Open Access Article
Open Access ArticleExpanding the functionality of proteins with genetically encoded dibenzo[b,f][1,4,5]thiadiazepine: a photo-transducer for photo-click decoration†
Qin
Xiong
,
Tingting
Zheng
,
Xin
Shen
,
Baolin
Li
,
Jielin
Fu
,
Xiaohu
Zhao
,
Chunxia
Wang
and
Zhipeng
Yu
 *
*
Key Laboratory of Green Chemistry and Technology of Ministry of Education, College of Chemistry, Sichuan University, 29 Wangjiang Road, Chengdu, 610064, P. R. China. E-mail: zhipengy@scu.edu.cn
First published on 28th February 2022
Abstract
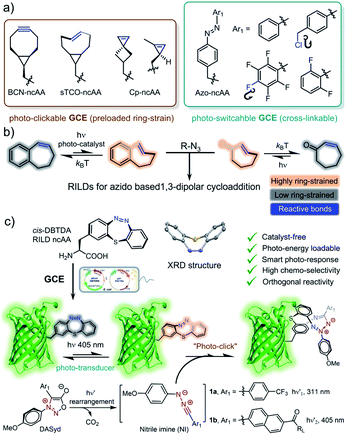
Genetic incorporation of novel noncanonical amino acids (ncAAs) that are specialized for the photo-click reaction allows the precisely orthogonal and site-specific functionalization of proteins in living cells under photo-control. However, the development of a ![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) ing-strain
ing-strain ![[i with combining low line]](https://www.rsc.org/images/entities/i_char_0069_0332.gif) n situ
n situ ![[l with combining low line]](https://www.rsc.org/images/entities/char_006c_0332.gif) oadable
oadable ![[d with combining low line]](https://www.rsc.org/images/entities/char_0064_0332.gif) ipolarophile (RILD) as a genetically encodable reporter for photo-click bioconjugation with spatiotemporal controllability is quite rare. Herein, we report the design and synthesis of a photo-switchable
ipolarophile (RILD) as a genetically encodable reporter for photo-click bioconjugation with spatiotemporal controllability is quite rare. Herein, we report the design and synthesis of a photo-switchable ![[d with combining low line]](https://www.rsc.org/images/entities/char_0064_0332.gif) i
i![[b with combining low line]](https://www.rsc.org/images/entities/char_0062_0332.gif) enzo[b,f][1,4,5]
enzo[b,f][1,4,5]![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif) hia
hia![[d with combining low line]](https://www.rsc.org/images/entities/char_0064_0332.gif) iazepine-based
iazepine-based ![[a with combining low line]](https://www.rsc.org/images/entities/char_0061_0332.gif) lanine (DBTDA) ncAA, together with the directed evolution of a pyrrolysyl-tRNA synthetase/tRNACUA pair (PylRS/tRNACUA), to encode the DBTDA into recombinant proteins as a RILD in living E. coli cells. The fast-responsive photo-isomerization of the DBTDA residue can be utilized as a converter of photon energy into ring-strain energy to oscillate the conformational changes of the parent proteins. Due to the photo-activation of RILD, the photo-switching of the DBTDA residue on sfGFP and OmpC is capable of promoting the photo-click ligation with diarylsydnone (DASyd) derived probes with high efficiency and selectivity. We demonstrate that the genetic code expansion (GCE) with DBTDA benefits the studies on the distribution of decorated OmpC-DBTD on specific E. coli cells under a spatiotemporal resolved photo-stimulation. The GCE for encoding DBTDA enables further functional diversity of artificial proteins in living systems.
lanine (DBTDA) ncAA, together with the directed evolution of a pyrrolysyl-tRNA synthetase/tRNACUA pair (PylRS/tRNACUA), to encode the DBTDA into recombinant proteins as a RILD in living E. coli cells. The fast-responsive photo-isomerization of the DBTDA residue can be utilized as a converter of photon energy into ring-strain energy to oscillate the conformational changes of the parent proteins. Due to the photo-activation of RILD, the photo-switching of the DBTDA residue on sfGFP and OmpC is capable of promoting the photo-click ligation with diarylsydnone (DASyd) derived probes with high efficiency and selectivity. We demonstrate that the genetic code expansion (GCE) with DBTDA benefits the studies on the distribution of decorated OmpC-DBTD on specific E. coli cells under a spatiotemporal resolved photo-stimulation. The GCE for encoding DBTDA enables further functional diversity of artificial proteins in living systems.
Introduction
With many advances over the last two decades, click chemistry1 has been increasingly exploited in the development of therapeutic agents,2 the functionalization of materials,3 and labelling biomacromolecules in living systems.4 Taking into account its unique spatiotemporal controllability,5 photo-click chemistry6 has had a wide impact on a broad range of interdisciplinary research because of its utilization of photon energy7 as well as the advantages of non-invasive manipulation.8 To fulfill the stringent requirements for the purpose of bioorthogonal ligation, great efforts have been made to improve the photo-click strategy,9 including the photo-induced 1,3-dipolar cycloaddition of tetrazole-alkene/alkyne systems,10 photo-click chemistry between diarylsydnone (DASyd) and alkenes/alkynes,11 light-triggered Diels–Alder reactions,12 and photo-mediated strain promoted azide–alkyne cycloaddition (photo-SPAAC).13 However, the diversity of bioorthogonal reporters dedicated to photo-click chemistry is quite limited (Scheme 1a).14 This predicament has been addressed by the development of![[r with combining low line]](https://www.rsc.org/images/entities/char_0072_0332.gif) ing-strain
ing-strain ![[i with combining low line]](https://www.rsc.org/images/entities/i_char_0069_0332.gif) n situ
n situ ![[l with combining low line]](https://www.rsc.org/images/entities/char_006c_0332.gif) oadable
oadable ![[d with combining low line]](https://www.rsc.org/images/entities/char_0064_0332.gif) ipolarophiles (RILDs)15 to revive the concept of photo-activation (Scheme 1b).
ipolarophiles (RILDs)15 to revive the concept of photo-activation (Scheme 1b).
Recently, our group reported a study regarding the photo-isomerization of a cyclic azobenzene, ![[d with combining low line]](https://www.rsc.org/images/entities/char_0064_0332.gif) i
i![[b with combining low line]](https://www.rsc.org/images/entities/char_0062_0332.gif) enzo[b,f]-[1,4,5]
enzo[b,f]-[1,4,5]![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif) hia
hia![[d with combining low line]](https://www.rsc.org/images/entities/char_0064_0332.gif) iazepine (DBTD), and its ability to serve as a dipolarophile in cycloaddition with the nitrile imine generated from the photolysis of DASyd (Scheme 1c).16 In fact, DBTD containing a cyclic azobenzene motif could swiftly respond to a programmable photo-stimulation sequence, transforming photon energy into ring-strain energy to accelerate the cycloaddition rate in situ (k2, up to 1.6 ± 0.16 × 105 M−1 s−1). The excellent photo-switching and anti-fatigue performances of DBTD demonstrate a next-generation RILD reporter that is ultra-durable during complex photo-operations. However, its incorporation into proteins has been restricted within chemical approaches previously.
iazepine (DBTD), and its ability to serve as a dipolarophile in cycloaddition with the nitrile imine generated from the photolysis of DASyd (Scheme 1c).16 In fact, DBTD containing a cyclic azobenzene motif could swiftly respond to a programmable photo-stimulation sequence, transforming photon energy into ring-strain energy to accelerate the cycloaddition rate in situ (k2, up to 1.6 ± 0.16 × 105 M−1 s−1). The excellent photo-switching and anti-fatigue performances of DBTD demonstrate a next-generation RILD reporter that is ultra-durable during complex photo-operations. However, its incorporation into proteins has been restricted within chemical approaches previously.
Genetic code expansion (GCE) with noncanonical amino acids (ncAAs, Scheme 1a) has emerged as an extremely powerful tool to incorporate artificial functionalities into engineered proteins in living systems.17 Harbouring the unique chemical or physical features of exogenous ncAAs at specific residues, the encoded proteins have been exploited for mapping protein–protein interactions (proximity dependent photo-crosslinking),18 probing and tuning protein structures19 and the construction of artificial conjugates on demand.20 In particular, it is of great interest to utilize GCE to encode RILDs (Scheme 1c) because its exceptional biocompatibility and the photo-chemical energy transduction can enhance the chemoselectivity of the photo-click chemistry.16 Therefore, the spatiotemporal resolving power of photo-control could be elevated to establish the precise decoration of key residues on proteins of interest (POI) in 3D space to gain high-order functionalization.21 Inheriting the characteristics of a T-type (another ![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif) hermal stimulus) photo-switch,22a the genetically encoded DBTD residue itself is also well-suited for regulating the micro-environment on a designated site of proteins with fast photo-response.22b
hermal stimulus) photo-switch,22a the genetically encoded DBTD residue itself is also well-suited for regulating the micro-environment on a designated site of proteins with fast photo-response.22b
Adding DBTD onto desired residues via amber codon suppression could equip the parent proteins with both photo-switch and photo-clickable features (Scheme 1c). To adapt to the hydrophobic pocket of the Methanosarcina aminoacyl-tRNA synthetase17d with one molecule of adenosine monophosphate (AMP), we designed a compact DBTD–alanine ncAA (DBTDA, Scheme 1c) that is amenable to producing DBTDA tRNA for genetic incorporation. However, there are still two challenges to overcome before realizing spatiotemporally photo-click ligation for visualizing or manipulating biological processes with superior resolution. First, the structure of DBTDA in the thermostable cis-configuration resembles a unique “bowl” shaped structure (Scheme 1c), which requires a diversified codon library to evolve the tRNA synthetase to recognize this “exotic” substrate. Second, optimization of the structure of DASyd probes is also essential to achieve high photo-reactivity and chemoselectivity toward DBTD. Through rational design of the codon library in combination with directed evolution toward the Methanosarcina tRNA synthetase in the E. coli system, we were able to express superfolder green fluorescent proteins (sfGFPs) with the DBTDA incorporated on three individual sites. By encoding the DBTDA into the extracellular domain of the outer membrane porin C protein (OmpC), we also evaluated the efficiency and selectivity of the DASyd-DBTD photo-click reaction on living E. coli cells by employing a trifunctional DASyd conjugate. Thus, the distribution of over-expressed OmpC on E. coli cells of interest could be captured in vitro in 3D space by implementing a designated illumination, which offers a protocol for exploring their dynamic processes in living systems. The successful incorporation of DBTDA also provides a chemical transducer for photochemical energy conversion23 on key loci of proteins, which could be further exploited in the patterning of proteins in biomaterials24 and the enrichment of protein molecular machines25 for synthetic biology,26 as well as protein photo-pharmaceuticals for precision medicine.27 The xolographic fabrication of artificial protein copolymers28 can also be envisioned because DASyd and DBTDA can be photo-activated separately.
Results and discussion
Genetic encoding of DBTDA and identification
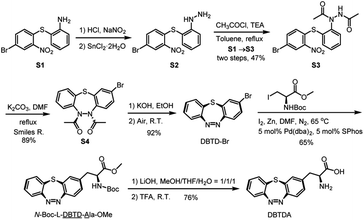
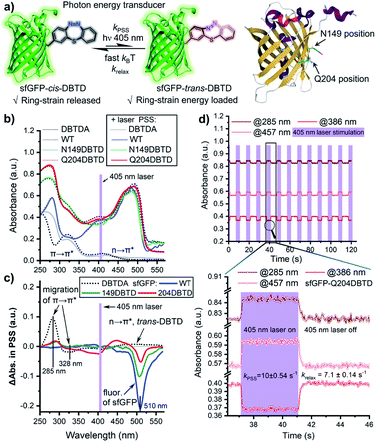
In order to obtain a sufficient amount of DBTDA for the directed evolution of the aminoacyl-tRNA synthetase, Smiles rearrangement29 of diacetyl-thiodiphenylhydrazine (S3, Scheme 2) was considered to be a key step to produce the seven-membered cyclic key intermediate, 2-bromo-5,6-diacetyl-5,6-dihydrodi-benzo[b,f][1,4,5]thiadiazepine (S4, diacetyl 2H-DBTD-Br) in gram scale, which is the precursor for DBTD-Br. After the consecutive deprotection and oxidation of S4 in air, we were able to afford DBTD-Br, and subsequently utilized Negishi coupling30 with a N-Boc-β-iodo-L-alanine-OMe derived organozinc reagent to harvest the desired N-Boc-L-DBTD-Ala-OMe, followed by hydrolysis of the ester and deprotection of N-Boc to furnish the DBTDA ncAA (Scheme 2).To genetically encode the DBTDA, we chose the pyrrolysyl-tRNA synthetase/tRNA(PylRS/tRNA) pair from Methanosarcina species31 as the evolving target because of its high efficiency32 in nonsense codon suppression, flexibility of substrates and full orthogonality in both prokaryotes and eukaryotes (Fig. 1a). It has been proven to be an extraordinary GCE system, and a wide array of ncAAs, e.g., p-benzoyl-L-phenyl-alanine (pBpa)33 and photo-switchable and clickable azobenzene–alanines (Scheme 1a),34 have been incorporated in both E. coli and mammalian cells via MmPylRS mutants. However, PylRS variants capable of encoding a seven-membered cyclic azobenzene framework have yet to be developed because of its wide-bodied and bowl-shaped morphology in the cis-form. Therefore, we have adopted a rationally designed library from pBpaRS mutations33 to evolve the MmPylRS. The goal of these mutations is to create an appropriate breadthwise space for the rigid and nonplanar macrocyclic DBTD that is orthogonal to other encodable ncAAs. Structural analyses of the MmPylRS binding pocket (PDB code: 2ZIM)35 for DBTDA clarified the constitution of the library, which includes a series of key residues subjected to either NNK (N = A, T, G or C; K = T or G) site saturation or degenerate-restricted mutagenesis (Tables S1 and S2 ESI†). To our delight, after two rounds of iterative selection of this library with both positive and negative cycles, a distinctive MmPylRS mutant carrying five-residue mutations (Fig. 1b) was obtained in the best consensus, and was named MmDBTDRS, exhibiting the effective incorporation of DBTDA into sfGFP (Fig. 1b). In order to verify the incorporation efficiency and fidelity of MmDBTDRS, we chose three characteristic sites on sfGFP to introduce DBTDA for evaluating its intact expression level. These sites include the N149 and Q204 residues on the outside of the β-barrel, which are in proximity to the sfGFP chromophore and exposed to the solvent,36 and the K3 site on the N-terminus flexible loop.37 The expression levels of sfGFP-K3DBTD, sfGFP-N149DBTD and sfGFP-Q204DBTD were found to be 2.7, 2.9 and 3.0 mg L−1, respectively, after purification. SDS-PAGE and deconvoluted LC-MS analysis (Fig. 1c) also confirmed the correct molecular weights, in agreement with theoretical predictions (Fig. 1d and S4–S9, ESI†). Furthermore, sfGFP-Q204DBTD was digested by trypsin for LC-MS/MS fingerprinting analysis, which confirmed the fidelity and site-specificity of the MmDBTDRS/tRNA pair, with the successful incorporation of DBTDA into the designated TAG-codon position (Fig. 1e and S1, ESI†). To enhance the incorporation efficiency of DBTDA in E. coli, we optimized the concentrations of DBTDA, as well as the pH of the culture medium, during the expression process (Fig. S2, ESI†). Under the optimized conditions (2 mM DBTDA, pH = 8.5), the purified sfGFP-Q204DBTD could be harvested with a yield of 5.7 mg L−1.
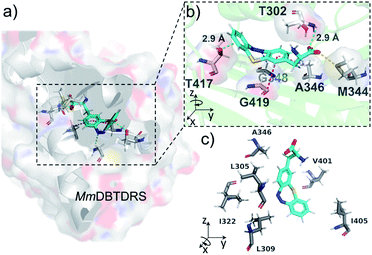
To understand the binding features of DBTDA involved in the protein–substrate interactions for the evolved MmDBTDRS synthetase, we utilized CHARMM-based molecular docking based on the crystal structures of both MmOmeRS (PDB code: 3QTC)38 and cis-DBTD (CCDC code: 1905279).16a All of the five residue mutations to the MmPylRS (Fig. 1b) and the putative conformation of L-DBTDA were applied to the active pocket with semi-flexible settings. Therefore, an optimal docking conformation for the MmDBTDRS-DBTDA complex was converged (Fig. 2a) with a binding free energy of −44.2 kcal mol−1. Indeed, DBTDA in the favourable “bowl” shaped energy pose was fitted into the expected binding pocket as the ligand for aminoacylation toward APM. The replacement of W348 and N346 residues with G and A, respectively, reserve a breadthwise space to accommodate the protruding cyclic azobenzene moiety on the DBTDA. The docking simulation also revealed that the DBTDA embeds into a bent binding pocket surrounded by hydrophobic residues, including L305, A346, V401 and I322 (Fig. 2c). As reported previously,38 T302 is located in the substrate-gating site of MmDBTDRS to form a hydrogen bond (2.9 Å, Fig. 2b) with the α-carboxyl acceptor on the amino acid backbone, stabilizing the polar subunit of DBTDA for acylation. Intriguingly, a lone pair(lp)–π interaction takes place on top of the distal phenyl of DBTDA toward the lone pair electrons of the oxygen atom on the T417 residue (2.9 Å), which might play an important role in keeping the distal phenyl in the right dihedral angle instead of adopting a π–π interaction (Fig. 2b). Compared with another plausible docking posture in which the azo group of DBTDA is flipped and facing outward to the V401 residue in the active pocket (Fig. S3 ESI†), the binding free energy of the displayed posture is advantageous, −44.2 vs. −33.3 kcal mol−1 (for the latter posture, Fig. S3, ESI†).
Photo-induced ring-strain energy loading of DBTD on proteins
The photo-isomerization performance of a molecular machine might be sensitive to its immediate chemical environment, including the photo-stationary state (PSS) and photo-switching kinetics. These are important factors that have to be considered when GCE is utilized to generate protein mutants for the photo-regulation of functions.39 To assess the impact of the protein micro-environment on the photo-switching efficiency of DBTD residues, the UV–Vis spectral changes of the two mutants (Q204 and N149) after the DBTD residues were equilibrated to PSS were investigated, using wild-type sfGFP (wt-sfGFP) and free DBTDA as controls (Fig. 3a). Under continuous 405 nm laser photo-stimulation to switch the DBTD residue, the wt-sfGFP did not exhibit any significant absorbance variation in the 250–430 nm region, but a fluorescence peak (510 nm, shown in the Δabsorbance spectra, Fig. 3c) could be observed because the presence of a 405 nm laser also excites the fluorophore of sfGFP simultaneously (Fig. 3b and c). In contrast, free DBTDA showed characteristic π → π* transition changes at both 285 and 328 nm, accompanied by a weak n → π* transition which moved to 450–550 nm under laser irradiation. Intriguingly, mutations of the native amino acid (either Q204 or N149) for DBTDA on sfGFP had an almost identical effect on the absorbance spectral changes around 285 nm (π → π*) after photo-stimulation, in which the PSS was estimated to be higher than 12% for sfGFP-Q204-trans-DBTD (Fig. S10, ESI†). But varying degrees of effects from 310 to 550 nm were observed, predominantly on the emission intensity around 510 nm (Fig. 3c). Notably, the Q204DBTD mutant had the most significant effect on the excitation spectral changes from 310–470 nm and there was a sharp suppression of the emission intensity (red line, Fig. 3c), in which the Q204 residue was reported to have a synergistically structural integration with the chromophore of sfGFP.40 Therefore, sfGFP-Q204DBTD was chosen to study the dynamic response of the photo-switchable DBTD in the protein environment under a programmable photo-stimulation sequence (Fig. 3d). Under an intermittent photo-stimulus, the absorbance signal responded precisely with the input showing no observable decay, outputting a nearly square-signal pattern at three different wavelengths with distinct phases (for wt-sfGFP as the control, see Fig. S11, ESI†). The ground-state of sfGFP-cis-DBTD approaches the PSS with a half-life of 70 ms (kPSS, Fig. 3d) under continuous photo-stimulation, and the energy-loaded “metastable” sfGFP-trans-DBTD relaxed back to the ground-state with a half-life of 98 ms (krelax, Fig. 3d) in the dark (lower panel of Fig. 3d and S12, ESI†). Compared with free DBTD (kPSS = 5.7 s−1, krelax = 3.8 s−1),16 the outer-surface environment at the Q204 position of sfGFP did not hinder the fast photo-response of the DBTD residue to transduce the photon energy into the ring-strain energy, which would accelerate the photo-click ligation for residue-specific labelling.Selective protein functionalization via photo-click chemistry
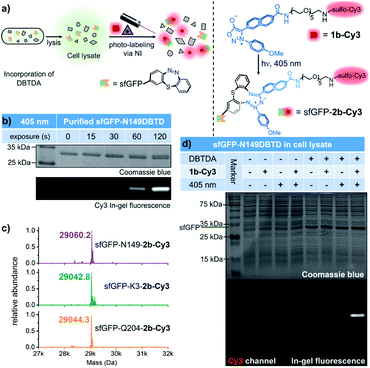
With the benefits of the DASyd-DBTD photo-click reaction with excellent reactivity and selectivity,16 we attempted to evaluate the site-specificity of this type of photo-labelling toward the DBTDA encoded sfGFP. Our initial study on the photo-labelling of purified sfGFP–DBTD with DASyd 1a under irradiation with 311 nm light (capable of the photolysis of 1a, 10.8 mW cm−2) for 2 min suggested an efficient photo-conjugation, giving the resultant proteins with yields in the order of N149 (85%) > K3 (71%) > Q204 (52%), detected by deconvoluted LC-MS (Fig. S13–S15 ESI†). To visualize this covalent modification via in-gel (SDS-PAGE) fluorescence analysis, we utilized 1b-Cy3 (Fig. 4a), a sulfo-Cy3 modified DASyd probe, for in vitro protein conjugation on the N149DBTD site to demonstrate the temporal control (Fig. 4b). The SDS-PAGE imaging analysis showed that a Cy3 fluorescence band correlated to sfGFP size was observed for the sample even within 15 s irradiation time via a 405 nm LED, and the labelling yield can be controlled in a time-dependent manner. Under optimized conditions, three sfGFP-DBTD mutants after photo-labelling by 3 min irradiation were also subjected to profiling by deconvoluted LC-MS, displaying almost quantitative efficiencies (Fig. 4c and S16–S18, ESI†) with site-specificity (for LC-MS/MS analysis, Fig. S19, ESI†). After subsequent purification to remove small molecules, the molar extinction coefficient (ε) ratio of the fluorophore of sfGFP (485 nm) and the characteristic absorption of the Cy3 (552 nm) can be measured to determine the yield of the labelled protein via UV–Vis spectral analysis, which resulted in 94 ± 1.7% yield (Fig. S20 ESI†), consistent with the result from the deconvoluted MS analysis. Through tracking the fluorescent bands in SDS-PAGE, we further analysed the bio-stability of the purified sfGFP–2b-Cy3 under biologically relevant conditions, e.g., in PBS or in a plasma mimicking environment with 5 mM GSH or even after further 405 nm LED irradiation. The purified sfGFP–2b-Cy3 was quite stable, without any degradation in PBS (98% retained), and 75% of the intact protein retained in the presence of GSH at 37 °C within 24 hours (Fig. S21 ESI†). Under further 405 nm light exposure for 5 min, 89% and 81% sfGFP–2b-Cy3 was conserved in PBS and 5 mM GSH, respectively (Fig. S21 ESI†). Even though there was slight protein degradation, the fluorescence of Cy3 remained tagged to the degraded components without being released as free Cy3, indicating that the covalent linkage built by the photo-click reaction is not damaged along with the degradation (Fig. S21 ESI†). These results validated that this photo-click chemistry is an effective tool for photo-controllable ligation toward DBTD-bearing proteins. To expand this photo-chemistry in a live-cell mimicking environment, we treated E. coli cell lysates over-expressing sfGFP-N149DBTD with the 1b-Cy3 probe, and implemented the ligation under the optimized conditions, followed by resolving via SDS-PAGE (Fig. 4d). Gratifyingly, only cell lysate containing the DBTD reporter on sfGFP exhibited an exclusive and strong fluorescent band after 405 nm illumination, which also matched the size of sfGFP. In contrast, there was rarely any fluorescent band detected in lanes for the samples either without DBTD on the proteins or under control conditions. Coomassie blue staining confirmed the fluorescence signal overlapping with the over-expressed sfGFP band (Fig. 4d). The photo-labelling experiments in cell lysate revealed the excellent selectivity of the DASyd-DBTD photo-click chemistry, capable of targeting the DBTD residues on a specific site in a complex of proteins.Since sfGFP-DBTD could be over-expressed in the living BL21(DE3) strain, we also attempted to evaluate intracellular bioorthogonal conjugation by utilizing a live-cell permeable probe, DASyd-Fl (photolysis under 373 nm LED, 38.5 mW cm−2) toward DBTDA residues under the excitation of both 373 nm and 405 nm LED (Fig. S22 ESI†). After the photo-click procedure inside living cells, SDS-PAGE analysis of the cell lysate revealed the successful labelling of the sfGFP-DBTD as a fluorescence band with the right size was detected. However, there were other stronger bands detected in the gel at the same time, which were likely attributed to the undesired binding of the DASyd-Fl dye to native proteins or to lipid phase components in the living E. coli cells. Therefore, the decent bioorthogonality of the DASyd-DBTD photo-click system with the anionic 1b-Cy3 probe is more suitable for us to study the dynamics of proteins in a single living cell when a spatiotemporal irradiation of submicron resolution could be exerted.
Spatiotemporally controlled and selective fluorescence labelling of the DBTDA encoded proteins on E. coli cells
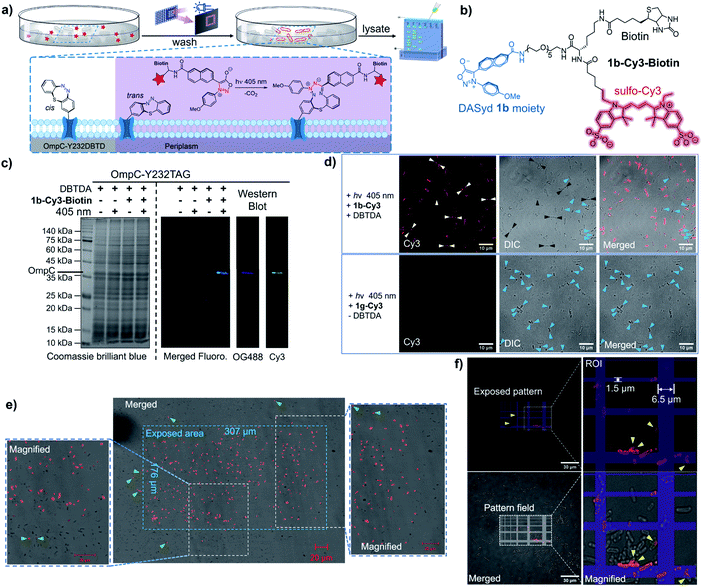
OmpC, known as a member of the outer membrane proteins (OMPs) family in Gram-negative bacteria, serves as a passive diffusion porin for transporting small molecules in the homotrimer form,41 and its expression level is regulated by external osmolarity.42 As a pharmacological target against antibiotic resistance,43 whether OmpC participates in the transport of other essential small molecules to support bacterial survival, as well as its dynamic distribution on cells, is yet to be discovered.Encouraged by the specific labelling of DBTD-bearing sfGFP, we were able to exploit the advantages of the DASyd-DBTD photo-click chemistry in the functionalization of OmpC-Y232DBTD44 on living E. coli cells under photo-control with high precision (Fig. 5a). The Y232 mutation site was chosen according to previous studies45 in which its location on one of the eight external loops of OmpC was pointed out. Live BL21(DE3) cells over-expressing either intact OmpC-Y232DBTD (2 mM DBTDA) or truncated OmpC (without DBTDA) via transformation with both pET-OmpC-Y232TAG and pEvol-MmDBTDRS were subjected to photo-click ligation with the 1b-Cy3 conjugate via exposure to the 405 nm LED for 5 min in PBS. The decorated cells were subsequently lysed, and the lysates were resolved by an in-gel fluorescence assay to investigate the selectivity of the photo-conjugation (Fig. S23 ESI†). To our delight, we could observe a prominent Cy3 fluorescence band of the correct OmpC size only in the lane where the prerequisites for the photo-click reaction were met, rather than those lanes under control conditions, indicating that the excellent chemo-selectivity of the photo-click chemistry is broadly applicable.
For the purpose of immunofluorescence labelling, we also constructed a biotinylated DASyd probe, 1b-Cy3-Biotin (Fig. 5b), which integrates three functionalities and displays photo-reactivity akin to 1b-Cy3. Therefore, multi-colored fluorescence Western blot (WB) imaging was performed to verify the chemical selectivity of the photo-click chemistry by staining the lysate PVDF membrane with the NeutrAvidin™-OG-488 probe for recognizing the photo-ligated biotin motif on OmpC (Fig. 5c). Encouragingly, the exclusive and overlapped bands shown in both the Cy3 channel and OG-488 channel reflected that the photo-conjugation was specifically targeted to the desired OmpC-Y232DBTD in the context of living cells.
After immobilization in agarose gels for the purpose of imaging, only intact BL21(DE3) cells over-expressing OmpC-Y232DBTD showed strong Cy3 fluorescence on the outer membrane,46 which was clarified by laser scanning confocal microscopic imaging (59% of the total cells were strongly labelled, Fig. 5d). Cells expressing truncated OmpC cannot be photo-labelled. Intriguingly, although most of the photo-labelled bacterial cells showed a uniform outer membrane fluorescence morphology, individual cells exhibited unique distributions of OmpC in either bipolar or multi-spotted shapes on the surface of cells (white arrows, Fig. 5d), which suggests the potential aggregation of the intact OmpC. Notably, inclusion bodies could be imaged when truncated OmpC was accumulated inside the E. coli cells either with very poor amber stop codon suppression efficiency (Fig. 5d upper row, blue arrows) or in the absence of DBTDA (Fig. 5d lower row, blue arrows).46
To demonstrate the spatial resolving power of the DASyd-DBTD photo-click chemistry, we projected a localized illumination in a rectangular shape (307 μm × 176 μm) via an embedded digital micromirror device (DMD) in the microscopic system to the cells over-expressing OmpC-Y232DBTD after treatment with 1b-Cy3-Biotin (Fig. 5e). The resulting images indicated that most of the E. coli cells inside the exposed area emitted the Cy3 fluorescence signal, whereas cells outside the area scarcely showed any signals. A few cells outside the stimulated area but close to the boundary were also photo-labelled, but the fluorescence intensity was relatively weak (Fig. 5e, green arrows). These results suggested that the genetically encoded DBTDA could potentially be utilized to unravel the functionalities of OmpC on single cells by more sophisticated irradiation in regions of interest (ROI). Thus, we projected a grid-patterned photo-stimulation via the DMD (405 nm LED, minimal grid line width = 0.44 μm) onto a cluster of E. coli cells over-expressing OmpC-Y232DBTD after treatment with 5 μM 1b-Cy3-Biotin (Fig. 5f) to preliminarily assess the spatial resolution. Encouragingly, the fluorescently labeled cells were mainly located within the grid lines of the stimulation pattern, although there was some offset labelling, presumably caused by either light scattering (the focal plane of the 100× objective is very thin) or diffusion of the photo-generated NI intermediate. Due to the varying expression level of GCE in encoding the DBTDA,47 about 40% of the total cells on the grid lines were strongly photo-labelled (Fig. 5f). We also found “U-shaped” subcellular fluorescence signals (Fig. 5f, yellow arrows) corresponding to half-segments of the bacterial outer membrane, which suggest that the spatial resolution of this photo-stimulation, together with the photo-click system, could be higher than the size of an E. coli cell (length < 3 μm, width ≈ 1 μm).48
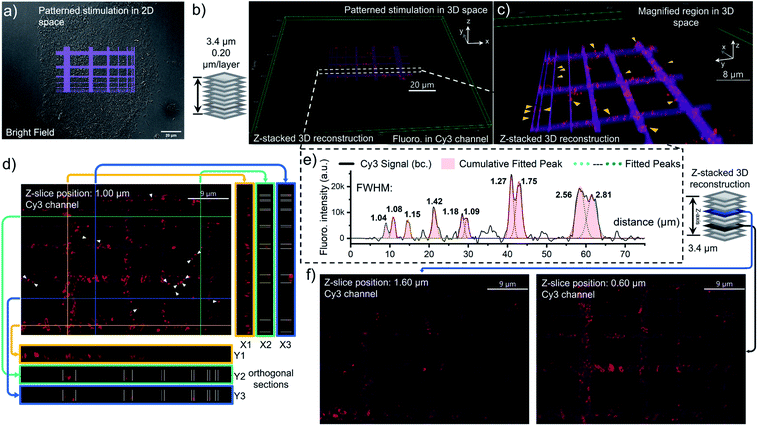
To better characterize the distribution of OmpC on E. coli cell via photo-click chemistry in 3D space, we projected the same grid-patterned photo-stimulation (via the DMD, 405 nm LED) onto clusters of cells over-expressing OmpC-Y232DBTD after treatment with 5 μM 1b-Cy3-Biotin (Fig. 6a, for other groups of imaging results as replications, see S24 and S25, ESI†). After washing, an 18-slice fluorescence tomogram covering the region of the photo-stimulated cells was acquired with a Z-stepping of 0.20 μm and reconstructed into a 3D Z-stacking model (Fig. 6b). The volumetric rendering allowed us to understand in more detail the distribution of OmpC on the outer membrane. As a result, the pattern of Cy3 fluorescence on E. coli cells was mostly overlapped with the photo-stimulation pattern in 3D space (Fig. 6b and c). Although, the scattering of light (multilayer structure of bacterial clusters) and diffusion of the NI intermediate resulted in some diffused fluorescence labelling at the edges of the stimulated area, depending on the Z-axial position and expression level of OmpC-DBTD of individual cells (Fig. 6d, f and S24, ESI†). It is noteworthy that subcellular segments on specific E. coli cells could be precisely distinguished at the edge of the photo-illumination lines, which were highlighted in the magnified 3D scene (yellow arrows, Fig. 6c). Furthermore, the cross-section slices in both the transverse and longitudinal direction also supported that the labelled OmpC is localized on the outer membrane because the typical hollow ring-shape of the Cy3 signal could be observed (Fig. 6d). In fact, Z-sliced images picked from the tomogram revealed that some of the Cy3 fluorescence signals distributed granularly on the outer membrane, presumably due to aggregation of the over-expressed OmpC (Fig. 6d and f), especially for few filamented cells (Fig. S25, ESI†).49 We also noticed that some of E. coli cells displayed enriched OmpC at both poles of the rod shape (white arrows, Fig. 6d and S24d, ESI†). In particular, some of them even appeared in pairs with a pole-to-pole spliced form. Therefore, it is possible that the pole distribution of OmpC might be related to the division of the bacterial cell (enlarged image in Fig. S24e, ESI†).50 Nevertheless, it is evident that there was some off-site and diffused Cy3 labelling shown in the imprinted fluorescence pattern (Fig. 6d and f). In addition, the spatial resolution of this photo-click approach on E. coli cells was assessed by the full width at half maximum (FWHM) of the imprinted Cy3 signal peak across the photo-stimulation line, which was determined to be as low as 1.1 μm on average (Fig. 6e). Considering that readily available LEDs were used as the light sources and DMD & epi-fluorescence microscopy were utilized as the photo-stimulation and imaging platform, respectively, the spatial resolution of 1.1 microns is decent.
Subsequently, we further employed an excitation pattern of a dot array via the DMD to evaluate the spatial-resolution of the photo-click labelling on OmpC at the single-cell level. The dot array was programmed to contain single-pixel excitations (the irradiation spot is roughly 135 nm per pixel on the specimen, Fig. S26, ESI†) to possibly demonstrate the finest resolution of the current photo-stimulation system. Surprisingly, the photo-stimulation of single-pixel dots could selectively label individual cells (Fig. S26, ESI†) in designated clusters of E. coli cells, albeit with some background labelling. Further study using laser (even femtosecond laser) scanning technology for stimulation and imaging is underway for a higher spatial resolution.
To be able to further track the dynamics of the photo-labelled OmpC on living cells, we also compared the survival of the BL21 strain before and after the photo-click reaction. The colony counting suggested that the photo-labelling of the over-expressed OmpC-DBTD protein on the outer membrane would cause a significantly weakened viability (10% survival rate, Fig. S27 ESI†). Meanwhile, applying the photo-stimulation (405 nm) alone would not lead to a bactericidal effect (78% survival rate, slight phototoxicity). As a result, the introduction of larger-sized dye conjugates on the outer membrane domain (Y232) of OmpC might alter the porin nature of OmpC into an accumulated hybrid conjugate that is possibly interfering with essential cellular processes.46 These data collectively suggested that the genetic incorporation of DBTDA in combination with the DASyd-DBTD photo-click chemistry enables spatiotemporal OmpC decoration on living cell membranes, offering the potential of triggering protein-mediated bactericidal activity selectively under photo-control. However, the incorporation sites of DBTDA can be expanded and screened to diversify the photo-functionalization of target proteins in living systems.
Conclusions
We have developed a novel ncAA bearing DBTD motif as a photo-switchable RILD reporter, and also evolved an MmDBTDRS/tRNACUA pair to allow its GCE incorporation into recombinant proteins site-specifically in E. coli systems. The incorporation efficiency of DBTDA into proteins was optimized and the binding model of DBTDA to the evolved synthetase was clarified by molecular docking to shed light on the design of synthase evolution for the incorporation of DBTDA derivatives. The successful incorporation of DBTDA facilitates the exploration of the photo-transducing performance of the DBTD residue on proteins, and also the photo-click conjugation between DBTD-encoded proteins and fluorescent DASyd probes in living cells with an excellent reaction rate, chemoselectivity and site-specificity. Moreover, an outer membrane porin C protein on E. coli cells was chosen to unravel its features and distribution characteristics via photo-click labelling in harmony with the genetically encoded DBTDA on its extracellular domain. By employing the DBTD as a RILD reporter together with the photo-generated nitrile imine, the spatial decoration of OmpC is now approaching the single-cell level. Encoding the DBTD reporter with both fast photo-response and photo-click features on proteins in a site-specific manner holds great potential for developing high-order manipulation of functionalized proteins via photon energy transduction in widespread application scenarios.Data availability
Additional figures as described in the main text, all experimental procedures, synthetic procedures, and copies of spectral data of the compounds and processes, computational details are available in the ESI.†Author contributions
Qin Xiong performed most of the experimental work in this manuscript and drafted the manuscript. Tingting Zheng assisted in the synthesis of chemical reagents and also in protein expression, purification and characterization. Xin Shen assisted in the photo-switching experiments. Baolin Li synthesized DASyd 1a and 1b. Jielin Fu performed the construction of pET-sfGFP-XTAG plasmids. Xiaohu Zhao participated in the analysis of 1H and 13C NMR spectra. Chunxia Wang revised the manuscript. Zhipeng Yu synthesized the fluorescent probe 1b-Cy3-Biotin, conceived of the project, analyzed the data and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Yanhong Liu at the Comprehensive Training Platform of Specialized Laboratory, College of Chemistry, for her assistance with the confocal image collection. We are grateful to Dr Pengchi Deng at the Analytical & Testing Center, SCU for help with NMR spectrum acquisition for the PSS. Financial support was provided by the National Natural Science Foundation of China (22001181 and 22077090) and the Fundamental Research Funds for the Central Universities (20826041D4117) and Institutional Research Fund from Sichuan University (2020SCUNL105). We also thank Dr Wenshe Liu (Texas A&M) and Dr Qing Lin (SUNY-UB) for generously providing us the plasmids, pEvol-PylT-PylRS and pET-sfGFP-Q204TAG. We also appreciate Dr Lunzhi Dai (State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University) for help with the LC-MS/MS analysis.Notes and references
- (a) H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004 CrossRef CAS; (b) C. R. Becer, R. Hoogenboom and U. S. Schubert, Angew. Chem., Int. Ed., 2009, 48, 4900 CrossRef CAS PubMed.
- P. Thirumurugan, D. Matosiuk and K. Jozwiak, Chem. Rev., 2013, 113, 4905 CrossRef CAS PubMed.
- W. Xi, T. F. Scott, C. J. Kloxin and C. N. Bowman, Adv. Funct. Mater., 2014, 24, 2572 CrossRef CAS.
- (a) H. Wang, M. Gauthier, J. R. Kelly, R. J. Miller, M. Xu, W. D. O’Brien Jr and J. Cheng, Angew. Chem., Int. Ed., 2016, 55, 5452 CrossRef CAS PubMed; (b) K. Lang, L. Davis, J. Torres-Kolbus, C. Chou, A. Deiters and J. W. Chin, Nat. Chem., 2012, 4, 298 CrossRef CAS PubMed.
- M. A. Tasdelen and Y. Yagci, Angew. Chem., Int. Ed., 2013, 52, 5930 CrossRef CAS PubMed.
- B. D. Fairbanks, L. J. Macdougall, S. Mavila, J. Sinha, B. E. Kirkpatrick, K. S. Anseth and C. N. Bowman, Chem. Rev., 2021, 121, 6915 CrossRef CAS PubMed.
- (a) C. E. Hoyle and C. N. Bowman, Angew. Chem., Int. Ed., 2010, 49, 1540 CrossRef CAS; (b) S. Chatani, C. J. Kloxin and C. N. Bowman, Polym. Chem., 2014, 5, 2187 RSC.
- (a) R. A. Olson, A. B. Korpusik and B. S. Sumerlin, Chem. Sci., 2020, 11, 5142 RSC; (b) N. A. Repina, T. McClave, H. J. Johnson, X. Bao, R. S. Kane and D. V. Schaffer, Cell Rep., 2020, 31, 107737 CrossRef CAS PubMed.
- G. S. Kumar and Q. Lin, Chem. Rev., 2021, 121, 6991 CrossRef CAS PubMed.
- (a) Y. Wang, W. Song, W. J. Hu and Q. Lin, Angew. Chem., Int. Ed., 2009, 48, 5330 CrossRef CAS PubMed; (b) Z. Yu, T. Y. Ohulchanskyy, P. An, P. N. Prasad and Q. Lin, J. Am. Chem. Soc., 2013, 135, 16766 CrossRef CAS PubMed; (c) Y. Wu, G. Guo, J. Zheng, D. Xing and T. Zhang, ACS Sens., 2019, 4, 44 CrossRef CAS PubMed; (d) S. Jiang, X. Wu, H. Liu, J. Deng, X. Zhang, Z. Yao, Y. Zheng, B. Li and Z. Yu, ChemPhotoChem, 2020, 4, 327 CrossRef CAS.
- (a) L. Zhang, X. Zhang, Z. Yao, S. Jiang, J. Deng, B. Li and Z. Yu, J. Am. Chem. Soc., 2018, 140, 7390 CrossRef CAS; (b) X. Zhang, X. Wu, S. Jiang, J. Gao, Z. Yao, J. Deng, L. Zhang and Z. Yu, Chem. Commun., 2019, 55, 7187 RSC.
- (a) S. Arumugam and V. V. Popik, J. Am. Chem. Soc., 2011, 133, 5573 CrossRef CAS PubMed; (b) S. Arumugam, S. V. Orski, J. Locklin and V. V. Popik, J. Am. Chem. Soc., 2012, 134, 179 CrossRef CAS PubMed.
- A. A. Poloukhtine, N. E. Mbua, M. A. Wolfert, G.-J. Boons and V. V. Popik, J. Am. Chem. Soc., 2009, 131, 15769 CrossRef CAS PubMed.
- (a) J. Wang, W. Zhang, W. Song, Y. Wang, Z. Yu, J. Li, M. Wu, L. Wang, J. Zang and Q. Lin, J. Am. Chem. Soc., 2010, 132, 14812 CrossRef CAS PubMed; (b) Y. Tian, M. P. Jacinto, Y. Zeng, Z. Yu, J. Qu, W. R. Liu and Q. Lin, J. Am. Chem. Soc., 2017, 139, 6078 CrossRef CAS PubMed; (c) Z. Yu, Y. Pan, Z. Wang, J. Wang and Q. Lin, Angew. Chem., Int. Ed., 2012, 51, 10600 CrossRef CAS PubMed; (d) Y. Wang, W. Song, W. J. Hu and Q. Lin, Angew. Chem., Int. Ed., 2009, 48, 5330 CrossRef CAS; (e) Z. Yu and Q. Lin, J. Am. Chem. Soc., 2014, 136, 4153 CrossRef CAS.
- (a) K. Singh, C. J. Fennell, E. A. Coutsias, R. Latifi, S. Hartson and J. D. Weaver, Chem, 2018, 4, 124 CrossRef CAS; (b) H. Yang, T. Zeng, S. Xi, S. Hu, Y. Wu and Y. Tang, Green Chem., 2020, 22, 7023 RSC.
- (a) J. Gao, Q. Xiong, X. Wu, J. Deng, X. Zhang, X. Zhao, P. Deng and Z. Yu, Commun. Chem., 2020, 3, 29 CrossRef CAS; (b) J. Deng, X. Wu, G. Guo, X. Zhao and Z. Yu, Org. Biomol. Chem., 2020, 18, 5602 RSC.
- (a) L. Davis and J. W. Chin, Nat. Rev. Mol. Cell Biol., 2012, 13, 168 CrossRef CAS PubMed; (b) A. Dumas, L. Lercher, C. D. Spicer and B. G. Davis, Chem. Sci., 2015, 6, 50 RSC; (c) J. W. Chin, Nature, 2017, 550, 53 CrossRef CAS; (d) J. M. Kavran, S. Gundllapalli, P. O’Donoghue, M. Englert, D. Söll and T. A. Steitz, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11268 CrossRef CAS PubMed.
- (a) T.-A. Nguyen, M. Cigler and K. Lang, Angew. Chem., Int. Ed., 2018, 57, 14350 CrossRef CAS; (b) W. Hu, Y. Yuan, C.-H. Wang, H.-T. Tian, A.-D. Guo, H.-J. Nie, H. Hu, M. Tan, Z. Tang and X.-H. Chen, Chem, 2019, 5, 2955 CrossRef CAS; (c) H. Mori and K. Ito, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 16159 CrossRef CAS.
- (a) J. C. Jackson, J. T. Hammill and R. A. Mehl, J. Am. Chem. Soc., 2007, 129, 1160 CrossRef CAS PubMed; (b) S. E. Cellitti, D. H. Jones, L. Lagpacan, X. Hao, Q. Zhang, H. Hu, S. M. Brittain, A. Brinker, J. Caldwell, B. Bursulaya, G. Spraggon, A. Brock, Y. Ryu, T. Uno, P. G. Schultz and B. H. Geierstanger, J. Am. Chem. Soc., 2008, 130, 9268 CrossRef CAS; (c) L. Wang, J. Zhang, M.-J. Han, L. Zhang, C. Chen, A. Huang, R. Xie, G. Wang, J. Zhu, Y. Wang, X. Liu, W. Zhuang, Y. Li and J. Wang, Angew. Chem., Int. Ed., 2021, 60, 11143 CrossRef CAS.
- (a) J. H. Mills, S. D. Khare, J. M. Bolduc, F. Forouhar, V. K. Mulligan, S. Lew, J. Seetharaman, L. Tong, B. L. Stoddard and D. Baker, J. Am. Chem. Soc., 2013, 135, 13393 CrossRef CAS PubMed; (b) Y. Fu, J. Huang, Y. Wu, X. Liu, F. Zhong and J. Wang, J. Am. Chem. Soc., 2021, 143, 617 CrossRef CAS PubMed; (c) J. Wang, Y. Liu, Y. Liu, S. Zheng, X. Wang, J. Zhao, F. Yang, G. Zhang, C. Wang and P. R. Chen, Nature, 2019, 569, 509 CrossRef CAS PubMed.
- (a) V. Klippenstein, L. Mony and P. Paoletti, Trends Biochem. Sci., 2018, 43, 436 CrossRef CAS PubMed; (b) J.-Y. Kang, D. Kawaguchi, I. Coin, Z. Xiang, D. D. M. O’Leary, P. A. Slesinger and L. Wang, Neuron, 2013, 80, 358 CrossRef CAS PubMed; (c) S. Liu, C. Lin, Y. Xu, H. Luo, L. Peng, X. Zeng, H. Zheng, P. R. Chen and P. Zou, Nat. Chem., 2021, 13, 472 CrossRef CAS PubMed.
- (a) S. Crespi, N. A. Simeth, M. D. Donato, S. Doria, C. N. Stindt, M. F. Hilbers, F. L. Kiss, R. Toyoda, S. Wesseling, W. J. Buma, B. L. Feringa and W. Szymański, Angew. Chem., Int. Ed., 2021, 60, 25290 CrossRef CAS PubMed; (b) M. Bose, D. Groff, J. Xie, E. Brustad and P. G. Schultz, J. Am. Chem. Soc., 2006, 128, 388 CrossRef CAS PubMed.
- Q.-Q. Zhou, Y.-Q. Zou, L.-Q. Lu and W.-J. Xiao, Angew. Chem., Int. Ed., 2019, 58, 1586 CrossRef CAS PubMed.
- (a) C. Guo, H. Kim, E. M. Ovadia, C. M. Mourafetis, M. Yang, W. Chen and A. M. Kloxin, Acta Biomater., 2017, 56, 80 CrossRef CAS; (b) M. A. Wijdeven, C. Nicosia, A. Borrmann, J. Huskens and F. L. van Delft, RSC Adv., 2014, 4, 10549 RSC; (c) A. D. de Araújo, J. M. Palomo, J. Cramer, M. Köhn, H. Schröder, R. Wacker, C. Niemeyer, K. Alexandrov and H. Waldmann, Angew. Chem., Int. Ed., 2006, 45, 296 CrossRef PubMed.
- A. M. Hartley, H. L. Worthy, S. C. Reddington, P. J. Rizkallah and D. D. Jones, Chem. Sci., 2016, 7, 6484 RSC.
- Y. Wang, X. Chen, W. Cai, L. Tan, Y. Yu, B. Han, Y. Li, Y. Xie, Y. Su, X. Luo and T. Liu, Angew. Chem., Int. Ed., 2021, 60, 10040 CrossRef CAS PubMed.
- X. Ji, Z. Pan, B. Yu, L. K. De La Cruz, Y. Zheng, B. Ke and B. Wang, Chem. Soc. Rev., 2019, 48, 1077 RSC.
- M. Regehly, Y. Garmshausen, M. Reuter, N. F. König, E. Israel, D. P. Kelly, C.-Y. Chou, K. Koch, B. Asfari and S. Hecht, Nature, 2020, 588, 620 CrossRef CAS PubMed.
- (a) A. A. Levy, H. C. Rains and S. Smiles, J. Chem. Soc., 1931, 3264 RSC; (b) C. Corral, J. Lissavetzky and G. Quintanilla, J. Org. Chem., 1982, 47, 2214 CrossRef CAS.
- Z. Yao, X. Wu, X. Zhang, Q. Xiong, S. Jiang and Z. Yu, Org. Biomol. Chem., 2019, 17, 6777 RSC.
- (a) G. Srinivasan, C. M. James and J. A. Krzycki, Science, 2002, 296, 1459 CrossRef CAS PubMed; (b) S. M. Hancock, R. Uprety, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2010, 132, 14819 CrossRef CAS PubMed; (c) P. R. Chen, D. Groff, J. Guo, W. Ou, S. Cellitti, B. H. Geierstanger and P. G. Schultz, Angew. Chem., Int. Ed., 2009, 48, 4052 CrossRef CAS PubMed; (d) S. Greiss and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 14196 CrossRef CAS PubMed; (e) A. Bianco, F. M. Townsley, S. Greiss, K. Lang and J. W. Chin, Nat. Chem. Biol., 2012, 8, 748 CrossRef CAS PubMed.
- M. Pott, M. J. Schmidt and D. Summerer, ACS Chem. Biol., 2014, 9, 2815 CrossRef CAS PubMed.
- V. K. Lacey, G. V. Louie, J. P. Noel and L. Wang, ChemBioChem, 2013, 14, 2100 CrossRef CAS PubMed.
- (a) C. Hoppmann, V. K. Lacey, G. V. Louie, J. Wei, J. P. Noel and L. Wang, Angew. Chem., Int. Ed., 2014, 53, 3932 CrossRef CAS PubMed; (b) C. Hoppmann, I. Maslennikov, S. Choe and L. Wang, J. Am. Chem. Soc., 2015, 137, 11218 CrossRef CAS PubMed.
- J. M. Kavran, S. Gundllapalli, P. Donoghue, M. Englert, D. Söll and T. A. Steitz, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11268 CrossRef CAS PubMed.
- (a) S. C. Reddington, E. M. Tippmann and D. Dafydd Jones, Chem. Commun., 2012, 48, 8419 RSC; (b) T. M. Roberts, F. Rudolf, A. Meyer, R. Pellaux, E. Whitehead, S. Panke and M. Held, Sci. Rep., 2016, 6, 28166 CrossRef CAS PubMed.
- J.-D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2006, 24, 79 CrossRef PubMed.
- J. K. Takimoto, N. Dellas, J. P. Noel and L. Wang, ACS Chem. Biol., 2011, 6, 733 CrossRef CAS PubMed.
- S. C. Reddington, A. J. Baldwin, R. Thompson, A. Brancale, E. M. Tippmannk and D. D. Jones, Chem. Sci., 2015, 6, 1159 RSC.
- H. L. Worthy, H. S. Auhim, W. D. Jamieson, J. R. Pope, A. Wall, R. Batchelor, R. L. Johnson, D. W. Watkins, P. Rizkallah, O. K. Castell and D. D. Jones, Commun. Chem., 2019, 2, 83 CrossRef.
- Z. Xu and S. Y. Lee, Appl. Environ. Microbiol., 1999, 65, 5142 CrossRef CAS PubMed.
- L. A. Pratt, W. Hsing, K. E. Gibson and T. J. Silhavy, Mol. Microbiol., 1996, 20, 911 CrossRef CAS PubMed.
- (a) I. Calderón, S. R. Lobos, H. A. Rojas, C. Palomino, L. H. Rodríguez and G. C. Mora, Infect. Immun., 1986, 52, 209 CrossRef PubMed; (b) Y.-F. Liu, J.-J. Yan, H.-Y. Lei, C.-H. Teng, M.-C. Wang, C.-C. Tseng and J.-J. Wu, Infect. Immun., 2012, 80, 1815 CrossRef CAS PubMed.
- S. V. Mayer, A. Murnauer, M.-K. von Wrisberg, M.-L. Jokisch and K. Lang, Angew. Chem., Int. Ed., 2019, 58, 15876 CrossRef CAS PubMed.
- C. D. Spicer, T. Triemer and B. G. Davis, J. Am. Chem. Soc., 2012, 134, 800 CrossRef CAS PubMed.
- (a) K. Kipper, E. G. Lundius, V. Ćurić, I. Nikić, M. Wiessler, E. A. Lemke and J. Elf, ACS Synth. Biol., 2017, 6, 233 CrossRef CAS PubMed; (b) A. B. Lindner, R. Madden, A. Demarez, E. J. Stewart and F. Taddei, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3076 CrossRef CAS PubMed.
- M. B. Elowitz, A. J. Levine, E. D. Siggia and P. S. Swain, Science, 2002, 297, 1183 CrossRef CAS PubMed.
- G. Reshes, S. Vanounou, I. Fishov and M. Feingold, Biophys. J., 2008, 94, 251 CrossRef CAS PubMed.
- A. Rokney, M. Shagan, M. Kessel, Y. Smith, I. Rosenshine and A. B. Oppenheim, J. Mol. Biol., 2009, 392, 589 CrossRef CAS PubMed.
- J. D. Wang and P. A. Levin, Nat. Rev. Microbiol., 2009, 7, 822 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Containing details on experimental procedures, spectra properties, and characterization of all new compounds. See DOI: 10.1039/d1sc05710c |
| This journal is © The Royal Society of Chemistry 2022 |