Open Access Article
Open Access ArticleMulti-coloured fluorescent sensing toolbox for selective detection of nitroxyl in vitro and ex vivo†
V.
Staikopoulos‡
ab,
X.
Zhang‡
bc,
B. P.
Pullen
abd,
P.
Reineck
 be,
A. K.
Vidanapathirana
abd,
S. M.
Lee
a,
J.
Liu
a,
C.
Bursill
abd,
M. R.
Hutchinson
ab and
A. D.
Abell
be,
A. K.
Vidanapathirana
abd,
S. M.
Lee
a,
J.
Liu
a,
C.
Bursill
abd,
M. R.
Hutchinson
ab and
A. D.
Abell
 *bc
*bc
aAdelaide Medical School, University of Adelaide, Adelaide, South Australia 5005, Australia
bAustralian Research Council (ARC) Centre of Excellence for Nanoscale BioPhotonics, Adelaide Medical School, University of Adelaide, Adelaide, South Australia 5005, Australia
cDepartment of Chemistry, University of Adelaide, Adelaide, South Australia 5005, Australia. E-mail: andrew.abell@adelaide.edu.au
dVascular Research Centre, Lifelong Health Theme, South Australian Health and Medical Research Institute (SAHMRI), Adelaide, South Australia 5000, Australia
eSchool of Science, RMIT University, Melbourne, Victoria, 3001 Australia
First published on 3rd February 2022
Abstract
Methods for the endogenous detection of nitroxyl (azanone; HNO), the reduced and protonated derivative of nitric oxide (NO), are required to define its cardiovascular function and its key role in chronic pain. This study reports the design, synthesis and biological evaluation of 3 super-bright, highly sensitive, specific and non-cytotoxic arylphosphine-based fluorescent nitroxyl sensors that enable the detection of endogenous nitroxyl in vitro and ex vivo. The presence of endogenous nitroxyl is observed in the murine microglial cell line (BV2), primary human coronary artery endothelial cells (HCAECs) and rat cardiomyocyte cell line (H9C2) by spectroscopy and/or confocal microscopy following the addition of relevant stimulants. Furthermore, the presence of endogenous nitroxyl is detected in rat blood samples. The novel sensor 3 (rhodol 4 on a 2-(diphenylphosphanyl)benzoate backbone) is the most sensitive in detecting endogenous nitroxyl following stimulation, in the presence of biological media such as HBSS and DMEM. This then provides a validated tool for detecting nitroxyl in biological systems.
Introduction
The one-electron reduction of nitric oxide to nitroxyl (HNO) occurs in both eukaryote and mammalian biology. HNO plays a role in many pharmacological effects and physiological functions such as potent vasodilation,1–4 increased cardiac output,5–7 altered post-ischemic myocardial injury,8–10 and treatment of alcoholism,11,12 and may act as an anti-nociceptive modulator of neuropathic and inflammatory pain.13–15 While the chemical properties and reactivity of HNO and NO are distinct, they do act on similar biological targets (thiols, thiol protein and metalloproteins). However, the mechanisms of these interactions and the resulting products are different.16–20 Due to the challenges associated with directly detecting HNO, the basis for the biological role of HNO in the aforementioned conditions has been established using donor species such as Angeli's salt and Piloty's acid. The mechanisms of endogenous production of HNO have only been speculated upon based on aqueous solution chemical reactions and the use of scavengers.21 Despite the growing body of evidence supporting the biochemical importance of HNO, it remains unclear if it is generated in vitro and the associated mechanism involved.A reliable method for detecting HNO in vitro is required to help answer these questions. The use of fluorescent probes for detecting reactive nitrogen species in vitro and in vivo shows much promise due to high sensitivity, ease of use, non-invasive nature, and good spatiotemporal resolution. In addition, fluorescent probes are available in a wide range of wavelengths, which provide opportunities to visualise multiple targets simultaneously with multiple probes of different colours. These probes are thus ideal candidates for detecting HNO in a complex biological environment. Several classes of HNO-sensitive fluorescent probes have been reported,22 which are based on the reaction of nitroxyl with chemical species, such as copper(II), nitroxide, 2-mercapto-2-methylpropionic acid or arylphosphine. Several existing HNO probes have drawbacks that limit their use in biological sensing. For example, Cu-based probes23–25 lack selectivity for HNO over other biological reductants such as ascorbate or glutathione. This can cause interference, thus limiting their application in biological studies. Arylphosphine-based probes are highly selective for HNO over other cellular reductants and have given rise to several probes for the detection of nitroxyl.27–33 However, none of these arylphosphine probes are capable of detecting endogenously produced HNO, possibly due to the low concentration of HNO present in living systems. Thus a more sensitive, stable and biocompatible multi-coloured sensing tool kit is required for the application of arylphosphine probes in real-time quantitative measurement of biologically produced HNO.
Here we report a new, highly sensitive and biostable tri-arylphosphine-based fluorescent chemosensor 3 (Scheme 1) for selective detection of HNO in cells and demonstrate its use in imaging exogenous and endogenous HNO in two types of cell-lines, BV2 and HCAEC. This sensor, together with two other highly selective sensors (1 and 2, Scheme 1),28,34 provides a multi-coloured sensing toolkit for HNO with applicability over a wide range of common wavelengths (480–560 nm) used in biological sensing studies under physiological pH. This study demonstrates the ability of these sensors to detect exogenous and endogenous derived HNO in mouse, rat and human cells and rat blood. These chemo-sensors are able to detect HNO in a range of aqueous buffers and cellular media without the use of an organic cosolvent, thus enabling sensing in biological samples that are vulnerable to the toxicity of organic solvents. In addition, they show low cytotoxicity to cell lines, such as BV2 (mouse microglia) and HCAEC (primary human coronary arterial endothelial cells) cells. Sensor 3 was able to detect HNO when added to fresh rat blood and maintained a stable fluorescent signal over time following freeze–thaw cycles. The excellent biocompatibility of these new arylphosphine-based HNO sensors makes them suitable for a wide range of biological studies.
Results – chemistry
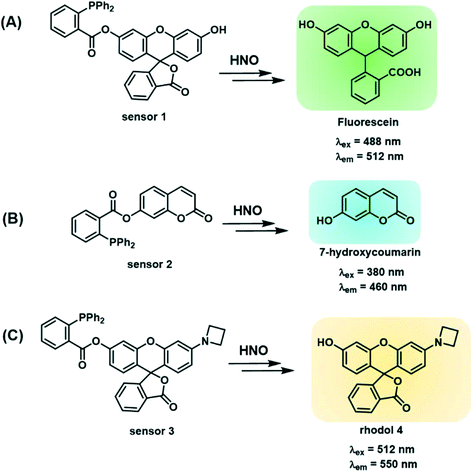
Design and synthesis of sensors 1–3
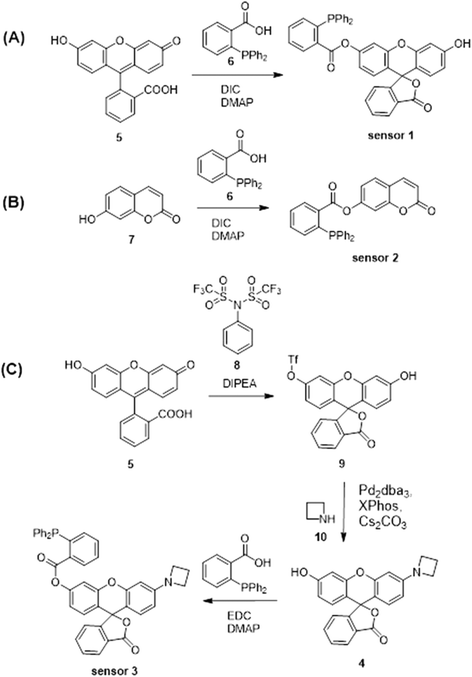
Sensors 1–3 consist of a triarylphosphine group conjugated with three distinctive fluorophores, fluorescein, coumarin and a mono-substituted rhodol 4, to selectively detect intracellular HNO (Scheme 1A–C). The conjugated probes are non-fluorescent.26 The reaction with HNO yields an aza-ylide intermediate and leads to subsequent ester aminolysis to the original fluorophores and a measurable increase in fluorescence by using fluorescence spectroscopy and confocal microscopy.27 The reaction is bio-orthogonal as phosphines are abiotic and essentially unreactive toward other biomolecules inside or near cells. The abovementioned fluorophores were selected to provide a wide range of emission wavelengths (460–550 nm) commonly used in sensing studies. Importantly, the three sensors encompass wavelength ranges commonly used in chemical sensing, with sensor 2 emitting in the blue range (λem = 460 nm), sensor 1 in the green range (λem = 512 nm) and sensor 3 in the yellow range (λem = 550 nm). These fluorophores were chosen due to their good fluorescent response to biological stimuli and excellent stability in biological environments.58–60 In particular, sensor 3 reacts with HNO to give highly fluorescent rhodol 10, which has high biostability to allow detection of subtle changes in concentrations of HNO in cells and tissues. In addition, this fluorophore emits at λem = 550 nm, where biological samples typically have weaker autofluorescence relative to the turn-on brightness of the sensor, as demonstrated in this study. This ensures that sensing of HNO occurs with minimal background fluorescence resulting in enhanced sensitivity. Together these sensors form a multi-coloured fluorescent sensing tool kit that can be used in conjunction with a broad range of biosensing techniques including green-fluorescent protein, fluorescent antibodies, nucleus stain and mitochondria stain.Sensors 1–3 were synthesised as shown in Scheme 2. Briefly, fluorescein5 or 7-hydroxycoumarin7 was esterified on reaction with 2-(diphenylphosphino) benzoic acid6 to give sensors 1 and 2 in 44% and 28% yield, respectively. Sensor 3 was synthesised from fluorescein.5 Fluorescein was protected as a triflate,9 which was subsequently coupled with azetidine10 under the catalysis of Pd2dba3 and XPhos to give rhodol 4. Esterification of rhodol 4 with 2-(diphenylphosphino) benzoic acid6 gave sensor 3 in 8% yield. All sensors were purified by HPLC.
Spectroscopic characterisation of sensors 1–3
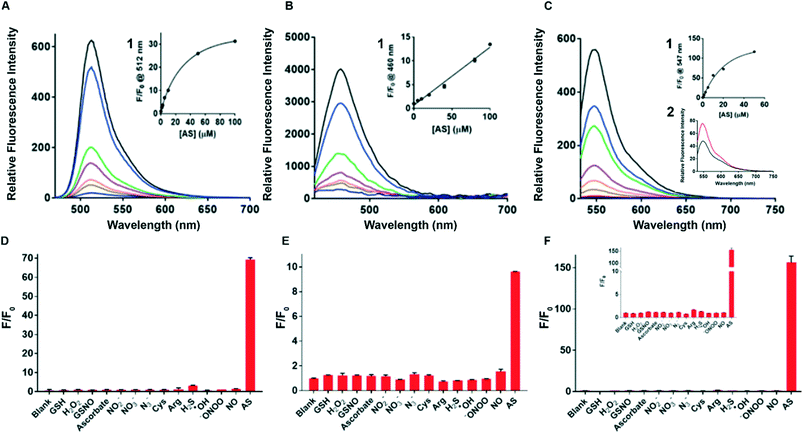
The spectroscopic properties of sensors 1–3 were assessed in water. First, the fluorescence of sensors 1–3 with and without added Angeli's salt (AS) was determined. Sensor 3 (λex = 512 nm and λem max = 550 nm) provides a 120-fold increase in fluorescence upon addition of 100 μM AS, the largest fold-increase in fluorescent response amongst all three sensors (Fig. 1C). In comparison, sensors 1 and 2 gave a 30-fold and 12-fold increase in an analogous experiment (Fig. 1A and B). Sensors 1 and 2 gave rise to typical fluorescence emission profiles of fluorescein and coumarin, respectively, with λex = 488 nm and λem max = 512 nm for sensor 1, and λex = 380 nm and λem max = 460 nm for sensor 2. (Fig. 1A and B). Standard curves of sensors 1–3 with added Angeli's salt (0–100 μM) were generated (Fig. 1 insert 1) to allow quantification of HNO. The fluorescence of sensors 1–3 increased with increasing concentration of AS, with sensors 1 and 3 following a trend of one-phase association and sensor 2 following a linear trend. The quantum yield of each sensor was determined in the presence of 100 μM Angeli's salt (ESI† Fig. S3A–C). All three sensors demonstrated very high quantum yields (Φsensor1 = 0.9, Φsensor2 = 0.85, Φsensor3 = 0.77) with sensor 1 being the highest (ESI† Fig. S3A). This is expected as the reaction of sensor 1 with HNO gives fluorescein, which has a reported quantum yield of 0.93 in 0.1 M NaOH. The high quantum yield ensures the high sensitivity of these chemo-sensors for HNO detection in biological studies. The limit of detection of sensor 3 with Angeli's salt was determined to be 100 nM (Fig. 1C insert 2), the lowest of the three sensors reported herein, and to our best knowledge is together with Dong et al.35 one of the lowest reported in the literature. Overall, sensor 3 provides an optimal sensing profile with a combined low detection limit and large fluorescence turn-on in response to HNO. Thus, sensor 3 is the optimal tool assessed here for detecting HNO levels in biological environments.Stability, selectivity profiles & kinetics
The photo and chemical stability of the sensors in common biological buffers (PBS and HBSS) and cell media (DMEM, DMEM serum-free (SF), DMEM phenol-red free (PRF) and DMEM serum-free and phenol-red free (SF PRF)) were investigated to examine their suitability for use in cell-based experiments. The sensor was dissolved in buffer or medium containing 1% DMSO followed by the addition of NaOH (final concentration 0.5 mM) or AS (100 μM, in 10 mM NaOH). The sample was incubated for 10 min in the dark and the resulting fluorescence was measured by spectroscopy, with the results shown in ESI† Fig. S4(A–C). All three sensors retain their sensing ability in the tested buffers and media, with sensor 3 providing the highest fold change in the fluorescent signal. PBS buffer gave a similar sensing profile to sensors in water, with sensors 1, 2 and 3 giving a 32-fold, 7-fold and 65-fold increase in fluorescence upon addition of AS, respectively. Incubation in HBSS resulted in a decreased fluorescence response in sensors 1 (22-fold) and 2 (6-fold) while enhancing the response of sensor 3 (75-fold) when compared to PBS response. Incubation in cell media resulted in a decrease in sensitivity for all three sensors. Sensor 3 is shown to be the most compatible with cell media, with the retained ability to produce a 40-fold fluorescence response in DMEM PRF. In comparison, sensors 1 and 2 gave lower fluorescence response (12-fold and 4-fold) in DMEM-related media. This demonstrates that sensor 3 is optimal for cell-based experiments.Next, sensors 1–3 were assayed against a range of biologically relevant RNS (NO, NO2−, NO3−, N3−, ONOO−), amino acids (Cys, Arg), small molecules (ascorbate, H2S, GSH, GSNO), and ROS (H2O2, OH) to define selectivity profiles and determine species specificity. All three sensors clearly show good selectivity for AS (Fig. 1D–F), with sensor 3 showing the optimal selectivity and being non-reactive to all species tested. Importantly, the sensors display an excellent selectivity for AS over all other tested RNS, NO in particular. This is critical as RNS always co-exist in biological environments and thus the ability to detect HNO amongst a complex mixture of RNS is highly desirable. Both sensor 1 and sensor 2 showed minimal affinity to arginine. This is important as arginine is the precursor of RNS and thus the ability to distinguish between these species is critical to accurate sensing of intracellular HNO. Sensor 1 is also found to react with H2S to a limited extent, which is consistent with the literature. However, due to the low concentration of H2S present in cells, the weak affinity to H2S is not expected to disrupt the applicability of sensor 1 in biosensing. Collectively, sensor 3 demonstrates excellent fluorescence turn-on response to AS, the highest fold change in fluorescence, low detection limit, high compatibility with cell media and excellent selectivity.
The kinetic profiles of the sensors were next investigated to examine their stability in water and rate of reaction with AS. Briefly, a solution of each sensor was prepared using the above conditions and mixed with AS (200 μM) in 10 mM NaOH. The fluorescence of each sample was continuously monitored over 60 min (ESI† Fig. S5A–C). Sensor 1 reacts with AS faster than sensors 2 and 3, taking 10 min to reach the saturation of the fluorescence signal, with sensors 2 and 3 taking more than 10 min. Rapid reaction with HNO (such as sensor 1) is required if one is to measure a reactive analyte such as HNO, which is oxidised rapidly to the corresponding reactive nitrogen species (RNS) in biological environments.36 At a concentration of 1 μM, sensor 1 showed a 67-fold increase in fluorescence compared to 0 μM, while sensor 2 showed a 19-fold increase in fluorescence. Sensor 3 gave the highest fluorescence increase with a 368-fold increase (ESI† Fig. S5D–F). Interestingly, at 100 nM concentration, sensor 1 showed a 5.5-fold increase in fluorescence compared to 0 μM, sensor 2 showed a 3-fold increase in fluorescence and sensor 3 showed a 38-fold increase. The high turn-on fluorescence of sensor 3 makes it more suitable for measuring low concentrations of endogenous HNO. This data was used to determine the optimal concentration of the sensor and incubation time to use in our subsequent in vitro work. Sensor 3 was selected for subsequent cell-based studies based on these results, unless otherwise stated.
Results – application in biological systems
Stability over time of sensors 1–3 in biological buffer
The fluorescence stability of all 3 sensors in biological buffer (HBSS) was assessed in cell-free media using spectroscopy. Angeli's salt (AS) was used as a donor of HNO and tested at concentrations ranging from 0 to 200 μM (ESI† Fig. S6). All 3 sensors were tested at 2.5, 5 and 10 μM and measured for up to an hour. Sensors 1 and 2 display a similar ‘turn-on’ fluorescent profile across all concentrations of the probe and donor, and the starting baseline increased in proportion to the sensor concentration. Sensor 2 appeared to be the least stable in AS concentration ranges between 100 and 200 μM. The time taken for the fluorescence signal to peak before plateauing was 20 minutes for sensors 1 and 2, and 10 minutes for sensor 3, making it the fastest responder of all 3 probes. Together with the kinetic data and fluorescent characteristics in biological buffers and media (ESI† Fig. S4) we were able to determine a concentration range (1–3 μM) and incubation time (at least 10 min) to proceed with future biological experiments.Detecting HNO in cell-free MesoEndo culture media used for HCAEC
The stability of sensor 3 in HCAEC buffer (MesoEndo cell growth medium) was investigated to examine its suitability for use in cell-based experiments. Briefly, sensor 3 (3 μM) was added to MesoEndo cell growth medium followed by the addition of AS (200 μM) and different concentrations of L-cysteine (0, 1, 10, 100, 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 and 1000 μM) and then read on a spectrophotometer after 15 min (ESI† Fig. S7). The data show that the fluorescence signal is higher in the presence of sensor 3 when compared to media only controls (pre-AS, **p < 0.0001; AS 200 μM, #p < 0.0001). Furthermore, the presence of L-cysteine reduces the AS derived HNO fluorescent signal in cell-free media when compared to sensor only (*p < 0.0001). This indicates that sensor 3 is suitable to be used for HCAEC experiments.
500 and 1000 μM) and then read on a spectrophotometer after 15 min (ESI† Fig. S7). The data show that the fluorescence signal is higher in the presence of sensor 3 when compared to media only controls (pre-AS, **p < 0.0001; AS 200 μM, #p < 0.0001). Furthermore, the presence of L-cysteine reduces the AS derived HNO fluorescent signal in cell-free media when compared to sensor only (*p < 0.0001). This indicates that sensor 3 is suitable to be used for HCAEC experiments.
HNO sensors 1, 2 and 3 are not cytotoxic to mouse microglial or human coronary artery cells
Following the evaluation of sensors 1–3 in biological media samples, a trypan blue dye exclusion test and/or MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) colorimetric assay were conducted to evaluate the changes in cell viability and metabolic activity in response to sensors 1–3 (ESI† Fig. S8). BV2 cells were incubated with 1–10 μM of sensor 1, 2 or 3, for either 1.5 hours for the cell viability assay using trypan blue (ESI† Fig. S8A–C) or up to 48 hours with MTT (ESI† Fig. S8D–I). The results indicate that all three sensors have low toxicity towards living cells at concentrations below 10 μM, up to 48 hours of exposure. Therefore, this further qualifies our concentration range of 1–3 μM for sensor 3, which was used for subsequent spectroscopy and confocal imaging studies, unless otherwise described.Endogenous release of HNO via iNOS enzyme following LPS stimulation of BV2 cells
Previously, the literature described the use of an NO donor followed by sodium ascorbate to convert NO to HNO as a model of endogenous stimulation.56 In the present study, we chose to use a physiologically relevant stimulant, lipopolysaccharide (LPS), in BV2 cells that is known to drive reactive nitrogen species pathways.57 To determine the mechanism deriving the endogenous signal within BV2 cells, spectroscopy experiments were carried out using chronic 24 hour treatment of stimuli LPS in the presence or absence of iNOS enzyme inhibitor, 1400 W (ESI† Fig. S9A). Twenty-four-hour treatment with LPS (500 ng ml−1) (4196 ± 489 RFU) significantly increased the fluorescence of sensor 3 when compared to ‘HNO only’ controls (2721 ± 142 RFU, **p < 0.05). Furthermore, pre-incubation with 1400 W (1861 ± 64 RFU) significantly prevented the HNO signal compared to both LPS (***p < 0.001) and the ‘HNO only’ control (**p < 0.01) which suggests that the homeostatic baseline fluorescent levels observed in the HNO only control may also be derived from iNOS production. Control data (ESI† Fig. S9B) indicates that sensor 3 is specific to HNO as AS (200 μM) significantly increased the RFU signal of sensor 3 (28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 589 ± 4636 RFU) (***p < 0.001), which was diminished by L-cysteine pre-treatment (8912 ± 175 RFU). This data suggests that the endogenous HNO signal in LPS treated BV2 cells is derived from iNOS enzymatic activity and that LPS is a good candidate for endogenous stimulation of HNO.
589 ± 4636 RFU) (***p < 0.001), which was diminished by L-cysteine pre-treatment (8912 ± 175 RFU). This data suggests that the endogenous HNO signal in LPS treated BV2 cells is derived from iNOS enzymatic activity and that LPS is a good candidate for endogenous stimulation of HNO.
Sensor 3 can detect exogenous and endogenous HNO in both rodent and human cell types
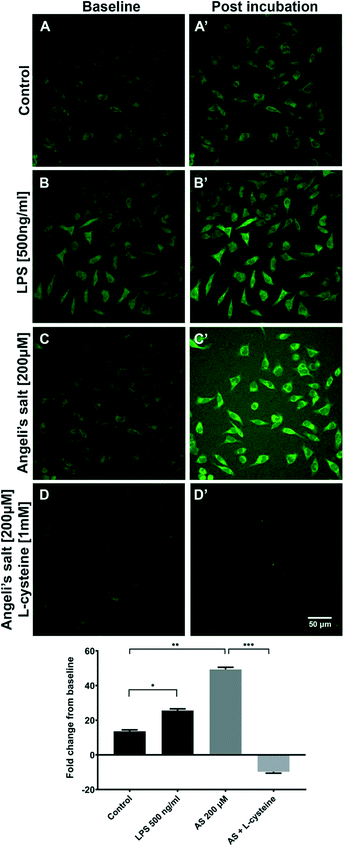
As sensor 3 was shown to be the brightest and most stable in biological media among all 3 probes, this sensor was selected to carry-out the following experiments, unless otherwise specified. The presence of both exogenous and endogenous HNO in BV2, HCAEC and H9C2 cells was examined by spectroscopy and/or confocal microscopy.Confocal microscopy imaging of BV2 cells was used to assess the uptake of sensor 3 and detect both exogenous and endogenous derived HNO. BV2 cells were incubated with 1 μM of sensor 3 for 30 min prior to confocal imaging (Fig. 2). The data shows that acute LPS (endogenous) treatment (*p < 0.0001) and HNO exogenous donor AS (**p < 0.0001) both increased the fluorescence signal compared to the control (sensor 3 only) (control: 13.59 ± 0.86 RFU; LPS: 25.58 ± 0.96 RFU; AS: 49.26 ± 1.2). Pre-treatment with the HNO scavenger, L-cystine (1 mM), was able to prevent the AS derived signal (LC: −9.718 ± 0.8 RFU; ***p < 0.0001), indicating that the sensor 3 probe is specific to HNO. Together, this data suggests that sensor 3 is sensitive enough to detect the endogenous HNO signal from BV2 cells using confocal microscopy.
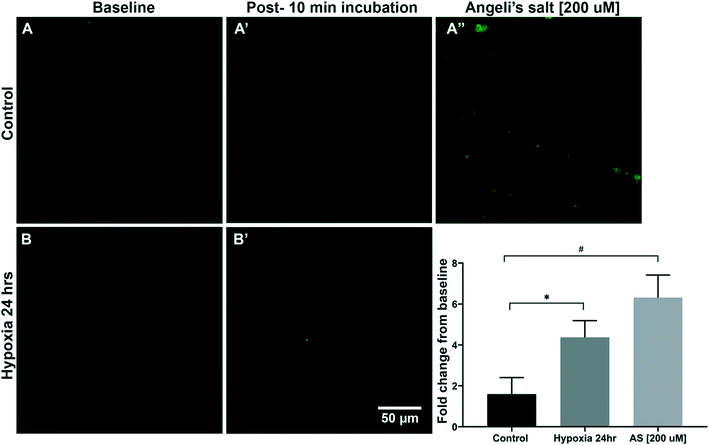
Confocal microscopy imaging of human coronary arterial endothelial cells (HCAECs) was then used to assess the uptake of sensor 3 and detect endogenous HNO following a 24 h incubation period in either normoxic or hypoxic solution (1% O2). HCAECs were imaged on the confocal microscope prior to and 15 min following the addition of sensor 3 (1 μM) (Fig. 3). The data showed that cells incubated in low O2 levels (hypoxic) yielded a 4.3-fold difference (*p < 0.05) and cells incubated in normal O2 levels (control) gave a 1.5-fold change, both compared to the baseline. Furthermore, the addition of the HNO donor AS (200 μM) gave a 6.3-fold change when compared to the control (#p < 0.05). These data suggest that sensor 3 is sensitive enough to detect the endogenous HNO signal from human arterial wall cells using confocal microscopy.
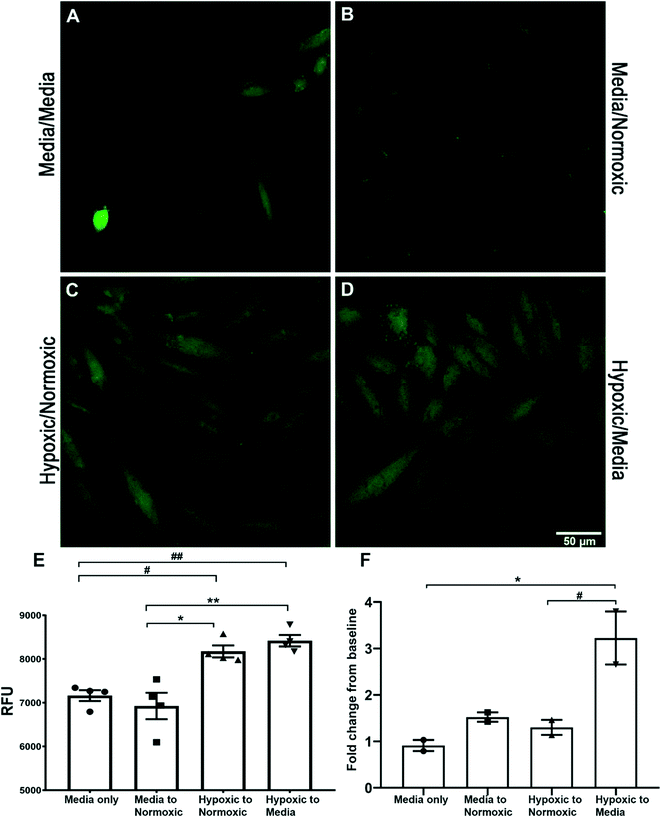
Both spectroscopy and confocal microscopy were used to explore the potential of sensor 1 (2 μM) to also detect the endogenous HNO signal in vitro, the fluorescent signal derived from rat cardiomyocyte (H9C2) treated cells (Fig. 4). HNO has been implicated as a factor in the damaged generated in cardiac tissue following ischemic reperfusion. Confocal microscopy imaging of cardiomyocyte cells was used to assess the uptake of sensor 1 and detect both exogenous (AS) and endogenous (hypoxic) derived HNO. H9C2 cells pre-treated with hypoxic solution followed by either normoxic solution (8175 ± 136 RFU) (Fig. 4C) or medium (8420 ± 130 RFU) (Fig. 4D) showed a significantly increased intracellular fluorescent signal when compared to cells pre-treated with media followed by either a second media exchange (7164 ± 124 RFU) (hypoxic/normoxic v media/media, #p < 0.05; hypoxic/media v media/media, ##p < 0.01) (Fig. 4A) or normoxic solution (6926 ± 303 RFU) (hypoxic/normoxic v media/normoxic, **p < 0.01; hypoxic/media v media/normoxic, ***p < 0.001) (Fig. 4B) (as shown in graph 4E). Spectroscopy was used to further determine the presence of endogenous HNO in supernatant samples taken from H9C2 cells. An initial fluorescent reading was taken from the supernatant of cells treated with either media, normoxic or hypoxic solution. The solution was then replaced with a second treatment of either media or normoxic buffer and a second fluorescent reading was taken from the supernatant after 1–2 minutes and compared to the initial reading (Fig. 4F; see Table S1 in ESI† data for treatment details). Only cells pre-treated with hypoxic solution followed by normoxic solution showed a significant fold-change increase (3.22 ± 0.57-fold) in the fluorescent signal when compared to media/media treatment only (0.91 ± 0.12-fold) (*p < 0.05) and media/normoxic treatment (1.3 ± 0.16-fold) (#p < 0.05). This data suggests that after a period of hypoxia, HNO is generated and released by H9C2 cells following the reintroduction of oxygen. This data supports the use of sensor 1 for the detection of endogenous HNO in H9C2 cells using both confocal microscopy and spectroscopy.
Sensor 3 is taken up into BV2 cell mitochondria
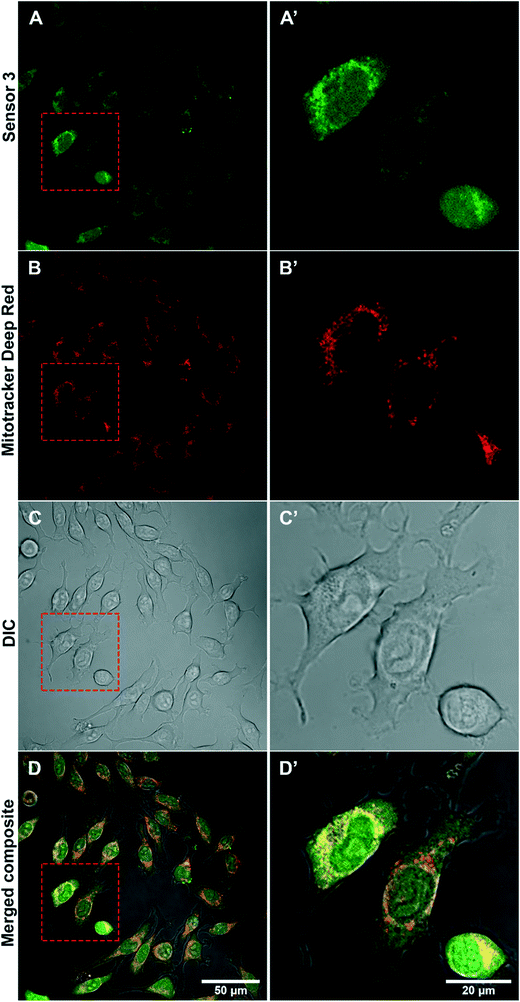
Sub-cellular localisation of sensor 3 within AS treated BV2 cells was determined by confocal microscopy using a mitochondria specific stain (Fig. 5). Confocal imaging determined that AS activated sensor 3 appears more punctate around the nucleus of cells (Fig. 5A and A′) in a similar location to Mitotracker Deep Red (Fig. 5B and B′). Overlay of the 2 channels shows the co-localisation of the signal (in yellow) (Fig. 5D and D′) suggesting that sensor 3 is taken up predominately by mitochondria within BV2 cells.Sensor 3 can detect HNO in rodent cardiac blood
Spectroscopy was used to detect the presence of extracellular HNO in plasma and/or serum of rat cardiac blood using sensor 3 (10 μM) with or without EDTA (0.5 M) (ESI† Fig. S10). We observed a 150-fold higher mean fluorescence value in the EDTA-free serum (ESI† Fig. S10A) containing sensor 3 compared to the PBS control (no sensor 3, *p < 0.0001) and a 114-fold higher mean fluorescence value in the plasma (with EDTA, ESI† Fig. S10B) containing sensor 3 compared to the PBS control (no sensor 3, *p < 0.0001) suggesting a minimal contribution of the blood (serum or plasma) to the basal non-specific reading. However, the presence of EDTA did reduce the fluorescence signal of sensor 3 by 30% when compared to the EDTA-free sensor 3 sample which may highlight a potential interference between EDTA and sensor 3 fluorescent read-out. We noted a further increase in the mean fluorescence value of a second reading, 30 min following the addition of AS (200 μM) to either serum with sensor 3 (14% increase, #p < 0.0001) or to plasma with sensor 3 (12% increase, &p < 0.001) indicating that the active sensor remained present throughout the whole freeze–thaw process. The presence of the HNO scavenger L-cysteine gave opposing results depending on the presence or absence of the chelator EDTA. In EDTA-free blood, L-cysteine gave a concentration dependent reduction in mean fluorescence values when compared to sensor 3 only (LC 100 μM; 18%, LC 500 μM; 21%, LC 1000 μM; 42%, ***p < 0.0001). This decrease was also observed in the presence of AS with L-cysteine decreasing the mean fluorescence values by 15%, 26% & 41% for 100, 500 & 1000 μM, respectively (###p < 0.0001). However, the presence of EDTA with L-cysteine resulted either in unchanged or increased mean fluorescence values. Higher concentrations of L-cysteine (500 & 1000 μM) lead to an 11% and 19% increase, respectively, when compared to sensor 3 (^p < 0.0001). This increase was also observed in the presence of AS, with higher concentrations of L-cysteine (500 & 1000 μM) leading to a 6% ($p < 0.05) and 15% (%p < 0.0001) increase, respectively, when compared to sensor 3 with AS. This observation indicates that the presence of EDTA can potentially interfere with the fluorescent signal of sensor 3 measured from blood samples and hence further assessment of other anti-coagulant chelators used for routine clinical blood collection is required.Discussion
Previous studies have shown the distinct chemical and physiological properties of the one-electron reduced, protonated form of NO, nitroxyl.16,21,37,38 Endogenous production of nitroxyl is suggested to occur via several possible mechanisms, such as NOS activation in the absence of the cofactor tetrahydrobiopterin (BH4),39 hydroxylamine (NH2OH) oxidation by a variety of heme proteins, reduction of NO to NO− by cytochrome c,40 or by reaction with either ubiquinol,41 manganese superoxide dismutase,42 or xanthine oxidase.43 Furthermore, potential mechanisms of HNO generation in microglial cells include: iNOS enzyme, reduction of hydroxylamine (NH2OH) to HNO via myeloperoxidase (MPO), the conversion of S-nitrosothiols to HNO by dithiols40 or by the interaction between H2S and NO.44 However, there is only indirect evidence to support the endogenous generation of HNO in vitro and in vivo and therefore the development of analytical tools to assess this is paramount. Fluorescent probes are highly sensitive, simple to use, non-invasive, have good spatiotemporal resolution and can be developed in a wide range of wavelengths, thus making them excellent candidates for detecting HNO. Many fluorescent probes have been developed over recent years based on different chemical reactions to HNO. However, the arylphosphine-based probes have demonstrated the highest selectivity and the least interference from other potential reducing agents in biological systems.22Here we have demonstrated the generation of a suite of bright, highly sensitive and very stable triarylphosphine-based HNO fluorescent probes, with the detection limit of sensor 3 being the lowest reported to our knowledge. Using LPS or hypoxia challenge, we have observed the presence of endogenous HNO in BV2, HCAEC and H9C2 cells using sensors 3 and 1, by both spectroscopy and confocal microscopy. We determined that sensor 2 did not provide a sufficient fold change in the signal in the presence of biological media and was below the sensitivity required to detect endogenous changes in this study. Furthermore, the wavelength required to excite sensor 2 falls in the UV range which can be detrimental to living cells when exposed for long periods of time and is therefore not an ideal candidate for biological use. We therefore focused on the use of sensors 3 and 1 for biological experiments. The cytotoxicity of the sensors was assessed on BV2 cells and HCAECs. HCAECs are derived from the inner lining of the coronary blood vessels that supply the heart. These cells did not demonstrate any changes in the cell viability or metabolism, in the presence of up to 10 μM of sensors 1–3, for longer periods (up to 48 hours) of exposure, which is a positive attribute for future translational applications. However, the use of the arylphosphine-based sensors is limited to samples in physiological pH as the fluorescence response can be pH dependent. In addition, the sensitivity of probes 1–3 is sufficient for in vitro detection of HNO; however, further research into a fluorescent backbone with a lower baseline fluorescence level is needed to improve the sensitivity for in vivo studies.
Our study found that all the sensors can detect HNO generated by AS in biological buffers and media, with the HBSS buffer and DMEM phenol red-free (DMEM-PRF) media yielding the best fold increase for sensor 3. This is important for live imaging studies which require the cells to be imaged in their conditioned media and stimulated to produce the endogenous HNO signal. Many studies have reported the generation of HNO probes that can detect intracellular signals via the use of HNO donors such as Angeli's salt,32,35 following the uptake of the probe and therefore require the removal of excess probe by washing. This method may be misleading in determining the sensitivity of the probe, as the timing and concentration of the endogenous signal may be significantly slower and lower than what is produced by AS, and hence below the limit of detection for many probes.
Our study detected increases in HNO fluorescence of sensor 3 with acute LPS (10–15 minutes) treatment of BV2 cells which was not present in control cells. However, the mechanism of HNO production following such a short time exposure to LPS is yet to be elucidated and requires further investigation. LPS has been shown to increase iNOS expression and release of NO in BV2 cells.45,46 Further to this, studies suggest that iNOS can produce HNO in the absence of co-factor BH4,39 which implicates iNOS as a potential source of HNO in vivo. However, as the measure of BH4 was beyond the scope of this study, we are unable to speculate its involvement in our findings.
The exposure of HCAECs to hypoxic conditions was used as an in vitro model of endothelial dysfunction. Both NO and HNO are known to be associated with endothelial vasodilator mechanisms,47 which are disrupted in oxidative stress, hypoxia, hypertension and ischemic heart disease.48 Demonstration of the specific role of HNO in these mechanisms is limited by the lack of HNO sensors37 and quantifiable fluorescent markers to study the synthesis and function of endogenous HNO. In the current study, we demonstrated the application of sensor 3 to report an increase in the levels of HNO under hypoxic conditions. To our knowledge, this is the first study to report the presence of endogenous HNO in primary HCAECs. It has been well established that HCAECs can produce reactive nitrogen species via enzyme eNOS49 but the role of HNO in endothelial cells has previously focused on the therapeutic effects of exogenous applications.50 Experiments were also carried out using sensor 1 to measure levels of endogenous HNO using a cellular model of ischemic-reperfusion injury. H9C2 cells showed increased intracellular HNO fluorescence following the replacement of oxygenated medium and/or buffer, after a period of hypoxia. This is the first study, to our knowledge, that demonstrates endogenous production of HNO in myocardial-like cells. Previous studies have identified a dual role of HNO in myocardial ischemia–reperfusion, whereby administration of AS just prior to reperfusion causes an increase in myocardial injury;8 however pre-ischemic administration is protective.9,51 Further investigation is required to validate this model and understand the role the endogenous HNO may be having following reperfusion.
We were able to localise HNO sensor 3 to the mitochondria of the BV2 cells via the use of live cell stain, Mitotracker Deep Red. The localisation of HNO probes within mitochondria has been previously reported52,53 and supports our findings. Further to this, the generation of ROS and RNS has been suggested to also be localised to the mitochondria of cells.52–54
Furthermore, sensor 3 is able to detect HNO fluorescent signals in both serum and plasma fractions of blood samples. Samples were collected via cardiac puncture directly into a syringe pre-loaded with sensor 3. Importantly, once the sensor was added to blood samples, fluorescence counts were relatively unaffected in serum/plasma by snap-freezing, confirming the stability of sensor 3 during freeze–thaw processing. This highlights the potential for future clinical translation using this sensor. Furthermore, we observed that after initial fluorescence readings were taken from sensor 3 samples, the addition of AS still resulted in increased fluorescence values indicating that the active sensor remained present in the plasma sample throughout the entire process of freeze–thawing. However, a limiting factor may be the presence of the commonly used chelator, EDTA, which showed a potential interference in the fluorescence signal output of sensor 3, when combined with the HNO scavenger, L-cysteine. In the absence of EDTA, we observed that L-cysteine was able to reduce (scavenge) the HNO fluorescent signal in blood, as noted in BV2 cells and previous literature reports. Further studies are required to understand the chemical interactions between these two chelators, which may cloud the output reading of blood HNO readings in a clinical setting.
Conclusion
This is the first report of synthesis, characterisation and biological evaluation of sensor 3 (rhodol 4 on a 2-(diphenylphosphanyl)benzoate backbone). The validation of these super bright, highly specific and stable HNO fluorescent sensors (sensors 1–3) in biological systems provides evidence to support the use of these tools to further our understanding of the role of endogenous nitroxyl. Since the first fluorogenic nitroxyl probe was synthesised in 2007 by Tennyson et al.,55 there have been a plethora of subsequent HNO probes designed and synthesised. However, this is the first study to report such an extensive validation in multiple biological systems using both spectroscopy and confocal microscopy for detection. As such, we were able to demonstrate the presence of endogenous HNO in mouse, rat and human cell lines, as well as rat blood samples. These triacylphosphine-based sensors described here show excellent potential for detecting HNO in vitro and in vivo in future studies and furthering our understanding of the role of this endogenously produced reactive nitrogen species.Methods
For general, chemical and biological methods and characterization data see the ESI† Methods section.For NMR spectra and HPLC traces of the compounds in this article, see the ESI.†
Author contributions
V. S. carried out experiments for Fig. 2, 4, 5, S5, S6 and S8–S10† and contributed to the project concept (biology), analysis of all biological figures and manuscript preparation; X. Z. carried out the creation of HNO sensors 1, 2 & 3 and contributed to Scheme 1, Fig. 1, S1, S2 and S4† project concept (chemistry) and manuscript preparation; B. P. carried out experiments for Fig. 3, S7 and S8† and contributed to manuscript preparation; A. K. V. contributed to the study design for Fig. 3, S7 and S8† and manuscript preparation; J. L. carried out experiments for Fig. S6;† S. M. L. carried out experiments for Fig. 4; P. R. carried out experiments for Fig. S3† (quantum yield); C. B., M. R. H. & A. D. A. contributed towards the design and development of the synthesis of the probes and manuscript preparation.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
This work was funded the Australian Research Council Centre of Excellence for Nanoscale Biophotonics CE140100003. The authors would like to acknowledge Ms Ashley Grant for her contribution to the confocal work in Fig. 5.References
- J. M. Fukuto, K. Chiang, R. Hszieh, P. Wong and G. Chaudhuri, The pharmacological activity of nitroxyl: a potent vasodilator with activity similar to nitric oxide and/or endothelium-derived relaxing factor, J. Pharmacol. Exp. Ther., 1992, 263(2), 546–551 CAS.
- A. Ellis, C. G. Li and M. J. Rand, Differential actions of L-cysteine on responses to nitric oxide, nitroxyl anions and EDRF in the rat aorta, Br. J. Pharmacol., 2000, 129, 315–322 CrossRef CAS PubMed.
- J. C. Wanstall, T. K. Jefery, A. Gambino, F. Lovren and C. R. Triggle, Vascular smooth muscle relaxation mediated by nitric oxide donors: a comparison with acetylcholine, nitric oxide and nitroxyl ion, Br. J. Pharmacol., 2001, 134(3), 463–472 CrossRef CAS PubMed.
- K. L. Andrews, N. G. Lumsden, J. Farry, A. M. Jefferis, B. K. Kemp-Harper and J. P. Chin-Dusting, Nitroxyl: a vasodilator of human vessels that is not susceptible to tolerance, Clin. Sci., 2015, 129(2), 179–187 CrossRef CAS PubMed.
- N. Paolocci, W. F. Saavedra, K. M. Miranda, C. Martignani, T. Isoda, J. M. Hare, M. G. Espey, J. M. Fukuto, M. Feelisch, D. A. Wink and D. A. Kass, Nitroxyl anion exerts redox-sensitive positive cardiac inotropy in vivo by calcitonin gene-related peptide signaling, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(18), 10463–10468 CrossRef CAS PubMed.
- D. A. Wink, K. M. Miranda, T. Katori, D. Mancardi, D. D. Thomas, L. Ridnour, M. G. Espey, M. Feelisch, C. A. Colton, J. M. Fukuto, P. Pagliaro, D. A. Kass and N. Paolocci, Orthogonal properties of the redox siblings nitroxyl and nitric oxide in the cardiovascular system: a novel redox paradigm, Am. J. Physiol., 2003, 285(6), H2264–H2276 CAS.
- H. N. Sabbah, C. G. Tocchetti, M. Wang, S. Daya, R. C. Gupta, R. S. Tunin, R. Mazhari, E. Takimoto, N. Paolocci, D. Cowart, W. S. Colucci and D. A. Kass, Nitroxyl (HNO) a Novel Approach for the Acute Treatment of Heart Failure, Circ.: Heart Failure, 2013, 6(6), 1250–1258 CAS.
- X. L. Ma, F. Gao, G. L. Liu, B. L. Lopez, T. A. Christopher, J. M. Fukuto, D. A. Wink and M. Feelisch, Opposite effects of nitric oxide and nitroxyl on post-ischemic myocardial injury, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14617–14622 CrossRef CAS PubMed.
- P. Pagliaro, D. Mancardi, R. Rastaldo, C. Penna, D. Gattullo, K. M. Miranda, M. Feelisch, D. A. Wink, D. A. Kass and N. Paolocci, Nitroxyl affords thiol-sensitive myocardial protective effects akin to early preconditioning, Free Radical Biol. Med., 2003, 34, 33–43 CrossRef CAS.
- R. Takahira, K. Yonemura, Y. Fujise and A. Hishida, Dexamethazone attenuates neutrophil infiltration in the rat kidney in ischemia/reperfusion injury: the possible role of nitroxyl, Free Radical Biol. Med., 2001, 31, 809–815 CrossRef CAS.
- H. T. Nagasawa, E. G. DeMaster, B. Redfern, F. N. Shirota and D. J. W. Goon, Evidence for nitroxyl in the catalase-mediated bioactivation of the alcohol deterrent agent cyanamide, J. Med. Chem., 1990, 33(12), 3120–3122 CrossRef CAS.
- E. G. DeMaster, B. Redfern and H. T. Nagasawa, Mechanisms of inhibition of aldehyde dehydrogenase by nitroxyl, the active metabolite of the alcohol deterrent agent cyanimide, Biochem. Pharmacol., 1998, 55(12), 2007–2015 CrossRef CAS.
- D. T. Longhi-Balbinot, A. C. Rossaneis, F. A. Pinho-Ribeiro, M. M. Bertozzi, F. Q. Cunha, J. C. Alves-Filho, T. M. Cunha, J. P. S. Peron, K. M. Miranda, R. Casagrande and W. A. Verri Jr., The nitroxyl donor, Angeli's salt, reduces chronic constriction injury-induced neuropathic pain, Chem.-Biol. Interact., 2016, 256, 1–8 CrossRef CAS.
- L. Staurengo-Ferrari, A. C. Zarpelon, D. T. Longhi-Balbinot, M. Marchesi, T. M. Cunha, J. C. Alves-Filho, F. Q. Cunha, S. H. Ferreira, R. Casagrande, K. M. Miranda and W. A. Verri, Nitroxyl inhibits overt pain-like behavior in mice: Role of cGMP/PKG/ATP-sensitive potassium channel signaling pathway, Pharmacol. Rep., 2014, 66(4), 691–698 CrossRef CAS PubMed.
- A. C. Zarpelon, G. R. Souza, T. M. Cunha, I. R. S. Schivo, M. Marchesi, R. Casagrande, P. Pinge-Filho, F. Q. Cunha, S. H. Ferreira, K. M. Miranda and W. A. Verri Jr., The nitroxyl donor, Angeli's salt, inhibits inflammatory hyperalgesia in rats, Neuropharmacology, 2013, 71, 1–9 CrossRef CAS PubMed.
- J. M. Fukuto, M. D. Bartberger, A. S. Dutton, N. Paolocci, D. A. Wink and K. N. Houk, The physiological chemistry and biological activity of nitroxyl (HNO): the neglected, misunderstood and enigmatic nitrogen oxide, Chem. Res. Toxicol., 2005, 18, 790–801 Search PubMed.
- J. M. Fukuto, C. L. Bianco and T. A. Chavez, Nitroxyl (HNO) signaling, Free Radical Biol. Med., 2009, 47, 1318–1324 CrossRef CAS PubMed.
- J. M. Fukuto, S. J. Carrington, D. J. Tantillo, J. G. Harrison, L. J. Ignarro, B. A. Freeman, A. Chen and D. A. Wink, Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species, Chem. Res. Toxicol., 2012, 25, 769–793 Search PubMed.
- J. M. Fukuto, C. J. Cisneros and R. L. Kinkade, A comparison of the chemistry associated with the biological signaling and actions of nitroxyl (HNO) and nitric oxide (NO), J. Inorg. Biochem., 2013, 118, 201–208 CrossRef CAS PubMed.
- J. M. Fukuto, A recent history of nitroxyl chemistry, pharmacology and therapeutic potential, Br. J. Pharmacol., 2019, 176, 135–146 CrossRef CAS PubMed.
- K. M. Miranda, The chemistry of nitroxyl (HNO) and implications in biology, Coord. Chem. Rev., 2005, 249, 433–455 CrossRef CAS.
- R. Smulik-Izydorczyk, K. Dębowska, J. Pięta, R. Michalski, A. Marcinek and A. Sikora, Fluorescent probes for the detection of nitroxyl (HNO), Free Radical Biol. Med., 2018, 128, 69–83 CrossRef CAS PubMed.
- J. Rosenthal and S. J. Lippard, Direct detection of nitroxyl in aqueous solution using a tripodal copper(II) BODIPY complex, J. Am. Chem. Soc., 2010, 132, 5536–5537 CrossRef CAS PubMed.
- M. Royzen, J. J. Wilson and S. J. Lippard, Physical and structural properties of [Cu(BOT1)Cl]Cl, a fluorescent imaging probe for HNO, J. Inorg. Biochem., 2013, 118, 162–170 CrossRef CAS PubMed.
- Y. Zhou, K. Liu, J. Y. Li, Y. Fang, T. C. Zhao and C. Yao, Visualization of nitroxyl in living cells by a chelated copper(II) coumarin complex, Org. Lett., 2011, 13, 1290 CrossRef CAS PubMed.
- J. A. Reisz, E. B. Klorig, M. W. Wright and S. B. King, Reductive phosphine-mediated ligation of nitroxyl (HNO), Org. Lett., 2009, 11, 2719–2721 CrossRef CAS PubMed.
- A. K. Kawai, N. Ieda, K. Aizawa, T. Suzuki, N. Miyata and H. Nakagawa, A reductant resistant and metal-free fluorescent probe for nitroxyl applicable to living cells, J. Am. Chem. Soc., 2013, 135(34), 12690–12696 CrossRef PubMed.
- G. J. Mao, X. B. Zhang, X. L. Shi, H. W. Liu, Y. X. Wu, L. Y. Zhou, W. Tan and R. Q. Yu, A highly sensitive and reductant-resistant fluorescent probe for nitroxyl in aqueous solution and serum, Chem. Commun., 2014, 50(43), 5790–5792 RSC.
- C. Liu, H. Wu, Z. Wang, C. Shao, B. Zhu and X. Zhang, A fast-response, highly sensitive and selective fluorescent probe for the ratio-metric imaging of nitroxyl in living cells, Chem. Commun., 2014, 50(45), 6013–6016 RSC.
- K. B. Zheng, W. Y. Lin, D. Cheng, H. Chen, Y. Liu and K. Y. Liu, A two-photon fluorescent turn-on probe for nitroxyl (HNO) and its bioimaging application in living tissues, Chem. Commun., 2015, 51(26), 5754–5757 RSC.
- X. T. Jing, F. B. Yu and L. X. Chen, Visualization of nitroxyl (HNO) in vivo via a lysosome-targetable near-infrared fluorescent probe, Chem. Commun., 2014, 50(91), 14253–14256 RSC.
- Z. Miao, J. A. Reisz, S. M. Mitroka, J. Pan, M. Xian and S. B. King, A selective phosphine based fluorescent probe for nitroxyl in living cells, Bioorg. Med. Chem. Lett., 2015, 25(1), 16–19 CrossRef CAS PubMed.
- H. Zhang, R. Liu, Y. Tan, W. H. Xie, H. Lei, H. Y. Cheung and H. Sun, A FRET-based ratiometric fluorescent probe for nitroxyl detection in living cells, ACS Appl. Mater. Interfaces, 2015, 7(9), 5438–5443 CrossRef CAS PubMed.
- D. Zhang, W. Chen, Z. Miao, Y. Ye, Y. Zhao, S. B. King and M. A. Xian, A reductive ligation based fluorescent probe for S-nitrosothiols, Chem. Commun., 2014, 50(37), 4806–4809 RSC.
- B. Dong, X. Kong and W. Lin, Reaction-Based Fluorescent Probes for the Imaging of Nitroxyl (HNO) in Biological Systems, ACS Chem. Biol., 2018, 13, 1714–1720 CrossRef CAS PubMed.
- Z. Miao and S. B. King, Recent advances in the chemical biology of nitroxyl (HNO) detection and generation, Nitric Oxide, 2016, 57, 1–14 CrossRef CAS PubMed.
- B. K. Kemp-Harper, Nitroxyl (HNO): A Novel Redox Signaling Molecule, Antioxid. Redox Signal., 2011, 14(9), 1609–1613 CrossRef CAS PubMed.
- J. M. Fukuto and S. J. Carrington, HNO Signaling Mechanisms, Antioxid. Redox Signal., 2011, 14(9), 1649–1657 CrossRef CAS PubMed.
- C. C. Wei, Z. Q. Wang, C. Hemann, R. Hille and D. J. Stuehr, A Tetrahydrobiopterin Radical Forms and then Becomes Reduced during N-Hydroxyarginine Oxidation by Nitric-oxide Synthase, J. Biol. Chem., 2003, 278, 46668–46673 CrossRef CAS PubMed.
- C. U. Choe, J. Lewerenz, C. Gerloff, T. Magnus and S. Donzelli, Nitroxyl in the Central Nervous System, Antioxid. Redox Signaling, 2011, 14, 1699–1711 CrossRef CAS PubMed.
- J. J. Poderoso, M. C. Carreras, F. Schopfer, C. L. Lisdero, N. A. Riobo, C. Guilivi, A. D. Boveris and E. Cadenas, Free Radical Biol. Med., 1999, 26, 925–935 CrossRef CAS PubMed.
- V. Niketic, S. Stojanovic, A. Nikolic, M. Spasic and A. M. Michelson, Exposure of Mn and FeSODs, but not Cu/ZnSOD, to NO leads to nitrosonium and nitroxyl ions generation which cause enzyme modification and inactivation: an in vitro study, Free Radical Biol. Med., 1999, 27, 992–996 CrossRef CAS PubMed.
- M. Saleem and H. Ohshima, Xanthine oxidase converts nitric oxide to nitroxyl that inactivates the enzyme, Biochem. Biophys. Res. Commun., 2004, 315(2), 455–462 CrossRef CAS PubMed.
- Q. C. Yong, L. F. Hu, S. Wang, D. Huang and J. S. Bian, Hydrogen sulfide interacts with nitric oxide in the heart: possible involvement of nitroxyl, Cardiovasc. Res., 2010, 88(3), 482–491 CrossRef CAS PubMed.
- S. S. Kim, P. J. Kong, B. S. Kim, D. H. Sheen, S. Y. Nam and W. Chun, Inhibitory action of minocycline on lipopolysaccharide-induced release of Nitric Oxide and Prostaglandin E2 in BV2 microglial cells, Arch. Pharmacal Res., 2004, 27(3), 314–318 CrossRef CAS PubMed.
- J. M. Pocock and A. C. Liddle, Microglial signalling cascades in neurodegenerative disease, Prog. Brain Res., 2001, 132, 555–565 CAS.
- J. L. Favaloro and B. K. Kemp-Harper, The nitroxyl anion (HNO) is a potent dilator of rat coronary vasculature, Cardiovasc. Res., 2007, 73(3), 587–596 CrossRef CAS PubMed.
- B. M. Wynne, H. Labazi, R. C. Tostes and R. C. Webb, Nitroxyl Anion Mediates Relaxation in Mesenteric Arteries from Angiotensin II Hypertensive Mice, Curr. Vasc. Pharmacol., 2018, 16(1), 93–101 CAS.
- S. Y. Ma, J. Z. Xiang, J. L. Wu, Y. X. Ma and B. R. Hu, Endogenous nitric oxide mediate lipoteichoic acid induced preconditioning on reoxygenation injury of cultured human coronary artery endothelial cells, Yaoxue Xuebao, 2005, 40(4), 316–321 CAS.
- K. L. Andrews, A. K. Samplson, J. C. Irvine, W. A. Shihata, D. L. Michell, N. G. Lumsden, C. Lim, O. Huet, G. R. Drummond, B. K. Kemp-Harper and J. P. Chin-Dusting, Nitroxyl (HNO) reduced endothelial and monocyte activation and promotes M2 macrophage polarization, Clin. Sci., 2016, 130(18), 1629–1640 CrossRef CAS PubMed.
- J. C. Irvine, R. H. Ritchie, J. L. Favaloro, K. L. Andrews, R. E. Widdop and B. K. Kemp-Harper, Nitroxyl (HNO): the Cinderella of the nitric oxide story, Trends Pharmacol. Sci., 2008, 29(12), 601–608 CrossRef CAS PubMed.
- M. Ren, B. Deng, K. Zhou, J. Y. Wang, X. Kong and W. Lin, A targetable fluorescent probe for imaging exogenous and intracellularly formed nitroxyl in mitochondria in living cells, J. Mater. Chem. B, 2017, 5, 1954–1961 RSC.
- X. Gong, X. F. Yang, Y. Zhong, Y. Chen and Z. Li, A mitochondria-targetable near-infrared fluorescent probe for imaging nitroxyl (HNO) in living cells, Dyes Pigm., 2016, 131, 24–32 CrossRef CAS.
- K. Sunwoo, K. N. Bobba, J. Y. Lim, T. Park, A. Podder, J. S. Heo, S. G. Lee, S. Bhuniya and J. S. Kim, A iorthogonal ‘turn-on’ fluorescent probe for tracking mitochondrial nitroxyl formation, Chem. Commun., 2017, 53, 1723 RSC.
- A. G. Tennyson, L. Do, R. C. Smith and S. J. Lippard, Selective fluorescence detection of nitroxyl over nitric oxide in buffered aqueous solution using a conjugated metallopolymer, Polyhedron, 2007, 26, 4625–4630 CrossRef CAS.
- A. Loas, R. J. Radford, A. D. Liang and S. J. Lippard, Solid-phase synthesis provides a modular, lysine-based platform for fluorescent discrimination of nitroxyl and biological thiols, Chem. Sci., 2015, 6, 4131–4140 RSC.
- O. I. Han, K. W. Kim, J. H. Ryu and W. K. Kim, p38 mitogen-activated protein kinase mediates lipopolysaccharide, not interferon-γ, −induced inducible nitric oxide synthase expression in mouse BV2 microglial cells, Neurosci. Lett., 2002, 325(1), 9–12 CrossRef PubMed.
- P. D. D. M. Rajasekar, Recent Development in Fluorescein derivatives, J. Mol. Struct., 2020, 1224, 129085 CrossRef.
- J. B. Grimm, B. P. English, J. Chen, J. P. Slaughter, Z. Zhang, A. Revyakin, R. Patel, J. J. Macklin, D. Normanno, R. H. Singer, T. Lionnet and L. D. Lavis, A general method to improve fluorophores for live-cell and single-molecule microscopy, Nat. Methods, 2015, 12(3), 244–250 CrossRef CAS PubMed.
- J. Ma, Y. Xu, X. Kong, Y. Wei, D. Meng and Z. Zhang, A coumarin-based biosensor for human hepatocellular carcinoma diagnosis with enhanced brightness and water solubility, Mater. Chem. Front., 2021, 5(20), 7548–7557 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2sd00006g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |