Open Access Article
Open Access ArticleHeterologous expression, molecular studies and biochemical characterization of a novel alkaline esterase gene from Bacillus thuringiensis for detergent industry
Asma Zafar *a,
Ziaur Rahmana,
Hira Mubeena,
Javairia Makhdoomb,
Javeria Tariqa,
Mahjabeena,
Zulqurnain Alia,
Attia Hamid
*a,
Ziaur Rahmana,
Hira Mubeena,
Javairia Makhdoomb,
Javeria Tariqa,
Mahjabeena,
Zulqurnain Alia,
Attia Hamid c,
Eeza Shafiquea and
Muhammad Nauman Aftabc
c,
Eeza Shafiquea and
Muhammad Nauman Aftabc
aFaculty of Science and Technology, University of Central Punjab, Lahore, Pakistan. E-mail: asma.zafar@ucp.edu.pk; Tel: +92-3006485797
bFaisalabad Medical University, Faisalabad, Pakistan
cInstitute of Industrial Biotechnology, Govt. College University, Lahore, Pakistan
First published on 30th November 2022
Abstract
Present study was aimed to clone and express the esterase encoding gene from Bacillus thuringiensis in E. coli BL21. Purification of recombinant esterase enzyme was achieved up to 48.6 purification folds by ion exchange chromatography with specific activity of 126.36 U mg−1. Molecular weight of esterase enzyme was 29 kDa as measured by SDS-PAGE. Purified esterase enzyme showed stability up to 90% at 90 °C and remained stable in a wide pH range (8–11). Molecular docking strengthens the experimental results by showing the higher binding energy with p-NP-butyrate. Enzyme activity was found to be reduced by EDTA but enhanced in the presence of other metal ions. Enzyme activity was reduced with 1% SDS, PMSF, and urea but organic solvents did not show considerable impact on it even at higher concentrations. Purified recombinant esterase was also found to be compatible with commercial laundry detergents and showed very good stability (up to 90%). All these properties proved the esterase enzyme from B. thuringensis a significant addition in detergent industry.
Introduction
The global consumption of daily life products such as food, feed, textile, detergents, paper, chemicals, fuel, etc. has increased daily due to the worldwide increase in population. To fulfill the demands and for improvements in economies, the industries consume an excessive quantity of raw material and energy resulting in huge amounts of waste production, which ultimately leads to the destruction of the environment and quality of life.1,2 There is an urgent need to explore the sources which can reduce environmental impact of production processes in order to protect the environment while producing the products without compromising the quality.3 Industries are taking great interest in those technologies through which they can produce good quality products without damaging the environment while utilizing fewer resources.4 Among these technologies, the use of enzymes in industrial processes either to replace or supplement conventional methods is one of the promising tools to move the industries towards sustainable development.5–8One of the major classes of hydrolases encompasses lipolytic enzymes (esterases & lipases) which are involved in both hydrolysis as well as the synthesis of ester compounds.9,10 Lipolytic enzymes are broadly distributed in plants, microorganisms and animals. True lipases enzymes mediate the hydrolysis and synthesis of moderately long-chain acylglycerols, whereas esterases enzymes catalyze glycerol esters with acyl chains of fewer than 10 carbon atoms.11 Another major difference among esterases and true lipases is the phenomenon of interfacial activation which is only detected for lipases. Both enzymes are also found to be stable in organic solvents.12 Esterases have gained particular interest owing to their affinity toward both natural as well synthetic substrate reactions. The selectivity of natural substrate creates a novel approach for setting up more eco-friendly reactions for the synthesis of value-added by-products.13 Microbial esterases have a wide range of substrate specificity, do not require a cofactor, have stereo and high area specificity, and have important uses in different industries like food industries, detergent, pharmaceutical, dairy, beauty products and physiological domains.14
Recently, the use of esterase enzyme along with other enzymes in detergent formulations has gotten a lot of interest because surfactants, corrosion inhibitors, builders, optical brighteners, bleaching agents, foam regulators and enzymes are the most common minor additives in detergents.15 For more than a century, enzymes like esterase, protease, amylase, cellulase, mannanase, and pectinase have been employed in laundry detergents. Many firms are now manufacturing enzyme-based detergents that are excellent at eliminating stains from a variety of sources, including proteins, oils, and carbohydrates, while also conserving water and energy.16 Because enzymes are produced from renewable sources, adding them into detergent formulations means that fewer chemicals are used and that people are exposed to less hazardous substances and solvents. Furthermore, the excellent cleaning impact of the enzymes removes the need for repeated washing to remove stubborn impurities such as oil stains and enables full cleaning of the clothes even during a short washing cycle.17 In this way, the use of enzymes in detergents provides eco-friendly and cost-effective cleaning without compromising washing effectiveness.
Heterologous expression of industrially important proteins is considered an advantageous tool to get a high yield of enzymes for industrial usage. Escherichia coli is one of the most important hosts for the production of heterologous proteins due to its genetic modification, rapid growth on cheap medium and the availability of cloning tools.18 BL21 (DE3) strain of E. coli is genetically modified by deficient in amino acids of Lon protease as well as OmpT protease which are responsible to degrade foreign proteins. In addition to these genetic modifications, naturally occurring hsdSB mutation in the genome of E. coli BL21 (DE3) prevent plasmid loss in this strain.19 E. coli BL21 (DE3) is also a well-known strain for the T7 expression system because the λDE3 prophage gene has been inserted into the genome of this bacterium and it also contains the T7 RNAP gene under lac UV5 promoter. pET expression vectors contain the T7 promoter system and are frequently utilized for the heterologous expression of proteins.20–24
In order to explore the enzymes used in detergent formulation having improved characteristics, the present study was conducted by using Bacillus thuringiensis. The soil bacterium B. thuringiensis is a Gram-positive, spore-forming rod-shaped and aerobic-to-facultative, which also grows anaerobically. This genus contains great potential for biotechnological applications.25 Taking into account the behavior of this potent candidate towards biotechnological applications, this study was aimed to clone, express, purify and characterize a novel alkaline esterase gene from B. thuringensis for utilization in detergent industry.
Materials and methods
Bacterial strains and plasmids
The strain of Bacillus thuringiensis was obtained from the Department of Mycology and Plant Pathology's First Fungal Culture Bank of University of the Punjab, Lahore, Pakistan. The expression strain E. coli BL21 (DE3) was acquired from the Biotechnology Department of Govt. College University Lahore, Pakistan for the expression of esterase gene in pET-21c vector. Both of these strains were grown in Lysogeny broth (0.5% yeast extract, 1% trypton and 1% NaCl) and stored at 4 °C.Extraction of genomic DNA and gene amplification
Genomic DNA from B. thuringiensis was extracted using the method given by Kronstad et al.26 To amplify the esterase gene, primers were designed using Primer III software. In the forward primer, NdeI restriction site was introduced and in reverse primer BamHI was inserted. The primer's sequences for the amplification of esterase gene are mentioned below:Est F′: 5′ CAT ATG ACC CGA CGG CAA AAT TAC 3′.
Est R′: 5′ GGA TCC CCA TTC GGA ATA GTC TTT 3′.
Esterase gene of B. thuringiensis was amplified through PCR. Reaction mixture contained, 1 μL of forward primer, 1 μL of reverse primer, 12.5 μL of master mix (Thermo Scientific™), 1 μL of DNA template and 9.5 μL of double distilled water. Initial denaturation was performed at 95 °C for 5 min, annealing at 56 °C for 30 s and extension at 72 °C for 30 s for 35 cycles following the final extension step at 72 °C for 20 min. For the purification of amplified esterase gene product, Gel Extraction kit (Thermo Scientific) was used.
Cloning of esterase gene in pET-21c vector
In the presence of 2× tango buffer, the purified amplified product of the esterase gene of B. thuringiensis as well as pET-21c were double restricted with BamHI and NdeI. Thermo Scientific GeneJET DNA purification kit was used to purify both the products. Amplified double restricted esterase gene of B. thuringiensis was ligated with pET-21c using DNA ligase enzyme. Ligated product was transformed in competent cell of E. coli BL21 (DE3) prepared by chemical method described by Cohen et al.27 Positive clones containing gene of interest were selected restriction of recombinant plasmid and also by colony PCR.Expression of recombinant esterase gene
To evaluate the expression of the recombinant esterase gene, ampicillin (100 μg mL−1) containing LB broth was used. Overnight grown transformed E. coli BL21 cells were diluted to 1% in LB broth containing ampicillin and incubated at 37 °C to raise the optical density of the culture to the range 0.4 to 0.6 at 600 nm followed by IPTG (0.5 mM) induction at 37 °C for 4 h. Cell pellet was separated by centrifugation at 6000 × g for 10 min. The pellet was dissolved in 5 mL of Tris–HCl (50 mM, pH 7.5) buffer and sonicated to lyse the cell. The cell lysate was separated by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C. The expression of the cloned esterase gene was determined using SDS-PAGE following the protocol given by Laemmli28 in intracellular and extracellular fractions.
000 × g for 10 min at 4 °C. The expression of the cloned esterase gene was determined using SDS-PAGE following the protocol given by Laemmli28 in intracellular and extracellular fractions.
Quantification assay
To measure esterase enzyme activity, p-nitrophenyl butyrate (pNPC4) was used as a substrate. A 20 mM of substrate stock solution was prepared. Reaction mixture contained, 100 μL purified esterase enzyme and 100 μL of substrate followed by an incubation at 40 °C. Reaction was stopped after 15 min by cooling the sample on ice and amount of produced p-nitrophenol after reaction was determined spectrophotometrically at 405 nm. The amount of enzyme that released 1 mol of p-nitrophenol per min was determined as one unit of esterase activity.Under standard assay conditions, activity of enzyme is defined as “the quantity of enzyme used to release 1 μmol of p-nitrophenol from substrate”.
Estimation of total proteins
The Bradford reagent was used to quantify total proteins. Total 6 mL reaction mixture was prepared by adding 100 μL of enzyme sample, 900 μL of phosphate buffer and 5 mL of Bradford reagent. In addition, a control with 1 mL of phosphate buffer and 5 mL of Bradford reagent was used. Absorbance of the reaction mixture was measured at 595 nm in order to determine the total protein contents in the mixture.Optimization of recombinant esterase expression
To gain the efficient expressions of cloned esterase gene, many variables were optimized e.g. temperature of incubation, time of induction, pH of medium and inducer concentrations. The maximum expression was obtained by optimizing the temperature ranges of 16–42 °C, pH range of 4.0–9.0, IPTG concentration (0.1–0.6 mM), optical density ranges (0.4–0.8) at the time of induction and time of induction 1–6 h.Enzyme purification
Purification of recombinant esterase enzyme was achieved in two steps. Partial purification was obtained using ammonium sulphate precipitation and final purification using ion exchange chromatography. Partial purification of recombinant esterase enzyme was achieved by saturating it with ammonium sulphate and precipitating it. Ammonium sulphate was added to cell lysate with constant stirring to equilibrate. Stirring was continuous until the 40% saturation of solution was achieved. To pellet out the cell proteins, this mixture was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min. Pellet obtained was dissolved in 50 mM sodium citrate buffer (pH: 7.5) and checked for enzyme activity as well as total protein contents.
000 × g for 10 min. Pellet obtained was dissolved in 50 mM sodium citrate buffer (pH: 7.5) and checked for enzyme activity as well as total protein contents.
Ion exchange chromatography
Final purification of esterase enzyme was used for ion exchange chromatography after dialysis and filtration. The enzyme was passed down to a Bio-scale Mini Unosphere anion exchange column that was pre-equilibrated with phosphate buffer of pH 7.5 (50 mM). Rate of flow was maintained at 1.5 mL min−1. For elution of bounded proteins, 50 mM phosphate buffer (pH 7.5) with 0–1 M NaCl gradient was used. The eluted fractions were pooled, checked for enzymatic activity and subjected to SDS-PAGE to evaluate the purity of esterase enzyme.Molecular weight determination
Purified recombinant esterase enzyme was run on SDS-PAGE for the determination of molecular mass. A broad range (10–230 kDa) ColorPlus pre-stained protein ladder was used for confirmation of exact size.Characterization of esterase enzyme
Impact of EDTA and metal ions
Purified esterase enzyme was treated with EDTA and different metal ions (Ni2+, Mg2+, Ca2+, Cu2+, K1+, and NH41+) ranging from 1 to 10 mM for 1 h at room temperature to observe how they influence the stability of the esterase enzyme. After 1 h of incubation, the remaining enzymatic activity was determined using standard conditions.Effects of inhibitors and surfactants
To investigate the effect of different surfactants and inhibitors on esterase enzyme treatment was carried out at room temperature for 1 h with several inhibitors such as SDS, Tween 20, PMSF, Tween 80, and urea (1–3%). The residual enzymatic activity was evaluated using standard conditions.Substrate specificity
Different substrate like p-NP-butyrate, methyl sinapinate, methyl ferulate and methyl caffeate were used against purified recombinant esterase enzyme to check its specificity towards substrate.Effect of organic solvent
The purified esterase enzyme was incubated with 10–30% (v/v) acetone, ethanol, isopropanol, methanol, and n-butanol for 1 h at room temperature to determine the impact of organic solvents on it. After 1 h, the activity of enzyme was evaluated using optimum parameters.Structure prediction and molecular docking analysis
Protein sequence of esterase enzyme was deduced by utilizing gene sequence through translation tool of expasy [https://expasy.org/tools/dna.html]. 3D structural model was obtained using deduced amino acid sequence through SWISSMODEL. Low energy structural model was selected by comparing five deduced 3D structural models of esterase protein through superimposing. Binding sites of the low energy protein model for p-NP-butyrate, methyl sinapinate, methyl ferulate and methyl caffeate were recognized using AutoDock Vina software.29 Protein structure was prepared by using AutoDockTools30 by adding hydrogen atoms and charges while by using Discovery Studio,31 3D atomic coordinates of ligands were proposed. The grid root was prepared using AutoDockTools for all possible protein–ligand interactions.Compatibility with commercial laundry detergents
Compatibility of esterase enzyme was checked with commercial laundry detergents (SurfExcel, Bright, Ariel), 10 mg mL−1 of each detergent was incubated at 90 °C for an hour in order to inactivate the endogenous enzymes. Cooled fractions of each detergent were incubated with 200 units of purified esterase enzyme at 50 °C for 1 h and enzymatic activity was checked using enzyme assay conditions.Washing efficiency of purified esterase enzyme for oil-stained cloth
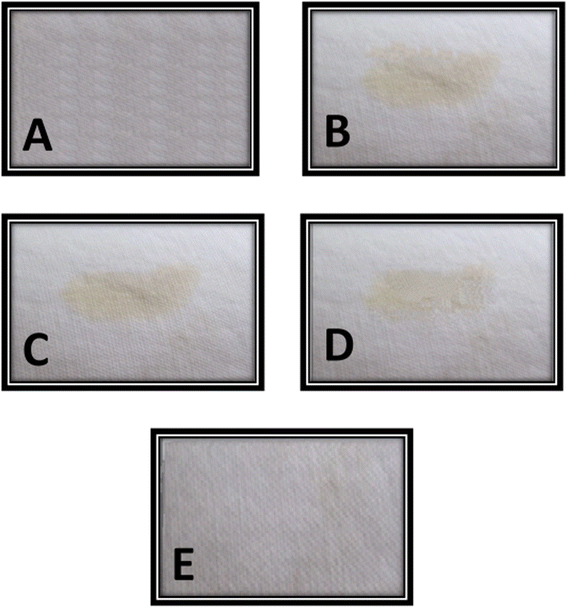
Washing efficiency of purified esterase enzyme was checked in combination of commercial laundry detergent (Bright) for the removal of oil stain from cotton cloth. Five piece of cotton cloth (6 cm × 6 cm) were taken among which four pieces were stained with oil containing solution and one piece without any treatment was used as a positive control. Stained cloth pieces were allowed to air dry at room temperature. Wash performance of purified recombinant esterase enzyme was evaluated by different combinations (i) simple distilled water (ii) distilled water with 10 mg mL−1 detergent (iii) distilled water with 200 U esterase enzyme (iv) distilled water with 10 mg mL−1 detergent and 200 U esterase enzyme. In shaking incubator, the reaction mixtures were incubated for 30 min at 50 °C. Afterwards, all cloth pieces were thoroughly rinsed with distilled water, air dried and observed for the wash efficiency of recombinant purified esterase.Results
Expression of esterase gene of B. thuringiensis in pET-21c(+)
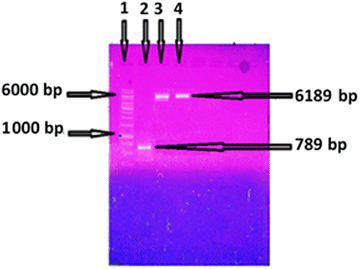
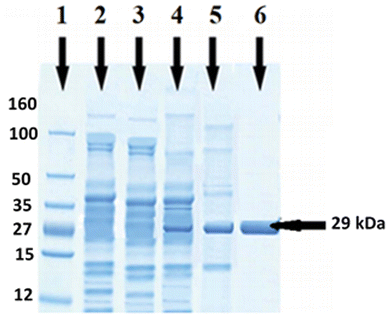
Genomic DNA of B. thuringiensis was isolated following the protocol of Kronstad et al.26 Amplification of esterase gene of B. thuringiensis was carried out using a pair of primer in a thermocycler. An amplicon of 789 bp of esterase gene was obtained after PCR (Fig. 1). Ligation of amplified product of esterase gene and expression vector pET-21c was attained using T4 DNA ligase and transformed into expression strain E. coli BL21 (DE3). Positive clones were confirmed by restriction of plasmid containing esterase gene and colony PCR. A single band of 6189 bp on agarose gel (Fig. 1) indicated that the esterase gene had been successfully ligated with pET-21c and transformed in E. coli BL21. Gene expression was checked by SDS-PAGE in both extra and intracellular samples after induction with IPTG. A well distinct band of 29 kDa was detected on SDS-PAGE in intracellular fraction (Fig. 3), which confirmed the expression of esterase gene of B. thuringiensis in E. coli. The activity of the esterase enzyme was checked in intracellular as well as extracellular samples. In intracellular fraction, the activity of esterase enzyme was calculated 3.75 U mL−1 while in extracellular fraction, the negligible activity of enzyme was found.Optimization of esterase gene expression
Following factors were studied in order to obtain the maximum expression of the cloned esterase gene of B. thuringiensis in E. coli BL21 (DE3).pH of medium and incubation temperature
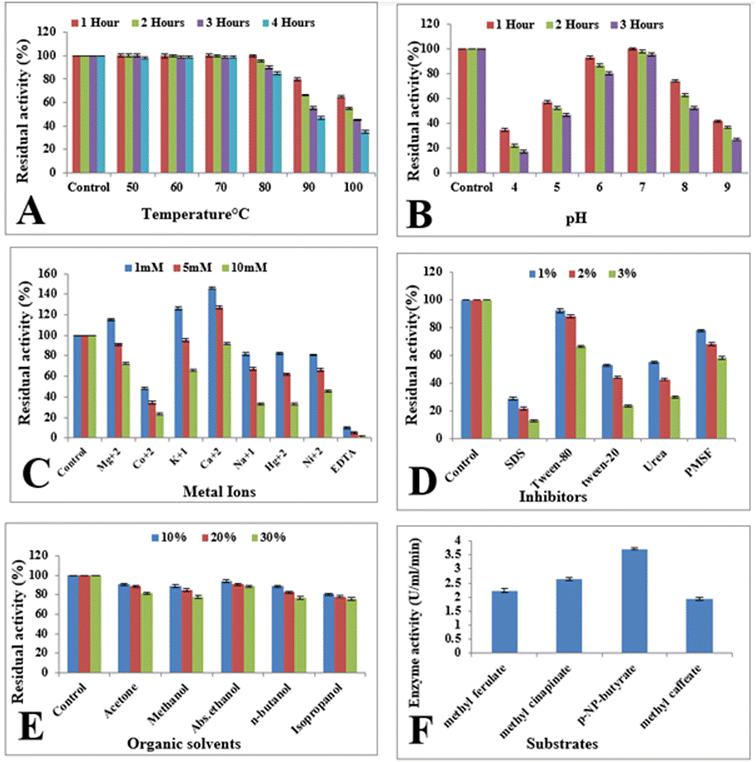
E. coli BL21 (DE3) containing recombinant vector was cultivated at temperatures ranging from 16 to 42 °C in order to check the influence of incubation temperature on the expression of the cloned esterase gene. The gene expression was evaluated in term of enzyme activity. At 22 °C, the maximum activity (2.2 U mL−1) of esterase enzyme was obtained (Fig. 2A). Enzyme activity was found to be decreased upon rise as well as reduction of temperature. At 16 °C, 30 °C, 37 °C and 42 °C, the enzyme activity was 0.93 U mL−1, 1.95 U mL−1, 1.87 U mL−1 and 1.75 U mL−1, respectively (Fig. 2A). The influence of media pH on the expression of esterase gene was determined by using a range of pH (4 to 9) of media. Maximum expression (2.73 U mL−1) of cloned gene was obtained in the culture medium with pH 7.0 (Fig. 2B). The activity of enzyme was found to be diminished at low and high pH. The activity of recombinant esterase enzyme was observed as 1.25 U mL−1, 1.68 U mL−1, 2.15 U mL−1, 1.75 U mL−1, and 0.53 U mL−1 in the culture media having pH 4.0, 5.0, 6.0, 8.0, 9.0, 10.0 and 11.0, respectively, as shown in Fig. 2B.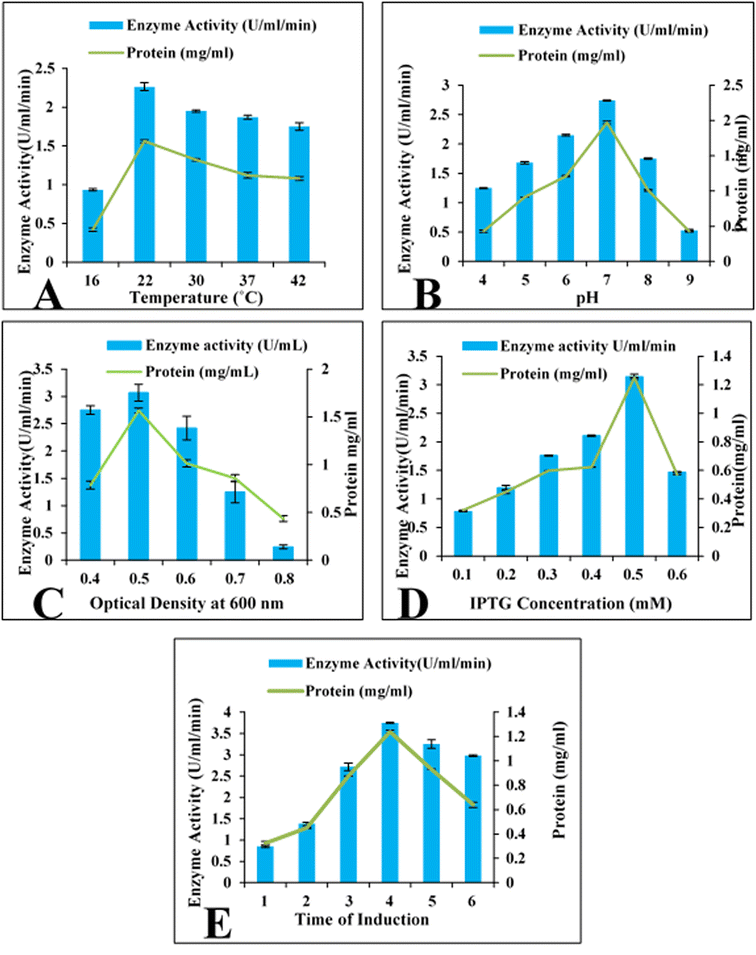 | ||
| Fig. 2 Optimization of cloned esterase gene expression. (A) Incubation temperature (B) pH of the medium (C) optical density (D) IPTG concentration (mM) (E) time of induction (hours). | ||
Optical density of culture
Evaluation of the influence of optical density of bacterial culture on the expression of cloned esterase gene was estimated. Maximum expression (3.01 U mL−1) of esterase gene was observed when induction was given at 0.5 optical density at 600 nm. Decrease expression of gene was observed at elevated optical densities as shown in Fig. 2C. At 0.6, 0.7 and 0.8 optical densities at the time of induction, the activity of esterase enzyme was 2.41 U mL−1, 1.25 U mL−1 and 0.24 U mL−1 (Fig. 2C). Similarly, induction at 0.4 optical density the esterase enzymatic activity was 2.74 U mL−1 as shown in Fig. 2C.Induction time and IPTG concentrations
The effect of the concentration of IPTG on the expression of the esterase gene was also investigated. E. coli BL21 (DE3) harbouring the esterase gene was incubated with 0.1 to 0.6 mM IPTG for 4 h. Maximum activity of enzyme (3.14 U mL−1) was obtained with 0.5 mM IPTG (Fig. 2D). With other concentration of IPTG like 0.1, 0.2, 0.3, 0.4, and 0.6 mM, the esterase enzyme activity was 0.78 U mL−1, 1.19 U mL−1, 1.76 U mL−1, 2.11 U mL−1 and 1.47 U mL−1 (Fig. 2D). To investigate the impact of induction duration, the incubation of recombinant E. coli BL21 culture was done for 1–6 h at 22 °C after induction. It was found that the cloned esterase gene was maximally expressed (3.75 U mL−1) after 4 h of induction period as shown in Fig. 2E. The activity of esterase enzyme was 0.85 U mL−1, 1.38 U mL−1, 2.72 U mL−1, 3.25 U mL−1 and 2.98 U mL−1 after 1, 2, 3, 5 and 6 h of incubation period after induction with IPTG (Fig. 2E).Purification of recombinant esterase enzyme
Ammonium sulphate precipitation method was used for partial purification of recombinant esterase enzyme. Ion exchange chromatography was carried out for further purification of recombinant esterase enzyme. Bio-scale Mini Unospheres pre-packed columns were used for this purpose. Specific activity and purification fold of recombinant enzyme was 126.36 U mg−1, and 48.60 times, respectively after purification, as represented in Table 1.| Purification steps | Esterase activity (U) | Total protein (mg) | Specific activity (U mg−1) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|
| Crude enzyme | 1250 | 465 | 2.6 | 1 | 100 |
| (NH4)2SO4 precipitation (70%) | 765 | 14.56 | 52.33 | 20.13 | 61.2 |
| Ion exchange chromatography | 925 | 7.32 | 126.36 | 48.6 | 74.0 |
Estimation of molecular mass
To determine the molecular mass of purified esterase enzyme SDS-PAGE was used. A well define protein band of 29 kDa was observed in purified intracellular fraction as shown in Fig. 3 while in controls (wild E. coli and un-induced vector containing the esterase gene) no protein band was observed at the same position (Fig. 3).Characterization of purified esterase enzyme
Effect of metal ions and EDTA
The residual activity of esterase enzyme was determined after 1 h pre-incubation of recombinant enzyme in the presence of 1–10 mM EDTA as well metal ions (Hg2+, K1+, Mg2+, Ca2+, Co2+, Na1+, and Ni2+) at room temperature to explore the impact of metal ions and EDTA on it. Results indicated that enzyme activity was enhanced up to 145.8% with the addition of 1 mM Ca2+ as shown in Fig. 4C, while with 1 mM Mg2+ and K1+, the activity of recombinant esterase enzyme was enhanced up to 115% and 126%, respectively. Higher concentration (10 mM) of metal ions seems to slightly inhibit the enzyme activity. On the other hand, EDTA resulted in almost complete inhibition of enzyme activity as shown in Fig. 4C. With the addition of 1 mM EDTA, the activity of enzyme was decreased up to 90% while 5 mM and 10 mM EDTA found to be completely inhibit the enzyme activity as shown in Fig. 4C.Effect of surfactants and inhibitors
To determine the effect of surfactants and inhibitors on recombinant esterase enzyme, purified enzyme was pre-incubated with different concentrations of (1–3%) of SDS, Tween 20, urea, Tween 80, and PMSF at room temperature for 1 h. Remaining activity of recombinant esterase enzyme was calculated after 1 h. Enzyme activity was slightly affected by 1% Tween-80 while 3% Tween-80 resulted in 66% residual activity after 1 h of incubation. Similarly, with 3% Tween-20, PMSF, and urea the activity of recombinant esterase enzyme was observed as 23%, 30% and 58%, respectively as shown in Fig. 4D. Influence of SDS on the activity of esterase enzyme was found to be severe and 3% SDS reduced the activity of enzyme up to 12% as shown in Fig. 4D.Effect of organic solvents
Pre-incubation of recombinant esterase was carried out with 10–30% organic solvents (ethanol, methanol, isopropanol, acetone, n-butanol) for 1 h at room temperature in order to determine their influence on the enzyme activity. The esterase enzyme was shown to be very stable in the presence of organic solvents with higher concentrations (30%). With 30% ethanol, methanol, acetone, isopropanol, and n-butanol, the calculated enzyme activity after 1 h of incubation was 88%, 77%, 75%, 81% and 77%, respectively as shown in Fig. 4E.Substrate specificity
The esterase enzyme was checked for specificity towards its specific substrate by determining its activity against 1.2% p-NP-butyrate, methyl sinapinate, methyl ferulate and methyl caffeate. The maximum esterase enzyme activity (3.25 U mL−1) was attained against p-NP-butyrate. With other substrates like methyl sinapinate, methyl ferulate and methyl caffeate, the enzyme activity was 2.6 U mL−1, 2.2 U mL−1 and 1.92 U mL−1 as shown in Fig. 4F.Molecular docking analysis
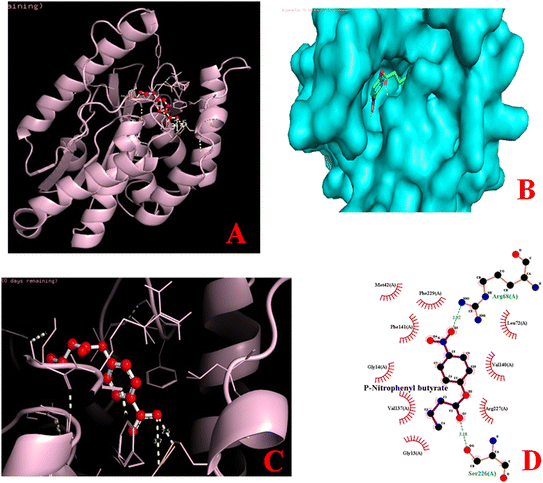
Catalytic behavior of esterase enzyme was elucidated by molecular docking analysis. The 3D structures of protein and ligands (p-NP-butyrate, methyl sinapinate, methyl ferulate, methyl caffeate) were utilized for this purpose. Docking studies with different substrates revealed that 3D structure of esterase contains a specific catalytic cleft for substrate binding (Fig. 5) where the binding was made possible with the help of various interactions such as alkyl bonds, van der Waals forces, and hydrogen bonding. Binding affinities with p-NP-butyrate, methyl sinapinate, methyl ferulate and methyl caffeate were −6.1 kcal mol−1, −6.2 kcal mol−1, −6.1 kcal mol−1 and −6.1 kcal mol−1, respectively. Molecular interaction of enzyme–substrate complexes by ligplot analysis showed that catalytic cleft of esterase enzyme contains different amino acid residue involved in the catalysis of different substrates but the arginine68 and arginine159 are the common amino acid residues involved the catalysis of all above mentioned substrates. Molecular docking simulations performed for p-NP-butyrate indicated that arginine68 and serine226 of esterase enzyme are involved in the enzyme–substrate binding while for binding of methyl sinapinate the amino acid residues were arginine159, glutamine202 and tyrosine191 as shown in Fig. 5. The amino acid residues involved in the binding of methyl ferulate with esterase enzyme were tyrosine191, arginine159 and serine12 but for methyl caffeate binding only arginine68 was involved as shown in Fig. 5.Application of esterase enzyme in detergent industry
Purified recombinant enzyme in this study showed a very good stability and compatibility with commercial laundry detergents. Enzyme showed maximum compatibility and stability in the presence of Bright laundry detergent and showed up to 90% residual activity after 60 min of incubation at 50 °C. However, a slight decrease in enzyme activity was observed after the treatment with other two detergents (SurfExcel & Ariel). Purified esterase enzyme showed 82% residual activity with 10 mg mL−1 SurfExcel and 78% residual activity with 10 mg mL−1 Ariel detergent after 60 min of incubation at 50 °C as shown in Table 2. Washing efficiency of purified recombinant esterase enzyme was determined in combination of 10 mg mL−1 laundry detergent (Bright) and it was observed that oil stained was efficiently removed with combined treatment of esterase enzyme and detergent after 30 min of incubation at 50 °C as shown in Fig. 6.| Sr. no. | Supplements | Residual activity |
|---|---|---|
| 1 | Control | 100% |
| 2 | Bright | 90% |
| 3 | SurfExcel | 82% |
| 4 | Ariel | 78% |
Discussion
Characterization of industrially important enzymes from various sources such as plants and microorganisms, is one of the most important needs for developing powerful biocatalysts for a variety of applications. The strategies for successful expression of genes into E. coli, could be beneficial for the enzymes used in industrial processes for their cost-effective production using cheap sources.32 Present study concerned with expression of an esterase gene from B. thuringiensis into E. coli BL21 (DE3) was carried out using pET-21c as an expression vector and purification and characterization of recombinant enzyme was accomplished for industrial utilization.Amplified fragment of esterase gene from B. thuringiensis was cloned and expressed in pET-21c. The pET expression systems are specifically designed for the expression of recombinant proteins in E. coli.33 Previous reports are present on the use of pET expression systems for the successful expression of recombinant proteins.34–38 Expression of cloned esterase gene as well as molecular weight determination of recombinant protein was evaluated by SDS-PAGE as shown in Fig. 3. SDS-PAGE is most useful method to obtain the high-resolution separation of proteins present in mixture.39 Evaluation of cloned gene expression in E. coli BL21 (DE3) is also reported by utilizing SDS-PAGE.40–43
Optimization of cloned gene expression was carried out by determining the optimal conditions (incubation temperature, pH of the culture medium, inducer concentration, optical density of the culture at the time of induction, induction period) for the maximum production of esterase enzyme. Maximum expression of cloned esterase gene (3.75 U mL−1) was found when culture with optical density of 0.5 at 600 nm was induced with 0.5 mM IPTG for four hours at 22 °C in LB medium with pH 7. Recombinant enzyme production and yield known to be influenced by above mentioned condition.44 Optimization of these variables is important because heterologous expression of genes generates metabolic burden on the host cell resulted in reduction of cell biomass, low expression of gene, as well as plasmid instability.45–47 Optimization of foreign gene expression is also reported in previous studies.48–50 Soumya et al.51 reported a cloned esterase gene from Bacillus subtilis E9 into E. coli using pTac Bs-est vector, the overexpression of which was attained by IPTG induction by optimizing the time required for over expression of the gene and culture media. Similarly, Fan et al.52 optimized different parameters for the maximum production of a recombinant esterase enzyme from E. coli and reported maximum production with LB medium having pH 5.5, with 5 h pre-induction period and 32 h after induction period at 23 °C.
Purification of recombinant esterase enzyme in this study was attained up to 48.6-fold with 78% recovery using anion exchange chromatography. Ion exchange chromatography is one of the most effectual methods for the purification of charged molecules. This technique has been successfully employed for the purification of different recombinant proteins for various purposes.53–57 The molecular mass of purified recombinant esterase enzyme was 29 kDa as analyzed by SDS-PAGE (Fig. 3). Previous study reported the molecular weight of esterase enzyme from B. gelatini KACC 12197 as 42 kDa.32 Brod et al.58 also reported the molecular weight of a recombinant esterase enzyme from Lactobacillus plantarum as 38 kDa by using SDS-PAGE.
Determination of the stability of esterase enzyme was evaluated by checking various characters like thermostability, pH stability, stability in the presence of organic solvents, effect of different inhibitors and surfactants on its stability and its specificity towards different substrates, were studied as shown in Fig. 4. Thermostability is an important parameter of enzyme's study, specifically for those which are employed in industrial sectors as temperature fluctuations during industrial process might affect the activity of enzyme and high temperature could be result in denaturation of the enzyme. Purified recombinant esterase enzyme in this study was found to be very stable at high temperature (up to 80 °C) for 1 h and retained its activity up to 95% as shown in Fig. 4A. However, prolonged exposure (4 h) to high temperature (100 °C) seems to denature the enzyme and almost 90% reduction in enzyme activity was observed. Various reports are present on the determination of thermostability of esterase enzyme from various microorganisms. Hamid et al.59 reported an esterase enzyme from Aspergillus niger with 56% activity at 80 °C after 4 h of incubation. While Tang et al.60 studied a glucuronoyl esterase from Thielavia terrestris and reported its thermostability at 55 °C. Rong et al.61 cloned and characterize the novel acetyl xylan esterase which showed a high stability across a broader range of temperature. Hou et al.62 reported a thermostable esterase which showed stability across a wide temperature range and retained more than 60% activity between 4 °C and 60 °C after 24 hours of incubation.
Enzyme stability at broad pH range is another factor which greatly influence the utilization of enzyme in industrial applications as change in pH, affects the shape and structure of the proteins by ionization of atoms, molecules and amino acids leading to the inhibition of enzyme activity. Esterase enzyme was found to be stable at broad pH range (4–11) as shown in Fig. 4B. The enzyme retained 74% activity at pH 4.0 and 55% activity at pH 11.0 after 1 h of incubation at room temperature. Zhang et al.,63 Adıgüzel64 and Ayna et al.65 also reported esterase enzymes which were active at pH range 4.0–8.0, 4.0–10.0 and 4.0 to 9.0, respectively.
Effect of metal ions on the activity and stability of recombinant esterase enzyme was checked in order to determine its metal ion dependency to become active. Ca2+ and Mg2+ was seemed to enhance the enzyme's residual activity, resulting in increased stability at high temperatures. The Ca2+ and Mg2+ ions were appeared to make the esterase conformation more stable. Ca2+ is expected to have a structural function inside the protein as a substitute for a component of the catalytic domain. Reports on Ca2+ dependency of esterases for structural and functional stability are present.66,67 However, Rodríguez et al.68 reported an alkaline esterase from bovine rumen which was inhibited by metal ions. Lee et al.69 described about an esterolytic enzyme, the activity of which was enhanced in the presence of Mg2+ and Ca2+. In another report, Zhang et al.70 purified an esterase enzyme from B. licheniformis with enhanced enzymatic activity in the presence of various metal ions such as Zn2+, Mn2+, Mg2+.
With respect to the effect of detergents, the results in this study are in accordance to the previous report.71 The residual activity of esterase enzyme in this study was not significantly affected by Tween 80 even at higher concentration whereas urea and SDS inhibited the residual activity of esterase as shown in Fig. 4D. Among inhibitors, PMSF decreased the residual activity of esterase enzyme which may be due to the alkylation of serine hydroxyl group.72 Reduced enzyme activity and stability reflects that histidine and serine may play a role in the enzyme's catalytic activity and substrate binding. Hamid et al.59 also reported the reduction in enzyme activity in the presence of SDS. Effect of organic solvents on enzyme's stability and activity was also determined in order for its utilization in industrial applications. The purified esterase enzyme found to be resistant in the presence of organic solvents used in this study (acetone, methanol, ethanol, n-butanol, isopropanol) even at higher concentration as shown in Fig. 4E. Similar characteristics were found in thermostable carboxylesterase from hyperthermophilic Thermotoga maritima.73 Zhu et al.74 also investigated the effect of several organic solvents on esterase enzyme from Geobacillus sp. JM6 and discovered that esterase enzyme remained stable at a concentration of around 10%. Like previously reported esterases from different sources, enzyme in this study was found to be more specific towards p-NP-butyrate as shown in Fig. 4F.64,75,76 These results are also in accordance to the in silico protein–ligand interaction. Molecular docking simulations performed for p-NP-butyrate indicated that specific bonding of esterase enzyme in this study with p-NP-butyrate occurred at arginine68 and serine226 of esterase as shown in Fig. 5.
Above mentioned characters of purified recombinant esterase enzyme give a privilege to this enzyme for efficient utilization in detergent industry. Wide pH range and good temperature stability offer the advantages for industrial utility of microbial enzymes. Other important characteristics like metal resistant as well as organic solvents and detergent resistant are the good attributes of the enzyme for detergent industry.77 Good compatibility (90%) of esterase enzyme with commercial detergent (Bright) also provide an advantage for its utilization in detergent manufacturing.44,78 Purified recombinant esterase enzyme's utilization along with detergent efficiently removed the oil stain from cotton cloth as shown in Fig. 6 which proved it as a potential candidate for the production of eco-friendly and skin protecting detergents at industrial level.
Conclusion
Modern research work is going to explore more and more efficient industrial candidates in order to sustain the industries for the fulfilment of present and future demands. In this regard, an esterase gene from B. thuringiensis was cloned and successfully expressed in the E. coli system to explore its enzymatic properties and utilization in detergent industry. Cloned esterase showed high thermostability and broad pH range with an excellent tolerance against organic solvents and increased activity in the presence of metal ions. On the basis of all parameters studied in this research work, recombinant alkaline esterase enzyme could be utilized in detergents industry in order to promote the industry in eco-friendly environment.Author contributions
All authors contributed in the present research work following is the specific contribution of authors: Asma Zafar: resources (provision of reagent), formal analysis, methodology. Ziaur Rahman: original manuscript writing, methodology, conceptualization. Javairia Makhdoom: data interpretation. Javeria Tariq: manuscript writing, and reviewing. Mahjabeen: discussion writing. Zulqurnain Ali: rephrasing. Attia Hamid: data interpretation. Eeza Shafique: experimental work. Muhammad Nauman Aftab: supervision (writing – reviewing and editing).Conflicts of interest
Authors declare that they have no financial and non-financial interests associated within present research work.Acknowledgements
Present research work is a part of ongoing research project at University of Central Punjab, Lahore, Pakistan and no specific funding is available for that project.References
- R. Woodward, The organisation for economic co-operation and development (OECD), Routledge, 2009, DOI:10.4324/9780203875773.
- P. Koller and L. Cashman, Implementing EC environmental law. Compliance promotion and enforcement by the European Commission, J. Eur. Environ. Plan. Law, 2009, 6, 1–12 Search PubMed.
- C. Benoît-Norris, G. Vickery-Niederman, S. Valdivia, J. Franze, M. Traverso, A. Ciroth, B. Mazijn, A. Ciroth and B. Mazijn, Introducing the UNEP/SETAC methodological sheets for subcategories of social LCA, Int. J. Life Cycle Assess., 2011, 16, 682–690, DOI:10.1007/s11367-011-0301-y.
- K. R. Jegannathan and P. H. Nielsen, Environmental assessment of enzyme use in industrial production–a literature review, J. Clean. Prod., 2013, 42, 228–240, DOI:10.1016/j.jclepro.2012.11.005.
- U. T. Bornscheuer and R. J. Kazlauskas, Hydrolases in organic synthesis: regio- and stereoselective biotransformations, John Wiley & Sons, 2006, DOI:10.1002/3527607544.
- R. Hofer, Sustainable solutions for modern economies, Royal Society of Chemistry, 2009, 10.1039/9781847552686.
- R. Wohlgemuth, The locks and keys to industrial biotechnology, New Biotechnol., 25, 204–213, DOI:10.1016/j.nbt.2009.01.002.
- O. K. Othmer, Encyclopedia of chemical technology, Watcher, 2021 Search PubMed.
- X. Fan, X. Niehus and G. Sandoval, Lipases as biocatalyst for biodiesel production, Lipases Phospholipases, 2012, pp. 471–483, DOI:10.1007/978-1-61779-600-5_27.
- L. Casas-Godoy, S. Duquesne, F. Bordes, G. Sandoval and A. Marty, Lipases: an overview, Lipases Phospholipases, 2012, pp. 3–30, DOI:10.1007/978-1-61779-600-5_1.
- P. Chandra, R. Singh and P. K. Arora, Microbial lipases and their industrial applications: a comprehensive review, Microb. Cell Fact., 2020, 19, 1–42, DOI:10.1186/s12934-020-01428-8.
- U. T. Bornscheuer, Microbial carboxyl esterases: classification, properties and application in biocatalysis, FEMS Microbiol. Rev., 2002, 26, 73–81, DOI:10.1111/j.1574-6976.2002.tb00599.x.
- J. A. Bååth, S. Mazurkewich, J.-C. N. Poulsen, L. Olsson, L. L. Leggio and J. Larsbrink, Structure–function analyses reveal that a glucuronoyl esterase from Teredinibacter turnerae interacts with carbohydrates and aromatic compounds, J. Biol. Chem., 2019, 294, 6635–6644, DOI:10.1074/jbc.RA119.007831.
- H. P. Sharma, H. Patel and S. Sharma, Enzymatic extraction and clarification of juice from various fruits—a review, Trends Post Harvest Technol., 2014, 2, 1–14, DOI:10.1080/10408398.2014.977434.
- B. Tyagi, C. D. Chudasama and R. V. Jasra, Determination of structural modification in acid activated montmorillonite clay by FT-IR spectroscopy, Spectrochim. Acta, Part A, 2006, 64, 273–278, DOI:10.1016/j.saa.2005.07.018.
- S. Gürkök, Microbial enzymes in detergents: a review, Int. J. Sci. Eng. Res., 2019, 10, 75–81 Search PubMed.
- F. Hasan, A. A. Shah, S. Javed and A. Hameed, Enzymes used in detergents: lipases, Afr. J. Biotechnol., 2010, 9, 4836–4844, DOI:10.4314/AJB.V9I31.
- B. Mesa-Pereira, M. C. Rea, P. D. Cotter, C. Hill and R. P. Ross, Heterologous expression of biopreservative bacteriocins with a view to low cost production, Front. Microbiol., 2018, 9, 1654, DOI:10.3389/fmicb.2018.01654.
- G. L. Rosano and E. A. Ceccarelli, Recombinant protein expression in Escherichia coli: advances and challenges, Front. Microbiol., 2014, 5, 172, DOI:10.3389/fmicb.2014.00172.
- A. Zafar, S. Aftab, M. N. Aftab, H. Mukhtar and M. A. Saleem, Purification and Characterization of cloned Endo-1, 4-β-Glucanase gene of Bacillus licheniformis for saccharification of plant biomass, Pak. J. Bot., 2019, 51, 957–966, DOI:10.30848/PJB2019-3(27).
- A. Zafar, M. N. Aftab, A. Manzoor, I. Iqbal, Z. Uddin, A. Kaleem, A. R. Khooharo and M. A. Saleem, Purification and Characterization of a recombinant β-Xylosidase from Bacillus licheniformis ATCC 14580 into E. coli Bl21, Pak. J. Pharm. Sci., 2018, 31, 2755–2762 CAS.
- M. N. Aftab, A. Zafar and A. R. Awan, Expression of thermostable β-xylosidase in Escherichia coli for use in saccharification of plant biomass, Bioengineered, 2017, 8, 665–669, DOI:10.1080/21655979.2016.1267884.
- I. Iqbal, M. Aftab, M. Afzal and A. Kaleem, Characterization of Geobacillus stearothermophilus protease for detergent industry, Rev. Mex. Ing. Quim., 2020, 19, 267–279, DOI:10.24275/rmiq/Bio1647.
- A. Saima, N. A. Muhammad, M. J. Muhammad, Z. Asma and I. Irfana, Cloning and expression of endo-1, 4β-glucanase gene from Bacillus licheniformis ATCC 14580 into Escherichia coli BL21 (DE 3), Afr. J. Biotechnol., 2012, 11, 2846–2854, DOI:10.5897/AJB11.2902.
- C. A. Mazzucotelli, M. d. R. Moreira and M. R. Ansorena, Statistical optimization of medium components and physicochemical parameters to simultaneously enhance bacterial growth and esterase production by Bacillus thuringiensis, Can. J. Microbiol., 2015, 62, 24–34 CrossRef PubMed.
- J. Kronstad, H. Schnepf and H. Whiteley, Diversity of locations for Bacillus thuringiensis crystal protein genes, J. Bacteriol., 1983, 154, 419–428, DOI:10.1128/jb.154.1.419-428.1983.
- S. N. Cohen, DNA cloning: a personal view after 40 years, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15521–15529, DOI:10.1073/pnas.1313397110.
- U. K. Laemmli, Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature, 1970, 227, 680–685, DOI:10.1038/227680a0.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31, 455–461 CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- D. S. Biovia, Discovery Studio modeling environment, San Diego, 2016 Search PubMed.
- J. Kim, L. Deng, E. Hong and Y. Ryu, Cloning and characterization of a novel thermostable esterase from Bacillus gelatini KACC 12197, Protein Expression Purif., 2015, 116, 90–97, DOI:10.1016/j.pep.2015.08.009.
- F. W. Studier and B. A. Moffatt, Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes, J. Mol. Biol., 1986, 189, 113–130, DOI:10.1016/0022-2836(86)90385-2.
- J. Sun, H. Wang, W. Lv, C. Ma, Z. Lou, H. Yao and Y. Dai, Cloning and expression of a thermostable β-1, 3-1, 4-glucanase from Bacillus amyloliquefaciens ATCC 23350, Ann. Microbiol., 2012, 62, 1235–1242, DOI:10.1007/s13213-011-0366-7.
- I. Iqbal, M. N. Aftab, M. Afzal, A. Ur-Rehman, S. Aftab, A. Zafar, Z. Ud-Din, A. R. Khuharo, J. Iqbal and I. Ul-Haq, Purification and characterization of cloned alkaline protease gene of Geobacillus stearothermophilus, J. Basic Microbiol., 2015, 55, 160–171, DOI:10.1002/jobm.201400190.
- A. Zafar, M. N. Aftab, S. Aftab and I. Iqbal, Cloning, purification and characterization of a highly thermostable amylase gene of Thermotoga petrophila into Escherichia coli, Appl. Biochem. Biotechnol., 2016, 178, 831–848, DOI:10.1007/s12010-015-1912-8.
- R. Zeng, Q. Hu, X.-Y. Yin, H. Huang, J.-B. Yan, Z.-W. Gong and Z.-H. Yang, Cloning a novel endo-1, 4-β-d-glucanase gene from Trichoderma virens and heterologous expression in E. coli, AMB Express, 2016, 6, 1–7, DOI:10.1186/s13568-016-0282-0.
- A. Zafar, M. N. Aftab, A. Asif, A. Karadag, L. Peng, H. U. Celebioglu, M. S. Afzal, A. Hamid and I. Iqbal, Efficient biomass saccharification using a novel cellobiohydrolase from Clostridium clariflavum for utilization in biofuel industry, RSC Adv., 2021, 11, 9246–9261, 10.1039/D1RA00545F.
- A. B. Nowakowski, W. J. Wobig and D. H. Petering, Native SDS-PAGE: high resolution electrophoretic separation of proteins with retention of native properties including bound metal ions, Metallomics, 2014, 6, 1068–1078, 10.1039/c4mt00033a.
- C. L. Kielkopf, W. Bauer and I. L. Urbatsch, Expression of cloned genes in E. coli using IPTG-inducible promoters, Cold Spring Harb. Protoc., 2021, 2021, pdb.prot102137, DOI:10.1101/pdb.prot102137.
- M. M. Barbosa, A. I. Kanno, G. C. Barazzone, D. Rodriguez, V. Pancakova, M. Trentini, E. L. Faquim-Mauro, A. P. Freitas, M. I. Khouri and J. Lobo-Silva, Robust immune response induced by Schistosoma mansoni TSP-2 antigen coupled to bacterial outer membrane vesicles, Int. J. Nanomed., 2021, 16, 7153, DOI:10.2147/IJN.S315786.
- N. Ahmed, B. Afroze, R. Abbas, M. A. Khan, M. Akram, S. Tahir, S. Bakht, A. Munir and A. A. Shahid, Method for efficient soluble expression and purification of recombinant human interleukin-15, Protein Expression Purif., 2021, 177, 105746, DOI:10.1016/j.pep.2020.105746.
- F. Yan, X. Ye, C. Li, P. Wang, S. Chen and H. Lin, Isolation, purification, gene cloning and expression of antifungal protein from Bacillus amyloliquefaciens MG-3, Food Chem., 2021, 349, 129130, DOI:10.1016/j.foodchem.2021.129130.
- A. Zafar, S. Yousaf, M. Aftab, A. Hamid, J. Wattoo, A. Masood and H. Mubeen, Purification and characterization of a novel pullulanase enzyme from Bacillus thuringiensis for detergent industry, Rev. Mex. Ing. Quim., 2022, 21, Bio2699, DOI:10.24275/rmiq/Bio2699.
- R. Bellaouchi, H. Abouloifa, Y. Rokni, A. Hasnaoui, N. Ghabbour, A. Hakkou, A. Bechchari and A. Asehraou, Characterization and optimization of extracellular enzymes production by Aspergillus niger strains isolated from date by-products, J. Genet. Eng. Biotechnol., 2021, 19, 1–8, DOI:10.1186/s43141-021-00145-y.
- W. E. Bentley, R. H. Davis and D. S. Kompala, Dynamics of induced CAT expression in E. coli, Biotechnol. Bioeng., 1991, 38, 749–760, DOI:10.1002/bit.260380709.
- W. E. Bentley, N. Mirjalili, D. C. Andersen, R. H. Davis and D. S. Kompala, Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria, Biotechnol. Bioeng., 1990, 35, 668–681, DOI:10.1002/bit.260350704.
- B. R. Glick, Metabolic load and heterologous gene expression, Biotechnol. Adv., 1995, 13, 247–261, DOI:10.1016/0734-9750(95)00004-A.
- A. Fazaeli, A. Golestani, M. Lakzaei, S. S. R. Varaei and M. Aminian, Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1, PLoS One, 2019, 14, e0212217, DOI:10.1371/journal.pone.0212217.
- M. Mühlmann, E. Forsten, S. Noack and J. Büchs, Optimizing recombinant protein expression via automated induction profiling in microtiter plates at different temperatures, Microb. Cell Fact., 2017, 16, 1–12, DOI:10.1186/s40643-014-0010-z.
- P. Soumya and J. Kochupurackal, An Esterase with Increased Acetone Tolerance from Bacillus subtilis E9 over Expressed in E. coli BL21 Using pTac Bs-est Vector, Mol. Biotechnol., 2022, 1–11 Search PubMed.
- G. Fan, Y. Zhu, Z. Fu, B. Sun, C. Teng, R. Yang and X. Li, Optimization of fermentation conditions for the production of recombinant feruloyl esterase from Burkholderia pyrrocinia B1213, 3 Biotech, 2020, 10, 1–15 CrossRef PubMed.
- M. Gutiérrez-González, C. Farías, S. Tello, D. Pérez-Etcheverry, A. Romero, R. Zúñiga, C. H. Ribeiro, C. Lorenzo-Ferreiro and M. C. Molina, Optimization of culture conditions for the expression of three different insoluble proteins in Escherichia coli, Sci. Rep., 2019, 9, 1–11, DOI:10.1038/s41598-019-53200-7.
- C. T. Carvalho, S. D. Oliveira Júnior, W. B. Lima, F. G. Medeiros, A. L. O. Leitão, J. M. Dantas, E. S. Santos, G. R. Macêdo and F. C. Sousa Júnior, Recovery of β-galactosidase produced by Kluyveromyces lactis by ion-exchange chromatography: Influence of pH and ionic strength parameters, An. Acad. Bras. Cienc., 2022, 94, e20200752, DOI:10.1590/0001-3765202220200752.
- A. K. El-Sayed, M. I. Abou-Dobara, A. A. El-Fallal and N. F. Omar, Heterologous expression, purification, immobilization and characterization of recombinant α-amylase AmyLa from Laceyella sp. DS3, Int. J. Biol. Macromol., 2019, 132, 1274–1281, DOI:10.1016/j.ijbiomac.2019.04.010.
- A. Taddia, M. Rito-Palomares, K. Mayolo-Deloisa and G. Tubio, Purification of xylanase from Aspergillus niger NRRL3 extract by an integrated strategy based on aqueous two-phase systems followed by ion exchange chromatography, Sep. Purif. Technol., 2021, 255, 117699, DOI:10.1016/j.seppur.2020.117699.
- P. Chai, X. Pu, J. Li, X. Xia, J. Ge, A. Luo, H. Su, W. Zhang and J. Ma, Expression and purification of tetanus toxin fragment C in Escherichia coli BL21 (DE3), Protein Pept. Lett., 2020, 27, 1132–1140, DOI:10.2174/0929866527666200528113327.
- F. C. A. Brod, J. Vernal, J. B. Bertoldo, H. Terenzi and A. C. M. Arisi, Cloning, expression, purification, and characterization of a novel esterase from Lactobacillus plantarum, Mol. Biotechnol., 2010, 44, 242–249, DOI:10.1007/s12033-009-9232-2.
- A. Hamid, Z. Hussain, M. Tayyab, A. Zafar, M. Nawaz, S. Ali, A. Rehman and M. Aftab, Production and characterization of a thermostable extracellular esterase from Aspergillus niger, Rev. Mex. Ing. Quim., 2021, 20, 839–852, DOI:10.24275/rmiq/Bio2034.
- J. Tang, L. Long, Y. Cao and S. Ding, Expression and characterization of two glucuronoyl esterases from Thielavia terrestris and their application in enzymatic hydrolysis of corn bran, Appl. Microbiol. Biotechnol., 2019, 103, 3037–3048, DOI:10.1007/s00253-019-09662-w.
- Z. Rong, Y.-Y. Huo, S.-L. Jian, Y.-H. Wu and X.-W. Xu, Characterization of a novel alkaline esterase from Altererythrobacter epoxidivorans CGMCC 1.7731T, Prep. Biochem. Biotechnol., 2018, 48, 113–120 CrossRef CAS PubMed.
- Y. Hou, Z. Yang, Y. Yin, Z. Meng, J. Wang, T. Zhao and Q. Yang, Expression of an alkaline feruloyl esterases from thermophilic Chaetomium thermophilum and its boosting effect on delignification of pulp, Enzyme Microb. Technol., 2021, 150, 109859 CrossRef CAS PubMed.
- S.-B. Zhang, L. Wang, Y. Liu, H.-C. Zhai, J.-P. Cai and Y.-S. Hu, Expression of feruloyl esterase A from Aspergillus terreus and its application in biomass degradation, Protein Expression Purif., 2015, 115, 153–157, DOI:10.1016/j.pep.2015.08.015.
- A. O. Adıgüzel, Production and characterization of thermo-, halo-and solvent-stable esterase from Bacillus mojavensis TH309, Biocatal. Biotransform., 2020, 38, 210–226, DOI:10.1080/10242422.2020.1715370.
- C. Ayna, Y. Kolcuoğlu, F. Öz, A. Colak and N. S. Ertunga, Purification and characterization of a pH and heat stable esterase from Geobacillus sp. TF17, Turk. J. Biochem., 2013, 38, 329–336, DOI:10.5505/tjb.2013.36035.
- T. C. Leung, Z. Qu, W. Nong, J. H. Hui and S. M. Ngai, Proteomic Analysis of the Venom of Jellyfishes Rhopilema esculentum and Sanderia malayensis, Mar. Drugs, 2020, 18, 655, DOI:10.3390/md18120655.
- N. Noby, H. Saeed, A. M. Embaby, I. V. Pavlidis and A. Hussein, Cloning, expression and characterization of cold active esterase (EstN7) from Bacillus cohnii strain N1: a novel member of family IV, Int. J. Biol. Macromol., 2018, 120, 1247–1255, DOI:10.1016/j.ijbiomac.2018.07.169.
- M. C. Rodríguez, I. Loaces, V. Amarelle, D. Senatore, A. Iriarte, E. Fabiano and F. Noya, Est10: a novel alkaline esterase isolated from bovine rumen belonging to the new family XV of lipolytic enzymes, PLoS One, 2015, 10, e0126651, DOI:10.1371/journal.pone.0126651.
- H. Y. Lee, D. Y. Cho, I. Ahmad, H. M. Patel, M. J. Kim, J. G. Jung, E. H. Jeong, M. A. Haque and K. M. Cho, Mining of a novel esterase (est3S) gene from a cow rumen metagenomic library with organosphosphorus insecticides degrading capability: catalytic insights by site directed mutations, docking, and molecular dynamic simulations, Int. J. Biol. Macromol., 2021, 190, 441–455 CrossRef CAS PubMed.
- M. Zhang, W. Lai, Y. Zhu, S. Chen, K. Zhou, X. Ao, L. He, Y. Yang, L. Zou and A. Liu, Purification and characterization of a novel cypermethrin-hydrolyzing esterase from Bacillus licheniformis B-1, J. Food Sci., 2021, 86, 1475–1487 CrossRef CAS PubMed.
- G. López, J. Chow, P. Bongen, B. Lauinger, J. Pietruszka, W. R. Streit and S. Baena, A novel thermoalkalostable esterase from Acidicaldus sp. strain USBA-GBX-499 with enantioselectivity isolated from an acidic hot springs of Colombian Andes, Appl. Microbiol. Biotechnol., 2014, 98, 8603–8616, DOI:10.1007/s00253-014-5775-7.
- R. S. Labow, E. Meek, L. A. Matheson and J. P. Santerre, Human macrophage-mediated biodegradation of polyurethanes: assessment of candidate enzyme activities, Biomaterials, 2002, 23, 3969–3975, DOI:10.1016/s0142-9612(02)00137-0.
- S. Kakugawa, S. Fushinobu, T. Wakagi and H. Shoun, Characterization of a thermostable carboxylesterase from the hyperthermophilic bacterium Thermotoga maritima, Appl. Microbiol. Biotechnol., 2007, 74, 585–591, DOI:10.1007/s00253-006-0687-9.
- Y. Zhu, W. Zheng, H. Ni, H. Liu, A. Xiao and H. Cai, Molecular cloning and characterization of a new and highly thermostable esterase from Geobacillus sp. JM6, J. Basic Microbiol., 2015, 55, 1219–1231, DOI:10.1002/jobm.201500081.
- Y. Fang, S. Wang, S. Liu and Y. Jiao, Discovery a novel organic solvent tolerant esterase from Salinispora arenicola CNP193 through genome mining, Int. J. Biol. Macromol., 2015, 80, 334–340, DOI:10.1016/j.ijbiomac.2015.06.045.
- Y.-J. Zhang, C.-S. Chen, H.-T. Liu, J.-L. Chen, Y. Xia and S.-J. Wu, Purification, identification and characterization of an esterase with high enantioselectivity to (S)-ethyl indoline-2-carboxylate, Biotechnol. Lett., 2019, 41, 1223–1232, DOI:10.1007/s10529-019-02727-w.
- P. M. d. Souza and P. d. O. Magalhães, Application of microbial α-amylase in industry-A review, Braz. J. Microbiol., 2010, 41, 850–861, DOI:10.1590/S1517-83822010000400004.
- I. Lahmar, H. El Abed, B. Khemakhem, H. Belghith, F. Ben Abdallah and K. Belghith, Optimization, purification, and starch stain wash application of two new α-amylases extracted from leaves and stems of Pergularia tomentosa, BioMed Res. Int., 2017, 2017, 6712742, DOI:10.1155/2017/6712742.
| This journal is © The Royal Society of Chemistry 2022 |