 0.5 MacFarland standard. Bacterial lawn was prepared by using a non-absorbent cotton swab which was dipped into prepared bacterial suspension and then spread over a Muller Hinton agar plate. 0.5 cm of blank antimicrobial susceptibility discs are placed at different position on agar plate maintaining a fixed distance. Then the prepared antibiotic suspensions were poured (with or without nanoparticles) on the discs. The Petri plates were incubated at 37 °C for 18 hours.
000g and suspended into normal physiological saline (0.85% NaCl). Turbidity of the bacterial suspensions were adjusted with 0.5 MacFarland standard. Bacterial lawn was prepared by using a non-absorbent cotton swab which was dipped into prepared bacterial suspension and then spread over a Muller Hinton agar plate. 0.5 cm of blank antimicrobial susceptibility discs are placed at different position on agar plate maintaining a fixed distance. Then the prepared antibiotic suspensions were poured (with or without nanoparticles) on the discs. The Petri plates were incubated at 37 °C for 18 hours.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio and incubated overnight. The cultures were then streaked on MH agar without any antibiotic/conjugate and incubate overnight to observe the bactericidal effect at different drug preparations.
100 ratio and incubated overnight. The cultures were then streaked on MH agar without any antibiotic/conjugate and incubate overnight to observe the bactericidal effect at different drug preparations.3. Results and discussions
3.1 Surface topographical analysis of MP@Tyramine-HNT
The surface topographical analysis of MP@Tyramine-HNT was investigated through the SEM and the surface chemistry was confirmed through FTIR analysis. Fig. 1(A) shows the SEM image of HNT which reveals nano size tube shape hematite (α-Fe2O3) particles are synthesized successfully. The length of the nanotube was approximately 200 to 300 nm and diameter of nanotube was less than 100 nm. Fig. 1(B) the SEM image of MP@Tyramine-HNT which represents the MP and tyramine were successfully incorporated on the surface HNT. To confirm the interaction of HNT, tyramine and MP, FTIR analysis of HNT, MP, tyramine and MP@Tyramine-HNT were done as shown in Fig. 1(C). The transmittance peak due to Fe–O vibration in the wavelength of 475 and 562 cm−1 is confirmed the presence of α-Fe2O3. The FTIR spectra of tyramine represents different peaks, such as 1417 and 1518 for tyramine oriented aromatic ring, 3333 cm−1 for –NH2 vibration and 3280 cm−1 for strong –OH vibration.25 While the FTIR spectra of MP shows a sharp peak at 1735.62 cm−1 due to CO elongation of –COOH and pyrrolidine ring of MP and at 665 cm−1 for flexion OH in COOH. After conjugation of MP, tyramine and HNT the peak at 1735.62 was shift into 1741 and formed CONH bond between MP and tyramine26 and unshifted peak in 475 and 562 cm−1 reveals successful incorporation of MP, tyramine and HNT.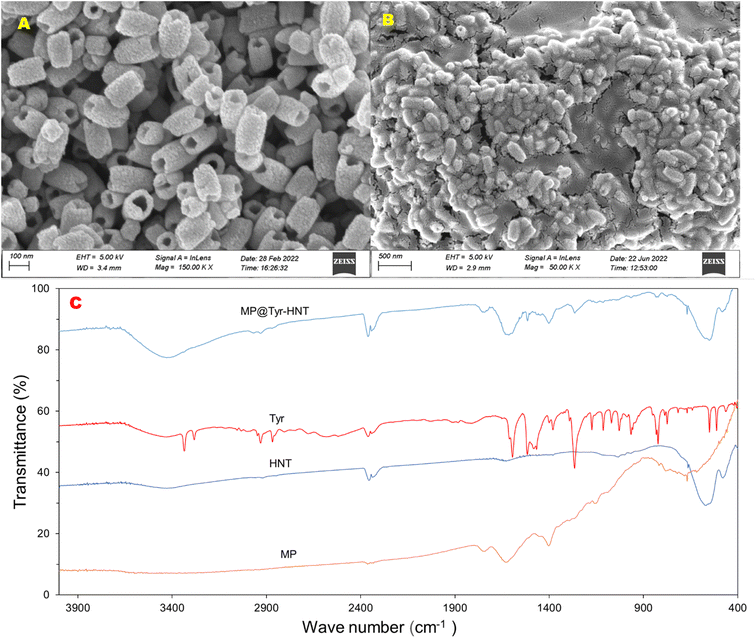 | ||
| Fig. 1 (A) SEM Of HNT, (B) MP@Tyramine-HNT and (C) FTIR spectra of MP, HNT, tyramine and MP@Tyramine-HNT. | ||
3.2 Effect of pH on antibiotic loading
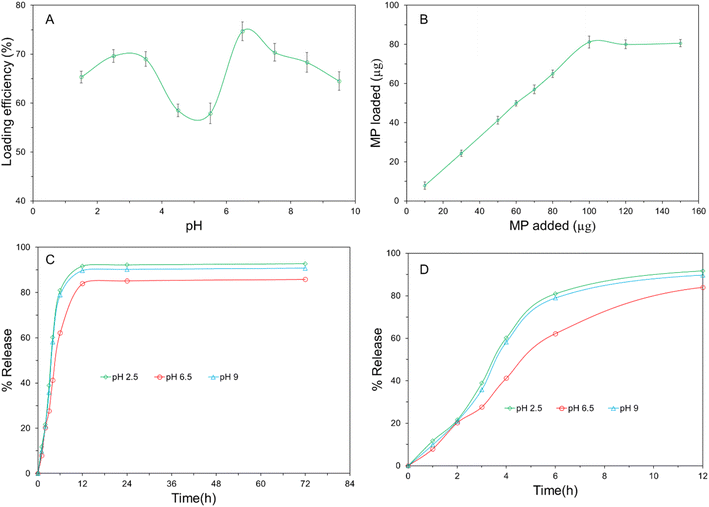
To examine the impact of pH on antibiotic loading efficiency, antibiotic efficiency analysis at different pH were performed. Fig. 2(A) shows drug loading efficiency of the surface of the proposed carrier at different pH and maximum antibiotic loading efficiency was observed at pH 6.5 that's means the nano carrier conjugated highest amount of MP at this pH which can be confirmed by observing the bipolar structure of MP. The antibiotic MP has an isoelectronic point of 6.336 which reveals carboxylic group of MP will be activated at a pH which is lower than 6.336 and produced positive charge.17 The thiol group of MP will be activated at a pH which is higher than the isoelectronic point and formed negative charge. The maximum number of carboxylic and thiol group of MP will be activated at pH 3.28 and 9.39 respectively. The probable cause of maximum adsorption found at pH 6.5 (between pH 3.28 and pH 9.39) is the better interaction between the –COOH group of MP and the –NH2 group of due to the presence of a maximum number of charge difference between them. The pH value greater than 6.5 and lower than 6.5, the electrostatic repulsion between MP and tyramine become higher, so the drug loading efficiency getting lower.3.3 Antibiotic encapsulation capacity analysis
The total amount of antibiotic conjugated with Tyramine-HNT was measured by performing UV-vis spectroscopy at a wavelength of 340 nm. Fig. 2(B) shows the amount of antibiotic loaded per milligram (mg) of Tyramine-HNT and total μg of MP loaded per mg of the nanocarrier was 81.2 ± 3 μg. The amount of antibiotic loaded on the surface of nano-carrier was increased with adding antibiotic but it became steady state after conjugating 81.2 μg. So, the antibiotic encapsulation capacity of Tyramine-HNT was 81.2 mg per gram of nanocarrier.3.4 Release profile analysis
To validify the practical application of Tyramine-HNT as an effective drug delivery agent, release profile of MP from the MP@Tyramine-HNT analysis is very important. For this case, a release profile analysis had been carried out at different pH (2.5, 6.5 and 9). Fig. 2(C) shows the pH dependent MP release profile from the nanocarrier at 37 °C. Initially, a vast release rate of MP was found at different pH. Almost 60 to 80% of MP was released from the MP@Tyramine-HNT in the first six hours at different pH and total 82 to 91% MP was released after 12 hours which is shown in Fig. 2(D). The release profile of MP revealed the dose of required to eradicate bacteria reduced and one dose of MP@Tyramine-HNT can kill the sufficient amount of K. pneumoniae in a day. The amount of MP released from MP@Tyramine-HNT was slower at pH 6.5 than higher and lower pH that means MP@Tyramine-HNT follows a pH-dependent release pattern. In acidic and basic condition, the antibiotic releasing rate was higher than that of pH 6.5 and almost 92.7% and 91.2% MP were released over 72 h releasing periods. The probable reason for this fast release pattern will be the loosening of the grafted MP@Tyramine from HNT bonding.The suitable release kinetic model of MP from MP@Tyramine-HNT was investigated by fitting the antibiotic release data in different kinetic models, such as zero-order, first order, Korsmeyer–Peppas and Higuchi kinetics models. Fig. 3 represents the different kinetic models for analysis of kinetic fitting and the kinetic rate constant (K) and correlation coefficient (R2) are listed in Table 1. The Table 1 reveals the release pattern follows the Higuchi kinetic model for releasing MP from MP@Tyramine-HNT.
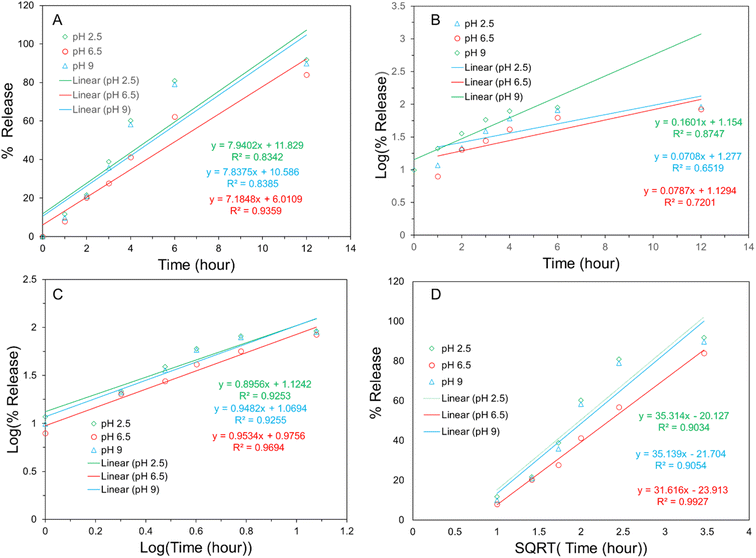 | ||
| Fig. 3 Kinetics study of antibiotic release profile (0 to 12 h): (A) zero order kinetic (B) first order kinetic (C) Korsmeyer–Peppas kinetic and (D) Higuchi kinetics. | ||
| Formulation | pH | Zero-order | First-order | Korsmeyer-Peppas | Higuchi | ||||
|---|---|---|---|---|---|---|---|---|---|
| K0 | R2 | K1 | R2 | KKP | R2 | KH | R2 | ||
| 2.5 | 11.829 | 0.83 | 1.154 | 0.87 | 1.124 | 0.93 | −20.127 | 0.91 | |
| MP@Tyramine-HNT | 6.5 | 6.011 | 0.94 | 1.129 | 0.65 | 0.976 | 0.96 | −21.704 | 0.99 |
| 9 | 10.586 | 0.84 | 1.277 | 0.72 | 1.069 | 0.93 | −23.913 | 0.91 | |
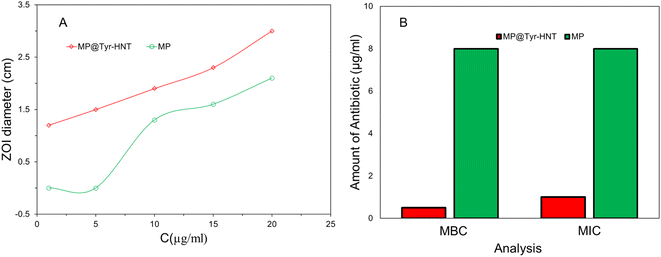
3.5 Antimicrobial activity of hematite/Tyramine-MP conjugated antibiotic
| Carrier matrix | Targeted bacteria | Antibiotic | Antibacterial properties | Release profile | Ref. | ||
|---|---|---|---|---|---|---|---|
| MIC (μg ml−1) | MBC (μg ml−1) | Percentage release over 24 h | Release kinetics | ||||
| ZnO/L-Cys | Salmonella typhimurium | Ceftizoxime | 5 | — | 86.7% | Korsmeyer–Peppas | 17 |
| GO@CoFe2O4@Ag | Escherichia coli, Staphylococcus aureus | Ciprofloxacin | 1.25, 2.5 | 1.25, 2.5 | 43% | — | 27 |
| Amine functionalized ZnO | Escherichia coli, Staphylococcus aureus, Klebsiella sp. | Ciprofloxacin | 10 | — | — | — | 28 |
| FAuNPs | Staphylococcus aureus, K. pnumoniae, Escherichia coli | Levofloxacin, Ciprofloxacin, Ciprofloxacin | 0.562, 0.281, 0.281 | 0.281, 0.14, 0.562 | — | — | 29 |
| Fe3O4@BSM | Escherichia coli, Staphylococcus aureus, Salmonella typhimurium | Cephalexin | — | — | 80% | Pseudo first order | 30 |
| Tyramine-HNT | K. pnumoniae | Meropenem | 1 | 0.5 | 85.2% | Higuchi | This work |
4. Conclusion
Nano drug delivery system is one of most promising way to address the disadvantage of conventional antibiotic base treatment strategy. In the present study, HNT and tyramine based an advanced nanodrug carrier had been fabricated and the fourth-generation beta lactam antibiotic conjugated with the nanocarrier. The conjugation of HNT, tyramine and MP were confirmed by different surface topographical analysis, such as SEM, ATR-FTIR. The amount of drug loading and drug loading efficiency of the nanocarrier drug was calculated by using UV-vis spectroscopy which reveals the nanocarrier capture the highest amount of MP at pH 6.5. The proposed nanocarrier showed lower MIC and MBC than the antibiotic only that's means the therapeutic efficiency of MP had been enhanced after conjugation with the drug carrier. The proposed nanodrug exhibited a pH sensitive release profile and controlled release manner in neutral pH. Thus, the as prepared nanocarrier will be a promising alternative of the conventional pneumonia treatment.Conflicts of interest
The authors declared no conflicts of interest.Acknowledgements
The work has been done with the financial support from Jashore University of Science & Technology, Jashore-7408, Bangladesh (Research Grant 2022-23).References
- S. Cao, X. Wu, J. Zhao and X. Jia, J. Nanosci. Nanotechnol., 2020, 20, 6063–6069 CrossRef CAS PubMed.
- S. J. Lam, N. M. O’Brien-Simpson, N. Pantarat, A. Sulistio, E. H. H. Wong, Y. Y. Chen, J. C. Lenzo, J. A. Holden, A. Blencowe, E. C. Reynolds and G. G. Qiao, Nat. Microbiol., 2016, 1, 16162 CrossRef CAS PubMed.
- M. Ye, J. Tu, J. Jiang, Y. Bi, W. You, Y. Zhang, J. Ren, T. Zhu, Z. Cao, Z. Yu, C. Shao, Z. Shen, B. Ding, J. Yuan, X. Zhao, Q. Guo, X. Xu, J. Huang and M. Wang, Front. Cell. Infect. Microbiol., 2016, 6, 1–12 CrossRef PubMed.
- N. Padmini, A. A. K. Ajilda, N. Sivakumar and G. Selvakumar, J. Basic Microbiol., 2017, 57, 460–470 CrossRef CAS PubMed.
- E. C. Rodríguez, S. Y. Saavedra, A. L. Leal, C. Álvarez, N. Olarte, A. Valderrama, S. I. Cuervo and J. Escobar, Biomédica Rev. del Inst. Nac. Salud, 2014, 34(suppl. 1), 224–231 Search PubMed.
- J. Reyes, A. C. Aguilar and A. Caicedo, Int. J. Gen. Med., 2019, 12, 437–446 CrossRef PubMed.
- G. Gebreyohannes, A. Nyerere, C. Bii and D. B. Sbhatu, Heliyon, 2019, 5, e02192 CrossRef PubMed.
- K. Hadinoto and W. S. Cheow, Colloids Surf. B Biointerfaces, 2014, 116, 772–785 CrossRef CAS PubMed.
- P. V. Baptista, M. P. McCusker, A. Carvalho, D. A. Ferreira, N. M. Mohan, M. Martins and A. R. Fernandes, Front. Microbiol., 2018, 9, 1–26 CrossRef PubMed.
- P. C. Kanth, S. K. Verma and N. Gour, Functionalized nanomaterials for biomedical and agriculture industries, INC, 2020 Search PubMed.
- S. Palit and C. M. Hussain, Functionalization of nanomaterials for industrial applications: recent and future perspectives, INC, 2020 Search PubMed.
- N. Gour, K. X. Ngo and C. Vebert-Nardin, Macromol. Mater. Eng., 2014, 299, 648–668 CrossRef CAS.
- E. Taylor and T. J. Webster, Int. J. Nanomed., 2011, 6, 1463–1473 CrossRef CAS PubMed.
- H. Zazo, C. I. Colino and J. M. Lanao, J. Control. Release, 2016, 224, 86–102 CrossRef CAS PubMed.
- L. Zhang, D. Pornpattananangkul, C.-M. Hu and C.-M. Huang, Curr. Med. Chem., 2010, 17, 585–594 CrossRef CAS PubMed.
- P. Walvekar, R. Gannimani and T. Govender, Eur. J. Pharm. Sci., 2019, 127, 121–141 CrossRef CAS PubMed.
- M. S. Bacchu, M. R. Ali, M. A. A. Setu, S. Akter and M. Z. H. Khan, Sci. Rep., 2021, 11, 1–11 CrossRef PubMed.
- K. Du, Y. Zhu, H. Xu and X. Yang, Prog. Chem., 2011, 23, 2287–2298 CAS.
- E. N. Taylor, K. M. Kummer, N. G. Durmus, K. Leuba, K. M. Tarquinio and T. J. Webster, Small, 2012, 8, 3016–3027 CrossRef CAS PubMed.
- M. Ashraf, I. Khan, M. Usman, A. Khan, S. S. Shah, A. Z. Khan, K. Saeed, M. Yaseen, M. F. Ehsan, M. N. Tahir and N. Ullah, Chem. Res. Toxicol., 2020, 33, 1292–1311 Search PubMed.
- V. Valdiglesias, N. Fernández-Bertólez, G. Kiliç, C. Costa, S. Costa, S. Fraga, M. J. Bessa, E. Pásaro, J. P. Teixeira and B. Laffon, J. Trace Elem. Med. Biol., 2016, 38, 53–63 CrossRef CAS PubMed.
- N. Malhotra, J. S. Lee, R. A. D. Liman, J. M. S. Ruallo, O. B. Villaflore, T. R. Ger and C. Der Hsiao, Molecules, 2020, 25, 1–26 Search PubMed.
- U. O. Häfeli, J. S. Riffle, L. Harris-Shekhawat, A. Carmichael-Baranauskas, F. Mark, J. P. Dailey and D. Bardenstein, Mol. Pharm., 2009, 6, 1417–1428 CrossRef PubMed.
- C. N. Jia, L. D. Sun, Z. G. Yan, Y. C. Pang, L. P. You and C. H. Yan, J. Phys. Chem. C, 2007, 111, 13022–13027 CrossRef CAS.
- M. Prokopijevic, O. Prodanovic, D. Spasojevic, G. Kovacevic, N. Polovic, K. Radotic and R. Prodanovic, Appl. Microbiol. Biotechnol., 2017, 101, 2281–2290 CrossRef CAS PubMed.
- L. M. D. A. C. Gaspar, A. C. S. Dórea, D. Droppa-Almeida, I. S. de Mélo Silva, F. E. Montoro, L. L. Alves, M. L. H. Macedo and F. F. Padilha, J. Nanoparticle Res., 2018, 20(11), 1–9 CrossRef CAS.
- M. Kooti, A. N. Sedeh, H. Motamedi and S. E. Rezatofighi, Appl. Microbiol. Biotechnol., 2018, 102, 3607–3621 CrossRef CAS PubMed.
- P. Patra, S. Mitra, N. Debnath, P. Pramanik and A. Goswami, Bull. Mater. Sci., 2014, 37, 199–206 CrossRef CAS.
- S. M. Pradeepa, S. Vidya, K. Mutalik, U. Bhat, P. Huilgol and K. Avadhani, Life Sci., 2016, 153, 171–179 CrossRef CAS PubMed.
- A. Rayegan, A. Allafchian, I. Abdolhosseini Sarsari and P. Kameli, Int. J. Biol. Macromol., 2018, 113, 317–328 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra05216d |
| This journal is © The Royal Society of Chemistry 2022 |