Open Access Article
Open Access ArticleTowards an integrative understanding of cancer mechanobiology: calcium, YAP, and microRNA under biophysical forces
Chenyu
Liang†
ab,
Miao
Huang†
ab,
Tianqi
Li†
bc,
Lu
Li†
bc,
Hayley
Sussman
d,
Yao
Dai
be,
Dietmar W.
Siemann
be,
Mingyi
Xie
*bcf and
Xin
Tang
 *ab
*ab
aDepartment of Mechanical & Aerospace Engineering, Herbert Wertheim College of Engineering (HWCOE), Gainesville, FL 32611, USA. E-mail: xin.tang@ufl.edu
bUF Health Cancer Center (UFHCC), Gainesville, FL 32611, USA
cDepartment of Biochemistry and Molecular Biology, College of Medicine (COM), Gainesville, FL 32611, USA. E-mail: mingyi.xie@ufl.edu
dDepartment of Radiation Oncology, COM, Gainesville, FL 32611, USA
eUF Genetics Institute (UFGI), University of Florida (UF), Gainesville, FL 32611, USA
fDepartment of Biomedical Engineering, College of Engineering (COE), University of Delaware (UD), Newark, DE 19716, USA
First published on 19th January 2022
Abstract
An increasing number of studies have demonstrated the significant roles of the interplay between microenvironmental mechanics in tissues and biochemical-genetic activities in resident tumor cells at different stages of tumor progression. Mediated by molecular mechano-sensors or -transducers, biomechanical cues in tissue microenvironments are transmitted into the tumor cells and regulate biochemical responses and gene expression through mechanotransduction processes. However, the molecular interplay between the mechanotransduction processes and intracellular biochemical signaling pathways remains elusive. This paper reviews the recent advances in understanding the crosstalk between biomechanical cues and three critical biochemical effectors during tumor progression: calcium ions (Ca2+), yes-associated protein (YAP), and microRNAs (miRNAs). We address the molecular mechanisms underpinning the interplay between the mechanotransduction pathways and each of the three effectors. Furthermore, we discuss the functional interactions among the three effectors in the context of soft matter and mechanobiology. We conclude by proposing future directions on studying the tumor mechanobiology that can employ Ca2+, YAP, and miRNAs as novel strategies for cancer mechanotheraputics. This framework has the potential to bring insights into the development of novel next-generation cancer therapies to suppress and treat tumors.
1. Introduction
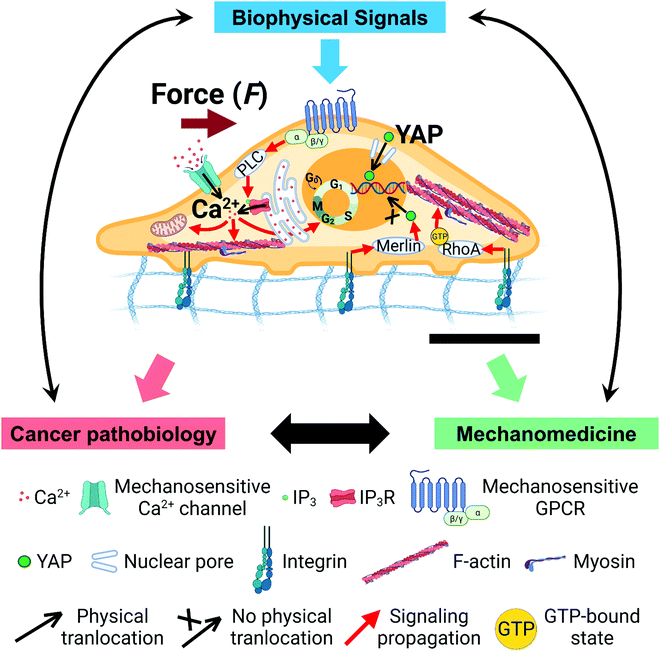
Cancer is the second leading cause of human death worldwide, accounting for 10 million deaths annually.1 90% of cancer deaths are the consequence of metastasis, the process of invasive tumor cells spreading from primary solid tumors to distant organs.2–6 The molecular mechanisms and parameters within primary tumors that regulate tumor progression and promote metastasis, however, are poorly understood. Such a knowledge gap in the understanding of tumor progression and prediction of metastasis onset is of serious concern. Increasingly studies from multiple disciplines, such as soft matter biophysics and mechanobiology, have begun to demonstrate the significant influences of biophysical microenvironments and signaling on tumor initiation, progression, and metastasis (Fig. 1).7–18 These influences are realized via mechanochemical transduction or mechanotransduction—the process in which cells sense and transduce extracellular biophysical stimuli into intracellular biochemical signals that elicit coherent biochemical responses and gene expression.14–16,19–29 Advances in the understanding of mechanotransduction have led to the design and development of new classes of pharmaceuticals, drug testing and delivery systems, wearable therapeutic devices, and engineered tissues that leverage biomechanics or target mechanobiology pathways to enable innovative combinatorial therapeutics.30–39In this review, we focus on reporting the interplay between biomechanical signals and three important biochemical effectors during tumor progression: (1) calcium ions (Ca2+), (2) yes-associated protein (YAP), and (3) microRNAs (miRNAs). Two other recent reviews discuss other types of biochemical effectors involved in cancer mechanobiology.7,31 During cancer progression, Ca2+ and YAP serve as critical signaling messengers to regulate gene-expression and cell functions, which are simultaneously modulated by miRNAs. However, how Ca2+, YAP, and miRNA interact with each other to influence the activities of cancer cells remains incompletely understood. Further, the functional interplay between mechanical signals and these three effectors, as well as the underpinning molecular mechanisms, are still elusive.
The goal of this review is to synergistically report (1) the recent advances in the understanding of these three effectors and their crosstalk with mechanotransduction pathways during cancer development, and (2) the mechanistic insights from soft matter and mechanobiology perspectives into the orchestrated functions of these three effectors. First, we introduce and discuss the current findings that demonstrate how biophysical forces induce calcium signaling in cancer cells and the identified molecular mechanisms (Section 3). Second, we address the functional responses of YAP to biophysical signals and the roles of cell cytoskeleton and nucleus in YAP mechanosensing (Section 4). Third, we discuss the functional interactions of miRNAs with Ca2+ and YAP, and the crosstalk between miRNAs and mechanical signaling in cancer (Section 5). We conclude by proposing promising future directions on the study of tumor mechanobiology using Ca2+, YAP, and miRNAs as potential targets, as well as novel strategies for cancer mechanotherapeutics (Section 6).
2. Mechanotransduction in cancer
All living cells and tissues in the human body experience biophysical forces from their micro- and macro-environments, including but not limited to tension, shear stress, compression, and fluid pressure.40–43 At the same time, cells actively generate and apply endogenous forces to their surroundings.31,41,44 The biophysical cues are sensed and transduced into intracellular biochemical and genetic signaling and further regulate specific cellular functions. This process is known as mechanochemical transduction or mechanotransduction (Fig. 1).14–16,19–21,25–27 Mechanotransduction involves a great number of mechanosensors or mechanosensitive biomolecules, such as integrin, cadherin, Piezo 1/2, G-protein-coupled receptor (GPCR), YAP/transcriptional co-activator with PDZ-binding motif (TAZ), Wnt, mitogen-activated protein kinase (MAPK)/extracellular-signal-regulated kinase (ERK), and phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT), which experience conformational and functional changes under mechanical stimuli.19,45–49 For example, cytoskeleton, such as filamentous (F)-actin, microtubules (MTs), and intermediate filaments (IFs), and their associated motor proteins, such as myosin II, act as molecular connectors to transmit forces directly from the extracellular environment, through the membrane mechanosensors, into the nucleus.31 Unlike diffusion-based chemical signal propagation, this force/deformation-based mechanical signal transmission can modulate the nucleus and affect gene activation within milliseconds.50–53 Dysregulated mechanotransduction often results in diseases.54–58Emerging studies have demonstrated the functional roles of mechanical cues in tumor progression at different stages (Fig. 1).7,9,11–17,59–62 Several aspects of the biophysical microenvironment in tumors are dramatically altered compared to their healthy counterparts, such as tissue stiffness,63 solid stress,64 interstitial fluid pressure,65 cell stiffness,66,67 cell contractility,68,69 and cell adhesion.70–72 These altered biophysical signals are transmitted into tumor cells via mechanotransduction and mediate cancer pathobiology. Understanding the molecular mechanisms of mechanotransduction in tumor development has inspired and enabled researchers to design and develop novel cancer therapies.40,73 In the following sections, we focus on three critical means of biochemical signaling in cancer: Ca2+, YAP, and miRNAs, and address their functions and crosstalk.
Ca2+ is a universal and indispensable signaling ion used by all eukaryotes.74–77 In tumor cells, Ca2+ regulates cellular activities and impact tumor progression including proliferation, metabolism, migration, epithelial–mesenchymal transition (EMT), and apoptosis.78–82 Emerging studies have demonstrated that Ca2+ signaling in cancer cells is affected by various mechanical stimuli including cyclic stretch,83 local membrane traction,84 fluid shear,85,86 and compression.87,88 Because the force-regulated Ca2+ signals have critical roles in cancer progression, the understanding of their underlying molecular mechanisms can provide insights into the development of novel cancer therapies. However, how mechanical stimuli convert into and regulate Ca2+ signals via mechanotransduction pathways has not been systematically studied.
YAP is a protein that can bind to transcription factors in the nucleus to regulate cellular functions.89,90 In cancer, expression and nucleus accumulation of YAP regulate tumor cell initiation, proliferation, migration, stemness, and chemoresistance.91–94 YAP's nucleus/cytoplasm distribution is sensitive to mechanical stimuli that cells experience, such as substrate stiffness, cytoskeleton tension, nuclear deformation, and extracellular mechanical tension/compression.95 However, how YAP responds to mechanical stimuli at molecular levels is still being actively investigated.96–99 Importantly, YAP activation is necessary in tumor initiation of squamous cell carcinoma and uveal melanoma100,101 and promotes the dissemination of circulating tumor cells.102 Aberrant YAP expression in different cancer types and its regulatory roles on the cancer progression necessitate the mechanistic dissections of how mechanical cues regulate YAP activity, which in turn contribute to the development of YAP-targeted anti-cancer mechano-medicine.
miRNAs are small non-coding RNAs that regulate gene-expression post-transcriptionally.103,104 Increasing evidence has demonstrated that miRNAs participate in the modulation of cancer-related pathways, from which diverse miRNAs have been used as diagnostic biomarkers and therapeutic targets/agents in anti-cancer treatments.105,106 How miRNAs can be potentially exploited for targeting calcium signaling, YAP activities, and mechanotransduction in cancer therapy is now being actively studied.
3. Calcium (Ca2+) signals in cancer
3.1 Significance of Ca2+ signals in tumor initiation, development, and progression
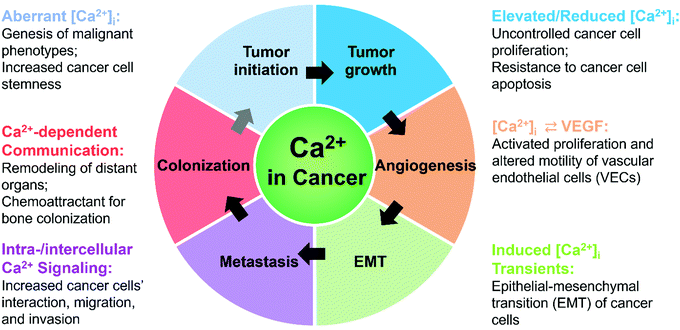
Calcium signals have pivotal roles in regulating cancer progression at different stages: tumor initiation, growth, angiogenesis, metastasis, and colonization (Fig. 2 and Table 1).79,107,108| Impact on cancer | Specific Ca2+ signal | Upstream | Downstream | Exact Influence | Cell type | In vitro/vivo? | Ref. |
|---|---|---|---|---|---|---|---|
| Initiation | Increased [Ca2+]cyt | Ca2+ influx via TRPV2 | Dysregulation of the Wnt/β-catenin signaling pathway | Induced Ca2+ influx inhibits stemness of cancer cells | Human bone osteosarcoma, breast, and colorectal adenocarcinoma | In vitro & in vivo | 117 |
| Elevated Ca2+ current | Overexpressed L- and T-type Ca2+ channels | AKT and ERK signaling pathways | Downregulation of Ca2+ channels reduces the expression of stemness markers | Ovarian cancer stem cells | In vitro & in vivo | 119 | |
| Growth | IP3R3-BKCa coupled Ca2+ signaling | ATP-induced ER Ca2+ release via IP3R3 | Activation of BKCa channel on plasma membrane | IP3R3/BKCa silencing impairs ATP-increased cell proliferation (arrested in G0/G1 phase) | Human breast cancer cells | In vitro | 121 |
| Spontaneous [Ca2+]cyt oscillations; 80% cancer cells showing oscillation vs. 30% non-cancer cells | Increased expression and activity of Orai1 | Downregulation of cdc2, Cyclin B1, and p27 | KD of Orai1 inhibits cell proliferation, migration, invasion, and tumor growth | Human esophageal squamous cell carcinoma | In vitro & in vivo | 122 | |
| Elevated [Ca2+]cyt | Ca2+ influx via Piezo1 | Akt/mTOR pathway; activation of CDK4 and cyclin D1 | Downregulation of Piezo1 suppresses cell proliferation and tumor growth | Human prostate cancer cells | In vitro & in vivo | 84 | |
| Decreased ATP-induced [Ca2+]cyt elevation | IP3R inhibition/silencing | Induced autophagy | IP3R inhibition induces cell death and suppresses tumor growth | Human breast cancer | In vitro & in vivo | 123 | |
| Reduced SOC current | Decreased Orai1 expression | Inhibited apoptosis-inducing pathways | Downregulation of Orai1 protects cells from apoptosis | Human prostate cancer cells | In vitro | 124 | |
| Enhanced ER-mitochondrial Ca2+ signaling | p53 binding to SERCA ⇒ ER Ca2+ overload ⇒ ER Ca2+ release | Mitochondrial Ca2+ overload ⇒ alteration of mitochondrial morphology | Tumor suppressor p53 induces apoptosis | Human colorectal, cervical, lung cancer cells | In vitro | 125 | |
| K-Ras G13D deletion-enhanced agonist-induced ER Ca2+ release | Remodeled IP3R expression and increased SERCA2b expression ⇒ increased ER Ca2+ content | Increased mitochondrial Ca2+ uptake | Deletion of K-RasG13D sensitizes cells to apoptosis | Human colorectal cancer cells | In vitro | 126 | |
| Increased ER Ca2+ release | BRCA1 binding to IP3R ⇒ regulation of IP3R function | Not through mitochondrial Ca2+ overload | Tumor suppressor BRCA1 is recruited for apoptosis | Human cervical, ovarian cancer cells | In vitro | 127 | |
| Angiogenesis | Elevated [Ca2+]cyt | Triclosan ⇒ TRPA1 | VEGF secretion | Triclosan stimulates epithelial cell proliferation | Human prostate cancer stromal cells | In vitro | 128 |
| Induced [Ca2+]cyt transients | Ca2+ influx via TRPV4 | Migration-related signaling | TRPV4 activation enhances migration of endothelial cells | Tumor-derived endothelial cells from human breast carcinomas | In vitro | 129 | |
| Reduced [Ca2+]cyt elevation | Decreased TRPV4 expression | High Rho activity | TRPV4 activation restores mechanosensitivity and inhibits migration of endothelial cells, and normalizes tumor vasculature | Tumor-derived endothelial cells from an adenocarcinoma mouse prostate model | In vitro & in vivo | 130 | |
| EMT | EGF/scratch-induced transient [Ca2+]cyt increase (2-fold higher)/Ca2+ wave | Ca2+ influx via TRPM7 (mechanosensitive) and other Ca2+ channels | STAT3 phosphorylation and vimentin expression; induction of Twist, N-cadherin, CD44/CD24 | Ca2+ signals are necessary for EGF/hypoxia-induced EMT (biomarkers) | Human breast cancer cells | In vitro | 135 |
| Intracellular Ca2+ elevation | Ca2+ influx via acid-sensing ion channels | Upregulation of RhoA activity | Inhibition of Ca2+ influx or intracellular Ca2+ chelation suppresses induced EMT | Pancreatic cancer cells | In vitro & in vivo | 133 | |
| Increased SOCE | Activation of STIM | EMT-related signaling | STIM-mediated SOCE facilitates induced EMT | Human breast cancer cells | In vitro | 134 | |
| Metastasis | Reduced [Ca2+]cyt elevation | Low TRPM7 activity | Inactivated RhoA/myosin-II and IQGAP1-Cdc42 | Reduced TRPM7-mediated Ca2+ influx inhibits shear flow sensing and facilitates intravasation | Human fibrosarcoma cells | In vitro & in vivo | 136 |
| [Ca2+]cyt elevation | Induced Ca2+ influx via TRPV4 | Accelerated actin dynamics and downregulated cytoskeleton-associated proteins at the cell cortex | Overexpression of TRPV4 increases cell invasiveness | Human breast cancer cells | In vitro & in vivo | 137 | |
| [Ca2+]cyt elevation | Ca2+ influx via Piezo1 | Protrusions of apical actin and expression of MMP-9 | Piezo1-mediated Ca2+ influx enhances cell invasion and matrix degradation | Human breast cancer cells | In vitro | 88 | |
| Sustained [Ca2+]cyt elevation | Induced ER Ca2+ release via IP3R3 | Migration-related signaling | Overexpression of IP3R3 enhances ATP-induced cell migration | Human breast cancer cells | In vitro | 138 | |
| Induced Ca2+ responses | GPCR/RTK ⇒ PLC ⇒ IP3R | Migration-related signaling | Caffeine inhibition of IP3R blocks glioblastoma invasion and increased survival of mouse model | Human glioblastoma | In vitro & in vivo | 139 | |
| [Ca2+]cyt elevation | ER Ca2+ release via IP3R | Promoted cortical actomyosin contractility | Nuclear envelope tension induced ER Ca2+ release facilitates cell transmigration through 3D matrix | Human cervical carcinoma cells | In vitro | 87 | |
| Spontaneous [Ca2+]cyt oscillations; 80% cancer cells showing oscillation vs. 30% non-cancer cells | Increased expression and activity of Orai1 | Increased expression of vimentin and Rac1; downregulation of E-cadherin | KD of Orai1 inhibits cell proliferation, migration, invasion, and tumor growth | Human esophageal squamous cell carcinoma | In vitro & in vivo | 122 | |
| Spontaneous periodic intracellular propagations of perimembrane Ca2+ waves (freq = 3 times min−1) | Low voltage-activated T-type Ca2+ channels and non-voltage-gated cation channels (on plasma membrane) | Migration-related signaling | Block of Ca2+ signals reduces cell motility and invasion | Human fibrosarcoma cells | In vitro | 140 | |
| Colonization | [Ca2+]cyt elevation | Ca2+ flow via gap junctions | Enriched NEAT and MEF2 activities | Ca2+ flow from osteogenic cells to cancer cells facilitates bone colonization | Human breast cancer cells | In vitro & in vivo | 145 |
During cancer initiation, altered calcium signals, due to aberrant expression and activities of Ca2+ channels/transporters, lead to abnormal cellular functions, such as defects in autophagy109 and resistance to apoptosis.110 Because autophagy functions as a tumor suppressor,111–113 both uncontrolled increases and decreases of cytoplasmic Ca2+ concentrations ([Ca2+]cyt) can cause defects in autophagy to break normal cellular homeostasis and favor cancerous phenotypes.109 In addition, p53-deficient cells fail to induce mitochondrial Ca2+ overload via endoplasmic reticulum (ER)-mitochondrial Ca2+ signaling and become apoptosis-resistant.110 This selection advantage of p53-deficiency favors survival of damaged cells, potentially resulting in cancer initiation. Moreover, altered [Ca2+]cyt that is mediated by a great number of Ca2+ channels and Ca2+-binding proteins contributes to increased cancer cell stemness and tumorigenesis.114–119
During tumor growth, increased expression and activities of Ca2+ channels in cancer cells raise [Ca2+]cyt to levels above those of healthy cells, and lead to uncontrolled, elevated cell proliferation by regulating downstream effectors in multiple key stages of the cell cycle.79,84,120–122 In addition, intracellular calcium signaling regulates tumor growth by modulating cancer cell death, partly through autophagy or mitochondrial Ca2+ overload.123–127 During angiogenesis in solid tumors, calcium signals regulate the proliferation and migration of vascular endothelial cells. Vascular endothelial growth factor (VEGF)- or basic fibroblast growth factor (BFGF)-induced increases of [Ca2+]cyt activates proliferation of vascular endothelial cells in solid tumors.79,128 Both elevated and reduced Ca2+ influxes can enhance the migration of tumor-derived endothelial cells (TECs) compared to normal endothelial cells (NECs), which results in abnormal tumor vasculature.86,129,130
EMT is the cellular process of acquiring mesenchymal features from epithelial cells, causing tumor cells to become invasive and metastatic.62,131,132 At the early stages of EMT and metastasis, calcium signals are required.133–135 During metastasis, diverse calcium signaling pathways are regulated by transient receptor potential (TRP) channels, inositol trisphosphate receptors (IP3Rs)/ryanodine receptors (RyRs), voltage-gated calcium channels (VGCCs), and store-operated Ca2+ entry (SOCE).81 These pathways (1) modulate local [Ca2+]cyt at distinct intracellular regions of cancer cells and (2) regulate Ca2+-dependent effectors for the formation or turnover of focal adhesions, thus facilitating migration.87,88,122,136–140 Calcium signaling including SOCE is proposed to remodel distant sites and facilitate the colonization of secondary tumors in new organs by assisting metastasized tumor cells to exploit growth factors embedded in the extracellular matrix (ECM).141 SOCE-enhanced secretion of VEGF and prostaglandins E2 from primary tumors may mobilize angiogenesis at distant organs to form pre-metastatic niches. In addition, Ca2+ itself serves as a chemoattractant of tumor cells for bone colonization.142–145
Readers are referred to Table 1 for the specific influence of altered Ca2+ signals on cancer development. Next, we will discuss how mechanical stimuli can influence and regulate [Ca2+]cytvia the interplay between mechanotransduction and Ca2+ signaling pathways.
3.2 Intra-/inter-cellular calcium responses induced by mechanical stimuli
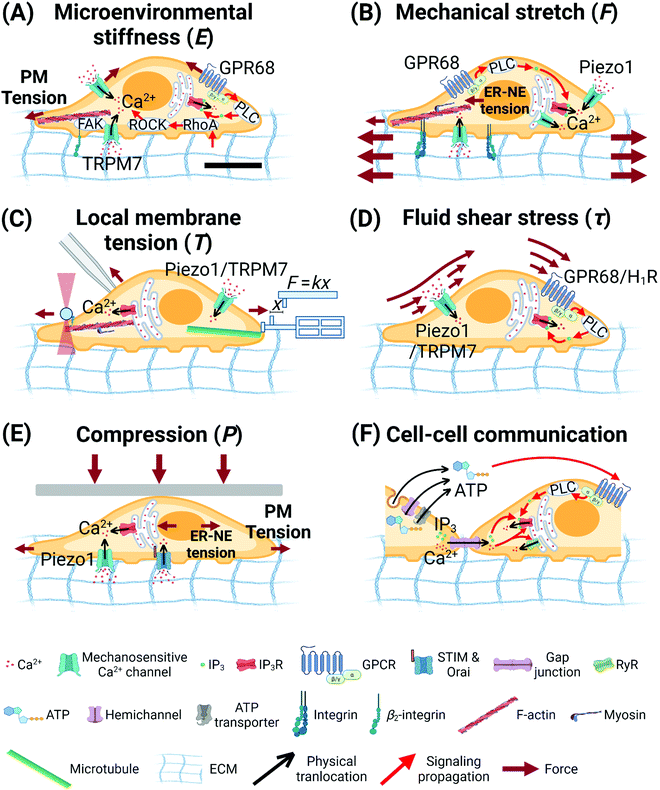 | ||
| Fig. 3 Biophysical cues regulate calcium signaling. (A) Stiff substrates enhance intracellular calcium signaling via RhoA/ROCK pathway,147 integrin/FAK/actin mechanotransduction,146,149 mechanosensitive ion channels,146,149–151 and mechanosensitive-GPR68-triggered Gq-PLC-IP3R pathway.152 Soft substrates enhance calcium oscillations in mouse fibroblasts in an F-actin-dependent manner.148 (B) Mechanical stretch from substrates enhances intracellular calcium signaling via actomyosin contraction,156,157 mechanosensitive ion channels,83,156,157 mechanosensitive GPR68,152 PLC-IP3R signaling pathway,156,157 and ER-NE tension.154 (C) Local membrane tension enhances intracellular calcium signaling via mechanosensitive ion channels84,158 and cytoskeletal-mechanotransduction-regulated ER calcium release.158 (D) Fluid shear stress enhances intracellular calcium signaling via mechanosensitive ion channels85,136 and mechanosensitive-GPCR-triggered Gq-PLC-IP3R pathway.86,159 (E) Mechanical compression enhances intracellular calcium signaling via mechanosensitive ion channels,88 ER-NE tension,87 and SOCE.162 (F) Mechanical stimuli regulate the expression and function of connexin-based gap junctions178–180 and hemichannel-/exocytosis-regulated ATP release,181,183–189 which are major pathways for intercellular propagation of calcium waves. The scale bar in (A) represents 5 μm length and applies to all other subfigures. | ||
In HMSCs, prolonged stretch triggers intracellular Ca2+ oscillations.156 This Ca2+ response is dependent on calcium influx via mechanosensitive Ca2+ channels on the plasma membrane, as well as the cytoskeleton, actomyosin contractility, and PLC activity. In HUVECs, vibrational stretch triggered global (80%) and local (20%) intracellular Ca2+ responses.157 The global [Ca2+]cyt increase is regulated by mechanosensitive Ca2+ channels on the plasma membrane, PLC-IP3R signaling pathway, and the resultant ER Ca2+ release, as well as F-actin assembly and actomyosin contractility. In the monolayer of human epidermal stem/progenitor cells (EPCs), cyclic stretch induces intracellular Ca2+ flashes.154 The underlying mechanism involves stretch-triggered nuclear deformation and ER-nuclear envelope (NE) tension, which causes Ca2+ release from the ER. Mechanically stretched human neuronal progenitor cells are more responsive to the activation of mechanosensitive GPR68 and show higher and faster elevation of [Ca2+]cyt compared to unstretched cells.152 Meniscus fibrochondrocytes (MFCs) (1) within the native tissues, (2) on aligned nanofibrous scaffolds, and (3) on silicone membranes, all show a baseline level of intracellular Ca2+ oscillations.155 Larger tensile deformation of all three types of substrates increases the population of cells that show intracellular Ca2+ oscillations, with the characteristics of a linear increase below 3% strain and a gradual plateau over 6%. The working mechanisms remain to be identified.
In HMSCs, laser-tweezer-induced tension at the plasma membrane triggers intracellular Ca2+ oscillations.158 The underlying mechanisms involve (1) Ca2+ influx via mechanosensitive TRPM7 channels, which is dependent on passive cytoskeletal support of F-actin and microtubules, and (2) ER Ca2+ release, which is dependent on cytoskeletal structure, actomyosin contractility, and TRPM7 activity. These data reveal that cytoskeleton indeed transmits mechanical signals from cell membrane into intracellular organelles and regulate Ca2+ signaling pathways.158
Intracellular ATP can be released to the extracellular milieu via (1) channels including hemichannels, maxi-anion channels and P2X7 receptors,169–172 (2) ATP-binding cassette (ABC) transporters,170,173,174 (3) exocytosis,169,170 and (4) lysis.170,174 ATP activates membrane (1) P2Y receptors and the downstream Gq-PLC-IP3R pathway to induce Ca2+ release from the ER,175,176 and (2) P2X receptors to trigger the influx of extracellular cations including Ca2+.176,177 In lung and prostate cancer cells, the mechanical-injury-triggered propagation of CWs in cultured epithelial layers is ATP-dependent.166 In non-contacted MCF-7 cells, the mechanically stimulated intercellular propagation of CWs involves extracellular ATP release.167 In HeLa cells, mechanically induced intercellular CWs involve both connexin-based gap junctions and extracellular ATP in a convoluted manner.168
The expression and function of connexin-hemichannel-formed gap junctions are responsive to mechanical stimuli, which further regulate intercellular communication.178–180 Certain connexins including Cx43 are sensitive to several types of mechanical stimuli, such as cyclic stretch, static tension, and shear stress. Moreover, ATP release can be stimulated by different types of mechanical stimuli including osmotic pressure, fluid shear stress, substrate stretch, compression, and injury.181,182 ATP-releasing connexin183,184 and pannexin185 hemichannels are mechanosensitive and further induces intercellular Ca2+ signaling.186,187 In addition, mechanical stretch,188 fluid shear stress,183,189 and injury181 induce exocytosis of ATP-containing vesicles in a Ca2+-dependent manner.
Overall, intra- and inter-cellular Ca2+ signaling can be triggered and enhanced by various types of mechanical stimuli, which have functional roles during tumor progression. We next review the current understanding of the molecular mechanisms of the mechano-regulated Ca2+ signaling pathways and the role of cytoskeletal proteins during this process.
3.3 Molecular mechanisms of the crosstalk between calcium signaling pathways and cytoskeletal proteins
In non-cancer cells, the actin cytoskeleton and its associated proteins regulate Ca2+ signaling.194,195 Actin cytoskeleton196 and cortical actin197,198 modulate Ca2+ flashes/waves during egg activation and fertilization.
In MDA-MB-231 human breast cancer cells, cyclic mechanical stretch induces a higher level of Piezo1-mediated Ca2+ influx than that in MCF10A normal human breast cells.83 In another study in MDA-MB-231 cells, Piezo1 is expressed in the cytoplasm including the plasma membrane, but in MCF10A cells, it is mainly expressed in the nuclear region, especially the nuclear envelope.201,202 This trait is likely to contribute to the different stretch-triggered Ca2+ responses between MDA-MB-231 cells and MCF10A cells. In the same study, expression of tropomyosin 2.1 (TPM2.1) is found in MCF10A cells but not in MDA-MB-231 cells, which is responsible for the different levels of stretch-induced Ca2+ influx in the two cell types.83 The data indicate that TPM2.1 regulates the expression location of Piezo1 in human breast cancer and normal cells.
In TECs from human breast carcinomas (BTECs), arachidonic acid (AA) treatment triggers actin remodeling and increases TRPV4 expression on the plasma membrane.129 Following pre-incubation of BTECs with AA, TRPV4 predominantly traffic from the cytoplasm to the cell membrane, demonstrating colocalization with the cortical actin in the cell periphery. In contrast, in the control untreated BTECs group, TRPV4 and actin mostly diffuse in the cytoplasm. The data indicate that the actin cytoskeleton interacts with TRPV4 channels and regulates the expression location of TRPV4 in BTECs.
In cancer cells, IP3Rs regulate cytoskeleton remodeling. In human breast cancer cells, IP3R3 mediates intracellular Ca2+ signaling and remodels profilin cytoskeleton via the ARHGAP18/RhoA/mDia1/FAK pathway.207 IP3R3 silencing causes oscillatory characteristics of [Ca2+]cyt signals after ATP administration or wound formation and alters the localization of F-actin and expression level of profilin. The remodeling of cytoskeletal proteins decreases cancer cell adhesion to collagen I-coated wells and induces rounded cell shape. However, how cytoskeletal proteins regulate IP3Rs and the downstream Ca2+ signaling in cancer is less known. To the best of our knowledge, only one study specifically reported that KRAS-induced actin-interacting protein (KRAP) is involved in the modulation of IP3R-regulated ER Ca2+ release in MCF7 breast cancer cells.208 Knockdown of KRAP attenuates the amplitude of ATP-induced Ca2+ release by 12–32% (peak response) in an ATP-concentration-dependent manner. KRAP is associated with IP3Rs in HCT116 colon cancer and HeLa cervical cancer cells, as well as in mouse liver and pancreas tissues.208 However, how KRAP functions in the IP3R-mediated Ca2+ signaling in those cancer cell lines and in vivo remains unclear.
In non-cancer cells, IP3Rs are directly regulated by or interact with cytoskeletal proteins,209 including but not limited to F-actin,210,211 protein 4.1N,212 myosin II,213,214 ankyrins,215–219 and microtubules.220 This evidence suggests that IP3Rs might be responsive to mechanical microenvironments via the cytoskeleton, and further influence the intracellular Ca2+ signaling. Indeed, in HMSCs, IP3R-regulated ER Ca2+ release in response to optical tweezer traction is dependent on cytoskeletal structure and actomyosin contractility but not IP3 level.158 Moreover, in human cervical carcinoma cells, IP3Rs release ER Ca2+ in response to ER-NE membrane tension, which further reinforces cortical actomyosin contractility to facilitate cancer cell transmigration through 3D collagen lattices and synthetic pores.87 These data suggest that in cancer cells, mechanical stimuli hold the potential to activate IP3Rs via cytoskeletal proteins and/or ER membrane tension to further induce Ca2+ signals.
PIP2, which produces IP3 following PLC cleavage, regulates actin-binding proteins including talin, gelsolin, ERM proteins (ezrin/radixin/moesin), formin, and actin-related protein 2/3 (ARP2/3) to mediate actin cytoskeleton dynamics.226–228 A myriad of actin-binding proteins interact with PIP2,229 including ERM proteins and myosin I,230 talin,228 formins and ARP2/3,227 and Coronin 1A.231
In summary, from our perspective, Ca2+ signals are instrumental at different stages of cancer progression. Various mechanical stimuli activate intra- and inter-cellular Ca2+ signaling pathways in cancer cells via mechanosensitive channels or through crosstalk with mechanotransduction pathways. Next, we introduce the functional roles and mechanisms of another mechanosensitive biochemical effector in cancer: YAP.
4. Yes-associated protein (YAP) in cancer
4.1 Significance of YAP in tumor progression
YAP (Yes-associated protein) and TAZ (Transcriptional co-activator with PDZ-binding motif) are two transcriptional co-activators in the Hippo pathway,234,235 and share ∼60% similarity in protein sequences.236 Binding with transcription factors in the nucleus, including YAP-TEA domains (TEADs), runt-related transcription factors (RUNXs), and p73, etc., YAP/TAZ regulate the transcription of genes including CTGF, IGFBP3, ITGB2, BIRC5, GLI2, and AXL, etc.,234 and regulate cell fates, functions (stemness, proliferation, apoptosis, migration, etc.), organ size, and homeostasis.234,237 Shuttling between nucleus and cytoplasm is an essential characteristic of YAP/TAZ because they only function when activated in the nucleus.90 The aberrant expression and nuclear accumulation of YAP/TAZ correlate with different cancers and at diverse tumour stages.90Recent studies show that, in response to biophysical signals, YAP/TAZ regulate the behaviors of both cancer cells and cancer-associated fibroblasts (CAFs) during tumor initiation, growth, and metastasis. For example, a stiff substrate (40 kPa) is needed when receptor tyrosine kinase (RTK)-Ras oncogenes transform normal cells into cancer cells through a YAP/TAZ-dependent mechano-transduction pathway.238 In CAFs, YAP activity is required to bridge biophysical signals from stretchable substrate and initiation of cytoskeleton remodeling, forming a self-reinforcing feed-forward loop.239 This important loop maintains CAFs’ phenotype and promotes tumor tissue stiffening, cancer cell growth, and invasion.239
In this review paper, we focus on the mechanobiology of YAP in cancer cells and their normal counterparts (Fig. 4). The fundamental biology of YAP/TAZ have recently been reviewed.90,94,234,240,241
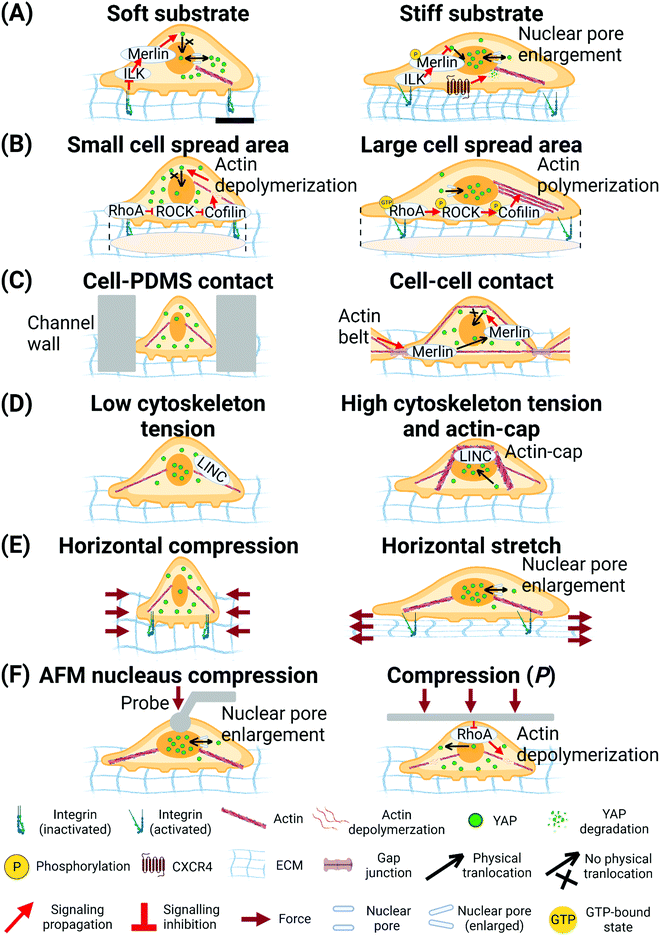 | ||
| Fig. 4 Biophysical cues regulate YAP translocation. (A) Soft substrate inhibits nuclear translocation of YAP through ILK/Merlin signalling.254 Stiff substrate inhibits Merlin function and enlarges nuclear pore size to facilitate nuclear translocation of YAP.96,254 (B) Cell spread area mediates RhoA-ROCK-Cofilin signalling to regulate actin polymerization and tension and translocation of YAP.98 (C) Cell-PDMS contact (confined micro-fluidic channel) induces compression on cells and inhibits nuclear translocation of YAP.265 Cell–cell contact increases actin belt tension and releases Merlin from gap junction to inhibit nuclear translocation of YAP.256 (D) LINC- complex-dependent actin-cap and peri-nuclear force facilitates nuclear translocation of YAP.257 (E) Horizontal compression of cells inhibits nuclear translocation of YAP. Horizontal stretching of cells enlarges nuclear pore size to facilitate nuclear translocation of YAP.96 (F) Local compression of cell nucleus by AFM probe enlarges nuclear pore size to facilitate nuclear translocation of YAP.96 Global compression of the cells by PDMS plate deactivates RhoA and triggers depolymerization of actin to inhibit nuclear translocation of YAP. The scale bar in (A) represents 5 μm length and applies to all other subfigures. | ||
4.2 Biophysical stimuli induce YAP responses
In both healthy and cancer cells, YAP and TAZ proteins respond to a broad range of biophysical stimuli, such as ECM mechanics (substrate stiffness and its heterogeneous patterns; material-type; dimensionality; geometry; topology; fiber directionality; and surface porosity), cellular mechanical states (cell spreading area; focal adhesion area; cytoskeleton tension or prestress; nuclear deformation; and cell shape), cell density, and extracellular mechanical stimuli (stretch; compression; pressure; and shear). Instructed by these biophysical stimuli, YAP responds differentially, represented by its translocation between the cell nucleus (N) and the cytoplasm (C), and translates the biophysical information into cell-specific transcriptional programmes. However, upon receiving the same biophysical stimuli or being in the same mechanical state, normal cells and cancer cells show distinct responses. For example, in normal cells, most studies show a positive correlation between the YAP nucleus/cytoplasm (N/C) ratio and cell spread area; a more spread cell shows a higher concentration of YAP in the nucleus and hence a higher N/C ratio,96,98,242 resulting in a higher proliferation rate.98 In contrast, in human breast cancer cells, YAP N/C ratio shows no notable correlation with cell spread area.243Conventional studies suggest that, prevailing in the evolutionarily conserved Hippo pathway, YAP and TAZ are regulated by biochemical cues and function in the nucleus to regulate cell fate and tissue homeostasis.237 Importantly, recent studies show that, in addition to biochemical cues, biophysical cues can independently regulate YAP's translocation from the cytoplasm to the nucleus through either the Hippo-dependent or -independent pathway. In the Hippo pathway, YAP/TAZ are phosphorylated by mammalian Ste20-like kinases 1/2 (MST1/2) and large tumor suppressor 1/2 (LATS1/2) and bind with the 14-3-3 protein and are retained in the cytoplasm.234 In both normal and cancer cells, substrate stiffness can regulate intracellular distribution of YAP through Hippo-dependent mechanisms.244,245 In the Hippo-independent case, extracellular biophysical cues regulate YAP translocation and bypass the Hippo pathway. For example, in the YAP-mutant cells that do not have Hippo-pathway-required interactions between YAP and LATS 1/2, substrate stiffness enhances nuclear YAP activity.98 Further, a study finds that modulation of biophysical cues can even dictate the Hippo pathway in regulating the YAP translocation.98
Importantly, emerging evidence suggests that, among all the mechano-sensitive components that participate in the regulation of YAP translocation, the nucleus can serve as a previously under-appreciated mechano-sensor that directly reads and translates biophysical cues into biochemical activities that regulate YAP translocation.96,97,246 However, the detailed molecular mechanism of how biophysical cues/states trigger, regulate, and maintain YAP translocation remains unclear at this time. Consequently, the potential mechanisms underpinning nuclear mechano-regulation remain an active area of research.
(1) In mammary epithelial cells and mouse embryonic fibroblasts (MEFs), YAP N/C ratio positively correlates with substrate stiffness.96,98 Cytoskeleton tension is necessary for substrate stiffness to regulate the translocation of YAP.98 The force-induced enlargement in nuclear pore size has been hypothesized to facilitate YAP nuclear translocation and is actively studied now.96
(2) Recent research on NIH 3T3 cells shows no correlation between YAP N/C ratio and substrate stiffness.246 Instead, YAP N/C ratio positively correlates with cellular traction force and nuclear deformation.246 The data suggest that while nuclear translocation of YAP, in most normal cells, positively correlates with substrate stiffness, substrate stiffness often leads to changes in downstream cellular behaviors such as cell spread area, traction, and nuclear shapes. It is these changes that directly regulate YAP translocation, rather than the substrate stiffness itself. We hypothesize that, in this experiment, substrate stiffness does not necessarily determine contractility. Therefore, if YAP translocation is regulated by contractility, the YAP N/C ratio shows correlation with contractility instead of substrate stiffness. To decouple these behaviors and reveal the true mechanotransduction mechanisms underlying YAP translocation, more studies are required.
In 3D culture milieu, positive correlation is observed between YAP N/C ratio and environmental stiffness.247,248 Importantly, in human liver organoids, signaling of integrin-mediated Src family kinases (SFKs) promotes YAP activity on stiff (1.3kPa vs. 0.3 kPa) 3D matrices.248 In contrast with 2D substrate findings, which suggest that YAP's response to extracellular biophysical stimuli necessitates cytoskeletal contractility,96,97,249 this alternative integrin-mediated SFK mechanism insinuates that YAP is not regulated through the conventional cytoskeletal tension or the downstream nuclear mechano-sensing in 3D substrates and in vivo. This possibility is supported by a recent finding which suggests that geometrical changes (including wrinkling) on nuclear envelopes in 2D/3D-cultured cells trigger diverse mechanisms to regulate YAP translocation.247
In this light, we hypothesize that a threshold of substrate stiffness may exist to determine the role of cytoskeletal tension in the regulation of YAP translocation. One recent report supports our hypothesis and shows that, in MEF cells, the unfolding of Talin by cytoskeletal tension and YAP nuclear translocation only occurs once substrate stiffness is larger than 5 kPa.250 Although the two results reported in human liver organoids248 and in MEF cells250 are obtained from different cell types on distinct 2D/3D substrates, we reason that the stiffness range within the organoid (1.3 kPa and 0.3 kPa) may not reach the hypothesized stiffness threshold (such as 5 kPa)250 to trigger the regulatory effects of cytoskeletal tension on YAP translocation. Our hypothesis can be evaluated by targeted disruption of the actin cytoskeleton, systematic characterization of YAP translocation, and real-time measurement of nuclear envelope geometry and tension in cells experiencing a range of environmental stiffness.
In certain cancer cell lines (pancreatic, brain, and liver), YAP N/C ratio is observed to show positive correlation with substrate stiffness,245,251–253 despite a few exceptions:99,254
(1) Brain cancer cells (on 10%- and 3%-acrylamide polyacrylamide (PAA) gels) and pancreatic cancer cells (on 1, 4, and 25 kPa PAA gel) show higher nucleus YAP localization on stiffer substrates.252,253 Human liver cancer cells show higher nucleus YAP localization on stiffer PAA gels (1.1 kPa vs. 400 Pa).245 Mechanistically, agrin and integrin sense the stiffness signals and trigger YAP translocation through two subsequent mechanisms including (a) the formation of actin stress fibers and (b) the diminishment of the Merlin function (Note: Merlin retains YAP in the cytoplasm through activation of the Hippo-pathway).245 Although cellular traction is not measured in this study, the observed formation of actin stress fibers, induced by agrin stiffness-sensing and nuclear translocation of YAP, shows the regulatory role of cytoskeletal tension on YAP translocation. In liver cancer cells, increased substrate stiffness triggers the increase in the expression of mechano-transducer C–X–C Motif Chemokine Receptor 4 (CXCR4) and maintains the positive correlation between YAP N/C ratio and substrate stiffness.251 These studies on liver cancer cells suggest that, even for the same cell type, multiple molecular pathways may co-regulate YAP translocation in response to ECM stiffness.
(2) Interestingly, in breast cancer cells, biphasic correlation is uncovered between YAP N/C ratio and substrate stiffness. YAP N/C ratio is lower than 1 on both soft (10 kPa) and stiff (57 kPa) PAA gel, while it is larger than 1 on intermediate stiff (38 kPa) PAA gel. Mechanistic studies suggest this translocation is regulated by Integrin Linked Kinase (ILK)/Merlin-controlled YAP nuclear transportation (Fig. 4A and Table 2).254 Specifically, ILK locates between integrin and actin and mediates the phosphorylation of Merlin to regulate YAP translocation. Supported by a series of functional results, the study concludes that the biphasic correlation between ILK and substrate stiffness causes the highest YAP nuclear translocation on substrates of intermediate stiffness. The mechanism by which ILK expression level is regulated by substrate stiffness and causes the YAP translocation is now under investigation.
| Mechanical stimulus | Normal/cancer | Cell type | Relation with nuclear translocation | Proposed mechanism or related protein | Ref. |
|---|---|---|---|---|---|
| Substrate Stiffness | Normal | Mammary epithelial cells | Positive | F-Actin-capping/severing proteins Cofilin, CapZ, and Gelsolin | 98 |
| MEF | Positive | Nuclear pore size increase | 96 | ||
| NIH 3T3 cells | No correlation | N/A | 246 | ||
| Cancer | Pancreatic cancer cell | Positive | N/A | 252 | |
| Liver cancer cell | Positive | Agrin/integrin mediated stiffness sensing and formation of stress fiber | 245 | ||
| Liver cancer cell | Positive | CXCR4 mediated YAP cytoplasmic degradation | 251 | ||
| Brain cancer cell, IOMM-Lee, (HKBMM) | Positive | Merlin mediated YAP cytoplasmic retension | 253 | ||
| Tumor repopulating cells (ovarian cancer cell line A2780, human MCF-7 breast cancer cell line and murine melanoma cell line B16-F1) | Positive (but on different substrate type) | Cdc42-mediated F-actin and Lats1 interactions | 406 | ||
| Breast cancer cell | Biphasic | ILK and Merlin mediated YAP cytoplasmic retension | 254 | ||
| Cell area | Normal | Mammary epithelial cells | Positive | F-Actin-capping/severing proteins Cofilin, CapZ, and Gelsolin | 98 |
| MSC | Positive | Rho/ROCK mediated actin polymerization | 242 | ||
| NIH 3T3 cells | (>∼1000 μm2) not related, (<∼1000 μm2) YAP in cytoplasm | Nuclear deformation | 246 | ||
| Cancer | Breast cancer (MDA-MB-231) | No correlation | N/A | 246 | |
| Cell traction | Normal | MEF | Positive | Nuclear pore size change | 96 |
| NIH 3T3 cells | Positive (not decoupled with nucleus deformation) | Nuclear deformation | 246 | ||
| No correlation (decoupled with nucleus deformation) | N/A | ||||
| Mesenchymal Stem Cells | Positive (when decrease traction) | Nuclear deformation | 97 | ||
| Perinuclear Traction | Normal | MEF | Positive | Actin-cap | 257 |
| Peri-cell traction | Normal | MEF | No correlation | N/A | 257 |
| normal cell (MDCKII) | Negative | Actin belt mediated Merlin | 256 | ||
| Fluid shear stress | Normal | zebrafish endothelial cells and human pulmonary artery endothelial cells | Positive | Cortical actin bundles release YAP from binding with angiomotin | 260 |
| Cancer | Human prostate cancer cells | Positive | Polymerization of F-actin | 261 | |
| Stretch | Normal | MEF | Positive (cyclic) | N/A | 263 |
| Mesenchymal stem cells | Positive | Nuclear deformation | 97 | ||
| Compression | Cancer | cervical cancer cell | Negative | F actin depolymerization and RhoA deactivation | 264 |
| Human fibrosarcoma HT1080 | Negative | Ca2+ dependent | 266 | ||
| Osteosarcoma, U2OS | Negative | N/A | 265 | ||
| Normal | MCF-10A | Negative | F actin depolymerization and RhoA deactivation | 264 | |
| MEF | Positive | Nuclear pore size increase | 96 |
In cancer cells, only one study has been conducted and shows no correlation between YAP N/C ratio and cell size.243 This study focused on single metastatic breast cancer cells (MDA-MB-231) and metastatic D3H2LN cells harvested from mouse lymph nodes in MDA-MB-231-injected mice.243 Further research on other cancer cell types needs to be conducted to verify if YAP is not correlated with cell area in all cancer cells.
In another study on brain cancer cell lines, the YAP N/C ratio is negatively correlated with cell density.253 Merlin expression shows a positive correlation with cell density, implying that cell density regulates YAP intracellular distribution through Hippo-dependent mechanisms in cancer cells. The potential roles of cytoskeletal structure and tension in cancer cells must be further investigated.
In the same type of MEF cells, perinuclear traction force and actin-cap are observed for the first time. Disrupting the linker of nucleoskeleton and cytoskeleton (LINC) complex (a transmembrane protein complex that locates on the nuclear envelope and connects the nuclear interior with the cytoskeleton) reduces perinuclear force, eliminates actin-cap formation, and shows the reduced YAP N/C ratio without influencing cell periphery traction (Fig. 4D and Table 2).257 This finding suggests that the perinuclear cytoskeletal tension and structure enable transmitting the force into the nucleus to regulate YAP translocation.
In the presence of cell–cell contact, the cortical actin tension regulates the translocation of YAP in a Hippo-dependent way. In normal MDCKII cells, increased actin belt tension (reflected by the amount of colocalized myosin-II and F-actin) negatively regulates the YAP N/C ratio by releasing Merlin from the adhesion junction to enhance the retention of YAP in the cytoplasm.256
Overall, in normal cells, YAP N/C ratio positively correlates with overall cell traction. However, the intracellular distribution of cytoskeletal tension, i.e., in the perinuclear and cell periphery regions, may have differential regulatory roles on YAP translocation. Additionally, cytoskeletal tension/structure and the LINC complex are needed for cells to sense extracellular biophysical stimuli and subsequently trigger YAP translocation. Third, nuclear deformation is positively correlated with YAP N/C ratio and traction force96,255 and needs to be decoupled from traction to determine if it is an independent regulatory effector. Fourth, unlike in normal cells, the correlation between cell traction and YAP translocation in cancer cells is still lacking.
To decouple the roles of nuclear deformation and cytoskeletal tension in YAP regulation under stretching, another study employed two drugs with distinct functions: ML7 and Y27632. ML7 is an inhibitor of myosin-II b and reduces the cytoskeletal tension but keeps the stress fibers and nuclear deformation. In the cells under cyclic stretching treated by ML7, YAP shows nuclear translocation. In contrast, Y27632 is an inhibitor of ROCK and eliminates the cytoskeletal tension as well as nuclear deformation. Cells treated by Y27632 under cyclic stretching show no YAP translocation into the nucleus.97 These results indicate that cytoskeletal contractility is not necessary in regulating YAP translocation. Instead, the force sensed by the nucleus is required to regulate YAP translocation. A recent study corroborates this indication. The study changes the nuclear stiffness through an up-regulation of Lamin A expression and observes that YAP N/C ratio correlates with nuclear deformation but not traction force.246 These results suggest that YAP translocation is regulated by nuclear deformation but not necessarily by the force transmitted through the LINC complex. However, Lamin A not only affects nuclear stiffness but also serves as the structural component that is downstream of the LINC complex and might affect the potential nuclear mechano-sensing through this route. Hence, how nuclear mechano-sensing regulates YAP translocation at the precise molecular level needs to be further investigated.
To address this question, we propose three potential approaches. First, one can achieve similar nuclear deformation in cells by different force transmission methods including stretching and compression and measuring the corresponding difference in YAP translocation. If the nuclear geometry regulates YAP translocation, then the YAP N/C ratio should be similar in cells that experience similar nuclear deformation regardless of the types of forces applied. Second, we can disrupt the cytoskeleton and directly apply force on the nucleus through either the LINC complex or other protein complexes, potentially with magnetic beads, followed by observing the differential relationship between YAP translocation and nuclear deformation. If force transmission through the LINC complex regulates YAP translocation, then the YAP N/C ratio should increase noticeably when forces are applied via the LINC complex but not via other protein complexes. Third, one can maintain the level of nuclear deformation without interfering with the nuclear force transmission using methods such as keeping Lamin A expression constant and stretching cells to increase the force transmitted into the nucleus through the LINC complex. If the nuclear deformation regulates YAP translocation, the YAP N/C ratio should remain stationary regardless of the magnitude of force transmitted into the nucleus. These strategies enable the decoupling of nuclear deformation from the force transmitted into the nucleus (through and not through LINC) and bring us closer to the discovery of the molecular underpins in YAP translocation.
Next, we discuss how YAP translocation responds to actively applied extracellular force.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dyne
dyne![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cm2 for 6 h) facilitates the polymerization of F-actin through ROCK–LIMK–cofilin signaling and triggers the nuclear translocation of YAP.261 In hepatocellular carcinomas, fluid shear stress (1.4 dyne cm2 for 2–8 h) triggers the nuclear YAP translocation in a F-actin-dependent way.262 Whether nuclear mechano-sensing and cortical actin tension are involved in this mechanism remains unclear.
cm2 for 6 h) facilitates the polymerization of F-actin through ROCK–LIMK–cofilin signaling and triggers the nuclear translocation of YAP.261 In hepatocellular carcinomas, fluid shear stress (1.4 dyne cm2 for 2–8 h) triggers the nuclear YAP translocation in a F-actin-dependent way.262 Whether nuclear mechano-sensing and cortical actin tension are involved in this mechanism remains unclear.
In normal cells, both static and cyclic stretching trigger nuclear translocation of YAP (Fig. 4E and Table 2).97,98,263 In MEFs, static stretching of the cell monolayer induces increased YAP N/C ratio.98 In MSCs, when the cytoskeletal contractility is inhibited by ML7 but the actin stress fibers are maintained, cyclic stretching can cause nuclear deformation and YAP nuclear translocation.97
Active compression on cells does not cause a universal trend on the regulation of YAP translocation. Compression force (1.5 nN), applied by AFM tips on the normal and cytoskeleton-disrupted MEF cells at the apical surface above the nucleus, triggers YAP nuclear translocation.96 In both HeLa (cervical cancer cell line) and MCF-10A (normal mammary epithelial cell line), compression (24 Pa) applied by a polydimethylsiloxane (PDMS) sheet causes F-actin depolymerization and YAP translocation into the cytoplasm (Fig. 4F).264 Similar to preceding research, deformations of the cell and the nucleus are not quantified in this study. In the osteosarcoma line, cells under narrow confinement from micro-fluidic devices show YAP cytoplasm translocation. However, the cells on the line patterns (width range: 5–50 μm) without confinement show no YAP translocation, even with large nucleus aspect ratio.265 This result implies that (1) the aspect ratio of the nucleus does not regulate YAP translocation, and (2) the real regulatory parameter of YAP translocation is influenced by force transmitted into the nucleus, instead of nuclear geometry. Compression on human fibrosarcoma cells inhibits RhoA activity through TRPV4 mediated Ca2+ currents and cause the cytoplasmic translocation of YAP.266
4.3 Summary of YAP mechano-transduction
The key understandings of the roles of YAP in mechano-transduction and the direct regulators of YAP are:(1) YAP acts as a mechano-transducer that transmits the extra- and intra-cellular biophysical cues into the cell nucleus and regulates cell functions through binding with transcription factors.
(2) YAP itself is unlikely to be a direct mechano-sensor that senses the biophysical cues. The mechano-sensors, such as integrin and potentially the cell nucleus, convert biophysical cues into chemical signals that are transmitted by YAP activation.
(3) Mechanistically, the mechano-regulation of YAP is believed to be mainly through the F-actin cytoskeletal tension and nuclear envelope mechanics.
(4) The nucleus is a promising mechano-sensor that can directly sense the biophysical signals.
How the nucleus senses the force and regulates YAP is being actively studied. Elosegui-Artola proposes that the size changes in nuclear pores, induced by nucleus flattening, regulate YAP translocation.96 However, because of the challenges in manipulating the size of nuclear pores in a controlled manner, this hypothesis is still under active investigation. In line with the finding that the nucleus is a direct mechano-sensor, we hypothesize that the combination of the LINC complex and nucleoskeleton may function as an alternative route to transmit force and regulate YAP. Our hypothesis is supported by a recent study that shows that, in the nucleus isolated out of the cell body, force transmission through the LINC complex and non-specific bindings into the nucleus triggers distinct changes in nuclear stiffness. Since the size changes in nuclear pores are unlikely to affect nuclear stiffness, we hypothesize that certain other mechano-sensitive underpins within the LINC complex and nucleoskeleton may respond to the force transmitted into the nucleus and alter nuclear mechanical states.
If the mechanisms of mechano-transduction through YAP are clear, it offers new opportunities to develop mechano-medicine for cancer treatment because of the important role of YAP in maining mechanical homeostasis.94 We propose:
(1) To reduce the possibility of tumor initiation in stiffened tissue, we can inhibit the stiffness sensing in normal cells through YAP translocation since YAP is required for RTK-Ras oncogenes to transform normal cells into tumor cells on stiff ECM.238 If the mechano-transduction through YAP is inhibited in normal cells within fibrosis tissue, which has higher stiffness and higher possibility for tumor initiation, the transformation of normal cells can be suppressed.
(2) To reduce tumor tissue stiffness and cancer cell extravasation, we can inhibit YAP-mediated mechano-transduction in CAFs because YAP activity is required for CAFs-dependent matrix stiffening, cell invasion, and angiogenesis.239
5. MicroRNA in cancer
5.1 miRNA biogenesis
MicroRNAs (miRNAs) are ∼22 nucleotide (nt) RNA, first discovered in Caenorhabditis elegans in 1993.267 Under most conditions, miRNAs interact with the 3' untranslated region (UTR) of target messenger RNAs (mRNAs) to cause mRNA deadenylation and decapping as well as to attenuate the translational output.268,269 In addition, multiple reports have demonstrated the capability of miRNAs to target protein-coding sequences (CDS) and 5′ UTR.270–272 The miRNA biogenesis can be classified into canonical and non-canonical pathways.In the canonical pathway, miRNAs are transcribed by RNA polymerase II (Pol II) (Fig. 5A). The Pol II-transcribed primary (pri-) miRNAs are capped and polyadenylated, harboring one or multiple hairpin structure(s), which contain the miRNA sequence.273,274 Processing of the pri-miRNAs is carried out in the nucleus by a heterotrimeric complex, Microprocessor, comprised of one molecule of the RNase III enzyme Drosha and two molecules of Digeorge critical region 8 (DGCR8; named Pasha in flies and nematodes).275–280 Drosha possesses two RNase III domains that each cleaves one strand of the stem in the pri-miRNA hairpin, which liberates a 60 nt to 70 nt stem-loop called a precursor (pre-) miRNA with a characteristic 3′ hydroxyl group (OH), overhangs of 2 nts, and a 5′ phosphate (P). The generated pre-miRNAs are exported to the cytoplasm by an exportin 5 (XPO5)/RanGTP complex and processed by another RNase III enzyme Dicer.281 As an endonuclease with two RNase III domains, Dicer functions in concert with trans-activation-responsive RNA-binding protein (named TRBP in mammals and Loquacious in flies).282 In this process, Dicer releases a dsRNA that is ∼22 base-pairs long from the stem of the pre-miRNA to the cleavage site contiguous to the apical loop and creates a mature miRNA duplex that interacts with the Argonaute (AGO) proteins.283,284 Afterwards, the AGO unwinds the RNA duplex and promotes the expulsion of the passenger strand to form the mature RNA-induced silencing complex (RISC).285 Depending on the origin from the hairpin arms, the mature miRNA is designated as either the 5p or the 3p miRNA. The initiated RISC then identifies a specific mRNA sequence by complementary base-pairing, resulting in translation inhibition and/or RNA degradation.286
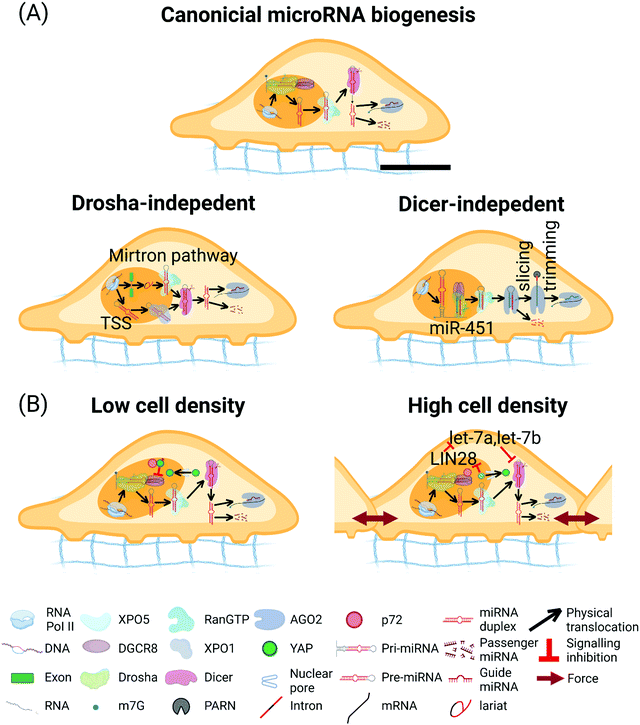 | ||
| Fig. 5 miRNA biogenesis pathway. (A) Canonical miRNA biogenesis pathway. In the nucleus, primary (pri-) miRNAs are transcribed by Pol II and then processed by the Microprocessor complex containing one Drosha and two DGCR8 to form pre-miRNAs.273 Pre-miRNAs are exported to the cytoplasm by the complex of XPO5/RanGTP. Subsequently, pre-miRNAs are cleaved by Dicer to form ∼22 nucleotides miRNA duplex. miRNA guide strand is then loaded into the AGO to form the RNA-induced silencing complex (RISC), the passenger strand is degraded. Drosha-independent miRNA biogenesis pathways. In the mirtron pathway, pre-miRNAs are spliced and debranched from the intron region of transcript, which bypass Drosha processing. After that, the intron-derived pre-miRNAs access the canonical miRNA pathway.293–295 In the transcription start site (TSS) miRNA biogenesis pathway, the 5′ end of the pre-miRNA hairpin intermediate contains an 7-methylguanosine (m7G)-Cap and the 5′ end of the pre-miRNA hairpin generated by transcription initiation directly, and the 3′ end generated by transcription termination. The Capped pre-miRNAs are exported by XPO1 protein and bypass Drosha processing. The 3p-capped miRNA is loaded onto the AGO complex, but the 5p-capped miRNA is degraded.296 Biogenesis of miR-451. pri-miR-451 is cleaved by the Microprocessor complex and bypass Dicer. Pre-miR-451 is directly loaded into AGO2, which cleaves the 3p arm of the hairpin. Poly-A specific ribonuclease (PARN) further trims the 5p arm to form miR-451.292 (B) Hippo-YAP signaling pathway affects the miRNA biogenesis. At low cell densities, activated YAP stays in the nucleus, sequestering p72 from Microprocessor and disrupting the miRNA biogenesis.380 At higher cell density, translocation of nuclear YAP/TAZ into cytoplasm inhibits LIN28, upregulates let-7a and let-7b, and represses the Dicer levels.381 The scale bar in (A) represents 5 μm length and applies to all other subfigures. | ||
Numerous non-canonical miRNA biogenesis pathways have been identified.287 These pathways take advantage of distinct combinations of the proteins engaged in the canonical pathway, namely Drosha, Dicer, XPO5, and AGO. Readers are referred to recent in-depth reviews for more information about non-canonical miRNA biogenesis pathways.288–300
5.2 Significance of miRNA regulation in cancer
During the last decade, convincing evidence has clarified that miRNA expression is dysregulated in human malignancies through diverse mechanisms, including miRNA biogenesis defect, miRNA gene mutation, and dysregulated transcriptional control or epigenetic modification of miRNA genes.301Microprocessor cleavage of the pri-miRNA is the initial processing step during miRNA biogenesis. Single-nucleotide substitution/deletion of the Microprocessor components Drosha and DGCR8 (15% of 534 Wilms tumors) is associated with diminished expression of mature let-7a and miR-200 family members.304
Considering the vital role of XPO5 in the nuclear export of pre-miRNAs, it is not surprising that downregulation of XPO5 causes decreased cellular proliferation, attenuated invasion, arrest of G1/S cell-cycle, and downregulation of pivotal oncogenic miRNAs (e.g., miR-21, miR-10b, miR-27, miR-182 and miR-155) in colorectal cancer (CRC) cells.305 Another example of dysregulation of XPO5 in cancer is that phosphorylation of XPO5 by hyper-activated ERK can repress the recruiting and exporting of pre-miRNA, which globally suppress miRNA biogenesis in hepatocellular carcinoma (HCC).306
Universal downregulation of miRNAs due to defective processing by Dicer is rising as a prevalent hallmark of cancer.307 In the DICER1 gene, somatic ‘hotspot’ mutations at the four catalytic residues in the RNase IIIb domain (D1709, E1705, E1813, D1810) and one catalytic residue in the RNase IIIa domain (G1809) were identified in ovarian sex cord-stromal tumors, pediatric tumors and endometrial tumors.308–310 Likewise, 15 RNase IIIb hotspot in uterine corpus endometrial carcinoma (UCEC) cases show down-regulation of specific 5p miRNAs.311
AGO2, the only slicing protein in the AGO family that cleave miRNA duplexes, plays a vital role in the accumulation of mature miRNAs.312 Acetylation, a novel post-translational modification (PTM) of AGO2, boosts cancer progression by specifically affecting miR-19b levels.313 Additionally, the AGO2 expression levels in HCC specimens are significantly higher in comparison to adjacent non-tumor liver.314
Amplification of miRNA genomic loci also exists. The miR-17–92 cluster is amplified in a variety of tumors, which resulted in the upregulation of the miRNAs, thus stimulating tumor development.319 Overexpression of miR-21, because of the amplification in 17q23–25, causes low expression of the tumor suppressor gene, phosphatase and tensin homolog (PTEN), in ovarian cancer.320 In fact, the upregulation of miR-21 has been revealed in numerous cancers, which has an effect in boosting drug resistance of cancer cells.321 Due to amplification of 3q26.2, a cancer-associated miRNA, miR-569, contributes to ovarian and breast cancer cell survival and proliferation.322
A high-resolution array-based assay in 227 specimens detected DNA copy number alterations in genomic loci consist of miRNA genes in ovarian cancer (37.1%), breast cancer (72.8%), and melanoma (85.9%).323 Genome-wide investigations revealed that 98 of 186 (52.5%) miRNA genes are in cancer-associated genomic regions or in fragile sites.324 In summary, abnormal miRNA expression in cancer cells could develop from the amplification or deletion of individual genomic regions containing the miRNA genes.
In addition, the MYC-miRNA feedback loop is indispensable for the development of HCC. miR-122 indirectly suppresses MYC expression by targeting Tfdp2 and E2f1. Furthermore, miR-148 directly targets the 3′ UTR of MYC and inhibits MYC, while miR-363 directly targets the 3′ UTR of ubiquitin-specific protease 28 (USP28) and indirectly destabilizes MYC.330
Another example is how p53 regulates miRNA abundance to exert its tumor suppressive activity.332p53 is one of the most ubiquitous tumor suppressors, whose mutation is detected in approximately 50% of human cancers.333p53 can induce the upregulation of miR-34a to prompt apoptosis, cell-cycle arrest and cell senescence through associating with the promoter of the miR-34a gene.334 As a feedback loop, miR-34a inhibits p53 expression by targeting sirtuin 1 (SIRT1), which is a negative regulator of p53 via deacetylation.335 Further, the miR-34 family inhibits tumor growth and progression by targeting regulatory factors including cyclin-dependent kinase 4/6 (CDK4/6), cyclin E2, and anti-apoptotic protein B-cell lymphoma 2 (BCL2), which are engaged in cell proliferation, the cell cycle, EMT, metastasis, and stemness.336 More studies revealed that p53 regulates the expression of a range of miRNAs, such as miR-605 miR-1246, miR-143 and miR-107, to perform its function.337–339
Overall, MYC and p53, two of the most comprehensively studied transcription factors, regulate miRNA expression. Other transcription factors and miRNA co-regulatory networks, such as E2Fs/miR-17/20 and PITX3/miR-133b, have been discovered in multiple tumors.340
Compared with healthy individuals, the methylation level of the nine CpGs of the miR-223 promoter was significantly lower in atherosclerotic cerebral infarction (ACI) patients but higher in carotid atherosclerotic patients.344 A total of seventeen miRNAs were upregulated higher than 3-fold after simultaneous treatment with DNA methylation and histone acetylation inhibitors. miR-127, one of 17 miRNAs located within a CpG island, is highly induced after treatment. Consistently, miR-127 is lowly expressed in the malignant cells, indicating that it is subject to epigenetic silencing.345 Further, decreased expression of miR-152/-137 and miR-34b/c is associated with DNA hypermethylation in endometrial, lung and gastric cells, respectively.346–349 The above evidence spotlights the intricate interpretation between miRNAs and the epigenetic architecture, revealing that abnormal DNA methylation and histone acetylation of miRNA genes can serve as biomarkers for cancer diagnosis and therapeutics.350
5.3 Mechanosensitive miRNA in cancers
Multiple mechanosensitive miRNAs (mechanomiRs) have been identified by miRNA microarray screening of either longitudinally or transversely stretched diaphragms from mice.351 Over the past few years, an increasing number of miRNAs have been reported to interact reciprocally with ECM proteins and regulate mechanotransduction via distinct mechanisms.352 miRNAs play a role in ECM regulation by directly targeting mRNAs that encode ECM proteins or by indirectly regulating the expression of genes that modulate the synthesis/degradation of ECM proteins (Table 3). Interestingly, different miRNAs from the miR-17–92 cluster are involved in both regulatory mechanisms.| microRNA | Function | Ref. | |
|---|---|---|---|
| Mechanotransduction | miR-17 | Repress the expression of fibronectin | 355 |
| miR-143 | Target the 3′ UTR of fibronectin type III domain | 357 | |
| let-7e-5p | Trigger muscle fibrosis by targeting the ECM proteins: Col1a1, Col1a2, Col3a1, Col24a1, Col27a1, Itga1, Itag4, Scd1, and Thbs1 | 351 | |
| miR-18a | Suppress PTEN via β-catenin stimulation of MYC-driven miR-18a and HOXA9 | 359 | |
| Calcium signaling targeting | miR-34a | Decreased Ca2+ influx | 364 |
| miR-195 | Regulate mitochondrial Ca2+ uptake by downregulating MICU1 | 375 | |
| miR-27a | Downregulate the ER-located Ca2+ transporter CACNA2D3 | 369 | |
| miR-28 | Downregulate TRPM7 | 371 | |
| miR-25 | Downregulate MCU | 374 | |
| Mechano-memory, crosstalk with YAP | miR-21 | Function as a mechanical memory keeper in myofibroblast activation and fibrogenesis | 392 |
| let-7a and let-7b | Downregulated let-7a and 7b expression rescues the miRNA biogenesis defects observed following TAZ/YAP knockdown | 381 | |
| miR-130a | Promote YAP-induced tumorigenesis and liver enlargement | 386 | |
| miR-130b | Target the MST1 and SAV1 resulting in Hippo signaling pathway inactivation | 387 |
Fibronectin (Fn) is a glycoprotein found in the ECM and the generation of active Fn fibers is required for collagen I matrix assembly. The ECM network is initially constructed by depositing Fn fibers, followed by collagen I fibers, which preferentially interact with the relaxed Fn in the ECM.353,354 miR-17, a member of miR-17–92 cluster, represses the expression of Fn which leads to reduced cell adhesion, migration, and proliferation.355 In addition, miR-143 can directly target the 3′ UTR of Fn type III domain containing 3A (FNDC3A) and repress its expression level. Therefore, upregulated miR-143 facilitates liver tumor cell invasion and metastasis, as local liver and distant lung metastasis were significantly reduced when miR-143 expression was suppressed.356,357 Another example is let-7e-5p, a member of the let-7 family, reported as mechanomiR, showing more than 1.5-fold downregulation in atrophic skeletal muscle; dysregulation of let-7e-5p may trigger muscle fibrosis by targeting the ECM proteins: Col1a1, Col1a2, Col3a1, Col24a1, Col27a1, Itga1, Itag4, Scd1, and Thbs.351
The miR-17–92 cluster can form an autoregulatory feedback loop with E2F transcription factors, thereby suppressing the expression of many tumor-associated proteins.358 Induced by increased stiffness in human and mouse tissue, miR-18a from the mi-17–92 cluster, leading to reduced levels of the tumor suppressor PTEN by base-pairing with the 3′ UTR of PTEN.359 Increased ECM stiffness could modulate PTEN suppression by directly suppressing PTEN via β-catenin stimulation of MYC-driven miR-18a and by indirectly reducing PTEN through the levels of homeobox A9 (HOXA9) regulation.359 In breast cancer, HOXA9 directly binds to the PTEN promoter to regulate its expression and inhibit the malignancy.359,360PTEN loss in stromal fibroblasts promotes ECM deposition and alignment independently from cancer cells’ presence, and this reorganization regulates cancer cell behavior.361 Therefore, stromal matrix stiffness controls cellular ECM deposition through the regulation of miRNA expression.
Furthermore, 122 miRNA families with their 73 mRNA targets which encode cytoskeleton-actin-matrix (CAM) proteins were identified in endothelial cells.362 The miRNA-CAM mRNA regulatory network is demonstrated to counteract the effects of ECM stiffness and promote mechanical stability of tissues.362
5.4 Molecular mechanism of the crosstalk between miRNA and Ca2+ signaling
In Section 3, we discussed that the intracellular Ca2+ signaling links to almost every cancer hallmark. Emerging studies have illustrated that miRNAs play a crucial role in regulating intracellular Ca2+ dynamics through the SOCE pathway, calcineurin/NFAT signaling, and Ca2+ ion channels (Table 3).In T cells, SOCE is the central pathway to modulate cellular activation, proliferation, apoptosis, and migration.363 In the human Jurkat T cell line, miR-34a overexpression significantly reduces calcium influx through targeting SOCE-related genes (ITPR1, ITPR3, CALM3, ATP2A2 and ATP2A3) and calcineurin/NFAT signaling related genes (RCAN1, PPP3R1 and NFATC4).364
miR-27a is involved in different regulatory functions in different types of cancer, and is upregulated in breast cancer,365 ovarian cancer,366 and prostate cancer.367 In breast carcinoma, ER-located Ca2+ transporter CACNA2D3 is frequently methylated and contributes to metastasis.368 In Mycobacterium tuberculosis (Mtb) infected peripheral blood mononuclear cells, miR-27a is abundantly expressed and contributes to autophagy inhibition through down-regulating ER Ca2+ signaling by directly targeting CACNA2D3.369 Thus, the study of miR-27a targeting CACNA2D3 in cancer metastasis may support the development of anti-metastasis therapeutic approaches.
TRPM7 forms a constitutively active Ca2+ permeable channel, which regulates diverse cellular processes in healthy and tumor cells.370 In glioblastomas, in addition to TRPM7's critical roles in regulating cell migration and invasion, an upregulated miR-28-5p expression results in a significant decrease in glioma cell proliferation and migration.371,372 Rap1b was reported to be a target of miR-28-5p and its expression level was downregulated. Therefore, it was demonstrated that TRPM7 targeting Rap1b signaling to suppress glioma cells’ proliferation and invasion by upregulating miR-28-5p expression.371
It is widely accepted that Ca2+ entry into the mitochondria is mediated by the activity of the mitochondrial calcium uniporter (MCU) complex, composed of the pore-forming subunit of the MCU channel together with several regulatory proteins. Abnormal changes in the expression of one or more members of the MCU complex have been associated with cancer-related phenotypes in HCC, breast cancer, colon cancer and pancreatic cancer.373 Oncogenic miR-25 is highly expressed in prostate and colon cancer. miR-25 induces the downregulation of MCU with subsequent decreases in mitochondrial Ca2+ uptake and reductions in the apoptotic process of prostate and colon cancer. Importantly, miR-25-dependent reduction of mitochondrial Ca2+ can be rescued by miR-25 inhibitor.374 In ovarian cancer, miR-195 contributes to regulating mitochondrial Ca2+ uptake in response to cytosolic Ca2+ concentration by repressing the mitochondrial calcium uptake 1 protein (MICU1).375 Therefore, miRNAs play crucial roles in modulating intracellular Ca2+ signals in different cancer stages and types. Overall, the interplay between miRNAs and Ca2+ signaling in tumor microenvironments will offer novel therapeutic targets for the progress of targeted metastasis.
5.5 Molecular mechanism of the crosstalk between miRNAs and YAP
During metastasis, the disseminating cancer cells experience alterations in the microenvironment of cell–cell and cell-matrix stiffness.376 These different mechanical cues can be remembered by cells for long- or short-term periods, influencing the tumor cell phenotype in cancer progression.377 The Hippo pathway regulates cell proliferation, apoptosis, and stemness in response to a wide range of extracellular and intracellular signals.378 YAP/TAZ have been investigated in cancer and stem cells as mechanosensors in response to mechanical stimulation.379 Metastatic tumor cells retain their “mechanical memory” to acclimate to a new surface with a different stiffness during migration. The tumor cells containing YAP translocation-dependent mechanical memory would lose the memory when YAP is depleted. Without YAP, cells migrate through the soft surface in the same way as through the stiff substrate. However, the roles of miRNAs in mechano-memory are poorly understood. Specifically, there exists a knowledge gap between miRNAs and Hippo-YAP/TAZ pathways in human malignancies.Dysregulation of the Hippo-YAP signaling pathway underlies various solid tumors, and misregulation of miRNAs is a common feature in human cancers. Recent advances show that the Hippo-YAP signaling pathway affects the miRNA biogenesis by regulating the Microprocessor-interacting protein p72 and Dicer expression in a cell-density-dependent manner. At higher cell density, YAP translocates from the nucleus into the cytoplasm, thereby allowing p72 to bind to the Microprocessor in the nucleus and leading to efficient miRNA biogenesis.380 In contrast, at low cell densities, YAP stays in the nucleus and is activated, thereby sequestering p72 from the Microprocessor and disrupting the miRNA biogenesis.380 Cell-density induced translocation of nuclear YAP/TAZ represses the Dicer levels.381 When nuclear YAP/TAZ are lost, levels of LIN28, a regulator of let7-a/b, is reduced. Lower LIN28 leads to let-7a and let-7b miRNAs accumulation, which down-regulates Dicer, resulting in decreased processing of pre-miRNA to mature miRNA (miR-23a, miR-22, miR-221, miR-24 and miR-21). Consistently, inhibition of let-7 rescues the miRNA biogenesis defects observed following YAP/TAZ knockdown (Fig. 5B and Table 3).381
The miR-130 family members, miR-130a and miR-130b, are located in chromosomes 11 and 22, respectively. Both miR-130a and miR-130b can mediate Hippo-YAP signaling in different cancers. Aberrant expression of miR-130a is observed in several types of cancer.382–385 miR-130a is significantly down-regulated in HCC.384 Conversely, miR-130a promotes YAP-induced liver tumorigenesis and liver enlargement in mice.386 miR-130a can be induced as a direct target of the TEAD transcription complex, and the loss of endogenous YAP/TAZ substantially represses the pri- and mature miR-130a level. Also, miR-130a could effectively target VGLL4, an inhibitor of YAP. Therefore, aberrant YAP activation alone is enough to lead to liver tumorigenesis in a normal tissue microenvironment. The inhibition of miR-130a reversed liver size enlargement induced by Hippo pathway inactivation and blocked YAP-induced tumorigenesis.386
miR-130b, another member of the miR-130 family, induces the glioblastoma cancer stem cell phenotype through the regulation of the YAP/TAZ signaling pathway.387 In the Hippo pathway, YAP/TAZ are phosphorylated and activated by kinase MST1/2 and LATS1/2 in mammals.388 In addition, MST1/2 can bind to and phosphorylate the adaptor protein SAV1 and form MST1/2-SAV1 interaction to phosphorylate LATS1/2.389 miR-130b is overexpressed in human glioblastoma and directly targets the MST1 and SAV1, resulting in the inactivation of the Hippo signaling pathway.387 Hence, understanding the role of miR-130b in glioblastoma pathogenesis may shed light on novel therapeutic strategies.
miR-21 is overexpressed in most tumor types and acts as an oncogene by targeting many tumor suppressor genes related to proliferation, apoptosis, and invasion.390,391 It has been demonstrated that miR-21 functions as a long-term mechanical memory keeper against different environmental mechanics, while YAP/TAZ primarily respond to acute changes of substrate mechanical cues in MSCs’ migration.392 In addition to MSCs, pancreatic cancer cells also commonly migrate through tissues of different stiffnesses during metastasis. Liver is the major metastatic site of pancreatic cancer. Metastatic niche in a softer environment presents a higher intrinsic resistance to gemcitabine monotherapy, a standard first-line treatment for patients with metastatic pancreatic cancer.393,394 In pancreatic cancer, YAP nuclear translocation and miR-21 expression mediate the mechanical memory in response to altered environmental stiffness.393 Meanwhile, environmental stiffness can influence the gemcitabine chemoresistance of soft-primed pancreatic cancer cells. These findings could shed light on how the regulation of miRNA expression affects tumor metastasis in patients, while miR-21 serve as a potential therapeutic target in metastatic tumor cells.
6. Conclusions and Outlook
In the past few decades, studies show that biophysical signals can regulate biochemical signaling in normal cells and cancer cells. Specifically, in response to biophysical inputs, (1) calcium-related ion channels and transporters mediate calcium signaling and interact with cytoskeletal proteins to regulate cellular function; (2) the mechanotransduction carried out by the cytoskeleton and nucleus mediate the YAP shuttling to trigger changes in the transcription of downstream genes, affecting cellular functions; (3) calcium signals trigger changes in cytoskeleton force to interact with YAP signaling; (4) the expression level of miRNAs can either change in response to ECM mechanics or directly regulate the gene expression of ECM proteins; (5) miRNAs target the gene expression of calcium-related transporters/channels to regulate calcium dynamics; and (6) modulated by cell density, the interactions between nuclear YAP and Microprocessors regulate the maturation of diverse miRNAs, mediating cell behaviors.Despite the significant advances in the understanding of mechanotransduction related to Ca2+, YAP, and miRNAs, several important questions are unanswered and being actively studied: (1) What are the direct mechano-sensor and molecular mechanism responsible for biophysical-signal-induced calcium and YAP signaling? (2) How does miRNA mediate mechanotransduction? (3) miRNA degradation can be induced by some target RNAs through a pathway called target-directed miRNA degradation (TDMD). Does mechanotransduction influence the target RNA expression levels and induce miRNA degradation through TDMD?
Most importantly, further mechanobiological studies, leveraging the in vivo imaging,395–399 CRISPR/Cas9 genome editing,400–402 and data science,29,403–405 could facilitate elucidating the roles and mechanisms of Ca2+/YAP/miRNA within mechanotranduction in vivo and empower the development of mechano-medicine for combinatorial cancer therapeutics.
7. Terminology of mechanics
Force (or load, F): physical interaction between two objects. It causes an object with mass (m) to accelerate (a), obeying the Newton's 2nd law F = ma.Stress (σ or τ): the internal force (F) per unit area (A) in continuum medium. It includes normal stress (σ) and shear stress (τ), σ or τ = F/A. For normal stress (tension and compression), the direction of force is perpendicular to the surface of area. For shear stress, the direction of force is parallel to the surface of area.
Strain (ε or γ): the change in length (Δl) of an object with respect to the initial length (l). It includes normal strain (ε) and shear strain (γ), ε or γ = Δl/l. For normal strain, the directions of Δl (elongation is positive and shortening is negative) and l are in parallel. For shear strain, the directions of Δl and l are perpendicular to each other. No rigid body rotation is included.
Tension (T): outward force (F) in the direction normal to the surface per unit area (A) that causes a positive normal stress and strain, T = F/A.
Compression (or pressure, P): inward force (F) in the direction normal to the surface per unit area (A) that causes a negative normal stress and strain, P = F/A.
Stiffness (k): the elastic resistance offered by an object to deformation (Δl) under an applied force (F), k = F/Δl.
Elastic modulus: a measure of the stiffness of an elastic object under an applied stress, defined as the slope of its stress–strain curve in the elastic deformation region. It includes Young's modulus (E) and Shear modulus (G), E = 2G(1 + v), where v is the Poisson's ratio.
Young's modulus (E): the ratio of tensile/compressive stress (σ) to normal strain (ε) in the linear elastic region of a material, E = σ/ε.
Shear modulus (G): the ratio of shear stress (τ) to shear strain (γ) of an elastic material, G = τ/γ.
Cell traction (or traction force): the force per unit area exerted by the cell on substrates.
Cell contractility: the capability of a cell to contract the microenvironment. It can be evaluated by traction.
Adhesion: the molecular attraction force in the area of contact between dissimilar particles or objects that tend to cling to each other.
Porosity (φ): the fraction of the void volume (VV) over the total volume (VT) in a material, φ = VV/VT.
Nomenclature
| ARP2/3 | Actin-related protein 2/3 |
| ATP | Adenosine-5′-triphosphate |
| ABC | ATP-binding cassette |
| AA | Arachidonic acid |
| AGO | Argonaute |
| ACI | Atherosclerotic cerebral infarction |
| AFM | Atomic force microscopy |
| BFGF | Basic fibroblast growth factor |
| BCL2 | B-cell lymphoma 2 |
| BRCA1 | Breast and ovarian cancer susceptibility protein 1 |
| Ca2+ | Calcium ion |
| CW | Ca2+ wave |
| CaMKII | Calcium/calmodulin-dependent protein kinase II |
| CICR | Calcium-induced calcium release |
| CalA | Calyculin A |
| CAF | Cancer-associated fibroblast |
| CLL | Chronic lymphocytic leukemia |
| CDS | Coding sequences |
| CRC | Colorectal cancer |
| Cx | Connexin |
| CXCR4 | C–X–C Motif Chemokine Receptor 4 |
| CDK4/6 | Cyclin-dependent kinase 4/6 |
| CytD | Cytochalasin D |
| [Ca2+]cyt | Cytoplasmic Ca2+ concentration |
| CAM | Cytoskeleton-actin-matrix |
| DRM | Detergent-resistant membrane |
| DGCR8 | Digeorge critical region 8 |
| ER | Endoplasmic reticulum |
| EGF | Epidermal growth factor |
| EPC | Epidermal stem/progenitor cell |
| EMT | Epithelial–mesenchymal transition |
| XPO | Exportin |
| ECM | Extracellular matrix |
| ERK | Extracellular-signal-regulated kinase |
| Fn | Fibronectin |
| FNDC3A | Fibronectin type III domain containing 3A |
| F-actin | Filamentous-actin |
| FAK | Focal adhesion kinase |
| G-actin | Globular-actin |
| GPCR | G-protein-coupled receptor |
| HCC | Hepatocellular carcinoma |
| HOXA9 | Homeobox A9 |
| HMSC | Human mesenchymal stem cell |
| HUVEC | Human umbilical vascular endothelial cell |
| OH | Hydroxyl group |
| IP3 | Inositol trisphosphate |
| IP3R | Inositol trisphosphate receptor |
| ILK | Integrin-linked kinase |
| IF | Intermediate filament |
| IEC | Intestinal epithelial cell |
| KRAP | KRAS-induced actin-interacting protein |
| LATS1/2 | Large tumor suppressor 1/2 |
| LINC | Linker of nucleoskeleton and cytoskeleton |
| LIMK | Lin11, Isl-1 and Mec-3 kinase |
| MST1/2 | Mammalian Ste20-like kinases 1/2 |
| MMP-9 | Matrix metallopeptidase 9 |
| MFC | Meniscus fibrochondrocyte |
| MSC | Mesenchymal stem cell |
| mRNA | Messenger RNA |
| miRNA | MicroRNA |
| mechanomiR | Mechanosensitive miRNA |
| MT | Microtubule |
| MCU | Mitochondrial calcium uniporter |
| MICU1 | Mitochondrial calcium uptake 1 protein |
| MAPK | Mitogen-activated protein kinase |
| MEF | Mouse embryonic fibroblast |
| Mtb | Mycobacterium tuberculosis |
| NEC | Normal endothelial cell |
| NE | Nuclear envelope |
| nt | Nucleotide |
| PTEN | Phosphatase and tensin homolog |
| P | Phosphate |
| PI3K | Phosphoinositide 3-kinase |
| PLC | Phospholipase C |
| PAA | Polyacrylamide |
| PARN | Poly-A specific ribonuclease |
| PDMS | Polydimethylsiloxane |
| Pol II | Polymerase II |
| PTM | Post-translational modification |
| pre-miRNA | Precursor miRNA |
| pri-miRNA | Primary miRNA |
| AKT | Protein kinase B |
| RhoA | Ras homolog family member A |
| ROCK | Rho-associated protein kinase |
| RTK | Receptor tyrosine kinase |
| RISC | RNA-induced silencing complex |
| RUNX | Runt-related transcription factor |
| RyR | Ryanodine receptor |
| SUN | Sad1p-UNC-84 |
| SERCA | Sarco/endoplasmic reticulum Ca2+ ATPase |
| SPINK1 | Serine peptidase inhibitor, Kazal type-1 |
| SIRT1 | Sirtuin 1 |
| SFK | Src family kinase |
| SOCE | Store-operated Ca2+ entry |
| STIM | Stromal interaction molecule |
| TDMD | Target-directed miRNA degradation |
| TG | Thapsigargin |
| TAZ | Transcriptional co-activator with PDZ-binding motif |
| TSS | Transcription start site |
| TRP | Transient receptor potential |
| TRPA | Transient receptor potential ankyrin |
| TRPC | Transient receptor potential canonical |
| TRPM | Transient receptor potential melastatin |
| TRPV | Transient receptor potential vanilloid |
| TPM | Tropomyosin |
| TEC | Tumor-derived endothelial cell |
| BTEC | TEC from human breast carcinomas |
| TNF | Tumor necrosis factor |
| TRAF6 | TNF receptor associated factor 6 |
| TRAIL | TNF-related apoptosis-inducing ligand |
| USP28 | Ubiquitin-specific protease 28 |
| UTR | Untranslated region |
| UCEC | Uterine corpus endometrial carcinoma |
| VEGF | Vascular endothelial growth factor |
| VGCC | Voltage-gated calcium channel |
| YAP | Yes-associated protein |
| TEAD | YAP-TEA domain |
| 2-APB | 2-Aminoethoxydiphenyl borate |
| m7G | 7-methylguanosine |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We sincerely apologize for not being able to cite all relevant works due to space constraints. This project is funded by UF Gatorade Award Start-up Package (X. T.) and Cancer Pilot Award from UFHCC (X. T. and D. S.). We genuinely appreciate the invaluable discussions with Dr Jonathan Licht (UFHCC), Dr Michael Sheetz (The University of Texas Medical Branch at Galveston), Dr Bo Zeng (Southwest Medical University, China), Dr Youhua Tan (Hong Kong Polytechnic University, China), Dr Marin Schwartz (Yale University), and Dr Sanjay Kumar (University of California at Berkley). We are grateful for the effective supports from all members of Tang's, Xie's, and Siemann's laboratories and all staff members of the MAE&UFHCC, UF.Notes and references
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram and A. Jemal, et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA Cancer J. Clin., 2021, 71(3), 209–249 CrossRef PubMed.
- American Cancer Society, Cancer Facts & Figures, American Cancer Society, Atlanta, 2021, https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html Search PubMed.
- M. Tanaka, S. S. Dykes and D. W. Siemann, Inhibition of the Axl pathway impairs breast and prostate cancer metastasis to the bones and bone remodeling, Clin. Exp. Metastasis, 2021, 38(3), 321–335 CrossRef CAS PubMed.
- D. Hanahan and R. Weinberg, Hallmarks of Cancer: The Next Generation, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- C. L. Chaffer and R. A. Weinberg, A perspective on cancer cell metastasis, Science, 2011, 331, 1559–1564 CrossRef CAS PubMed.
- A. Chambers, A. Groom and I. MacDonald, Metastasis: dissemination and growth of cancer cells in metastatic sites, Nat. Rev. Cancer, 2002, 2, 563–572 CrossRef CAS PubMed.
- H. T. Nia, L. L. Munn and R. K. Jain, Physical traits of cancer, Science, 2020,(6516), 370 Search PubMed.
- A. S. G. van Oosten, X. Chen, L. K. Chin, K. Cruz, A. E. Patteson and K. Pogoda, et al., Emergence of tissue-like mechanics from fibrous networks confined by close-packed cells, Nature, 2019, 573(7772), 96–101 CrossRef CAS PubMed.
- X. Tang, T. Kuhlenschmidt, J. Zhou, P. Bell, F. Wang and M. Kuhlenschmidt, et al., Mechanical force affects expression of an in vitro metastasis-like phenotype in HCT-8 cells, Biophys. J., 2010, 99(8), 2460–2469 CrossRef CAS PubMed.
- M. J. Paszek, N. Zahir, K. R. Johnson, J. N. Lakins, G. I. Rozenberg and A. Gefen, et al., Tensional homeostasis and the malignant phenotype, Cancer Cell, 2005, 8(3), 241–254 CrossRef CAS PubMed.
- A. Sontheimer-Phelps, B. A. Hassell and D. E. Ingber, Modelling cancer in microfluidic human organs-on-chips, Nat. Rev. Cancer, 2019, 19, 65–81 CrossRef CAS PubMed.
- B. A. Hassell, G. Goyal, E. Lee, A. Sontheimer-Phelps, O. Levy and C. S. Chen, et al., Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro, Cell Rep., 2017, 21(2), 508–516 CrossRef CAS PubMed.
- C. R. Pfeifer, C. M. Alvey, J. Irianto and D. E. Discher, Genome variation across cancers scales with tissue stiffness–An invasion-mutation mechanism and implications for immune cell infiltration, Curr. Opin. Syst. Biol., 2017, 2, 103–114 CrossRef PubMed.
- L. Chin, Y. Xia, D. Discher and P. Janmey, Mechanotransduction in cancer, Curr. Opin. Chem. Eng., 2016, 11, 77–84 CrossRef PubMed.
- X. Tang, T. B. Kuhlenschmidt, Q. Li, S. Ali, S. Lezmi and H. Chen, et al., A mechanically-induced colon cancer cell population shows increased metastatic potential, Mol. Cancer, 2014, 13(1), 1–15 CrossRef PubMed.
- Y. Tan, A. Tajik, J. Chen, Q. Jia, F. Chowdhury and L. Wang, et al., Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression, Nat. Commun., 2014, 5(1), 1–12 Search PubMed.
- M. W. Pickup, J. K. Mouw and V. M. Weaver, The extracellular matrix modulates the hallmarks of cancer, EMBO Rep., 2014, 15(12), 1243–1253 CrossRef CAS PubMed.
- J. Liu, Y. Tan, H. Zhang, Y. Zhang, P. Xu and J. Chen, et al., Soft fibrin gels promote selection and growth of tumorigenic cells, Nat. Mater., 2012, 11(8), 734–741 CrossRef CAS PubMed.
- N. Wang, Review of cellular mechanotransduction, J. Phys. D: Appl. Phys., 2017, 50, 233002 CrossRef PubMed.
- C. DuFort, M. Paszek and V. Weaver, Balancing forces: architectural control of mechanotransduction, Nat. Rev. Mol. Cell Biol., 2011, 12, 308–319 CrossRef CAS PubMed.
- C. P. Johnson, H. Y. Tang, C. Carag, D. W. Speicher and D. E. Discher, Forced unfolding of proteins within cells, Science, 2007, 317(5838), 663–666 CrossRef CAS PubMed.
- K. Amar, F. Wei, J. Chen and N. Wang, Effects of forces on chromatin, APL Bioeng., 2021, 5, 041503 CrossRef CAS PubMed.
- S. Nemec and K. A. Kilian, Materials control of the epigenetics underlying cell plasticity, Nat. Rev. Mater., 2021, 6, 69–83 CrossRef CAS.
- V. D. Tran and S. Kumar, Transduction of cell and matrix geometric cues by the actin cytoskeleton, Curr. Opin. Cell Biol., 2021, 68, 64–71 CrossRef CAS PubMed.
- P. A. Janmey, D. A. Fletcher, C. A. Reinhart-king, P. A. Janmey, D. A. Fletcher and C. A. Reinhart-king, Stiffness Sensing by Cells, Physiol. Rev., 2020, 100(2), 695–724 CrossRef PubMed.
- O. Chaudhuri, J. Cooper-White, P. A. Janmey, D. J. Mooney and V. B. Shenoy, Effects of extracellular matrix viscoelasticity on cellular behaviour, Nature, 2020, 584, 535–546 CrossRef CAS PubMed.
- H. H. Yu and J. A. Zallen, Abl and Canoe/Afadin mediate mechanotransduction at tricellular junctions, Science, 2020, 370(6520), eaba5528 CrossRef PubMed.
- T. P. Lele, R. B. Dickinson and G. G. Gundersen, Mechanical principles of nuclear shaping and positioning, J. Cell Biol., 2018, 217, 3330–3342 CrossRef CAS PubMed.
- D. E. Discher, L. Smith, S. Cho, M. Colasurdo, A. J. Garciá and S. Safran, Matrix Mechanosensing: From Scaling Concepts in’Omics Data to Mechanisms in the Nucleus, Regeneration, and Cancer, Annu. Rev. Biophys., 2017, 46, 295–315 CrossRef CAS PubMed.
- A. Singh, A. Tijore, F. Margadant, C. Simpson, D. Chitkara and B. C. Low, et al., Enhanced tumor cell killing by ultrasound after microtubule depolymerization, Bioeng Trans. Med., 2021, 6(3), e10233 CAS.
- F. Chowdhury, B. Huang and N. Wang, Cytoskeletal prestress: the cellular hallmark in mechanobiology and mechanomedicine, Cytoskeleton, 2021, 78, 249–276 CrossRef PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- B. Abar, A. Alonso-Calleja, A. Kelly, C. Kelly, K. Gall and J. L. West, 3D printing of high-strength, porous, elastomeric structures to promote tissue integration of implants, J. Biomed. Mater. Res., Part A, 2021, 109(1), 54–63 CrossRef CAS PubMed.
- A. Herland, B. M. Maoz, D. Das, M. R. Somayaji, R. Prantil-Baun and R. Novak, et al., Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips, Nat. Biomed. Eng., 2020, 4(4), 421–436 CrossRef CAS PubMed.
- R. Lanza, R. Langer, J. Vacanti and A. Atala, Principles of Tissue Engineering. Vol. 53, Elsevier Inc. 2020 Search PubMed.
- J. Guck, Some thoughts on the future of cell mechanics, Biophys. Rev., 2019, 11, 667–670 CrossRef PubMed.
- K. Naruse, Mechanomedicine, Biophys. Rev., 2018, 10(5), 1257–1262 CrossRef PubMed.
- J. M. Northcott, I. S. Dean, J. K. Mouw and V. M. Weaver, Feeling Stress: The Mechanics of Cancer Progression and Aggression, Front. Cell Dev. Biol., 2018, 6, 17 CrossRef PubMed.
- J. Ma, Y. Zhang, K. Tang, H. Zhang, X. Yin and Y. Li, et al., Reversing drug resistance of soft tumor-repopulating cells by tumor cell-derived chemotherapeutic microparticles, Cell Res., 2016, 26(6), 713–727 CrossRef CAS PubMed.
- S. W. Wong, S. Lenzini and J. W. Shin, Perspective: biophysical regulation of cancerous and normal blood cell lineages in hematopoietic malignancies, APL Bioeng., 2018, 2, 031802 CrossRef PubMed.
- K. H. Vining and D. J. Mooney, Mechanical forces direct stem cell behaviour in development and regeneration, Nat. Rev. Mol. Cell Biol., 2017, 18, 728–742 CrossRef CAS PubMed.
- B. Martinac, Mechanosensitive ion channels: molecules of mechanotransduction, J. Cell Sci., 2004, 117(Pt 12), 2449–2460 CrossRef CAS PubMed.
- D. E. Ingber, Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology, Circ. Res., 2002, 91, 877–887 CrossRef CAS PubMed.
- W. J. Polacheck and C. S. Chen, Measuring cell-generated forces: a guide to the available tools, Nat. Methods, 2016, 13, 415–423 CrossRef CAS PubMed.
- M. Sheetz, A tale of two states: normal and transformed, with and without rigidity sensing, Annu. Rev. Cell Dev. Biol., 2019, 35, 169–190 CrossRef CAS PubMed.
- T. J. Kirby and J. Lammerding, Emerging views of the nucleus as a cellular mechanosensor, Nat. Cell Biol., 2018, 20, 373–381 CrossRef CAS PubMed.
- S. Murthy, A. Dubin and A. Patapoutian, Piezos thrive under pressure: mechanically activated ion channels in health and disease, Nat. Rev. Mol. Cell Biol., 2017, 18, 771–783 CrossRef CAS PubMed.
- R. Janoštiak, A. C. Pataki, J. Brábek and D. Rösel, Mechanosensors in integrin signaling: the emerging role of p130Cas, Eur. J. Cell Biol., 2014, 93, 445–454 CrossRef PubMed.
- S. Chien, Mechanotransduction and endothelial cell homeostasis: the wisdom of the cell, Am. J. Physiol.: Heart Circ. Physiol., 2007, H1209–H1224 CrossRef CAS PubMed.
- F. Martino, A. R. Perestrelo, V. Vinarský, S. Pagliari and G. Forte, Cellular mechanotransduction: from tension to function, Front. Physiol., 2018, 9, 824 CrossRef PubMed.
- Z. Shi, Z. T. Graber, T. Baumgart, H. A. Stone and A. E. Cohen, Cell Membranes Resist Flow, Cell, 2018, 175(7), 1769–1779 CrossRef CAS PubMed.
- P. A. Janmey and R. T. Miller, Mechanisms of mechanical signaling in development and disease, J. Cell Sci., 2011, 124, 9–18 CrossRef CAS PubMed.
- N. Wang, J. D. Tytell and D. E. Ingber, Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus, Nat. Rev. Cell Biol., 2009, 10(1), 75–82 CrossRef CAS PubMed.
- I. Muhamed, F. Chowdhury and V. Maruthamuthu, Biophysical tools to study cellular mechanotransduction, Bioengineering., 2017, 4(1), 12 CrossRef PubMed.
- T. Marin, B. Gongol, Z. Chen, B. Woo, S. Subramaniam and S. Chien, et al., Mechanosensitive microRNAs - Role in endothelial responses to shear stress and redox state, Free Radical Biol. Med., 2013, 64, 61–68 CrossRef CAS PubMed.
- W. Sun, Li YS Julie, H Da Huang, J. Y. J. Shyy and S. Chien, MicroRNA: a master regulator of cellular processes for bioengineering systems, Annu. Rev. Biomed. Eng., 2010, 12, 1–27 CrossRef CAS PubMed.
- D. E. Jaalouk and J. Lammerding, Mechanotransduction gone awry, Nat. Rev. Mol. Cell Biol., 2009, 10, 63–73 CrossRef CAS PubMed.
- M. A. Wozniak and C. S. Chen, Mechanotransduction in development: a growing role for contractility, Nat. Rev. Mol. Cell Biol., 2009, 10, 34–43 CrossRef CAS PubMed.
- J. Jin, K. Tang, Y. Xin, T. Zhang and Y. Tan, Hemodynamic shear flow regulates biophysical characteristics and functions of circulating breast tumor cells reminiscent of brain metastasis, Soft Matter, 2018, 14(47), 9528–9533 RSC.
- J. Liu, Y. Tan, H. Zhang, Y. Zhang, P. Xu and J. Chen, et al., Soft fibrin gels promote selection and growth of tumorigenic cells, Nat. Mater., 2012, 11(8), 734–741 CrossRef CAS PubMed.
- Y. Wei and J. L.-S. Au, Role of tumour microenvironment in chemoresistance. Integration/Interaction of Oncologic Growth. Springer, 2005. pp. 285–321 Search PubMed.
- Y. Xin, K. Li, M. Yang and Y. Tan, Fluid shear stress induces emt of circulating tumor cells via jnk signaling in favor of their survival during hematogenous dissemination, Int. J. Mol. Sci., 2020, 21(21), 1–16 Search PubMed.
- J. Riegler, Y. Labyed, S. Rosenzweig, V. Javinal, A. Castiglioni and C. X. Dominguez, et al., Tumor elastography and its association with collagen and the tumor microenvironment, Clin. Cancer Res., 2018, 24(18), 4455–4467 CrossRef CAS PubMed.
- T. Stylianopoulos, J. D. Martin, V. P. Chauhan, S. R. Jain, B. Diop-Frimpong and N. Bardeesy, et al., Causes, consequences, and remedies for growth-induced solid stress in murine and human tumors, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(38), 15101–15108 CrossRef CAS PubMed.
- S. J. Lunt, A. Fyles, R. P. Hill and M. Milosevic, Interstitial fluid pressure in tumors: therapeutic barrier and biomarker of angiogenesis, Future Oncol., 2008, 4(6), 793–802 CrossRef PubMed.
- M. Lekka, K. Pogoda, J. Gostek, O. Klymenko, S. Prauzner-Bechcicki and J. Wiltowska-Zuber, et al., Cancer cell recognition - Mechanical phenotype, Micron, 2012, 43(12), 1259–1266, DOI:10.1016/j.micron.2012.01.019.
- S. E. Cross, Y.-S. Jin, J. Rao and J. K. Gimzewski, Nanomechanical analysis of cells from cancer patients, Nat. Nanotechnol., 2007, 2(12), 780–783 CrossRef CAS PubMed.
- D. B. Agus, J. F. Alexander, W. Arap, S. Ashili, J. E. Aslan and R. H. Austin, et al., A physical sciences network characterization of non-tumorigenic and metastatic cells, Sci. Rep., 2013, 3, 1449 CrossRef CAS PubMed.
- C. M. Kraning-Rush, J. P. Califano and C. A. Reinhart-King, Cellular traction stresses increase with increasing metastatic potential, PLoS One, 2012, 7(2), e32572 CrossRef CAS PubMed.
- C. Alibert, B. Goud and J.-B. Manneville, Are cancer cells really softer than normal cells?, Biol. Cell., 2017, 109(5), 167–189 CrossRef PubMed.
- K. A. Paschos, D. Canovas and N. C. Bird, The role of cell adhesion molecules in the progression of colorectal cancer and the development of liver metastasis, Cell Signalling, 2009, 21(5), 665–674 CrossRef CAS PubMed.
- U. Cavallaro and G. Christofori, Cell adhesion in tumor invasion and metastasis: loss of the glue is not enough, Biochim. Biophys. Acta, 2001, 1552(1), 39–45 CAS.
- M. C. Lampi and C. A. Reinhart-King, Targeting extracellular matrix stiffness to attenuate disease: from molecular mechanisms to clinical trials, Sci. Transl. Med., 2018, 10(422), eaao0475 CrossRef PubMed.
- H. Ren, X. Zhao, W. Li, J. Hussain, G. Qi and S. Liu, Calcium Signaling in Plant Programmed Cell Death, Cells., 2021, 10(5), 1089 CrossRef CAS PubMed.
- A. F. T. Arnsten, D. Datta and M. Wang, The genie in the bottle-magnified calcium signaling in dorsolateral prefrontal cortex, Mol. Psychiatry, 2021, 26(8), 3684–3700 CrossRef CAS PubMed.
- R. Peruzzo, R. Costa, M. Bachmann, L. Leanza and I. Szabò, Mitochondrial metabolism, contact sites and cellular calcium signaling: implications for tumorigenesis, Cancers, 2020, 12(9), 2574 CrossRef CAS PubMed.
- D. E. Schäffer, L. M. Iyer, A. M. Burroughs and L. Aravind, Functional Innovation in the Evolution of the Calcium-Dependent System of the Eukaryotic Endoplasmic Reticulum, Front. Genet., 2020, 11, 34 CrossRef PubMed.
- M. Trebak and J. P. Kinet, Calcium signalling in T cells, Nat. Rev. Immunol., 2019, 19, 154–169 CrossRef CAS PubMed.
- M. M. Xu, A. Seas, M. Kiyani, K. S. Y. Ji and H. N. Bell, A temporal examination of calcium signaling in cancer- from tumorigenesis, to immune evasion, and metastasis, Cell Biosci., 2018, 8(1), 1–9, DOI:10.1186/s13578-018-0223-5.
- M. Bittremieux, J. B. Parys, P. Pinton and G. Bultynck, ER functions of oncogenes and tumor suppressors: modulators of intracellular Ca2+ signaling, Biochim. Biophys. Acta, Mol. Cell Res., 2016, 1863, 1364–1378 CrossRef CAS PubMed.
- N. Prevarskaya, R. Skryma and Y. Shuba, Calcium in tumour metastasis: new roles for known actors, Nat. Rev. Cancer, 2011, 11(8), 609–618 CrossRef CAS PubMed.
- M. J. Berridge, P. Lipp and M. D. Bootman, The versatility and universality of calcium signalling, Nat. Rev. Mol. Cell Biol., 2000, 1, 11–21 CrossRef CAS PubMed.
- A. Tijore, M. Yao, Y. H. Wang, A. Hariharan, Y. Nematbakhsh and B. Lee Doss, et al., Selective killing of transformed cells by mechanical stretch, Biomaterials, 2021, 275 Search PubMed.
- Y. Han, C. Liu, D. Zhang, H. Men, L. Huo and Q. Geng, et al., Mechanosensitive ion channel Piezo1 promotes prostate cancer development through the activation of the Akt/mTOR pathway and acceleration of cell cycle, Int. J. Oncol., 2019, 55(3), 629–644 CAS.
- J. M. Hope, M. Lopez-Cavestany, W. Wang, C. A. Reinhart-King and M. R. King, Activation of Piezo1 sensitizes cells to TRAIL-mediated apoptosis through mitochondrial outer membrane permeability, Cell Death Dis., 2019, 10(11), 836–837 CrossRef PubMed.
- J. Xu, J. Mathur, E. Vessières, S. Hammack, K. Nonomura and J. Favre, et al., GPR68 Senses Flow and Is Essential for Vascular Physiology, Cell, 2018, 173(3), 762–775 CrossRef CAS PubMed.
- A. J. Lomakin, C. J. Cattin, D. Cuvelier, Z. Alraies, M. Molina and G. P. F. Nader, et al., The nucleus acts as a ruler tailoring cell responses to spatial constraints, Science, 2020,(6514), 370 Search PubMed.
- M. Luo, K. K. Y. Ho, Z. Tong, L. Deng and A. P. Liu, Compressive Stress Enhances Invasive Phenotype of Cancer Cells via Piezo1 Activation, bioRxiv, 2019, 513218 Search PubMed.
- K. Zhang, H. X. Qi, Z. M. Hu, Y. N. Chang, Z. M. Shi and X. H. Han, et al., YAP and TAZ Take Center Stage in Cancer, Biochemistry, 2015, 54(43), 6555–6566 CrossRef CAS PubMed.
- S. Piccolo, S. Dupont and M. Cordenonsi, The biology of YAP/TAZ: hippo signaling and beyond, Physiol. Rev., 2014, 94(4), 1287–1312 CrossRef CAS PubMed.
- B. J. Thompson, YAP/TAZ: Drivers of Tumor Growth, Metastasis, and Resistance to Therapy, BioEssays, 2020, 42, 1900162 CrossRef PubMed.
- F. Zanconato, M. Cordenonsi and S. Piccolo, YAP and TAZ: a signalling hub of the tumour microenvironment, Nat. Rev. Cancer, 2019, 19, 454–464 CrossRef CAS PubMed.
- X. Zhang, H. Zhao, Y. Li, D. Xia, L. Yang and Y. Ma, et al., The role of YAP/TAZ activity in cancer metabolic reprogramming, Molecular Cancer, 2018, 17, 134 CrossRef PubMed.
- F. Zanconato, M. Cordenonsi and S. Piccolo, YAP/TAZ at the Roots of Cancer, Cancer Cell, 2016, 29(6), 783–803 CrossRef CAS PubMed.
- T. Panciera, L. Azzolin, M. Cordenonsi and S. Piccolo, Mechanobiology of YAP and TAZ in physiology and disease, Nat. Rev. Mol. Cell Biol., 2017, 18(12), 758–770, DOI:10.1038/nrm.2017.87.
- A. Elosegui-Artola, I. Andreu, A. E. M. Beedle, A. Lezamiz, M. Uroz and A. J. Kosmalska, et al., Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores, Cell, 2017, 171(6), 1397–1410.e14 CrossRef CAS PubMed.
- T. P. Driscoll, B. D. Cosgrove, S. J. Heo, Z. E. Shurden and R. L. Mauck, Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells, Biophys. J., 2015, 108(12), 2783–2793 CrossRef CAS PubMed.
- M. Aragona, T. Panciera, A. Manfrin, S. Giulitti, F. Michielin and N. Elvassore, et al., A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors, Cell, 2013, 154(5), 1047–1059 CrossRef CAS PubMed.
- Q. Luo, J. Zhang, G. Lin, M. Huang, M. Tanaka and S. Lepler, et al., Automatic Multi-functional Integration Program (AMFIP) towards All-optical Mechanobiology Interrogation, BioRxiv, 2021, 1–22, DOI:10.1101/2021.03.31.437936.
- F. X. Yu, J. Luo, J. S. Mo, G. Liu, Y. C. Kim and Z. Meng, et al., Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP, Cancer Cell, 2014, 25(6), 822–830 CrossRef CAS PubMed.
- K. Schlegelmilch, M. Mohseni, O. Kirak, J. Pruszak, J. R. Rodriguez and D. Zhou, et al., Yap1 acts downstream of α-catenin to control epidermal proliferation, Cell, 2011, 144(5), 782–795 CrossRef CAS PubMed.
- D. C. Benjamin, J. H. Kang, B. Hamza, E. M. King, J. M. Lamar and S. R. Manalis, et al., YAP enhances tumor cell dissemination by promoting intravascular motility and reentry into systemic circulation, Cancer Res., 2020, 80(18), 3867–3879 CrossRef CAS PubMed.
- T. X. Lu and M. E. Rothenberg, MicroRNA, J. Allergy Clin. Immunol., 2018, 141(4), 1202–1207 CrossRef CAS PubMed.
- M. Ha and V. N. Kim, Regulation of microRNA biogenesis, Nat. Rev. Mol. Cell Biol., 2014, 15, 509–524 CrossRef CAS PubMed.
- M. Acunzo, G. Romano, D. Wernicke and C. M. Croce, MicroRNA and cancer - A brief overview, Adv. Biol. Regulation., 2015, 57, 1–9 CrossRef CAS PubMed.
- M. D. Jansson and A. H. Lund, MicroRNA and cancer. 6, Molecular Oncology, No longer published by Elsevier, 2012. pp. 590–610 Search PubMed.
- S. J. Roberts-Thomson, S. B. Chalmers and G. R. Monteith, The calcium-signaling toolkit in cancer: remodeling and targeting, Cold Spring Harbor Perspect. Biol., 2019, 11(8), 1–20 Search PubMed.
- G. R. Monteith, N. Prevarskaya and S. J. Roberts-Thomson, The calcium-cancer signalling nexus, Nat. Rev. Cancer, 2017, 17(6), 367–380, DOI:10.1038/nrc.2017.18.
- A. Kondratskyi, M. Yassine, K. Kondratska, R. Skryma, C. Slomianny and N. Prevarskaya, Calcium-permeable ion channels in control of autophagy and cancer, Front. Physiol., 2013, 4(October), 1–12 Search PubMed.
- M. Bittremieux and G. Bultynck, p53 and Ca2+ signaling from the endoplasmic reticulum: partners in anti-cancer therapies, Oncoscience, 2015, 2(3), 233–238 CrossRef PubMed.
- E. White, Deconvoluting the context-dependent role for autophagy in cancer, Nat. Rev. Cancer, 2012, 12, 401–410 CrossRef CAS PubMed.
- Z. J. Yang, C. E. Chee, S. Huang and F. A. Sinicrope, The role of autophagy in cancer: therapeutic implications, Mol. Cancer Ther., 2011, 10, 1533–1541 CrossRef CAS PubMed.
- D. Gozuacik and A. Kimchi, Autophagy as a cell death and tumor suppressor mechanism, Oncogene, 2004, 23, 2891–2906 CrossRef CAS PubMed.
- D. O’Reilly and P. Buchanan, Calcium channels and cancer stem cells, Cell Calcium, 2019, 81, 21–28 CrossRef PubMed.
- E. Terrié, V. Coronas and B. Constantin, Role of the calcium toolkit in cancer stem cells, Cell Calcium, 2019, 80, 141–151 CrossRef PubMed.
- A. H. L. Bong and G. R. Monteith, Calcium signaling and the therapeutic targeting of cancer cells, Biochim. Biophys. Acta, Mol. Cell Res., 2018, 1865(11), 1786–1794, DOI:10.1016/j.bbamcr.2018.05.015.
- Y. Yu, X. Yang, S. Reghu, S. C. Kaul, R. Wadhwa and E. Miyako, Photothermogenetic inhibition of cancer stemness by near-infrared-light-activatable nanocomplexes, Nat. Commun., 2020, 11, 4117 CrossRef CAS PubMed.
- R. Wei, W. W. Zhu, G. Y. Yu, X. Wang, C. Gao and X. Zhou, et al., S100 calcium-binding protein A9 from tumor-associated macrophage enhances cancer stem cell-like properties of hepatocellular carcinoma, Int. J. Cancer, 2021, 148(5), 1233–1244 CrossRef CAS PubMed.
- H. Lee, J. W. Kim, D. K. Kim, D. K. Choi, S. Lee and J. H. Yu, et al., Calcium channels as novel therapeutic targets for ovarian cancer stem cells, Int. J. Mol. Sci., 2020, 21(7), 2327 CrossRef CAS PubMed.
- H. L. Roderick and S. J. Cook, Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival, Nat. Rev. Cancer, 2008, 8(5), 361–375 CrossRef CAS PubMed.
- A. Mound, L. Rodat-Despoix, S. Bougarn, H. Ouadid-Ahidouch and F. Matifat, Molecular interaction and functional coupling between type 3 inositol 1,4,5-trisphosphate receptor and BKCa channel stimulate breast cancer cell proliferation, Eur. J. Cancer, 2013, 49(17), 3738–3751 CrossRef CAS PubMed.
- H. Zhu, H. Zhang, F. Jin, M. Fang, M. Huang and C. S. Yang, et al., Elevated Orai1 expression mediates tumor-promoting intracellular Ca2+ oscillations in human esophageal squamous cell carcinoma, Oncotarget, 2014, 5(11), 3455–3471 CrossRef PubMed.
- A. Singh, M. Chagtoo, S. Tiwari, N. George, B. Chakravarti and S. Khan, et al., Inhibition of Inositol 1,4,5-Trisphosphate Receptor Induce Breast Cancer Cell Death Through Deregulated Autophagy and Cellular Bioenergetics, J. Cell. Biochem., 2017, 118(8), 2333–2346 CrossRef CAS PubMed.
- M. Flourakis, V. Lehen’kyi, B. Beck, M. Raphaël, M. Vandenberghe and F. V. Abeele, et al., Orai1 contributes to the establishment of an apoptosis-resistant phenotype in prostate cancer cells, Cell Death Dis., 2010, 1(9), e75 CrossRef CAS PubMed.
- C. Giorgi, M. Bonora, G. Sorrentino, S. Missiroli, F. Poletti and J. M. Suski, et al., P53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(6), 1779–1784 CrossRef CAS PubMed.
- C. Pierro, S. J. Cook, T. C. F. Foets, M. D. Bootman and H. L. Roderick, Oncogenic K-Ras suppresses IP3-dependent Ca2+ release through remodelling of the isoform composition of IP3Rs and ER luminal Ca2+ levels in colorectal cancer cell lines, J. Cell Sci., 2014, 127(7), 1607–1619 CAS.
- S. C. Hedgepeth, M. I. Garcia, L. E. Wagner, A. M. Rodriguez, S. V. Chintapalli and R. R. Snyder, et al., The BRCA1 tumor suppressor binds to inositol 1,4,5-trisphosphate receptors to stimulate apoptotic calcium release, J. Biol. Chem., 2015, 290(11), 7304–7313 CrossRef CAS PubMed.
- S. Derouiche, P. Mariot, M. Warnier, E. Vancauwenberghe, G. Bidaux and P. Gosset, et al., Activation of TRPA1 channel by antibacterial agent triclosan induces vegf secretion in human prostate cancer stromal cells, Cancer Prev. Res., 2017, 10(3), 177–187 CrossRef CAS PubMed.
- A. Fiorio Pla, H. L. Ong, K. T. Cheng, A. Brossa, B. Bussolati and T. Lockwich, et al., TRPV4 mediates tumor-derived endothelial cell migration via arachidonic acid-activated actin remodeling, Oncogene, 2012, 31(2), 200–212 CrossRef CAS PubMed.
- R. K. Adapala, R. J. Thoppil, K. Ghosh, H. C. Cappelli, A. C. Dudley and S. Paruchuri, et al., Activation of mechanosensitive ion channel TRPV4 normalizes tumor vasculature and improves cancer therapy, Oncogene, 2016, 35(3), 314–322 CrossRef CAS PubMed.
- D. Ribatti, R. Tamma and T. Annese, Epithelial-Mesenchymal Transition in Cancer: A Historical Overview, Translational Oncology, 2020, 13, 100773 CrossRef PubMed.
- T. Brabletz, R. Kalluri, M. A. Nieto and R. A. Weinberg, EMT in cancer, Nat. Rev., 2018, 18(2), 128–134 CAS.
- S. Zhu, H. Y. Zhou, S. C. Deng, S. J. Deng, C. He and X. Li, et al., Asic1 and asic3 contribute to acidity-induced emt of pancreatic cancer through activating Ca2+/RhoA pathway, Cell Death Dis., 2017, 8(5), e2806 CrossRef CAS PubMed.
- S. Zhang, Y. Miao, X. Zheng, Y. Gong, J. Zhang and F. Zou, et al., STIM1 and STIM2 differently regulate endogenous Ca2+ entry and promote TGF-β-induced EMT in breast cancer cells, Biochem. Biophys. Res. Commun., 2017, 488(1), 74–80 CrossRef CAS PubMed.
- F. M. Davis, I. Azimi, R. A. Faville, A. A. Peters, K. Jalink and J. W. Putney, et al., Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent, Oncogene, 2014, 33(18), 2307–2316 CrossRef CAS PubMed.
- C. L. Yankaskas, K. Bera, K. Stoletov, S. A. Serra, J. Carrillo-Garcia and S. Tuntithavornwat, et al., The fluid shear stress sensor TRPM7 regulates tumor cell intravasation, Sci. Adv., 2021, 7(28), eabh3457 CrossRef CAS PubMed.
- W. H. Lee, L. Y. Choong, N. N. Mon, S. Lu, Q. Lin and B. Pang, et al., TRPV4 regulates breast cancer cell extravasation, stiffness and actin cortex, Sci. Rep., 2016, 6, 27903 CrossRef CAS PubMed.
- A. Mound, A. Vautrin-Glabik, A. Foulon, B. Botia, F. Hague and J. B. Parys, et al., Downregulation of type 3 inositol (1,4,5)-trisphosphate receptor decreases breast cancer cell migration through an oscillatory Ca2+ signal, Oncotarget, 2017, 8(42), 72324–72341 CrossRef PubMed.
- S. S. Kang, K. S. Han, B. M. Ku, Y. K. Lee, J. Hong and H. Y. Shin, et al., Caffeine-mediated inhibition of calcium release channel inositol 1,4,5-trisphosphate receptor subtype 3 blocks glioblastoma invasion and extends survival, Cancer Res., 2010, 70(3), 1173–1183 CrossRef CAS PubMed.
- J.-B. Huang, A. L. Kindzelskii, A. J. Clark and H. R. Petty, Identification of Channels Promoting Calcium Spikes and Waves in HT1080 Tumor Cells, Cancer Res., 2004, 64, 2482–2489 CrossRef CAS PubMed.
- P. Mo and S. Yang, The store-operated calcium channels in cancer metastasis: from cell migration, invasion to metastatic colonization, Front. Biosci., 2018, 23(7), 1241–1256 CAS.
- S. Das, P. Clézardin, S. Kamel, M. Brazier and R. Mentaverri, The CaSR in Pathogenesis of Breast Cancer: A New Target for Early Stage Bone Metastases, Front. Oncology, 2020, 10, 69 CrossRef PubMed.
- T. Yoneda and T. Hiraga, Crosstalk between cancer cells and bone microenvironment in bone metastasis, Biochem. Biophys. Res. Commun., 2005, 328, 679–687 CrossRef CAS PubMed.
- U. Harjes, A source of calcium, Nat. Rev. Cancer, 2019, 19, 3 CrossRef CAS PubMed.
- H. Wang, L. Tian, J. Liu, A. Goldstein, I. Bado and W. Zhang, et al., The Osteogenic Niche Is a Calcium Reservoir of Bone Micrometastases and Confers Unexpected Therapeutic Vulnerability, Cancer Cell, 2018, 34(5), 823–839 CrossRef CAS PubMed.
- T. J. Kim, L. Lei, J. Seong, J. S. Suh, Y. K. Jang and S. H. Jung, et al., Matrix Rigidity-Dependent Regulation of Ca2+ at Plasma Membrane Microdomains by FAK Visualized by Fluorescence Resonance Energy Transfer, Adv. Sci., 2019, 6(4), 1801290 CrossRef PubMed.
- T. J. Kim, J. Seong, M. Ouyang, J. Sun, S. Lu and P. H. Jun, et al., Substrate rigidity regulates Ca2+ oscillation via RhoA pathway in stem cells, J. Cell. Physiol., 2008, 218(2), 285–293 CrossRef PubMed.
- J. Lembong, B. Sabass, B. Sun, M. E. Rogers and H. A. Stone, Mechanics regulates ATP-stimulated collective calcium response in fibroblast cells, J. R. Soc., Interface, 2015, 12, 20150140 CrossRef PubMed.
- Y. Xu, D. Huang, S. Lü, M. Long and Y. Zhang, Mechanical features of endothelium regulate cell adhesive molecule-induced calcium response in neutrophils, APL Bioeng., 2019, 3(1), 016104, DOI:10.1063/1.5045115.
- S. O. Rahaman, L. M. Grove, S. Paruchuri, B. D. Southern, S. Abraham and K. A. Niese, et al., TRPV4 mediates myofibroblast differentiation and pulmonary fibrosis in mice, J. Clin. Invest., 2014, 124(12), 5225–5238 CrossRef PubMed.
- R. G. Scheraga, S. Abraham, K. A. Niese, B. D. Southern, L. M. Grove and R. D. Hite, et al., TRPV4 Mechanosensitive Ion Channel Regulates Lipopolysaccharide-Stimulated Macrophage Phagocytosis, J. Immunol., 2016, 196(1), 428–436 CrossRef CAS PubMed.
- W. C. Wei, F. Bianchi, Y. K. Wang, M. J. Tang, H. Ye and M. D. Glitsch, Coincidence Detection of Membrane Stretch and Extracellular pH by the Proton-Sensing Receptor OGR1 (GPR68), Curr. Biol., 2018, 28(23), 3815–3823 CrossRef CAS PubMed.
- N. E. Karagas and K. Venkatachalam, Roles for the Endoplasmic Reticulum in Regulation of Neuronal Calcium Homeostasis, Cells, 2019, 8(10), 1232 CrossRef CAS PubMed.
- M. M. Nava, Y. A. Miroshnikova, L. C. Biggs, D. B. Whitefield, F. Metge and J. Boucas, et al., Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage, Cell, 2020, 181(4), 800–817 CrossRef CAS PubMed.
- W. M. Han, S. J. Heo, T. P. Driscoll, M. E. Boggs, R. L. Duncan and R. L. Mauck, et al., Impact of cellular microenvironment and mechanical perturbation on calcium signalling in meniscus fibrochondrocytes, Eur. Cells Mater., 2014, 27, 321–331 CrossRef CAS PubMed.
- T. J. Kim, J. Sun, S. Lu, Y. X. Qi and Y. Wang, Prolonged mechanical stretch initiates intracellular calcium oscillations in human mesenchymal stem cells, PLoS One, 2014, 9(10), e109378 CrossRef PubMed.
- W. S. Nishitani, T. A. Saif and Y. Wang, Calcium signaling in live cells on elastic gels under mechanical vibration at subcellular levels, PLoS One, 2011, 6(10), e26181 CrossRef CAS PubMed.
- T. J. Kim, C. Joo, J. Seong, R. Vafabakhsh, E. L. Botvinick and M. W. Berns, et al., Distinct mechanisms regulating mechanical force-induced Ca2+ signals at the plasma membrane and the ER in human MSCs, eLife, 2015, 4, e04876 CrossRef PubMed.
- S. Erdogmus, U. Storch, L. Danner, J. Becker, M. Winter and N. Ziegler, et al., Helix 8 is the essential structural motif of mechanosensitive GPCRs, Nat. Commun., 2019, 10, 5784 CrossRef CAS PubMed.
- G. Seano, H. T. Nia, K. E. Emblem, M. Datta, J. Ren and S. Krishnan, et al., Solid stress in brain tumours causes neuronal loss and neurological dysfunction and can be reversed by lithium, Nat. Biomed. Eng., 2019, 3(3), 230–245 CrossRef CAS PubMed.
- M. E. Fernandez-Sanchez, S. Barbier, J. Whitehead, G. Bealle, A. Michel and H. Latorre-Ossa, et al., Mechanical induction of the tumorigenic beta-catenin pathway by tumour growth pressure, Nature, 2015, 523(7558), 92–95 CrossRef CAS PubMed.
- V. Venturini, F. Pezzano, F. C. Castro, H. M. Häkkinen, S. Jiménez-Delgado and M. Colomer-Rosell, et al., The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior, Science, 2020,(6514), 370 Search PubMed.
- D. Zhu, L. Feng, N. Feliu, A. H. Guse and W. J. Parak, Stimulation of Local Cytosolic Calcium Release by Photothermal Heating for Studying Intra- and Intercellular Calcium Waves, Adv. Mater., 2021, 33(24), 2008261 CrossRef CAS PubMed.
- J. T. Lock, I. Parker and I. F. Smith, Communication of Ca2+ signals via tunneling membrane nanotubes is mediated by transmission of inositol trisphosphate through gap junctions, Cell Calcium, 2016, 60(4), 266–272 CrossRef CAS PubMed.
- T. Fry, J. H. Evans and M. J. Sanderson, Propagation of intercellular calcium waves in C6 glioma cells transfected with connexins 43 or 32, Microsc. Res. Tech., 2001, 52(3), 289–300 CrossRef CAS PubMed.
- G. J. Block, G. D. DiMattia and D. J. Prockop, Stanniocalcin-1 Regulates Extracellular ATP-Induced Calcium Waves in Human Epithelial Cancer Cells by Stimulating ATP Release from Bystander Cells, PLoS One, 2010, 5(4), 1–11 CrossRef PubMed.
- W. Zhang, W. T. Couldwell, H. Song, T. Takano, J. H. C. Lin and M. Nedergaard, Tamoxifen-induced enhancement of calcium signaling in glioma and MCF-7 breast cancer cells, Cancer Res., 2000, 60(19), 5395–5400 CAS.
- K. Paemeleire, P. E. M. Martin, S. L. Coleman, K. E. Fogarty, W. A. Carrington and L. Leybaert, et al., Intercellular calcium waves in HeLa cells expressing GFP-labeled connexin 43, 32, or 26, Mol. Biol. Cell, 2000, 11(5), 1815–1827 CrossRef CAS PubMed.
- L. Leybaert and M. J. Sanderson, Intercellular Ca2+ waves: mechanisms and function, Physiol. Rev., 2012, 92(3), 1359–1392 CrossRef CAS PubMed.
- E. Beamer, M. Kuchukulla, D. Boison and T. Engel, ATP and adenosine—Two players in the control of seizures and epilepsy development, Prog. Neurobiol., 2021, 204, 102105 CrossRef CAS PubMed.
- A. Taruno, ATP release channels, Int. J. Mol. Sci., 2018, 19, 808 CrossRef PubMed.
- E. F. Diezmos, P. P. Bertrand and L. Liu, Purinergic signaling in gut inflammation: the role of connexins and pannexins, Front. Neurosci., 2016, 10, 311 Search PubMed.
- S. Kojima, Y. Ohshima, H. Nakatsukasa and M. Tsukimoto, Role of ATP as a key signaling molecule mediating radiation-induced biological effects, Dose-Response, 2017, 15(1), 1559325817690638 CAS.
- R. Z. Sabirov and Y. Okada, ATP release via anion channels, Purinergic Signalling, 2005, 1, 311–328 CrossRef CAS PubMed.
- I. von Kügelgen, Molecular pharmacology of P2Y receptor subtypes, Adv. Biochem. Pharmacol., 2021, 187, 114361 CrossRef PubMed.
- G. Burnstock, Purinergic signalling: from discovery to current developments, Exp. Physiol., 2014, 99(1), 16–34 CrossRef CAS PubMed.
- R. A. North, Molecular physiology of P2X receptors, Physiol. Rev., 2002, 82, 1013–1067 CrossRef CAS PubMed.
- A. Salameh and S. Dhein, Effects of mechanical forces and stretch on intercellular gap junction coupling, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 147–156 CrossRef CAS PubMed.
- J. E. Saffitz and A. G. Kléber, Effects of Mechanical Forces and Mediators of Hypertrophy on Remodeling of Gap Junctions in the Heart, Circ. Res., 2004, 94, 585–591 CrossRef CAS PubMed.
- P. P. Cherian, B. Cheng, S. Gu, E. Sprague, L. F. Bonewald and J. X. Jiang, Effects of Mechanical Strain on the Function of Gap Junctions in Osteocytes are Mediated through the Prostaglandin EP2 Receptor, J. Biol. Chem., 2003, 278, 44 CrossRef PubMed.
- N. Mikolajewicz, A. Mohammed, M. Morris and S. V. Komarova, Mechanically stimulated ATP release from mammalian cells: systematic review and meta-analysis, J. Cell Sci., 2018, 131(22), jcs223354 CrossRef PubMed.
- G. Burnstock and G. E. Knight, Cell culture: complications due to mechanical release of ATP and activation of purinoceptors, Cell Tissue Res., 2017, 370, 1–11 CrossRef CAS PubMed.
- D. C. Genetos, C. J. Kephart, Y. Zhang, C. E. Yellowley and H. J. Donahue, Oscillating fluid flow activation of gap junction hemichannels induces ATP release from MLO-Y4 osteocytes, J. Cell. Physiol., 2007, 212(1), 207–214 CrossRef CAS PubMed.
- L. Bao, F. Sachs and G. Dahl, Connexins are mechanosensitive, Am. J. Physiol.: Cell Physiol., 2004, 287(5), C1389–C1395 CrossRef CAS PubMed.
- L. Bao, S. Locovei and G. Dahl, Pannexin membrane channels are mechanosensitive conduits for ATP, FEBS Lett., 2004, 572(1–3), 65–68 CrossRef CAS PubMed.
- K. Diem, M. Fauler, G. Fois, A. Hellmann, N. Winokurow and S. Schumacher, et al., Mechanical stretch activates piezo1 in caveolae of alveolar type I cells to trigger ATP release and paracrine stimulation of surfactant secretion from alveolar type II cells, FASEB J., 2020, 34(9), 12785–12804 CrossRef CAS PubMed.
- H. Takada, K. Furuya and M. Sokabe, Mechanosensitive ATP release from hemichannels and Ca2+ influx through TRPC6 accelerate wound closure in keratinocytes, J. Cell Sci., 2014, 127(19), 4159–4171 CAS.
- N. Takahara, S. Ito, K. Furuya, K. Naruse, H. Aso and M. Kondo, et al., Real-time imaging of ATP release induced by mechanical stretch in human airway smooth muscle cells, Am. J. Respir. Cell Mol. Biol., 2014, 51(6), 772–782 CrossRef PubMed.
- K. Woo, A. K. Dutta, V. Patel, C. Kresge and A. P. Feranchak, Fluid flow induces mechanosensitive ATP release, calcium signalling and Cl-Transport in biliary epithelial cells through a PKCζ-dependent pathway, J. Physiol., 2008, 586(11), 2779–2798 CrossRef CAS PubMed.
- F. C. Tsai, G. H. Kuo, S. W. Chang and P. J. Tsai, Ca2+ signaling in cytoskeletal reorganization, cell migration, and cancer metastasis, BioMed Res. Int., 2015, 2015, 409245 Search PubMed.
- F. Li, N. Abuarab and A. Sivaprasadarao, Reciprocal regulation of actin cytoskeleton remodelling and cell migration by Ca2+ and Zn2+: role of TRPM2 channels, J. Cell Sci., 2016, 129(10), 2016–2029 CAS.
- L. Bastatas, D. Martinez-Marin, J. Matthews, J. Hashem, Y. J. Lee and S. Sennoune, et al., AFM nano-mechanics and calcium dynamics of prostate cancer cells with distinct metastatic potential, Biochim. Biophys. Acta, Gen. Subj., 2012, 1820(7), 1111–1120 CrossRef CAS PubMed.
- Q. Wang, A. J. Symes, C. A. Kane, A. Freeman, J. Nariculam and P. Munson, et al., A novel role for Wnt/Ca2+ signaling in actin cytoskeleton remodeling and cell motility in prostate cancer, PLoS One, 2010, 5(5), e10456 CrossRef PubMed.
- L. Santella, N. Limatola, F. Vasilev and J. T. Chun, Maturation and fertilization of echinoderm eggs: role of actin cytoskeleton dynamics, Biochem. Biophys. Res. Commun., 2018, 506(2), 361–371 CrossRef CAS PubMed.
- K. Lange and J. Gartzke, F-actin-based Ca signaling - A critical comparison with the current concept of Ca signaling, J. Cell. Physiol., 2006, 209, 270–287 CrossRef CAS PubMed.
- A. H. York-Andersen, R. M. Parton, C. J. Bi, C. L. Bromley, I. Davis and T. T. Weil, A single and rapid calcium wave at egg activation in Drosophila, Biol. Open., 2015, 4(4), 553–560 CrossRef PubMed.
- K. Kyozuka, J. T. Chun, A. Puppo, G. Gragnaniello, E. Garante and L. Santella, Actin cytoskeleton modulates calcium signaling during maturation of starfish oocytes, Dev. Biol., 2008, 320(2), 426–435 CrossRef CAS PubMed.
- A. Puppo, J. T. Chun, G. Gragnaniello, E. Garante and L. Santella, Alteration of the cortical actin cytoskeleton deregulates Ca2+ signaling, monospermic fertilization, and sperm entry, PLoS One, 2008, 3(10), e3588 CrossRef CAS PubMed.
- X. Chen, S. Wanggou, A. Bodalia, M. Zhu, W. Dong and J. J. Fan, et al., A feedforward mechanism mediated by mechanosensitive ion channel PIEZO1 and tissue mechanics promotes glioma aggression, Neuron, 2018, 100(4), 799–815.e7 CrossRef CAS PubMed.
- C. Pardo-Pastor, F. Rubio-Moscardo, M. Vogel-Gonzalez, S. A. Serra, A. Afthinos and S. Mrkonjic, et al., Piezo2 channel regulates RhoA and actin cytoskeleton to promote cell mechanobiological responses, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(8), 1925–1930 CrossRef CAS PubMed.
- A. Tijore, F. Margadant, M. Yao, A. Hariharan, C. A. Z. Chew and S. Powell, et al., Ultrasound-mediated mechanical forces selectively kill tumor cells, bioRxiv, 2020 DOI:10.1101/2020.10.09.332726.
- S. A. Gudipaty, J. Lindblom, P. D. Loftus, M. J. Redd, K. Edes and C. F. Davey, et al., Mechanical stretch triggers rapid epithelial cell division through Piezo1, Nature, 2017, 543(7643), 118–121 CrossRef CAS PubMed.
- M. J. Berridge, The inositol trisphosphate/calcium signaling pathway in health and disease, Physiol. Rev., 2016, 96(4), 1261–1296 CrossRef CAS PubMed.
- D. E. Clapham, Calcium Signaling, Cell, 2007, 131(6), 1047–1058 CrossRef CAS PubMed.
- J. B. Parys, G. Bultynck and T. Vervliet, IP3 Receptor Biology and Endoplasmic Reticulum Calcium Dynamics in Cancer, Prog. Mol. Subcell. Biol., 2021, 59, 215–237 CrossRef CAS PubMed.
- M. Bittremieux, R. M. La Rovere, H. Akl, C. Martines, K. Welkenhuyzen and K. Dubron, et al., Constitutive IP 3 signaling underlies the sensitivity of B-cell cancers to the Bcl-2/IP 3 receptor disruptor BIRD-2, Cell Death Differ., 2019, 26(3), 531–547 CrossRef CAS PubMed.
- A. Vautrin-Glabik, B. Botia, P. Kischel, H. Ouadid-Ahidouch and L. Rodat-Despoix, IP3R3 silencing induced actin cytoskeletal reorganization through ARHGAP18/RhoA/mDia1/FAK pathway in breast cancer cell lines, Biochim. Biophys. Acta, Mol. Cell Res., 2018, 1865(7), 945–958 CrossRef CAS PubMed.
- T. Fujimoto, T. Machida, T. Tsunoda, K. Doi, T. Ota and M. Kuroki, et al., KRAS-induced actin-interacting protein regulates inositol 1,4,5-trisphosphate-receptor-mediated calcium release, Biochem. Biophys. Res. Commun., 2011, 408(2), 214–217 CrossRef CAS PubMed.
- D. L. Prole and C. W. Taylor, Inositol 1,4,5-trisphosphate receptors and their protein partners as signalling hubs, J. Physiology, 2016, 594, 2849–2866 CrossRef CAS PubMed.
- K. Fukatsu, H. Bannai, T. Inoue and K. Mikoshiba, Lateral diffusion of inositol 1,4,5-trisphosphate receptor type 1 in Purkinje cells is regulated by calcium and actin filaments, J. Neurochem., 2010, 114(6), 1720–1733 CrossRef CAS PubMed.
- M. R. Turvey, K. E. Fogarty and P. Thorn, Inositol (1,4,5)-trisphosphate receptor links to filamentous actin are important for generating local Ca2+ signals in pancreatic acinar cells, J. Cell Sci., 2005, 118(5), 971–980 CrossRef CAS PubMed.
- K. Fukatsu, H. Bannai, S. Zhang, H. Nakamura, T. Inoue and K. Mikoshiba, Lateral diffusion of inositol 1,4,5-trisphosphate receptor type 1 is regulated by actin filaments and 4.1 N in neuronal dendrites, J. Biol. Chem., 2004, 279(47), 48976–48982 CrossRef CAS PubMed.
- M. C. Hours and L. Mery, The N-terminal domain of the type 1 Ins(1,4,5)P3 receptor stably expressed in MDCK cells interacts with myosin IIA and alters epithelial cell morphology, J. Cell Sci., 2010, 123(9), 1449–1459 CrossRef CAS PubMed.
- D. S. Walker, S. Ly, K. C. Lockwood and H. A. Baylis, A direct interaction between IP3 receptors and myosin II regulates IP3 signaling in C. elegans, Curr. Biol., 2002, 12(11), 951–956 CrossRef CAS PubMed.
- M. M. El Refaey and P. J. Mohler, Ankyrins and spectrins in cardiovascular biology and disease, Front. Physiol., 2017, 8, 852 CrossRef PubMed.
- P. J. Mohler, J. Q. Davis, L. H. Davis, J. A. Hoffman, P. Michaely and V. Bennett, Inositol 1,4,5-Trisphosphate Receptor Localization and Stability in Neonatal Cardiomyocytes Requires Interaction with Ankyrin-B, J. Biol. Chem., 2004, 279(13), 12980–12987 CrossRef CAS PubMed.
- L. Y. W. Bourguignon and H. Jin, Identification of the ankyrin-binding domain of the mouse T-lymphoma cell inositol 1,4,5-trisphosphate (IP3) receptor and its role in the regulation of IP3-mediated internal Ca2+ release, J. Biol. Chem., 1995, 270(13), 7257–7260 CrossRef CAS PubMed.
- L. Y. W. Bourguignon, H. Jin, N. Iida, N. R. Brandt and Z. She Hui, The involvement of ankyrin in the regulation of inositol 1,4,5- trisphosphate receptor-mediated internal Ca2+ release from Ca2+ storage vesicles in mouse T-lymphoma cells, J. Biol. Chem., 1993, 268(10), 7290–7297 CrossRef CAS.
- S. K. Joseph and S. Samanta, Detergent solubility of the inositol trisphosphate receptor in rat brain membranes. Evidence for association of the receptor with ankyrin, J. Biol. Chem., 1993, 268(9), 6477–6486 CrossRef CAS.
- É. Béliveau and G. Guillemette, Microfilament and microtubule assembly is required for the propagation of inositol trisphosphate receptor-induced Ca2+ waves in bovine aortic endothelial cells, J. Cell. Biochem., 2009, 106(2), 344–352 CrossRef PubMed.
- X. T. Hu, F. B. Zhang, Y. C. Fan, X. S. Shu, A. H. Y. Wong and W. Zhou, et al., Phospholipase c delta 1 is a novel 3p22.3 tumor suppressor involved in cytoskeleton organization, with its epigenetic silencing correlated with high-stage gastric cancer, Oncogene, 2009, 28(26), 2466–2475 CrossRef CAS PubMed.
- C. A. Sengelaub, K. Navrazhina, J. B. Ross, N. Halberg and S. F. Tavazoie, PTPRN 2 and PLC β1 promote metastatic breast cancer cell migration through PI (4,5)P 2-dependent actin remodeling, EMBO J., 2016, 35(1), 62–76 CrossRef CAS PubMed.
- G. Sala, F. Dituri, C. Raimondi, S. Previdi, T. Maffucci and M. Mazzoletti, et al., Phospholipase Cγ1 is required for metastasis development and progression, Cancer Res., 2008, 68(24), 10187–10196 CrossRef CAS PubMed.
- F. Vasilev, N. Limatola, D. R. Park, U. H. Kim, L. Santella and J. T. Chun, Disassembly of subplasmalemmal actin filaments induces cytosolic Ca2+ increases in astropecten aranciacus eggs, Cell. Physiol. Biochem., 2018, 48(5), 2011–2034 CrossRef CAS PubMed.
- I. Kovacevic, J. M. Orozco and E. J. Cram, Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Caenorhabditis elegans Spermatheca, PLoS Genet., 2013, 9(5), e1003510 CrossRef CAS PubMed.
- O. Fatunmbi, R. P. Bradley, S. K. Kandy, R. Bucki, P. A. Janmey and R. Radhakrishnan, A multiscale biophysical model for the recruitment of actin nucleating proteins at the membrane interface, Soft Matter, 2020, 16(21), 4941–4954 RSC.
- R. Bucki, Y. H. Wang, C. Yang, S. K. Kandy, O. Fatunmbi and R. Bradley, et al., Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation, J. Biol. Chem., 2019, 294(12), 4704–4722 CrossRef CAS PubMed.
- P. A. Janmey, R. Bucki and R. Radhakrishnan, Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: an update on possible mechanisms, Biochem. Biophys. Res. Commun., 2018, 506, 307–314 CrossRef CAS PubMed.
- B. Catimel, C. Schieber, M. Condron, H. Patsiouras, L. Connolly and J. Catimel, et al., The PI(3,5)P2 and PI(4,5)P2 interactomes, J. Proteome Res., 2008, 7(12), 5295–5313 CrossRef CAS PubMed.
- K. Tsujita and T. Itoh, Phosphoinositides in the regulation of actin cortex and cell migration, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2015, 1851, 824–831 CrossRef CAS PubMed.
- K. Tsujita, T. Itoh, A. Kondo, M. Oyama, H. Kozuka-Hata and Y. Irino, et al., Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of coronin 1A by phosphoinositides, J. Biol. Chem., 2010, 285(9), 6781–6789 CrossRef CAS PubMed.
- K. Vanoverberghe, V. Lehen’kyi, S. Thébault, M. Raphaël, F. Vanden Abeele and C. Slomianny, et al., Cytoskeleton Reorganization as an Alternative Mechanism of Store-Operated Calcium Entry Control in Neuroendocrine-Differentiated Cells, PLoS One, 2012, 7(9), e45615 CrossRef CAS PubMed.
- A. F. Vanden, L. Lemonnier, S. Thébault, G. Lepage, J. B. Parys and Y. Shuba, et al., Two types of store-operated Ca2+ channels with different activation modes and molecular origin in LNCaP human prostate cancer epithelial cells, J. Biol. Chem., 2004, 279(29), 30326–30337 CrossRef PubMed.
- W. Hong and K. L. Guan, The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway, Semin. Cell Dev. Biol., 2012, 23(7), 785–793, DOI:10.1016/j.semcdb.2012.05.004.
- M. Sudol, Yes-Associated Protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product, Oncogene, 1994, 9(8), 2145–2152 CAS.
- F. Kanai, P. A. Marignani, D. Sarbassova, R. Yagi, R. A. Hall and M. Donowitz, et al., TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins, EMBO J., 2000, 19(24), 6778–6791 CrossRef CAS PubMed.
- F. X. Yu and K. L. Guan, The Hippo pathway: regulators and regulations, Genes Dev., 2013, 27, 355–371 CrossRef CAS PubMed.
- T. Panciera, A. Citron, D. Di Biagio, G. Battilana, A. Gandin and S. Giulitti, et al., Reprogramming normal cells into tumour precursors requires ECM stiffness and oncogene-mediated changes of cell mechanical properties, Nat. Mater., 2020, 1–10 Search PubMed.
- F. Calvo, N. Ege, A. Grande-Garcia, S. Hooper, R. P. Jenkins and S. I. Chaudhry, et al., Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts, Nat. Cell Biol., 2013, 15(6), 637–646 CrossRef CAS PubMed.
- T. Moroishi, C. G. Hansen and K. L. Guan, The emerging roles of YAP and TAZ in cancer, Nat. Rev. Cancer, 2015, 15(2), 73–79 CrossRef CAS PubMed.
- B. Zhao, L. Li, Q. Lei and K. L. Guan, The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version, Genes Dev., 2010, 24, 862–874 CrossRef CAS PubMed.
- G. Nardone, J. Oliver-De La Cruz, J. Vrbsky, C. Martini, J. Pribyl and P. Skladal, et al., YAP regulates cell mechanics by controlling focal adhesion assembly, Nat. Commun., 2017, 8(1), 15321 CrossRef CAS PubMed.
- J. E. Sero and C. Bakal, Multiparametric Analysis of Cell Shape Demonstrates that β-PIX Directly Couples YAP Activation to Extracellular Matrix Adhesion, Cell Syst., 2017, 4(1), 84–96 CrossRef CAS PubMed.
- Z. Meng, Y. Qiu, K. C. Lin, A. Kumar, J. K. Placone and C. Fang, et al., RAP2 mediates mechanoresponses of the Hippo pathway, Nature, 2018, 560(7720), 655–660 CrossRef CAS PubMed.
- S. Chakraborty, K. Njah, A. V. Pobbati, Y. B. Lim, A. Raju and M. Lakshmanan, et al., Agrin as a Mechanotransduction Signal Regulating YAP through the Hippo Pathway, Cell Rep., 2017, 18(10), 2464–2479 CrossRef CAS PubMed.
- N. Koushki, A. Ghagre, L. K. Srivastava, C. Sitaras, H. Yoshie and C. Molter, et al., Lamin A redistribution mediated by nuclear deformation determines dynamic localization of YAP, BioRxiv, 2020 DOI:10.1101/2020.03.19.998708.
- B. D. Cosgrove, C. Loebel, T. P. Driscoll, T. K. Tsinman, E. N. Dai and S. J. Heo, et al., Nuclear envelope wrinkling predicts mesenchymal progenitor cell mechano-response in 2D and 3D microenvironments, Biomaterials, 2021, 270 Search PubMed.
- G. Sorrentino, S. Rezakhani, E. Yildiz, S. Nuciforo, M. H. Heim and M. P. Lutolf, et al., Mechano-modulatory synthetic niches for liver organoid derivation, Nat. Commun., 2020, 11(1), 1–10 CrossRef PubMed.
- S. Dupont, L. Morsut, M. Aragona, E. Enzo, S. Giulitti and M. Cordenonsi, et al., Role of YAP/TAZ in mechanotransduction, Nature, 2011, 474(7350), 179–183 CrossRef CAS PubMed.
- A. Elosegui-Artola, R. Oria, Y. Chen, A. Kosmalska, C. Perez-Gonzalez and N. Castro, et al., Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity, Nat. Cell Biol., 2016, 18(5), 540–548 CrossRef CAS PubMed.
- N. Yang, T. Chen, L. Wang, R. Liu, Y. Niu and L. Sun, et al., CXCR4 mediates matrix stiffness-induced downregulation of UBTD1 driving hepatocellular carcinoma progression via YAP signaling pathway, Theranostics, 2020, 10(13), 5790 CrossRef CAS PubMed.
- A. J. Rice, E. Cortes, D. Lachowski, B. C. H. Cheung, S. A. Karim and J. P. Morton, et al., Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells, Oncogenesis, 2017, 6(7), e352 CrossRef CAS PubMed.
- K. Tanahashi, A. Natsume, F. Ohka, K. Motomura, A. Alim and I. Tanaka, et al., Activation of Yes-Associated Protein in Low-Grade Meningiomas Is Regulated by Merlin, Cell Density, and Extracellular Matrix Stiffness, J. Neuropathol. Exp. Neurol., 2015, 74(7), 704–709 CrossRef CAS PubMed.
- X. Qin, X. Lv, P. Li, R. Yang, Q. Xia and Y. Chen, et al., Matrix stiffness modulates ILK-mediated YAP activation to control the drug resistance of breast cancer cells, Biochim. Biophys. Acta, Mol. Basis Dis., 2020, 1866(3), 165625 CrossRef CAS PubMed.
- N. Perez Gonzalez, J. Tao, N. D. Rochman, D. Vig, E. Chiu and D. Wirtz, et al., Cell tension and mechanical regulation of cell volume, Mol. Biol. Cell, 2018, 29(21), 0–0213 CrossRef PubMed.
- K. T. Furukawa, K. Yamashita, N. Sakurai and S. Ohno, The Epithelial Circumferential Actin Belt Regulates YAP/TAZ through Nucleocytoplasmic Shuttling of Merlin, Cell Rep., 2017, 20(6), 1435–1447 CrossRef CAS PubMed.
- J. Y. Shiu, L. Aires, Z. Lin and V. Vogel, Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction, Nat. Cell Biol., 2018, 20(3), 262–271 CrossRef CAS PubMed.
- J. Lee, A. Abdeen, X. Tang, T. Saif and K. Kilian, Geometric guidance of integrin mediated traction stress during stem cell differentiation, Biomaterials, 2015, 69, 174–183 CrossRef CAS PubMed.
- J. P. Califano and C. A. Reinhart-King, Substrate stiffness and cell area predict cellular traction stresses in single cells and cells in contact, Cell. Mol. Bioeng., 2010, 3(1), 68–75 CrossRef PubMed.
- H. Nakajima, K. Yamamoto, S. Agarwala, K. Terai, H. Fukui and S. Fukuhara, et al., Flow-Dependent Endothelial YAP Regulation Contributes to Vessel Maintenance, Dev. Cell, 2017, 40(6), 523–536.e6 CrossRef CAS PubMed.
- H. J. Lee, M. F. Diaz, K. M. Price, J. A. Ozuna, S. Zhang and E. M. Sevick-Muraca, et al., Fluid shear stress activates YAP1 to promote cancer cell motility, Nat. Commun., 2017, 8, 14122 CrossRef CAS PubMed.
- H. Yu, J. He, G. Su, Y. Wang, F. Fang and W. Yang, et al., Fluid shear stress activates YAP to promote epithelial–mesenchymal transition in hepatocellular carcinoma, Mol. Oncol., 2021, 15(11), 3164–3183 CrossRef PubMed.
- Y. Cui, F. M. Hameed, B. Yang, K. Lee, C. Q. Pan and S. Park, et al., Cyclic stretching of soft substrates induces spreading and growth, Nat. Commun., 2015, 6, 1–8 Search PubMed.
- J. Gao, L. He, L. Zhou, Y. Jing, F. Wang and Y. Shi, et al., Mechanical force regulation of YAP by F-actin and GPCR revealed by super-resolution imaging, Nanoscale, 2020, 12(4), 2703–2714 RSC.
- C. Rianna, M. Radmacher and S. Kumar, Direct evidence that tumor cells soften when navigating confined spaces, Mol. Biol. Cell, 2020, 31(16), 1726–1734 CrossRef CAS PubMed.
- L. He, J. Tao, D. Maity, F. Si, Y. Wu and T. Wu, et al., Role of membrane-tension gated Ca2+ flux in cell mechanosensation, J. Cell Sci., 2018, 131(4), jcs208470 CrossRef PubMed.
- R. C. Lee, R. L. Feinbaum and V. Ambros, The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14, Cell, 1993, 75(5), 843–854 CrossRef CAS PubMed.
- A. Eulalio, E. Huntzinger, T. Nishihara, J. Rehwinkel, M. Fauser and E. Izaurralde, Deadenylation is a widespread effect of miRNA regulation, RNA, 2009, 15(1), 21–32 CrossRef CAS PubMed.
- I. Behm-Ansmant, J. Rehwinkel, T. Doerks, A. Stark, P. Bork and E. Izaurralde, mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes, Genes Dev., 2006, 20(14), 1885–1898 CrossRef CAS PubMed.
- J. Zhang, W. Zhou, Y. Liu, T. Liu, C. Li and L. Wang, Oncogenic role of microRNA-532-5p in human colorectal cancer via targeting of the 5′UTR of RUNX3, Oncol. Lett., 2018, 15(5), 7215–7220 Search PubMed.
- K. Zhang, X. Zhang, Z. Cai, J. Zhou, R. Cao and Y. Zhao, et al., A novel class of microRNA-recognition elements that function only within open reading frames, Nat. Struct. Mol. Biol., 2018, 25(11), 1019–1027 CrossRef CAS PubMed.
- A. Helwak and D. Tollervey, Mapping the miRNA interactome by cross-linking ligation and sequencing of hybrids (CLASH), Nat. Protoc., 2014, 9(3), 711–728 CrossRef CAS PubMed.
- Y. Lee, M. Kim, J. Han, K. H. Yeom, S. Lee and S. H. Baek, et al., MicroRNA genes are transcribed by RNA polymerase II, EMBO J., 2004, 23(20), 4051–4060 CrossRef CAS PubMed.
- S. P. Kabekkodu, V. Shukla, V. K. Varghese, J. D’Souza, S. Chakrabarty and K. Satyamoorthy, Clustered miRNAs and their role in biological functions and diseases, Biol. Rev., 2018, 93(4), 1955–1986 CrossRef PubMed.
- S. Kim, M. R. Park, C. Choi, J. B. Kim and C. Cha, Synergistic control of mechanics and microarchitecture of 3D bioactive hydrogel platform to promote the regenerative potential of engineered hepatic tissue, Biomaterials, 2021, 270, 120688, DOI:10.1016/j.biomaterials.2021.120688.
- K. Kim, T. Duc Nguyen, S. Li and T. Anh Nguyen, SRSF3 recruits DROSHA to the basal junction of primary microRNAs, RNA, 2018, 24(7), 892–898 CrossRef CAS PubMed.
- E. Dardenne, M. PolayEspinoza, L. Fattet, S. Germann, M. P. Lambert and H. Neil, et al., RNA Helicases DDX5 and DDX17 Dynamically Orchestrate Transcription, miRNA, and Splicing Programs in Cell Differentiation, Cell Rep., 2014, 7(6), 1900–1913 CrossRef CAS PubMed.
- A. M. Denli, B. B. J. Tops, R. H. A. Plasterk, R. F. Ketting and G. J. Hannon, Processing of primary microRNAs by the Microprocessor complex, Nature, 2004, 432(7014), 231–235 CrossRef CAS PubMed.
- M. Landthaler, A. Yalcin and T. Tuschl, The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis, Curr. Biol., 2004, 14(23), 2162–2167 CrossRef CAS PubMed.
- C. R. Alarcón, H. Lee, H. Goodarzi, N. Halberg and S. F. Tavazoie, N6-methyladenosine marks primary microRNAs for processing, Nature, 2015, 519(7544), 482–485 CrossRef PubMed.
- M. T. Bohnsack, K. Czaplinski and D. Görlich, Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs, RNA, 2004, 10(2), 185–191 CrossRef CAS PubMed.
- T. P. Chendrimada, R. I. Gregory, E. Kumaraswamy, J. Norman, N. Cooch and K. Nishikura, et al., TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing, Nature, 2005, 436(7051), 740–744 CrossRef CAS PubMed.
- J. E. Park, I. Heo, Y. Tian, D. K. Simanshu, H. Chang and D. Jee, et al., Dicer recognizes the 5′ end of RNA for efficient and accurate processing, Nature, 2011, 475(7355), 201–205 CrossRef CAS PubMed.
- S. Iwasaki, M. Kobayashi, M. Yoda, Y. Sakaguchi, S. Katsuma and T. Suzuki, et al., Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes, Mol. Cell, 2010, 39(2), 292–299 CrossRef CAS PubMed.
- R. W. Carthew and E. J. Sontheimer, Origins and Mechanisms of miRNAs and siRNAs, Cell, 2009, 136, 642–655 CrossRef CAS PubMed.
- M. H. Jo, S. Shin, S. R. Jung, E. Kim, J. J. Song and S. Hohng, Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs, Mol. Cell, 2015, 59(1), 117–124 CrossRef CAS PubMed.
- T. Treiber, N. Treiber and G. Meister, Regulation of microRNA biogenesis and its crosstalk with other cellular pathways, Nat. Rev. Mol. Cell Biol., 2019, 20, 5–20 CrossRef CAS PubMed.
- M. Xie and J. A. Steitz, Versatile microRNA biogenesis in animals and their viruses, RNA Biol., 2014, 11, 673–681 CrossRef PubMed.
- S. K. Pong and M. Gullerova, Noncanonical functions of microRNA pathway enzymes – Drosha, DGCR8, Dicer and Ago proteins, FEBS Lett., 2018, 592, 2973–2986 CrossRef CAS PubMed.
- D. Cifuentes, H. Xue, D. W. Taylor, H. Patnode, Y. Mishima and S. Cheloufi, et al., A novel miRNA processing pathway independent of dicer requires argonaute2 catalytic activity, Science, 2010, 328(5986), 1694–1698 CrossRef CAS PubMed.
- S. Yang, T. Maurin, N. Robine, K. D. Rasmussen, K. L. Jeffrey and R. Chandwani, et al., Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(34), 15163–15168 CrossRef PubMed.
- M. Yoda, D. Cifuentes, N. Izumi, Y. Sakaguchi, T. Suzuki and A. J. Giraldez, et al., Poly(A)-specific ribonuclease mediates 3′-end trimming of argonaute2-cleaved precursor micrornas, Cell Rep., 2013, 5(3), 715–726 CrossRef CAS PubMed.
- E. Berezikov, W. J. Chung, J. Willis, E. Cuppen and E. C. Lai, Mammalian Mirtron Genes, Mol. Cell, 2007, 28(2), 328–336 CrossRef CAS PubMed.
- K. Okamura, J. W. Hagen, H. Duan, D. M. Tyler and E. C. Lai, The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila, Cell, 2007, 130(1), 89–100 CrossRef CAS PubMed.
- J. G. Ruby, C. H. Jan and D. P. Bartel, Intronic microRNA precursors that bypass Drosha processing, Nature, 2007, 448(7149), 83–86 CrossRef CAS PubMed.
- M. Xie, M. Li, A. Vilborg, N. Lee, M. D. Shu and V. Yartseva, et al., Mammalian 5′-capped microRNA precursors that generate a single microRNA, Cell, 2013, 155(7), 1568–1580 CrossRef CAS PubMed.
- N. Lemus-Diaz, R. R. Ferreira, K. E. Bohnsack, J. Gruber and M. T. Bohnsack, The human box C/D snoRNA U3 is a miRNA source and miR-U3 regulates expression of sortin nexin 27, Nucleic Acids Res., 2020, 48(14), 8074–8089 CrossRef CAS PubMed.
- E. S. Martens-Uzunova, M. Olvedy and G. Jenster, Beyond microRNA - Novel RNAs derived from small non-coding RNA and their implication in cancer, Cancer Lett., 2013, 340, 201–211 CrossRef CAS PubMed.
- D. Stribling, Y. Lei, C. M. Guardia, L. Li, C. J. Fields and P. Nowialis, et al., A noncanonical microRNA derived from the snaR-A noncoding RNA targets a metastasis inhibitor, RNA, 2021, 27(6), 694–709 CrossRef PubMed.
- S. Cheloufi, C. O. Dos Santos, M. M. W. Chong and G. J. Hannon, A dicer-independent miRNA biogenesis pathway that requires Ago catalysis, Nature, 2010, 465(7298), 584–589 CrossRef CAS PubMed.
- W. Si, J. Shen, H. Zheng and W. Fan, The role and mechanisms of action of microRNAs in cancer drug resistance, Clin. Epigenet., 2019, 11, 1–24 CrossRef PubMed.
- J. Bråte, R. S. Neumann, B. Fromm, A. A. B. Haraldsen, J. E. Tarver and H. Suga, et al., Unicellular Origin of the Animal MicroRNA Machinery, Curr. Biol., 2018, 28(20), 3288–3295 CrossRef PubMed.
- R. Kian, S. Moradi and S. Ghorbian, Role of components of MicroRNA machinery in carcinogenesis, Exp. Oncol., 2018, 40, 2–9 CrossRef CAS PubMed.
- A. L. Walz, A. Ooms, S. Gadd, D. S. Gerhard, M. A. Smith and J. M. GuidryAuvil, et al., Recurrent DGCR8, DROSHA, and SIX Homeodomain Mutations in Favorable Histology Wilms Tumors, Cancer Cell, 2015, 27(2), 286–297 CrossRef CAS PubMed.
- K. Shigeyasu, Y. Okugawa, S. Toden, C. R. Boland and A. Goel, Exportin-5 Functions as an oncogene and a potential therapeutic target in colorectal cancer, Clin. Cancer Res., 2017, 23(5), 1312–1322 CrossRef CAS PubMed.
- H. L. Sun, R. Cui, J. K. Zhou, K. Teng, Y. H. Hsiao and K. Nakanishi, et al., ERK Activation Globally Downregulates miRNAs through Phosphorylating Exportin-5, Cancer Cell, 2016, 30(5), 723–736 CrossRef CAS PubMed.
- J. Ramírez-Moya, L. Wert-Lamas, G. Riesco-Eizaguirre and P. Santisteban, Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness, Oncogene, 2019, 38(27), 5486–5499 CrossRef PubMed.
- A. M. Caroleo, M. A. De Ioris, L. Boccuto, I. Alessi, G. Del Baldo and A. Cacchione, et al., DICER1 Syndrome and Cancer Predisposition: From a Rare Pediatric Tumor to Lifetime Risk, Front. Oncology, 2021, 10, 2989 Search PubMed.
- T. Goulvent, I. Ray-Coquard, S. Borel, V. Haddad, M. Devouassoux-Shisheboran and M. C. Vacher-Lavenu, et al., DICER1 and FOXL2 mutations in ovarian sex cord-stromal tumours: a GINECO Group study, Histopathology, 2016, 68(2), 279–285 CrossRef PubMed.
- J. Chen, Y. Wang, M. K. McMonechy, M. S. Anglesio, W. Yang and J. Senz, et al., Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis, J. Pathol., 2015, 237(2), 215–225 CrossRef CAS PubMed.
- J. Vedanayagam, W. K. Chatila, B. A. Aksoy, S. Majumdar, A. J. Skanderup and E. Demir, et al., Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis, Nat. Commun., 2019, 10(1), 1–14 CrossRef CAS PubMed.
- M. M. Janas, B. Wang, A. S. Harris, M. Aguiar, J. M. Shaffer and Y. V. B. K. Subrahmanyam, et al., Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins, RNA, 2012, 18(11), 2041–2055 CrossRef CAS PubMed.
- H. Zhang, Y. Wang, J. Dou, Y. Guo, J. He and L. Li, et al., Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis, Oncogene, 2019, 38(9), 1410–1431 CrossRef CAS PubMed.
- N. Cheng, Y. Li and Z. G. Han, Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma, Hepatology, 2013, 57(5), 1906–1918 CrossRef CAS PubMed.
- Z. A. Syeda, S. S. S. Langden, C. Munkhzul, M. Lee and S. J. Song, Regulatory mechanism of microrna expression in cancer, Int. J. Mol. Sci., 2020, 21, 1723 CrossRef CAS PubMed.
- G. A. Calin, C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo and E. Noch, et al., Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(24), 15524–15529 CrossRef CAS PubMed.
- O. A. Kent, R. R. Chivukula, M. Mullendore, E. A. Wentzel, G. Feldmann and K. H. Lee, et al., Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway, Genes Dev., 2010, 24(24), 2754–2759 CrossRef CAS PubMed.
- L. P. Garo, A. K. Ajay, M. Fujiwara, G. Gabriely, R. Raheja and C. Kuhn, et al., MicroRNA-146a limits tumorigenic inflammation in colorectal cancer, Nat. Commun., 2021, 12(1), 1–16 CrossRef PubMed.
- V. Olive, I. Jiang and L. He, Mir-17-92, a cluster of miRNAs in the midst of the cancer network, Int. J. Biochem. Cell Biol., 2010, 42(8), 1348–1354 CrossRef CAS PubMed.
- Y. Hirata, N. Murai, N. Yanaihara, M. Saito, M. Saito and M. Urashima, et al., MicroRNA-21 is a candidate driver gene for 17q23-25 amplification in ovarian clear cell carcinoma, BMC Cancer, 2014, 14(1), 1–10 CrossRef CAS PubMed.
- S. R. Pfeffer, C. H. Yang and L. M. Pfeffer, The Role of MIR-21 in Cancer, Drug Dev. Res., 2015, 76(6), 270–277 CrossRef CAS PubMed.
- P. Chaluvally-Raghavan, F. Zhang, S. Pradeep, M. P. Hamilton, X. Zhao and R. Rupaimoole, et al., Copy Number Gain of hsa-miR-569 at 3q26.2 Leads to Loss of TP53INP1 and Aggressiveness of Epithelial Cancers, Cancer Cell, 2014, 26(6), 863–879 CrossRef CAS PubMed.
- L. Zhang, J. Huang, N. Yang, J. Greshock, M. S. Megraw and A. Giannakakis, et al., microRNAs exhibit high frequency genomic alterations in human cancer, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(24), 9136–9141 CrossRef CAS PubMed.
- G. A. Calin, C. Sevignani, C. D. Dumitru, T. Hyslop, E. Noch and S. Yendamuri, et al., Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(9), 2999–3004 CrossRef CAS PubMed.
- T. C. Chang, D. Yu, Y. S. Lee, E. A. Wentzel, D. E. Arking and K. M. West, et al., Widespread microRNA repression by Myc contributes to tumorigenesis, Nat. Genet., 2008, 40(1), 43–50 CrossRef CAS PubMed.
- Y. Li, P. S. Choi, S. C. Casey, D. L. Dill and D. W. Felsher, MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a Neoplastic state, Cancer Cell, 2014, 26(2), 262–272 CrossRef CAS PubMed.
- Z. Wang, S. Lin, J. J. Li, Z. Xu, H. Yao and X. Zhu, et al., MYC protein inhibits transcription of the MicroRNA cluster MC-let-7a-1–let-7d via noncanonical E-box, J. Biol. Chem., 2011, 286(46), 39703–39714 CrossRef CAS PubMed.
- S. Dey, J. J. Kwon, S. Liu, G. A. Hodge, S. Taleb and T. A. Zimmers, et al., MiR-29a is repressed by MYC in pancreatic cancer and its restoration drives tumor-suppressive effects via downregulation of LOXL2, Mol. Cancer Res., 2020, 18(2), 311–323 CrossRef CAS PubMed.
- B. Wang, S. H. Hsu, X. Wang, H. Kutay, H. K. Bid and J. Yu, et al., Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2, Hepatology, 2014, 59(2), 311–323 CrossRef PubMed.
- H. Han, D. Sun, W. Li, H. Shen, Y. Zhu and C. Li, et al., A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis, Hepatology, 2013, 57(6), 2378–2389 CrossRef CAS PubMed.
- X. Zhang, X. Chen, J. Lin, T. Lwin, G. Wright and L. C. Moscinski, et al., Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas, Oncogene, 2012, 31(24), 3002–3008 CrossRef CAS PubMed.
- J. Sargolzaei, T. Etemadi and A. Alyasin, The P53/microRNA network: a potential tumor suppressor with a role in anticancer therapy, Pharmacol. Res., 2020, 160, 105179 CrossRef CAS PubMed.
- J. Liu, C. Zhang, W. Hu and Z. Feng, Tumor suppressor p53 and its mutants in cancer metabolism, Cancer Lett., 2015, 356, 197–203 CrossRef CAS PubMed.
- G. Misso, M. T. Di Martino, G. De Rosa, A. A. Farooqi, A. Lombardi and V. Campani, et al., Mir-34: a new weapon against cancer?, Mol. Ther.–Nucleic Acids, 2014, 3, e195 CrossRef PubMed.
- M. Yamakuchi and C. J. Lowenstein, MiR-34, SIRT1 and p53: the feedback loop, Cell Cycle, 2009, 8, 712–715 CrossRef CAS PubMed.
- S. Z. Zheng, P. Sun, J. P. Wang, Y. Liu, W. Gong and J. Liu, MiR-34a overexpression enhances the inhibitory effect of doxorubicin on HepG2 cells, World J. Gastroenterol., 2019, 25(22), 2752 CrossRef CAS PubMed.
- T. Cooks, I. S. Pateras, L. M. Jenkins, K. M. Patel, A. I. Robles and J. Morris, et al., Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246, Nat. Commun., 2018, 9(1), 1–15 CrossRef CAS PubMed.
- P. Dong, Y. Xiong, S. J. B. Hanley, J. Yue and H. Watari, Musashi-2, a novel oncoprotein promoting cervical cancer cell growth and invasion, is negatively regulated by p53-induced miR-143 and miR-107 activation, J. Exp. Clin. Cancer Res., 2017, 36(1), 1–12 CrossRef PubMed.
- J. Xiao, H. Lin, X. Luo, X. Luo and Z. Wang, MiR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress, EMBO J., 2011, 30(3), 524–532 CrossRef CAS PubMed.
- G. Qin, S. Mallik, R. Mitra, A. Li, P. Jia and C. M. Eischen, et al., MicroRNA and transcription factor co-regulatory networks and subtype classification of seminoma and non-seminoma in testicular germ cell tumors, Sci. Rep., 2020, 10(1), 1–14 CrossRef PubMed.
- K. M. Taufiqul Arif, E. K. Elliot, L. M. Haupt and L. R. Griffiths, Regulatory mechanisms of epigenetic mirna relationships in human cancer and potential as therapeutic targets, Cancers, 2020, 12, 2922 CrossRef PubMed.
- M. Tomasetti, S. Gaetani, F. Monaco, J. Neuzil and L. Santarelli, Epigenetic Regulation of miRNA Expression in Malignant Mesothelioma: miRNAs as Biomarkers of Early Diagnosis and Therapy, Front. Oncology, 2019, 9, 1293 CrossRef PubMed.
- V. Bhatia, A. Yadav, R. Tiwari, S. Nigam, S. Goel and S. Carskadon, et al., Epigenetic silencing of miRNA-338-5p and miRNA-421 drives SPINK1-positive prostate cancer, Clin. Cancer Res., 2019, 25(9), 2755–2768 CAS.
- Z. Li, F. Yu, X. Zhou, S. Zeng, Q. Zhan and M. Yuan, et al., Promoter hypomethylation of microRNA223 gene is associated with atherosclerotic cerebral infarction, Atherosclerosis, 2017, 263, 237–243 CrossRef CAS PubMed.
- Y. Saito, G. Liang, G. Egger, J. M. Friedman, J. C. Chuang and G. A. Coetzee, et al., Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells, Cancer Cell, 2006, 9(6), 435–443 CrossRef CAS PubMed.
- W. Zhang, J. H. Chen, T. Shan, I. Aguilera-Barrantes, L. S. Wang and T. H. M. Huang, et al., miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation, Lab Investig., 2018, 98(11), 1397–1407 CrossRef CAS PubMed.
- L. Xu, F. Wang, X. F. Xu, W. H. Mo, Y. J. Xia and R. Wan, et al., Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells, Med. Oncol., 2011, 28(SUPPL. 1), 189–196 CrossRef CAS PubMed.
- T. Tsuruta, K. I. Kozaki, A. Uesugi, M. Furuta, A. Hirasawa and I. Imoto, et al., miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer, Cancer Res., 2011, 71(20), 6450–6462 CrossRef CAS PubMed.
- Z. Wang, Z. Chen, Y. Gao, N. Li, B. Li and F. Tan, et al., DNA hypermethylation of microRNA-34b/c has prognostic value for stage I non-small cell lung cancer, Cancer Biol. Ther., 2011, 11(5), 490–496 CrossRef CAS PubMed.
- P. D. C. Monroig and G. A. Calin, MicroRNA and Epigenetics: Diagnostic and Therapeutic Opportunities, Curr. Pathobiol. Rep., 2013, 1(1), 43–52 CrossRef PubMed.
- J. S. Mohamed, A. Hajira, M. A. Lopez and A. M. Boriek, Genome-wide mechanosensitive microRNA (MechanomiR) screen uncovers dysregulation of their regulatory networks in the mdm mouse model of muscular dystrophy, J. Biol. Chem., 2015, 290(41), 24986–25011 CrossRef CAS PubMed.
- J. D. Humphrey, E. R. Dufresne and M. A. Schwartz, Mechanotransduction and extracellular matrix homeostasis, Nat. Rev. Mol. Cell Biol., 2014, 15, 802–812 CrossRef CAS PubMed.
- K. E. Kubow, R. Vukmirovic, L. Zhe, E. Klotzsch, M. L. Smith and D. Gourdon, et al., Mechanical forces regulate the interactions of fibronectin and collagen i in extracellular matrix, Nat. Commun., 2015, 6, 1–11 Search PubMed.
- Y. C. Poh, J. Chen, Y. Hong, H. Yi, S. Zhang and J. Chen, et al., Generation of organized germ layers from a single mouse embryonic stem cell, Nat. Commun., 2014, 5, 1–12 Search PubMed.
- S. W. Shan, D. Y. Lee, Z. Deng, T. Shatseva, Z. Jeyapalan and W. W. Du, et al., MicroRNA MiR-17 retards tissue growth and represses fibronectin expression, Nat. Cell Biol., 2009, 11(8), 1031–1038 CrossRef CAS PubMed.
- Z. J. Rutnam, T. N. Wight and B. B. Yang, MiRNAs regulate expression and function of extracellular matrix molecules, Matrix Biol., 2013, 32, 74–85 CrossRef CAS PubMed.
- X. Zhang, S. Liu, T. Hu, S. Liu, Y. He and S. Sun, Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression, Hepatology, 2009, 50(2), 490–499 CrossRef CAS PubMed.
- Y. Sylvestre, V. De Guire, E. Querido, U. K. Mukhopadhyay, V. Bourdeau and F. Major, et al., An E2F/miR-20a autoregulatory feedback loop, J. Biol. Chem., 2007, 282(4), 2135–2143 CrossRef CAS PubMed.
- J. K. Mouw, Y. Yui, L. Damiano, R. O. Bainer, J. N. Lakins and I. Acerbi, et al., Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression, Nat. Med., 2014, 20(4), 360–367 CrossRef CAS PubMed.
- P. M. Gilbert, J. K. Mouw, M. A. Unger, J. N. Lakins, M. K. Gbegnon and V. B. Clemmer, et al., HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype, J. Clin. Invest., 2010, 120(5), 1535–1550 CrossRef CAS PubMed.
- A. J. Trimboli, C. Z. Cantemir-Stone, F. Li, J. A. Wallace, A. Merchant and N. Creasap, et al., Pten in stromal fibroblasts suppresses mammary epithelial tumours, Nature, 2009, 461(7267), 1084–1091 CrossRef CAS PubMed.
- A. Moro, T. P. Driscoll, L. C. Boraas, W. Armero, D. M. Kasper and N. Baeyens, et al., MicroRNA-dependent regulation of biomechanical genes establishes tissue stiffness homeostasis, Nat. Cell Biol., 2019, 21(3), 348–358 CrossRef CAS PubMed.
- S. Feske, Calcium signalling in lymphocyte activation and disease, Nat. Rev. Immunol., 2007, 7, 690–702 CrossRef CAS PubMed.
- C. Diener, M. Hart, D. Alansary, V. Poth, B. Walch-Rückheim and J. Menegatti, et al., Modulation of intracellular calcium signaling by microRNA-34a-5p, Cell Death Dis., 2018, 9(10), 1–13 CAS.
- S. U. Mertens-Talcott, S. Chintharlapalli, X. Li and S. Safe, The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells, Cancer Res., 2007, 67(22), 11001–11011 CrossRef CAS PubMed.
- Z. Li, S. Hu, J. Wang, J. Cai, L. Xiao and L. Yu, et al., MiR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells, Gynecol. Oncol., 2010, 119(1), 125–130 CrossRef CAS PubMed.
- C. E. Fletcher, D. A. Dart, A. Sita-lumsden, H. Cheng, P. S. Rennie and C. L. Bevan, Androgen-regulated processing of the oncomir MiR-27a, which targets Prohibitin in prostate cancer, Hum. Mol. Genet., 2012, 21(14), 3112–3127 CrossRef CAS PubMed.
- C. Palmieri, B. Rudraraju, M. Monteverde, L. Lattanzio, O. Gojis and R. Brizio, et al., Methylation of the calcium channel regulatory subunit α2δ-3 (CACNA2D3) predicts site-specific relapse in oestrogen receptor-positive primary breast carcinomas, Br. J. Cancer, 2012, 107(2), 375–381 CrossRef CAS PubMed.
- F. Liu, J. Chen, P. Wang, H. Li, Y. Zhou and H. Liu, et al., MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy, Nat. Commun., 2018, 9(1), 1–14 CrossRef CAS PubMed.
- P. Beesetty, K. B. Wieczerzak, J. N. Gibson, T. Kaitsuka, C. T. Luu and M. Matsushita, et al., Inactivation of TRPM7 kinase in mice results in enlarged spleens, reduced T-cell proliferation and diminished store-operated calcium entry, Sci. Rep., 2018, 8(1), 1–22 CAS.
- J. Wan, A. A. Guo, I. Chowdhury, S. Guo, J. Hibbert and G. Wang, et al., TRPM7 Induces Mechanistic Target of Rap1b Through the Downregulation of miR-28-5p in Glioma Proliferation and Invasion, Front. Oncol., 2019, 9, 1413 CrossRef PubMed.
- M. Liu, K. Inoue, T. Leng and S. Guo, Xiong Z gang. TRPM7 channels regulate glioma stem cell through STAT3 and Notch signaling pathways, Cell Signalling, 2014, 26(12), 2773–2781 CrossRef CAS PubMed.
- A. Vultur, C. S. Gibhardt, H. Stanisz and I. Bogeski, The role of the mitochondrial calcium uniporter (MCU) complex in cancer, Pflugers Arch., 2018, 470, 1149–1163 CrossRef CAS PubMed.
- S. Marchi, L. Lupini, S. Patergnani, A. Rimessi, S. Missiroli and M. Bonora, et al., Downregulation of the mitochondrial calcium uniporter by cancer-related miR-25, Curr. Biol., 2013, 23(1), 58–63 CrossRef CAS PubMed.
- G. Rao, S. K. D. Dwivedi, Y. Zhang, A. Dey, K. Shameer and R. Karthik, et al., Micro RNA -195 controls MICU 1 expression and tumor growth in ovarian cancer, EMBO Rep., 2020, 21(10), e48483 CrossRef CAS PubMed.
- J. Fares, M. Y. Fares, H. H. Khachfe, H. A. Salhab and Y. Fares, Molecular principles of metastasis: a hallmark of cancer revisited, Signal Transduction Targeted Ther., 2020, 5, 1–17 CrossRef PubMed.
- T. P. Lele, A. Brock and S. R. Peyton, Emerging Concepts and Tools in Cell Mechanomemory, Ann. Biomed. Eng., 2020, 48(7), 1–10 CrossRef PubMed.
- B. Zhao, K. Tumaneng and K. L. Guan, The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal, Nat. Cell Biol., 2011, 13, 877–883 CrossRef CAS PubMed.
- C. Yang, M. Tibbitt, L. Basta and K. Anseth, Mechanical memory and dosing influence stem cell fate, Nat. Mater., 2014, 13(6), 645–652 CrossRef CAS PubMed.
- M. Mori, R. Triboulet, M. Mohseni, K. Schlegelmilch, K. Shrestha and F. D. Camargo, et al., Hippo signaling regulates microprocessor and links cell-density-dependent mirna biogenesis to cancer, Cell, 2014, 156(5), 893–906 CrossRef CAS PubMed.
- S. G. Chaulk, V. J. Lattanzi, S. E. Hiemer, R. P. Fahlman and X. Varelas, The hippo pathway effectors TAZ/YAP regulate dicer expression and MicroRNA biogenesis through Let-7, J. Biol. Chem., 2014, 289(4), 1886–1891 CrossRef CAS PubMed.
- H. D. Zhang, L. H. Jiang, D. W. Sun, J. Li and Z. L. Ji, The role of miR-130a in cancer, Breast Cancer, 2017, 24, 521–527 CrossRef PubMed.
- J. Chen, D. Yan, W. Wu, J. Zhu, W. Ye and Q. Shu, Micro RNA-130a promotes the metastasis and epithelialmesenchymal transition of osteosarcoma by targeting PTEN, Oncol. Rep., 2016, 35(6), 3285–3292 CrossRef CAS PubMed.
- B. Li, P. Huang, J. Qiu, Y. Liao, J. Hong and Y. Yuan, MicroRNA-130a is down-regulated in hepatocellular carcinoma and associates with poor prognosis, Med. Oncol., 2014, 31(10), 230 CrossRef PubMed.
- X. C. Wang, L. L. Tian, H. L. Wu, X. Y. Jiang, L. Q. Du and H. Zhang, et al., Expression of miRNA-130a in nonsmall cell lung cancer, Am. J. Med. Sci., 2010, 340(5), 385–388 CrossRef PubMed.
- S. Shen, X. Guo, H. Yan, Y. Lu, X. Ji and L. Li, et al., A miR-130a-YAP positive feedback loop promotes organ size and tumorigenesis, Cell Res., 2015, 25(9), 997–1012 CrossRef CAS PubMed.
- G. Zhu, Y. Wang, M. Mijiti, Z. Wang, P. F. Wu and D. Jiafu, Upregulation of MIR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway, Biochem. Biophys. Res. Commun., 2015, 465(2), 194–199 CrossRef CAS PubMed.
- B. Zhao, L. Li and K. L. Guan, Hippo signaling at a glance, J. Cell Sci., 2010, 123, 4001–4006 CrossRef CAS PubMed.
- B. A. Callus, A. M. Verhagen and D. L. Vaux, Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation, FEBS J., 2006, 273(18), 4264–4276 CrossRef CAS PubMed.
- X. Pan, Z. X. Wang and R. Wang, MicroRNA-21: a novel therapeutic target in human cancer, Cancer Biol. Therapy, 2010, 10, 630–639 Search PubMed.
- M. L. Si, S. Zhu, H. Wu, Z. Lu, F. Wu and Y. Y. Mo, miR-21-mediated tumor growth, Oncogene, 2007, 26(19), 2799–2803 CrossRef CAS PubMed.
- C. Li, N. Talele, S. Boo, A. Koehler, E. Knee-Walden and J. Balestrini, et al., MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells, Nat. Mater., 2016, 16, 379–389 CrossRef PubMed.
- I. Carnevale, M. Capula, E. Giovannetti, T. Schmidt and S. Coppola, A mechanical memory of pancreatic cancer cells, bioRxiv, 2019, 730960 Search PubMed.
- H. A. Burris, M. J. Moore, J. Andersen, M. R. Green, M. L. Rothenberg and M. R. Modiano, et al., Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial, J. Clin. Oncol., 1997, 15(6), 2403–2413 CrossRef CAS PubMed.
- M. Momcilovic, A. Jones, S. T. Bailey, C. M. Waldmann, R. Li and J. T. Lee, et al., In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer, Nature, 2019, 575(7782), 380–384 CrossRef CAS PubMed.
- J. A. Katzenellenbogen, C. G. Mayne, B. S. Katzenellenbogen, G. L. Greene and S. Chandarlapaty, Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance, Nat. Rev. Cancer, 2018, 18, 377–388 CrossRef CAS PubMed.
- Y. Zhao, O. Bucur, H. Irshad, F. Chen, A. Weins and A. L. Stancu, et al., Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy, Nat. Biotechnol., 2017, 35(8), 757 CrossRef CAS PubMed.
- S. W. Hell, S. J. Sahl, M. Bates, X. Zhuang, R. Heintzmann and M. J. Booth, et al., The 2015 super-resolution microscopy roadmap, J. Phys. D: Appl. Phys., 2015, 48, 443001 CrossRef.
- Q. Luo, M. Huang, C. Liang, J. Zhang, G. Lin and S. Yu, et al., All-optical Mechanobiology Interrogation of Yes-associated Protein in Human Cancer and Normal Cells using a Multi-functional System, JoVE, 2021, 178, e62934 Search PubMed.
- J. Strecker, A. Ladha, Z. Gardner, J. L. Schmid-Burgk, K. S. Makarova and E. V. Koonin, et al., RNA-guided DNA insertion with CRISPR-associated transposases, Science, 2019,(6448), 364 Search PubMed.
- C.-H. Huang, K.-C. Lee and J. Doudna, Applications of CRISPR-Cas Enzymes in Cancer Therapeutics and Detection, Trends Cancer, 2018, 4(7), 499–512 CrossRef CAS PubMed.
- S. J. Patel, N. E. Sanjana, R. J. Kishton, A. Eidizadeh, S. K. Vodnala and M. Cam, et al., Identification of essential genes for cancer immunotherapy, Nature, 2017, 548(7669), 537–542 CrossRef CAS PubMed.
- J. G. Reiter, M. Baretti, J. M. Gerold, A. P. Makohon-Moore, A. Daud and C. A. Iacobuzio-Donahue, et al., An analysis of genetic heterogeneity in untreated cancers, Nat. Rev. Cancer, 2019, 19(11), 639–650 CrossRef CAS PubMed.
- J. Gao, J. F. Ward, C. A. Pettaway, L. Z. Shi, S. K. Subudhi and L. M. Vence, et al., VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer, Nat. Med., 2017, 23(5), 551–555 CrossRef CAS PubMed.
- L. B. Alexandrov, S. Nik-Zainal, D. C. Wedge, S. A. J. R. Aparicio, S. Behjati and A. V. Biankin, et al., Signatures of mutational processes in human cancer, Nature, 2013, 500(7463), 415–421 CrossRef CAS PubMed.
- Q. Jia, W. Zhou, W. Yao, F. Yang, S. Zhang and R. Singh, et al., Downregulation of YAP-dependent Nupr1 promotes tumor-repopulating cell growth in soft matrices, Oncogenesis, 2016, 5(4), e220 CrossRef CAS PubMed.
Footnote |
| † These authors contribute to the work equally. |
| This journal is © The Royal Society of Chemistry 2022 |