Open Access Article
Open Access ArticleIdentification of two-dimensional copper signatures in human blood for bladder cancer with machine learning†
Weichao
Wang‡
ah,
Xian
Liu‡
a,
Changwen
Zhang
b,
Fei
Sheng
b,
Shanjun
Song
c,
Penghui
Li
d,
Shaoqing
Dai
 e,
Bin
Wang
f,
Dawei
Lu
ah,
Luyao
Zhang
ah,
Xuezhi
Yang
ah,
Zhihong
Zhang
b,
Sijin
Liu
e,
Bin
Wang
f,
Dawei
Lu
ah,
Luyao
Zhang
ah,
Xuezhi
Yang
ah,
Zhihong
Zhang
b,
Sijin
Liu
 a,
Aiqian
Zhang
ah,
Qian
Liu
a,
Aiqian
Zhang
ah,
Qian
Liu
 *agh and
Guibin
Jiang
ah
*agh and
Guibin
Jiang
ah
aState Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, China. E-mail: qianliu@rcees.ac.cn
bDepartment of Urology, The Second Hospital of Tianjin Medical University, Tianjin Institute of Urology, Tianjin 300211, China
cNational Institute of Metrology, Beijing 100029, China
dTianjin University of Technology, Tianjin 300384, China
eFaculty of Geo-Information Science and Earth Observation (ITC), University of Twente, P.O. Box 217, 7500AE Enschede, The Netherlands
fInstitute of Reproductive and Child Health, National Health Commission's Key Laboratory of Reproductive Health, Peking University, Beijing 100191, China
gInstitute of Environment and Health, Jianghan University, Wuhan 430056, China
hUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 11th January 2022
Abstract
Currently, almost all available cancer biomarkers are based on concentrations of compounds, often suffering from low sensitivity, poor specificity, and false positive or negative results. The stable isotopic composition of elements provides a different dimension from the concentration and has been widely used as a tracer in geochemistry. In health research, stable isotopic analysis has also shown potential as a new diagnostic/prognostic tool, which is still in the nascent stage. Here we discovered that bladder cancer (BCa) could induce a significant variation in the ratio of natural copper isotopes (65Cu/63Cu) in the blood of patients relative to benign and healthy controls. Such inherent copper isotopic signatures permitted new insights into molecular mechanisms of copper imbalance underlying the carcinogenic process. More importantly, to enhance the diagnostic capability, a machine learning model was developed to classify BCa and non-BCa subjects based on two-dimensional copper signatures (copper isotopic composition and concentration in plasma and red blood cells) with a high sensitivity, high true negative rate, and low false positive rate. Our results demonstrated the promise of blood copper signatures combined with machine learning as a versatile tool for cancer research and potential clinical application.
Introduction
Cancer biomarkers are vital for cancer diagnosis, prognosis prediction, epidemiologic studies, and therapeutic interventions. Currently, almost all available cancer biomarkers are based on concentrations of specific compounds.1 However, the concentrations of biomarkers are easily interfered by other non-tumor sources, often leading to low sensitivity, poor specificity, and false positive or negative results.2 The natural stable isotopic composition of an element provides an independent dimension of information from the concentration. Most elements have multiple naturally occurring stable isotopes with relatively constant compositions, while some physical, chemical, or biological processes are able to separate isotopes to cause stable isotopic fractionation.3 Compared with the concentration, the natural stable isotopic composition can bear fingerprint information about the source or processes of an element, due to the isotopic homogeneity of a source and the unidirectional stable isotopic fractionation during a specific process.4 Thus, the stable isotopic composition has been widely used as a powerful tracer in geochemistry, archaeology, anthropology, and environmental forensics.4–6 However, in other fields, its power has been less realized.7In the last two decades, application of stable isotopic analysis in health research has also been explored.8–10 An increasing number of studies have reported the stable isotopic fractionation phenomena related to a variety of human diseases,11–13 revealing the possibility of using stable isotopic analysis as a diagnostic/prognostic tool.14 For example, the stable iron (Fe) isotopic composition in human blood was reported to differ between individuals and genders,15 and hereditary hemochromatosis was associated with enrichment of heavy Fe isotopes in blood.16 Stable calcium (Ca) isotope ratios in urine could rapidly reflect the changes in bone mineral balance and bone loss.17 The serum copper (Cu) of Wilson's disease patients was isotopically lighter than that of controls,10 and patients with liver conditions of different etiologies showed lower serum copper isotopic compositions than controls.18,19 With regard to cancers, it has been reported that copper and sulfur (S) were enriched in light isotopes in the blood of hepatocellular carcinoma (HCC) patients, and the tumor tissue had a heavier copper isotopic composition than the adjacent healthy tissue.20 A similar trend in blood or tissue was also observed with breast cancer, colorectal cancer, ovarian cancer, hematological malignancies, and oral squamous cell carcinoma.14,21–25 In addition, zinc (Zn) isotopic fractionation was observed with breast cancer, pancreatic cancer, and hematological malignancies.23,26,27 However, until now, the mechanisms for disease-related isotopic fractionation are still poorly understood.28 Most previous studies were limited to reporting the isotopic fractionation phenomena between patients and controls, but the actual clinical applications are still very scarce, partly due to the insufficient distinguishing effect between patients and controls by stable isotopic analysis. The specificity of the disease-related isotopic fractionation also needs further evaluation.
In this study, we investigated the variations of the copper concentration and copper isotopic ratio in the blood of bladder cancer (BCa) patients, and developed a machine learning model to classify BCa and non-BCa subjects. To the best of our knowledge, this work may represent the first example of combination of stable isotopic data and machine learning for clinical applications. BCa is one of the most common cancers,29 but its early diagnosis is still challenging partially due to the lack of reliable molecular biomarkers in clinical practice, though some tests have been available.30,31 We focus on copper, because copper plays vital roles in fundamental life processes (e.g., oxidative processes),32,33 and urinary cancers have been hypothesized to be closely associated with abnormal systemic copper distribution34 but the underlying mechanisms are still indistinct.35 Thus, stable copper isotopes (65Cu and 63Cu) are expected to provide a novel approach to understand the copper imbalance underlying the carcinogenic process. Note that inherent natural copper isotopes were used in this study, which is basically different from the artificial isotope labelling method.
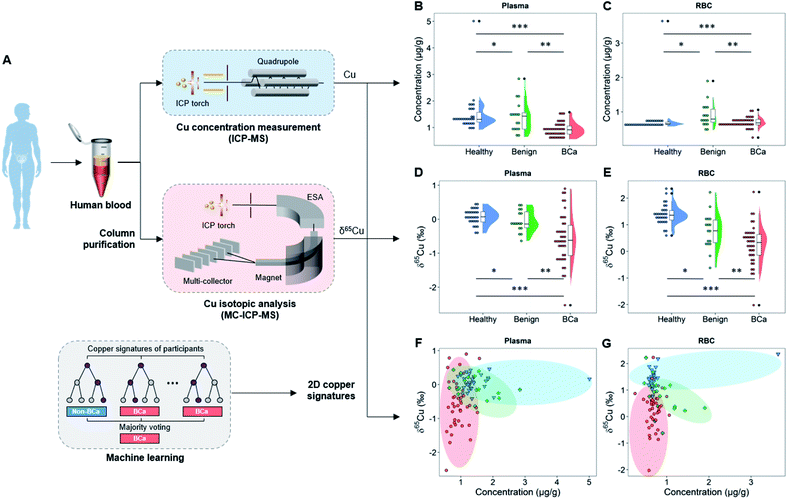
The copper concentration and copper isotopic ratio in blood were measured by inductively coupled plasma mass spectrometry (ICP-MS) and multi-collector ICP-MS (MC-ICP-MS) after chromatographical purification, respectively (Fig. 1A). Notably, to validate whether the copper isotopic fractionation was induced by BCa, we not only compared the copper isotopic ratio between BCa patients and healthy controls, but also examined other benign urinary diseases and studied the dependence of copper isotopes on the grades and stages of BCa. Based on the two-dimensional (2D) copper signatures in blood, i.e., the copper isotopic ratio and concentration in plasma and red blood cell (RBC), a machine learning model was developed to classify BCa and non-BCa subjects to reveal the potential role of this tool in cancer diagnosis and surveillance. To generalize the approach, we also validated the machine learning model with a previously published data set of HCC.
Results
Copper concentration variations in the blood of BCa patients
To realize the copper signatures related to BCa, we set three groups, i.e., the cancer group, age-matched healthy control, and benign urinary diseases (Tables S1–S8†). The cancer group includes BCa patients with different types, malignancy grades, and cancer stages. Note that men have a higher BCa morbidity than women36 and the BCa risk increases with age (most BCa cases are diagnosed in patients older than 50),37 and a young healthy control group was also included for reference. All study participants were recruited from the same region (Tianjin, China) to exclude the effects arising from geographical factors. The blood biochemical indexes of study participants are given in Tables S6–S8 and Fig. S1.† Here we chose plasma and RBC rather than urine as target media for copper isotopic analysis, because blood is the main medium of substance exchange between the tumor and the environment,38 and as two main components of blood, plasma and RBC have almost all of the blood copper burden, so the blood copper may directly reflect the disruption of copper metabolism by the tumor. In contrast, urine is not the main metabolic pathway of copper and the urinary copper concentration is too low to perform the isotopic analysis.39We first measured the copper concentration in blood. As shown in Fig. 1B, the mean copper concentration in plasma of BCa patients was lower than that of healthy and benign subjects (P < 0.05) but the difference was relatively small. The healthy and benign groups showed different copper concentrations in RBC, but the BCa group showed no significant difference from the healthy or benign groups (Fig. 1C). Noteworthily regarding the copper concentration in the blood of patients with urinary cancers, inconsistent trends have been obtained in the previous literature.40–44
Copper isotopic signatures of BCa in blood
We then measured the copper isotopic ratio in the plasma and RBC of study participants by MC-ICP-MS (see the Experimental section in the ESI for details†). Prior to isotopic analysis, the sample was purified by an anion exchange chromatographic method.45 The copper isotopic ratio (65Cu/63Cu) is expressed as the δ65Cu value (in ‰) relative to a standard material (ERM-AE633): | (1) |
A decrease in the δ65Cu value means a depletion of 65Cu (i.e., enrichment of 63Cu). The effect of the biological matrix on the copper isotopic analysis has been excluded to ensure a high precision (Fig. S2†).
As shown in Fig. 1D and E, we found that the BCa patients accumulated the light copper isotope (63Cu) in blood (δ65Cu value = −0.57 ± 0.73‰ in plasma and 0.25 ± 0.74‰ in RBC) compared with age-matched healthy (δ65Cu value = 0.07 ± 0.23‰ in plasma and 1.39 ± 0.38‰ in RBC) and benign control groups (δ65Cu value = −0.06 ± 0.31‰ in plasma and 0.76 ± 0.68‰ in RBC). For the healthy controls, the blood δ65Cu value was constrained in a relatively narrow range that was close to the literature data.20 Both benign and malignant diseases could lead to a broadening of the δ65Cu value range. Remarkably, in BCa patients, the blood δ65Cu value was significantly dispersed and 65Cu-depleted (Fig. 1D and E), suggesting that the incidence of BCa disrupted the copper homeostasis in the body. Compared with the copper concentration variations (Fig. 1B and C), the δ65Cu value showed larger discrepancies between BCa and control groups in both plasma and RBC (Δ65Cuplasma = 0.64‰ between BCa and healthy control; P < 0.0001), indicating that the blood copper isotope signature was more sensitive than the copper concentration in response to the cancer development. This result also revealed the significance of the blood copper isotopic ratio as a biomarker for BCa.
Notably, in plasma, only BCa (P < 0.0001), but not benign diseases (P > 0.05), could induce a significant copper isotopic fractionation compared to healthy control (Fig. 1D). Such a phenomenon was absent with the copper concentration or in RBC. Furthermore, by jointly using the copper concentration and δ65Cu value (Fig. 1F and G), we can better differentiate BCa, benign, and healthy controls. As shown in the 2D plot of copper signatures in plasma and RBC (Fig. 1F and G), the three groups are distributed in different zones as indicated by different colors. However, some overlaps were still observed, indicating that the distinguishing ability needs to be further improved.
Dependence of 2D copper signatures on cancer grades and stages
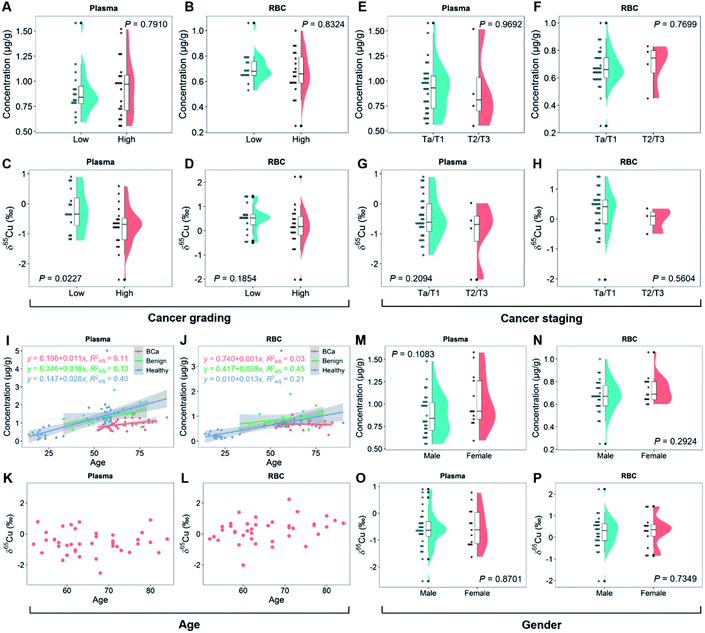
To clarify the relationship of copper isotopic fractionation with BCa, we then studied the 2D copper signatures of BCa patients grouped by cancer grading (high or low grade of the malignancy) and staging (Ta-T4 according to the TNM system).2,46 The cancer grades and stages were determined based on physical exams, biopsies, and imaging tests. As shown in Fig. 2A and B, high- and low-grade BCa showed no significant difference in the blood copper level (P > 0.7). By contrast, high-grade BCa was significantly 65Cu-depleted in plasma relative to low-grade (P < 0.05; Fig. 2C). This isotopic fractionation direction was consistent with that shown in Fig. 1D. Fig. 2A and C again suggest that the δ65Cu value should be a more sensitive BCa marker than the copper concentration. Furthermore, the cancer grade describes how malignant the cancer cells are. Thus, the result shown in Fig. 2C, i.e., the more malignant the tumor was, the larger the copper isotopic fractionation, evidenced that the significant copper isotopic fractionation was related to BCa.Regarding cancer staging, BCa was divided into two groups, superficial (Ta/T1) and muscle-invasive cancer (T2/T3), representing different degrees of tumor invasion and metastasis. In this study, the sample collection was mainly performed in the inpatient department, so only a limited number of T2/T3 patients were included (patients with T2 or a higher stage BCa were rarely subjected to surgical treatment). As shown in Fig. 2E–H, no difference in the blood copper level or δ65Cu value at different cancer stages was observed (P > 0.7 for the copper concentration and P > 0.2 for the δ65Cu value). Due to the small sample set of the T2/T3 group, it was difficult to draw any conclusions about the cancer staging yet. However, it was interesting to note that the plasma showed a tendency to be enriched in 63Cu in the T2/T3 group (Fig. 2G). More studies on this aspect are needed in the future.
It has been reported that age and gender may affect the copper concentration and δ65Cu value in human blood.21,47Fig. 2I and J show the relationship of age with the blood copper concentration in groups of BCa, benign, and healthy controls. In all groups, there was a clear linear positive correlation between the blood copper concentration and age. Interestingly, the slope of linearity decreased in the order of healthy > benign > BCa group. It thus appeared that the incidence of BCa could erase the effect of age on the blood copper. This result was also evidenced by the isotope data. In contrast to the previous report that the blood δ65Cu value decreased with age in healthy people,47 there was no correlation between the blood δ65Cu value and age of BCa patients (Fig. 2K and L). With regard to gender, it has been reported that women tend to have a lower blood δ65Cu value than men because of menstruation.48 However, in this study, the female BCa patients were mostly postmenopause women. This can explain why we did not find any difference in the blood copper concentration (Fig. 2M and N) or δ65Cu value between genders (Fig. 2O and P).
In addition, we have also performed a comprehensive correlation analysis taking into account multiple factors (including blood metals and biochemical indexes) (see Fig. S3†). Overall, only weak correlations between biochemical indexes and the blood copper concentration or δ65Cu value were observed (see Discussion in the ESI†).
Machine learning model for distinguishing BCa from non-BCa
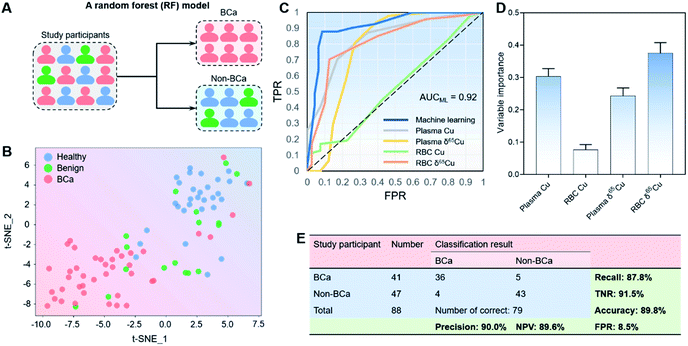
Since the subjects were difficult differentiate manually (Fig. 1F and G), we developed a machine learning model to further improve the distinguishing ability by combining all copper signatures. T-distributed stochastic neighbor embedding (t-SNE) is a non-linear technique for visualizing high-dimensional data by giving each sample data a location in a two- or three-dimensional map.49 As such, t-SNE could be used for automatically classifying and rationalizing the study participants according to copper-related variables that would otherwise have been difficult to differentiate manually.As shown in Fig. 3A, we first carried out the t-SNE dimensionality reduction process on data with all the four copper-related variables (i.e., the plasma copper concentration, RBC copper concentration, plasma δ65Cu value, and RBC δ65Cu value), and gained a conspicuous distinguishing result between BCa (n = 41) and non-BCa groups (benign plus healthy controls; n = 47). Since different groups could be visibly distinguished in the t-SNE map (Fig. 3B), we then developed a random forest (RF) model using the “leave-one-out” cross-validation (LOOCV) testing method50 to classify BCa and non-BCa groups (Fig. 3A). RF is an ensemble supervised learning method that fits a number of individual decision tree classifiers on sub-samples of the whole dataset. Such an ensemble strategy can reduce the risk of overfitting compared with single decision tree and is particularly suitable for the problems with low data volumes. The area under the receiver operator characteristic (ROC) curve (AUC) of the classification model reached 0.92 (Fig. 3C). Notably, we also performed the ROC analysis with single copper-related variables without RF classification (see also Fig. 3C). The results showed that the AUC of the machine learning model distinctly exceeded that without machine learning, clearly justifying the use of machine learning. The variable importance (VI) in the RF model was also analyzed. As shown in Fig. 3D, all the four variables contributed substantially to the modelling result, and the RBC δ65Cu value played the most important role in the RF model. The overall classification results are shown in Fig. 3E. For BCa and non-BCa subjects, the classification accuracy was 89.8%. A high precision (90.0%), high recall rate (also referred to as sensitivity, 87.8%), high TNR (true negative rate, also referred to as specificity, 91.5%), and low FPR (false positive rate, 8.5%) were achieved. Specifically, for high-grade BCa, the precision and recall rate could even reach 100% and 94.7%, respectively. These results indicated that the machine learning model had a strong distinguishing ability toward BCa.
Furthermore, to test the universality of the machine learning model, we also applied the algorithm to a previously published data set of HCC patients.20 We found that the machine learning model established here also worked well for classification of HCC and non-HCC subjects with accuracy, recall, TNR, and FPR of 87.5, 90.5, 84.2, and 15.8%, respectively (see Fig. S4, ESI†), thus suggesting the applicability of the machine learning model for different cancer types.
Discussion
Our results have demonstrated that 2D blood copper signatures offer a potentially new tool for BCa diagnosis, and the ROC analysis shows that machine learning plays a critical role in enabling the diagnosis (Fig. 3C). Currently, diagnosis and monitoring of BCa mainly rely on an invasive test, i.e., periodic cystoscopy. This test is reliable but rather uncomfortable for the patients and it is not exempt of comorbidities. Hence, there is an imperative need for reliable molecular biomarkers for clinical non-invasive BCa diagnosis. Noteworthily, the previous studies on disease-related isotopic analysis were largely limited to reporting stable isotopic ratio variations between patients and controls but seldom to realize the actual diagnosis, partly due to the insufficient distinguishing effect between patients and controls by only stable isotopic analysis. By contrast, we herein used 2D copper signatures integrating four copper-related variables by using the machine learning model. A prominent advantage offered by this approach is that it can provide a probability score for each subject for diagnosis, thus greatly improving the practicability of the stable isotopic analysis-based approach in clinical practice. The good performance of the machine learning model with the previously reported HCC data has also verified the universality of the approach (Fig. S4†).In addition, the dependence of the copper isotopic fractionation on the cancer grade of the tumor is also of potential merit for diagnosis. Remarkably, compared with the established biomarkers for BCa,31 this approach showed better or competitive performance with a high sensitivity, high TNR, and low FPR (see Fig. 3E and Table S9†). Despite that, it is also noteworthy that liver conditions of different etiologies showed a similar trend in the blood copper isotopic fractionation with cancers,18,19 suggesting that the specificity of the method needs further evaluation. Thus, in clinical practice, it is suggested to jointly use the 2D copper signatures with other existing diagnostic means to achieve a more reliable diagnosis.
Natural copper isotopic signatures also provide a novel point of perspective to reveal the molecular mechanisms underlying the carcinogenic process, assuming that cancer development may induce a specific isotopic fractionation effect of essential elements. The unique ability of stable isotopic signatures to identify the origins of elemental abnormity is particularly useful for cancer etiological research. Here, BCa is associated with a systemic significant depletion of 65Cu in blood, which could not be ascribed to the dietary intake because a typical human diet is 65Cu-enriched (δ65Cu value = ∼0.4‰).51 Therefore, the BCa-induced copper isotopic fractionation is deemed to result from the endogenous copper metabolic imbalance.
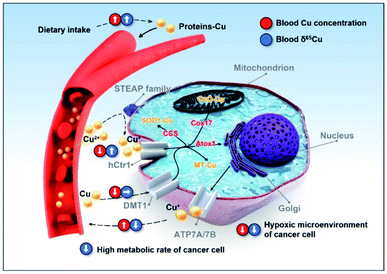
Bladder is a urinary organ, but the absorption, utilization, and excretion of copper mainly occur in the intestine and liver,32 so the copper isotopic fractionation should not be caused by direct changes in organ functions but rather impaired copper metabolism by cancer cells. The fractionation can be produced by equilibrium or kinetics processes.4 In an equilibrium process, a heavy isotope (65Cu) tends to be enriched in chemical species with higher oxidation states and stronger binding energies.52Fig. 4 shows the copper metabolic pathways in human cells and the possible factors linked to the copper isotopic fractionation. It is generally thought that the uptake and efflux of copper by cells are valence-dependent. The copper transport processes via hCtr1 (with Cu2+ being reduced to Cu+ by STEAP proteins as copper reductases) can induce the blood to be enriched in 65Cu2+ and the cells to be enriched in 63Cu+,51 and the amplitude of the copper isotopic fractionation is likely modulated by the copper reductases.53 It has been reported that hCtr1 is down-regulated and ATP7A is up-regulated in cancer cells,54–59 which can explain the lower δ65Cu value in the blood of BCa patients due to the lower cellular uptake of 63Cu+, leaving more 63Cu in blood (Fig. 1). However, the lower cellular copper uptake would lead to a higher copper level in blood, which however was not observed here (Fig. 1B and C). Therefore, our results imply the existence of additional copper uptake mechanisms that can compensate the lowered copper uptake in cancer cells. We consider that via divalent metal transporter 1 (DMT1) is a very likely mechanism, since its transport of copper does not involve copper reduction and thereby would not affect the copper isotopic fractionation induced by the Cu+ uptake (Fig. 4).60–62 This redox-independent process is normally less considered in the copper metabolism thus deserving more attention.
Some characteristics of cancer cells may aggravate the isotopic fractionation. The hypoxic tumor microenvironment is favorable for the enrichment of the heavy isotope (65Cu) in cancer cells.63,64 Furthermore, cancer cells have a relatively high metabolic rate for more material exchange,65 which may exacerbate the Cu+ uptake and excretion by cells, intensifying the enrichment of 63Cu in blood. Until now, the investigations on protein-associated copper isotopic fractionation are still very scarce. More in vitro and animal experiments are definitely needed in future studies to verify the cancer-related isotopic fractionation mechanisms.
It is interesting to note that the response of the blood δ65Cu value to the cancer development is more sensitive than the copper concentration (Fig. 1). A plausible explanation is that there are some regulators and sophisticated mechanisms to maintain the copper homeostasis in the human body,35e.g., the DMT1 pathway mentioned above.60 However, such mechanisms may not be able to correct the deviation in the δ65Cu value (i.e., no homeostatic mechanism toward copper isotopes). Thus, the cancer-induced deviation in the δ65Cu value is more prone to be accumulated and recorded in the human body. Furthermore, the copper isotopic fractionation in the human blood associated with different cancer types appears to be consistent (i.e., enriched in 63Cu),14,20–23,25 suggesting that there may be similar rules governing the copper metabolic imbalance in cancer development, notwithstanding more cancer types and larger cohort studies are required to verify this point.
Conclusions
In summary, we have identified the variations in the copper concentration and natural copper isotopic composition in the blood of BCa patients relative to benign and healthy controls, and such 2D copper signatures enabled the distinguishing of BCa and non-BCa by the aid of machine learning. We show that the combined use of multiple signatures and machine learning can greatly enhance the distinguishing capability between cancer and non-cancer. It is worth noting that such a good performance was achieved with only a single element. It is rational to expect that exploring and integrating more types of elemental signatures in the machine learning model will further enhance the reliability and practicability. It should also be noted that the sample size used in this study was relatively limited and so a larger sample set is needed to validate the method in future studies. The feasibility of the method for other types of cancers also needs to be explored. Overall, the machine learning-aided 2D elemental signatures provide a versatile label-free approach for molecular mechanism research and diagnosis of diseases. Considering that the stable isotopic signature is still a relatively new tool for health research, this work may represent an important step to push forward the application of stable isotopic chemistry in biomedicine.Ethical statement
All participants provided informed consent, and the study protocol was approved by the Ethics Committee of the Second Hospital of Tianjin Medical University (No. KY2018K082) and compliant with all relevant ethical regulations for studies involving human subjects.Data availability
All experimental procedures, supporting tables and figures are available in the ESI.†Author contributions
Q. Liu and W. Wang designed the research; W. Wang performed most of experiments; X. Liu and A. Zhang performed the machine learning calculations. C. Zhang, F. Sheng, S. Liu, and Z. Zhang collected the blood samples of patients; S. Song, P. Li, and B. Wang collected the blood samples of healthy subjects; D. Lu, L. Zhang, and X. Yang helped with the MC-ICP-MS measurements; S. Dai helped with the statistical analysis; G. Jiang supervised the project; W. Wang and Q. Liu analyzed the data; Q. Liu, W. Wang, and X. Liu wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 22050001, 21825403, 21976194, 92143301, and 22188102), the National Key Research and Development Program of China (2018YFA0901101, and 2020YFA0907500), the Strategic Priority Research Program of CAS (XDPB2003), and the Sanming Project of Medicine in Shenzhen (SZSM201811070). We would like to acknowledge the help from the working group of environmental exposure and human health of the China Cohort Consortium (http://chinacohort.bjmu.edu.cn/).Notes and references
- S. Gutman and L. G. Kessler, Nat. Rev. Cancer, 2006, 6, 565–571 CrossRef CAS PubMed.
- J. A. Ludwig and J. N. Weinstein, Nat. Rev. Cancer, 2005, 5, 845–856 CrossRef CAS PubMed.
- J. Hoefs, Stable Isotope Geochemistry, Springer Nature, 8th edn, 2018 Search PubMed.
- J. G. Wiederhold, Environ. Sci. Technol., 2015, 49, 2606–2624 CrossRef CAS PubMed.
- D. Lu, Q. Liu, T. Zhang, Y. Cai, Y. Yin and G. Jiang, Nat. Nanotechnol., 2016, 11, 682–686 CrossRef CAS PubMed.
- L. Yang, Mass Spectrom. Rev., 2009, 28, 990–1011 CrossRef CAS PubMed.
- Q. Liu, H. Hintelmann and G. Jiang, Natl. Sci. Rev., 2016, 3, 410 CrossRef.
- Y.-k. Tanaka and T. Hirata, Anal. Sci., 2018, 34, 645–655 CrossRef CAS PubMed.
- F. Albarede, P. Telouk, V. Balter, V. P. Bondanese, E. Albalat, P. Oger, P. Bonaventura, P. Miossec and T. Fujii, Metallomics, 2016, 8, 1056–1070 CrossRef CAS PubMed.
- M. Aramendia, L. Rello, M. Resano and F. Vanhaecke, J. Anal. At. Spectrom., 2013, 28, 675–681 RSC.
- Y. Anoshkina, M. Costas-Rodriguez, M. Speeckaert, W. Van Biesen, J. Delanghe and F. Vanhaecke, Metallomics, 2017, 9, 517–524 CrossRef CAS PubMed.
- L. Sauzeat, E. Bernard, A. Perret-Liaudet, I. Quadrio, A. Vighetto, P. Krolak-Salmon, E. Broussolle, P. Leblanc and V. Balter, iScience, 2018, 6, 264–271 CrossRef CAS PubMed.
- R. Grigoryan, M. Costas-Rodriguez, S. Van Laecke, M. Speeckaert, B. Lapauw and F. Vanhaecke, J. Anal. At. Spectrom., 2019, 34, 1514–1521 RSC.
- F. Vanhaecke and M. Costas-Rodríguez, View, 2021, 2, 20200094 CrossRef.
- T. Walczyk and F. von Blanckenburg, Science, 2002, 295, 2065–2066 CrossRef CAS PubMed.
- P. A. Krayenbuehl, T. Walczyk, R. Schoenberg, F. von Blanckenburg and G. Schulthess, Blood, 2005, 105, 3812–3816 CrossRef CAS PubMed.
- J. L. L. Morgan, J. L. Skulan, G. W. Gordon, S. J. Romaniello, S. M. Smith and A. D. Anbar, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9989–9994 CrossRef CAS PubMed.
- M. Costas-Rodriguez, Y. Anoshkina, S. Lauwens, H. Van Vlierberghe, J. Delanghe and F. Vanhaecke, Metallomics, 2015, 7, 491–498 CrossRef CAS PubMed.
- S. Lauwens, M. Costas-Rodriguez, H. Van Vlierberghe and F. Vanhaecke, Sci. Rep., 2016, 6, 30683 CrossRef CAS PubMed.
- V. Balter, A. N. da Costa, V. P. Bondanese, K. Jaouen, A. Lamboux, S. Sangrajrang, N. Vincent, F. Fourel, P. Telouk, M. Gigou, C. Lecuyer, P. Srivatanakul, C. Brechot, F. Albarede and P. Hainaut, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 982–985 CrossRef CAS PubMed.
- P. Telouk, A. Puisieux, T. Fujii, V. Balter, V. P. Bondanese, A.-P. Morel, G. Clapisson, A. Lamboux and F. Albarede, Metallomics, 2015, 7, 299–308 CrossRef CAS PubMed.
- B. Toubhans, A. T. Gourlan, P. Telouk, K. Lutchman-Singh, L. W. Francis, R. S. Conlan, L. Margarit, D. Gonzalez and L. Charlet, J. Trace Elem. Med. Biol., 2020, 62, 126611 CrossRef CAS.
- A. A. M. B. Hastuti, M. Costas-Rodriguez, A. Matsunaga, T. Ichinose, S. Hagiwara, M. Shimura and F. Vanhaecke, Sci. Rep., 2020, 10, 16389 CrossRef PubMed.
- G. W. Gordon, J. Monge, M. B. Channon, Q. Wu, J. L. Skulan, A. D. Anbar and R. Fonseca, Leukemia, 2014, 28, 2112–2115 CrossRef CAS PubMed.
- L. Lobo, M. Costas-Rodriguez, J. Carlos de Vicente, R. Pereiro, F. Vanhaecke and A. Sanz-Medel, Talanta, 2017, 165, 92–97 CrossRef CAS PubMed.
- F. Larner, L. N. Woodley, S. Shousha, A. Moyes, E. Humphreys-Williams, S. Strekopytov, A. N. Halliday, M. Rehkaemper and R. C. Coombes, Metallomics, 2015, 7, 112–117 CrossRef CAS PubMed.
- K. Schilling, F. Larner, A. Saad, R. Roberts, H. M. Kocher, O. Blyuss, A. N. Halliday and T. Crnogorac-Jurcevic, Metallomics, 2020, 12, 752–757 CrossRef CAS.
- F. Albarede, P. Telouk and V. Balter, Rev. Mineral. Geochem., 2017, 82, 851–885 CrossRef CAS.
- S. Wach, K. Weigelt, B. Michalke, V. Lieb, R. Stoehr, B. Keck, A. Hartmann, B. Wullich, H. Taubert and A. Chaudhri, J. Trace Elem. Med. Biol., 2018, 46, 150–155 CrossRef CAS PubMed.
- J. R. E. Wolfs, T. J. N. Hermans, E. L. Koldewijn and D. van de Kerkhof, Urol. Oncol., 2021, 39, 161–170 CrossRef PubMed.
- EAU, EAU Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and CIS), European Association of Urology, 2018 Search PubMed.
- A. Gupte and R. J. Mumper, Cancer Treat. Rev., 2009, 35, 32–46 CrossRef CAS PubMed.
- S. Ishida, P. Andreux, C. Poitry-Yamate, J. Auwerx and D. Hanahan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 19507–19512 CrossRef CAS PubMed.
- H. Mazdak, F. Yazdekhasti, A. Movahedian, N. Mirkheshti and M. Shafieian, Int. Urol. Nephrol., 2010, 42, 89–93 CrossRef CAS PubMed.
- B.-E. Kim, T. Nevitt and D. J. Thiele, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS PubMed.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- R. Madeb and E. M. Messing, Urol. Oncol., 2004, 22, 86–92 CrossRef PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57–70 CrossRef CAS.
- G. H. O. Rocha, C. Steinbach, J. R. Munhoz, M. A. O. Madia, J. K. Faria, D. Hoeltgebaum, F. Barbosa Jr, B. L. Batista, V. C. O. Souza, S. B. Nerilo, E. Bando, S. A. G. Mossini and P. Nishiyama, J. Trace Elem. Med. Biol., 2016, 35, 61–65 CrossRef CAS PubMed.
- W. I. Mortada, A. Awadalla, S. Khater, A. Ahmed, E. T. Hamam, M. El-zayat and A. A. Shokeir, Environ. Sci. Pollut. Res., 2020, 27, 15835–15841 CrossRef CAS PubMed.
- M. Kaba, N. Pirincci, M. B. Yuksel, I. Gecit, M. Gunes, H. Ozveren, H. Eren and H. Demir, Asian Pac. J. Cancer Prev., 2014, 15, 2625–2629 CrossRef PubMed.
- T. Golabek, B. Darewicz, M. Borawska, K. Socha, R. Markiewicz and J. Kudelski, Urol. Int., 2012, 89, 342–347 CrossRef CAS PubMed.
- J. T. Lim, Y. Q. Tan, L. Valeri, J. Lee, P. P. Geok, S. E. Chia, C. N. Ong and W. J. Seow, Environ. Int., 2019, 132, 105109 CrossRef CAS PubMed.
- S. Songchitsomboon, S. Komindr, A. Komindr, S. Kulapongse, O. Puchaiwatananon and U. Udomsubpayakul, J. Med. Assoc. Thailand, 1999, 82, 701–706 CAS.
- L. Van Heghe, E. Engstrom, I. Rodushkin, C. Cloquet and F. Vanhaecke, J. Anal. At. Spectrom., 2012, 27, 1327–1334 RSC.
- G. D. Chisholm, J. R. Hindmarsh, A. G. Howatson, J. N. Webb, A. Busuttil, T. B. Hargreave and J. E. Newsam, Br. J. Urol., 1980, 52, 500–505 CAS.
- K. Jaouen, M. Gibert, A. Lamboux, P. Telouk, F. Fourel, F. Albarede, A. N. Alekseev, E. Crubezy and V. Balter, Metallomics, 2013, 5, 1016–1024 CrossRef CAS.
- L. Van Heghe, O. Deltombe, J. Delanghe, H. Depypere and F. Vanhaecke, J. Anal. At. Spectrom., 2014, 29, 478–482 RSC.
- L. van der Maaten and G. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
- D. G. Beer, S. L. R. Kardia, C. C. Huang, T. J. Giordano, A. M. Levin, D. E. Misek, L. Lin, G. A. Chen, T. G. Gharib, D. G. Thomas, M. L. Lizyness, R. Kuick, S. Hayasaka, J. M. G. Taylor, M. D. Iannettoni, M. B. Orringer and S. Hanash, Nat. Med., 2002, 8, 816–824 CrossRef CAS PubMed.
- V. Balter, A. Lamboux, A. Zazzo, P. Telouk, Y. Leverrier, J. Marvel, A. P. Moloney, F. J. Monahan, O. Schmidt and F. Albarede, Metallomics, 2013, 5, 1470–1482 CrossRef CAS.
- T. Fujii, F. Moynier, M. Abe, K. Nemoto and F. Albarede, Geochim. Cosmochim. Acta, 2013, 110, 29–44 CrossRef CAS.
- J.-L. Cadiou, S. Pichat, V. P. Bondanese, A. Soulard, T. Fujii, F. Albarede and P. Oger, Sci. Rep., 2017, 7, 44533 CrossRef CAS PubMed.
- Y. Kita, A. Hamada, R. Saito, Y. Teramoto, R. Tanaka, K. Takano, K. Nakayama, K. Murakami, K. Matsumoto, S. Akamatsu, T. Yamasaki, T. Inoue, Y. Tabata, Y. Okuno, O. Ogawa and T. Kobayashi, Br. J. Cancer, 2019, 121, 1027–1038 CrossRef CAS PubMed.
- T. Yang, M. Chen, T. Chen and A. Thakur, Oncol. Lett., 2015, 10, 2584–2590 CrossRef CAS PubMed.
- V. Shanbhag, K. Jasmer-McDonald, S. Zhu, A. L. Martin, N. Gudekar, A. Khan, E. Ladomersky, K. Singh, G. A. Weisman and M. J. Petris, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 6836–6841 CrossRef CAS PubMed.
- I. M. Gomes, C. J. Maia and C. R. Santos, Mol. Cancer Res., 2012, 10, 573–587 CrossRef CAS PubMed.
- M. Azumi, H. Kobayashi, N. Aoki, K. Sato, S. Kimura, H. Kakizaki and M. Tateno, J. Urol., 2010, 183, 2036–2044 CrossRef CAS PubMed.
- A. K. Holzer, N. M. Varki, Q. T. Le, M. A. Gibson, P. Naredi and S. B. Howell, J. Histochem. Cytochem., 2006, 54, 1041–1049 CrossRef CAS PubMed.
- C. Lin, Z. Zhang, T. Wang, C. Chen and Y. J. Kang, Metallomics, 2015, 7, 1285–1289 CrossRef CAS PubMed.
- M. Arredondo and M. T. Nunez, Mol. Aspects Med., 2005, 26, 313–327 CrossRef CAS.
- R. O. Koch, H. Zoller, I. Theurl, P. Obrist, G. Egg, W. Strohmayer, W. Vogel and G. Weiss, Histol. Histopathol., 2003, 18, 1095–1101 CAS.
- M. R. Florez, M. Costas-Rodriguez, C. Grootaert, J. Van Camp and F. Vanhaecke, Anal. Bioanal. Chem., 2018, 410, 2385–2394 CrossRef CAS.
- V. P. Bondanese, A. Lamboux, M. Simon, J. E. Lafont, E. Albalat, S. Pichat, J.-M. Vanacker, P. Telouk, V. Balter, P. Oger and F. Albarede, Metallomics, 2016, 8, 1177–1184 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, supporting discussion, supporting tables and figures. See DOI: 10.1039/d1sc06156a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |