Open Access Article
Open Access ArticleModification of the antigenicity of cancer cells by conjugates consisting of hyaluronic acid and foreign antigens†
Soichi
Ogata
,
Reika
Tsuji
,
Atsushi
Moritaka
,
Shoya
Ito
and
Shinichi
Mochizuki
 *
*
Department of Chemistry and Biochemistry, The University of Kitakyushu, 1-1 Hibikino, Wakamatsu-ku, Kitakyushu, Fukuoka 808-0135, Japan. E-mail: mochizuki@kitakyu-u.ac.jp; Fax: +81-93-695-3572; Tel: +81-93-695-3203
First published on 20th July 2023
Abstract
Tumor-specific cytotoxic T-lymphocytes (CTLs) recognize tumor-associated antigens presented on major histocompatibility complex (MHC) class I molecules. However, it is difficult to induce potent CTLs by vaccination because the antigenicity is not so high, compared with that of foreign antigens derived from viruses and microbes. The affinity of binding to MHC class I molecules is proportional to the antigenicity of the antigen that they are presenting. Here, we prepared several conjugates consisting of hyaluronic acid (HA) as a carrier to cancer cells and ovalbumin (OVA) as a foreign protein and changed the antigens on cancer cells from intrinsic antigens to OVA fragments. The conjugate containing multiple HA and OVA molecules (100k4HA-3OVA) adopted a highly condensed structure and was well recognized by recombinant CD44 molecules in quartz crystal microbalance analysis and incorporated into cancer cells (CT26 cells). A mixture of CT26 cells treated with 100k4HA-3OVA and splenocytes including OVA-specific CTLs induced abundant secretion of IFN-γ into the supernatant. At 48 h after mixing with the CTLs, almost all CT26 cells had died. These results indicate that 100k4HA-3OVA is actively internalized into the cells through interaction between HA and CD44. Subsequently, CT26 cells present not only self-antigens, but also OVA fragments on MHC class I molecules and are recognized by OVA-specific CTLs. We thus succeeded in modifying the antigenicity from self- to non-self-antigens on cancer cells. Therefore, this foreign-antigen delivery using HA to cancer cells, followed by antigen replacement, could be used as a novel strategy for treating cancers.
1. Introduction
Expectations have been raised about the possibility of treating cancer by activating the naïve immune system by vaccination. Tumor-specific cytotoxic T-lymphocytes (CTLs) induced by immunization with tumor antigens and adjuvants are the primary cells for eradicating cancer cells.1–3 CTLs recognize and bind antigenic peptides that are presented on major histocompatibility complex (MHC) class I molecules and induce direct and indirect killing mechanisms to target cancer cells. Such tumor therapy using the immune system is expected to not only attack existing tumor cells, but also provide long-term memory against recurrence.4 Although cancer vaccines with tumor specificity and low side effects are anticipated to become alternatives to chemotherapy and radiotherapy, they have had only limited clinical success so far. One of the major reasons for this is assumed to be related to the low antigenicity of tumor-associated antigens (TAAs).5,6 In general, immunity acts strongly against foreign antigens derived from viruses and microbes. However, tumor cells are generated by the host itself and thus present self-antigens. It is thus difficult to induce fully activated CTLs targeting TAAs because most of these CTLs are eliminated during maturation due to negative selection at the thymus.7,8 As a result, immunological tolerance can be easily acquired, even after immunization with potent vaccines for tumor antigens. In addition, the high heterogeneity of tumor cells causes low efficacy of cancer vaccines.9 Tumor subpopulations do not necessarily express the cognate antigens on MHC class I molecules, resulting in treatment failure.10Vaccination with foreign antigens is advantageous for inducing strong immune responses. Compared with tumor antigens, foreign antigens can avoid negative selection because of the “non-self” antigenicity and have high affinity to MHC class I molecules.11,12 The binding affinity to MHC class I molecules is proportional to the antigenicity of the antigen that they are presenting. Therefore, if proteins with high antigenicity can be internalized into tumor cells, the degraded peptide can easily bind to host MHC class I molecules instead of the intrinsic antigens. MHC class II molecules are mainly expressed on macrophages, dendritic cells, and B cells,13 while MHC class I molecules are expressed on all nucleated cells.14 If the foreign antigenic proteins are administered and internalized into cells non-specifically, the cells would be targeted by the antigen-specific CTLs in the host. To propose a cancer vaccine strategy using foreign antigenic proteins, the development of carriers that specifically deliver the antigens to tumor cells is a primary requirement.
Hyaluronic acid (HA) is a linear high-molecular-weight polysaccharide consisting of alternating N-acetyl-β-D-glucosamine and β-D-glucuronate residues linked at 1–3 and 1–4 positions.15,16 HA is a component of extracellular matrices and is abundant in hydrated tissues such as the vitreous of the eye, articular cartilage, synovial fluid, and skin.17 Because HA possesses high biocompatibility and biodegradability, along with no toxicity, immunogenicity, or inflammatory effects, HA and its derivatives have been widely used as medical materials, such as for visco-supplementation, drug delivery, and tissue engineering.18–21 HA can specifically bind to various HA receptors on the cell surface such as CD4422,23 and receptors for HA-mediated motility (RHAMM).24 Cells from a variety of cancers, such as epithelial, ovarian, colon, and stomach, overexpress these HA-binding receptors.25 This background prompted the idea of taking advantage of HA for tumor-targeted drug delivery. HA has carboxylic acid (COOH) and alcohol (OH) functional groups in the repeating unit of polymers, making various chemical modifications possible. For example, a carboxylic acid group can be used for the formation of electrostatic complexes with positively charged polymers, but also for crosslinking with materials with an amino group through a dehydration reaction.26 Oxidation of hydroxy groups produces dialdehydes by opening the sugar rings.27 In addition, the reducing end of HA can also be used for reductive amination with materials having amino groups.20,28 A variety of chemically modified HAs have been extensively studied to deliver anti-tumor drugs, short-interfering RNA, and antisense DNA into tumor cells.
We previously prepared various conjugates consisting of HA and bovine serum albumin (BSA) by changing the mixing ratio between the carboxylic groups of HA and the amino groups of BSA.21 The resultant conjugates contained multiple HA and BSA molecules and showed almost the same size and morphology as HA, indicating that the conjugates adopted a highly condensed structure. The recombinant CD44 molecule recognized the conjugates more than it did HA before the conjugation. The conjugates showing a high affinity for CD44 were also effectively taken up at a high level by cancer cells expressing CD44. From these findings, we can conclude that CD44 preferentially recognized the prepared HA with a condensed structure and the same morphology as the original HA.
Based on the above framework, we initially hypothesized that foreign antigens attached to HA would be specifically delivered to cancer cells, leading to foreign peptide presentation on the cell surface (Fig. 1). As a proof of concept, Kim et al. reported that a conjugate made from HA and a foreign antigenic protein, ovalbumin (OVA), through reductive amination was taken up into CD44high tumor cells.29 The cells treated with the conjugate reacted with OVA-specific immune cells. In this study, we demonstrated the preparation of various conjugates consisting of HA and OVA through a dehydration reaction by changing the molecular weight of HA and the mixing ratio between HA and OVA and characterized them by precise analysis using multiangle light scattering (MALS). We also evaluated the uptake of the resultant conjugates into cancer cells and modified the antigenicity by changing the peptides presented on MHC class I and II molecules from intrinsic antigens to OVA in vitro.
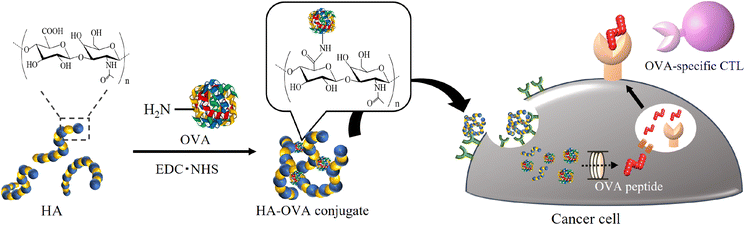 | ||
| Fig. 1 Schematic illustration showing antigen replacement from intrinsic antigens to the foreign and highly antigenic antigen OVA on cancer cells using an HA delivery system. | ||
2. Materials and methods
2.1. Materials
Hyaluronic acid (Mw = 1.0 × 106, determined by gel permeation chromatography (GPC) coupled to MALS analysis), and OVA were purchased from Sigma Aldrich (St Louis, MO). 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC·HCl) and N-hydroxysuccinimide NHS were purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan).2.2. Degradation of high-molecular-weight HA by sonication
High-molecular-weight HA was partially degraded by sonication to low-molecular-weight fragments. HA (40 mg) was dissolved in 40 mL of water and subjected to sonication using a QSONICA Q700 (Qsonica L.L.C., Newtown, CT). HA fragments with Mws of 5.0 × 105 and 1.0 × 105 were obtained by sonication at 12 W for 2 and 10 min, respectively.2.3. Preparation of HA-OVA conjugates
HA fragments obtained by treatment with sonication were conjugated to OVA through a condensation reaction using EDC and NHS reagents. HA, EDC and NHS were dissolved in 50 mL of 10 mM MES (pH 4.5) and incubated on ice. After 2 h, OVA was added to the mixture. The concentrations of each component in the reaction mixture are summarized in Table 1. After incubation at 37 °C for 24 h, the reaction mixtures were dialyzed with water using a Biotech CE Tubing (molecular weight cut-off of 3500) and freeze-dried. To remove unreacted OVA from freeze-dried samples and obtain HA-OVA conjugates with high purity, the samples were dissolved in distilled water and subjected to GPC. The fractions that were detected at an earlier elution time than the OVA fraction were collected, dialyzed with water, and freeze-dried.| Sample | Concentration in preparation | Conjugate | ||||||
|---|---|---|---|---|---|---|---|---|
| HA | OVA | EDC | NHS | M w/105 | HA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OVAa OVAa |
Code | ||
| M w/105 | Conc. (μM) | Conc. (μM) | Conc. (μM) | Conc. (μM) | ||||
| a Estimated from the relationship between the OVA concentration determined from protein quantification and the weight concentration of the conjugate. | ||||||||
| 1 | 1.0 | 2.0 | 4.0 | 1.0 | 1.7 | 1.5 | 1.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0 1.0 |
100k1HA-1OVA |
| 2 | 1.0 | 1.1 | 5.5 | 3.0 | 5.0 | 5.0 | 4.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 2.5 |
100k4HA-3OVA |
| 3 | 5.0 | 1.0 | 4.0 | 2.6 | 4.3 | 4.3 | 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.8 0.8 |
500k1HA-1OVA |
2.4. Determination of the free amino groups of OVA in a conjugate
The conjugates were dissolved in 50 μl of PBS (0.1–1 mg ml−1). Equal volumes of 0.1 M borate buffer (pH 8.5) and 0.1% trinitrobenzene sulfonic acid (TNBS) were added. After incubation at 40 °C for 1 h, the absorbance at 340 nm was measured with a UV spectrometer (V-630; JASCO Co., Ltd, Tokyo, Japan).30 The concentration of amino groups in the conjugate was determined from a standard curve constructed using a known concentration of OVA.2.5. Characterization of HA-OVA conjugates
GPC was carried out using a SHIMADZU Prominence 501 pumping system (SHIMADZU Co., Ltd, Kyoto, Japan) at a flow rate of 0.8 ml min−1 with an OHpak SB-806M column (Showa Denko Co., Ltd, Tokyo, Japan). Here, 10 mM phosphate buffer (pH 7.4) containing 0.1 M NaCl was used as a mobile phase. The eluate was detected using a reflective index (RI) detector (RI-501; Showa Denko), a UV detector (SPD-20A; SHIMADZU), and a MALS detector (DAWN HELEOS-II; Wyatt Technology Co., Santa Barbara, CA). RI and MALS signals were used to calculate the weight-averaged molecular weight (Mw) and z-averaged radius of gyration (Rg; 〈S2〉1/2z). The size and morphology of HA-OVA conjugates were also observed by dynamic light scattering (DLS) and transmission electron microscopy (TEM), respectively. After HA-OVA conjugates were dissolved in PBS at 0.5 mg ml−1, the sizes were determined using a DelsaMax PRO (Beckman Coulter, Fullerton, CA). For morphological observation, HA or HA-OVA conjugate solution diluted in deionized water was dropped on a carbon-coated copper grid, dried at room temperature, and observed using a JEM2100Plus operated at an accelerating voltage of 200 kV (JEOL Ltd, Tokyo, Japan).2.6. Interaction between the recombinant CD44 (rCD44) molecule and HA-OVA conjugates
The anti-His6 antibody (ThermoFisher Scientific, Waltham, MA) was immobilized on a gold substrate in a quartz crystal microbalance (QCM) sensor cell (AFFINIX QNμ; INITIUM, Inc., Tokyo, Japan) overnight. After washing the sensor cell with PBS three times, a His6-tagged rCD44 protein (10 μg ml−1) containing only the extracellular domain (Sino Biological, Beijing, China) in PBS was added to the sensor cell and incubated for 1 h at room temperature. After washing with PBS three times, the sensor cell was filled with 500 μl of PBS and HA or the HA–OVA conjugate was added at the indicated concentration, followed by measurement of the frequency change at 25 °C. The total mass of the adsorbed substrates on a QCM sensor was determined with Sauerbrey's equation (eqn (1)):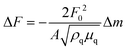 | (1) |
 | (2) |
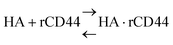
The dissociation constant (Kd) is given using eqn (3). Because HA is added in excess to rCD44 on the QCM sensor cell, at a steady state, [HA] = [HA]0 and [rCD44] = [rCD44]0 − [HA/rCD44] can be assumed. Eqn (3) can be converted to eqn (4) and (5).
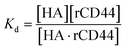 | (3) |
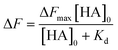 | (4) |
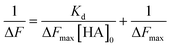 | (5) |
The Kd and the maximum frequency change (ΔFmax) can be obtained according to eqn (5).
2.7. Cell culture
CT26 cells (mouse colon carcinoma cells) were purchased from American Type Culture Collection (Manassas, VA), and cultured in RPMI-1640 medium (FUJIFILM Wako Pure Chemical Corporation) containing 10% FBS, 100 U ml−1 penicillin, and 0.1 mg ml−1 streptomycin.2.8. In vitro uptake of HA-OVA conjugates
CT26 cells were seeded at 2.0 × 105 cells per well in 12-well plates and supplemented with fluorescein isothiocyanate (FITC; FUJIFILM Wako Pure Chemical Corporation)-labelled OVA or HA-OVA conjugates at an OVA dose of 3 μg ml−1. After incubation for 24 h, the cells were washed with PBS three times and the fluorescence intensity of the cells was observed using a flow cytometer (CytoFLEX; Beckman Coulter). At the same time, the images of the cells were obtained using a BZ-X810 digital fluorescence microscope (Keyence, Osaka, Japan).2.9. Immune responses between CT26 cells treated with HA–OVA conjugates and splenocytes including OVA-specific CTLs
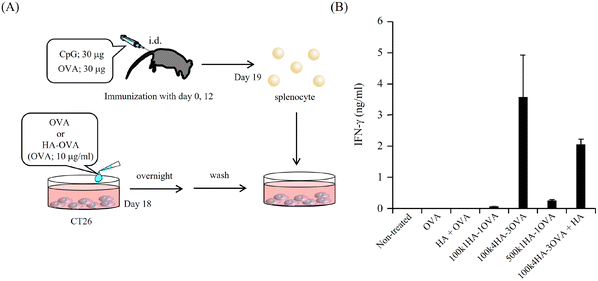
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of the University of Kitakyushu and approved by the Animal Ethics Committee of the University of Kitakyushu. Balb/c mice were purchased from Japan SLC, Inc. (Shizuoka, Japan). The mice were immunized with OVA (30 μg) and CpG-DNA (30 μg, K3 Et-FREE; Gene Design Co., Ltd, Osaka, Japan) at the base of the tail twice on days 0 and 12. On day 19, the splenocytes were collected by Histopaque gradient centrifugation. Briefly, excised spleen was minced and centrifuged at 1700 rpm for 5 min. After the filtration of the suspension through a 70 μm cell strainer and the centrifugation at 1700 rpm for 5 min, the pellet was resuspended in PBS and loaded on 100% Histopaque-1083 (Sigma Aldrich). The density gradient column was centrifuged at 1700 rpm for 30 min. The middle layer was collected. This layer contained an enriched fraction of splenocytes. On day 18, CT26 cells were seeded at 2.0 × 104 cells per well in 96-well plates. After cell adhesion, the cells were treated with OVA or HA-OVA conjugates (OVA concentration of 10 μg ml−1) and incubated overnight. The cells were washed with RPMI-1640 three times and supplemented with the collected splenocytes at 1.0 × 106 cells per well. After 24 h, the amount of IFN-γ secreted into the supernatant was measured with an IFN-γ Mouse ELISA Kit (ThermoFisher Scientific).2.10. Cytotoxicity assay
The cytotoxicity of CT26 cells was evaluated by measuring the activity of lactate dehydrogenase (LDH) released into the supernatant. After mixing CT26 cells (target cells; 2.0 × 104 cells per well) treated with the indicated HA-OVA conjugates and splenocytes (effector cells; 1.0 × 105 to 1.0 × 106 cells per well) obtained from mice immunized with OVA and CpG-DNA at the indicated effector cell/target cell (E/T) ratios, 24 or 48 h later the amount of LDH in the supernatants was determined using a cytotoxicity LDH assay kit-WST (DOJINDO, Kumamoto, Japan).2.11. Statistical analysis
All data are presented as mean ± SD. The statistical significance of differences between two groups was analyzed using two-tailed Student's t-test. Differences were considered statistically significant if the p value was less than 0.01.3. Results
3.1. Preparation of HA-OVA conjugates
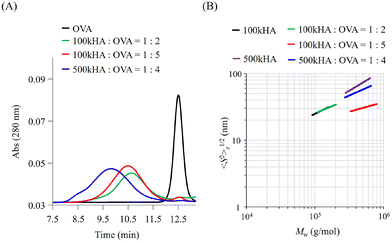
The molecular size of HA can be easily controlled by enzymatic or physical cleavage. In this study, we prepared two types of HA with different molecular weights (Mw = 1.0 × 105 and 5.0 × 105) by adjusting the duration of treatment with sonication for high-molecular-weight HA (Mw = 1.0 × 106). The obtained HA fragments were conjugated with OVA using a dehydration reaction between the carboxylic groups of HA and amino groups of OVA using EDC·HCl and NHS as condensation reagents. When the reaction mixtures of OVA and HA (Mw = 1.0 × 105; 100k) were subjected to GPC measurement, the peaks of OVA detected by UV absorbance at 280 nm were shifted to an earlier elution time (Fig. 2A). The elution times derived from the UV absorbance of OVA were almost the same irrespective of the mixing ratio between 100k HA and OVA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in Table 1). After the reaction of OVA with 500k HA at a molar ratio of 1
5 in Table 1). After the reaction of OVA with 500k HA at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, the elution time was shifted to an earlier time than that after the reaction with 100k HA (Fig. 2A). When the mixture of HA and OVA without condensation reagents was subjected to GPC measurement, the peak detected based on UV absorbance corresponded to that of the control OVA, which eluted at around 12.5 min (data not shown).
4, the elution time was shifted to an earlier time than that after the reaction with 100k HA (Fig. 2A). When the mixture of HA and OVA without condensation reagents was subjected to GPC measurement, the peak detected based on UV absorbance corresponded to that of the control OVA, which eluted at around 12.5 min (data not shown).
These results indicate that an earlier elution time of OVA after the reaction is attributed to the coupling with HA and a subsequent increase in the molecular size. The elution time depends on the coupling molecular weight of HA. The GPC profiles of the resultant conjugates also indicate the presence of no unreacted OVA, undesired aggregation, or degradation. The composition of each conjugate was estimated from the molecular weight and protein concentration determined by MALS and the bicinchoninic acid (BCA) protein assay, respectively, and coded as summarized in Table 1.
We can obtain information about the solution properties of the complexes from the plots of z-averaged Rg (〈S2〉1/2z) against Mw (Fig. 2B).21 100kHA and 500kHA are fitted by a straight line with slopes of 0.52 and 0.62, respectively, which correspond to a random coil swollen by the excluded volume effect. After conjugation with OVA, the slopes obtained from the double logarithmic plot suggest that the 100k1HA-1OVA conjugate adopts a random coil conformation like 100kHA. The straight line of the plot for 100k1HA-1OVA overlaps with that for 100kHA. Because OVA is less than 10 nm in size, the molecular size and conformation of 100kHA could not be affected by the attachment of one OVA molecule. From determination of the number of free amino groups in OVA, only 17% of amino groups in 100k1HA-1OVA are used for conjugation with carboxylic groups on HA (Table 2). This corroborates the assertion that the conformation of HA did not change after the conjugation with OVA. From DLS measurements, the hydrodynamic radii (Rh) of 100kHA and 100k1HA-1OVA were determined to be 17 nm and 21 nm, respectively (Fig. S1†). The relationship between Rg and Rh can also give us information regarding the internal structure and morphology.31,32 The rho ratio (ρ = Rg/Rh) of 0.78 corresponds to a homogeneous spherical conformation. The ρ values greater than 1 show characteristics of rod, random coil, and branched polymers. The ρ ratios of 100kHA and 100k1HA-1OVA were 1.8 and 1.4, respectively. Considering the TEM observations (Fig. S2†) together, 100kHA and 100k1HA-1OVA were revealed to adopt a random coil conformation. The 100k4HA-3OVA conjugate consists of multiple HA and OVA molecules and has a large molecular weight. However, the molecular size showed almost the same value at 25 nm as 100kHA and 100k1HA-1OVA (Fig. 2B and Table 2). The slope of 100k4HA-3OVA shows a value of 0.28, corresponding to a globular particle. Furthermore, the TEM image (Fig. S2†) and the ρ ratio of 0.74 determined in combination with Rh (Fig. S1†) indicate that 100k4HA-3OVA adopts a homogeneous sphere structure. From these findings, the high concentration of the condensation reagents in the reaction mixture induced intermolecular crosslinks, resulting in a highly folded condensed structure being adopted. The substitution rate of more than half of the amino groups in OVA also reveals the existence of an abundance of intermolecular crosslinking points in the conjugate (Table 2). Although 500k1HA-1OVA showed almost the same molecular weight and morphology as 500kHA, it became slightly smaller than it. However, in contrast to 100kHA, the excluded volume effect of 500kHA can interfere with intermolecular crosslinking. Once an amino group on OVA reacts, the HA with a large molecular size (numerous carboxy groups) surrounds the bound OVA. This promotes intramolecular crosslinking more than intermolecular crosslinking. As a result, the size of 500kHA slightly decreased via intramolecular crosslinking through OVA. We succeeded in acquiring various types of conjugates consisting of HA and OVA with different compositions, morphologies, and sizes.
| Code | 〈S2〉1/2z/nm | % Substitution of amino groups in OVAa | % Substitution of carboxylic groups in HAb | OVA wt% |
|---|---|---|---|---|
| a Determined from the number of free amino groups using TNBS. b Estimated from the number of used amino groups in OVA. | ||||
| 100k1HA-1OVA | 30 | 17 | 1.4 | 29 |
| 100k4HA-3OVA | 25 | 61 | 3.6 | 22 |
| 500k1HA-1OVA | 52 | 49 | 0.8 | 6.7 |
3.2. Interaction between the conjugates and the rCD44 molecule
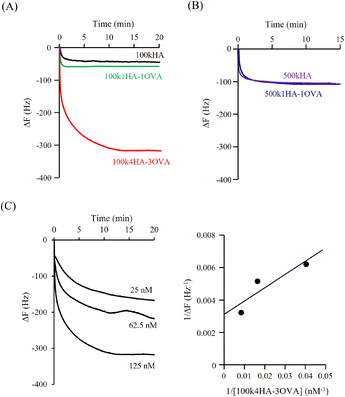
Fig. 3A shows the QCM frequency changes when 100kHA, 100k1HA-1OVA, or 100k4HA-3OVA at the same concentration of HA (50 μg ml−1) was added to the QCM cell. The addition of 100kHA and 100k1HA-1OVA showed decreases in the frequency, which was at almost the same level of around −50 Hz. In our previous study, the addition of HA to cells uncoated with rCD44 did not induce a decrease in the QCM frequency.21 Therefore, the frequency decreases observed in this study also reflect the specific binding of HA to rCD44. Although 100k1HA-1OVA showed a slightly larger decrease than 100kHA, this was due to it containing an OVA protein and having a heavier weight per molecule than 100kHA. The addition of 100k4HA-3OVA showed a decrease in the frequency greater than −300 Hz. Considering the weight % of OVA in the conjugate (Table 2), HA corresponding to the induction of a frequency change of −250 Hz was bound to rCD44 on the QCM cell. These results indicate that the conjugate adopting a highly condensed structure had higher affinity to rCD44 than the original HA. The relationship between 100kHA and 100k1HA-1OVA resembled that between 500kHA and 500k1HA-1OVA (Fig. 3B). The decreasing profiles of 500kHA and 500k1HA-1OVA completely overlapped, which was attributable to the negligible low weight % of OVA in 500k1HA-1OVA. Comparing 100kHA with 500kHA, the frequency change depended on the molecular weight of HA.Fig. 3C shows the QCM frequency changes when 100k4HA-3OVA was added at different concentrations. From eqn (5), the slope and intercept obtained from a reciprocal plot of ΔF and [guest] give us the equilibrium constant Kd.33 The determined Kd of 100k4HA-3OVA for rCD44 was 26 nM, while that of 100kHA for rCD44 was 145 nM (Fig. S3†). These results indicate that the affinity of 100kHA for rCD44 is enhanced by entanglements through inter-molecular crosslinking. Furthermore, despite the substitution rate of 4% of carboxylic groups for conjugation with amino groups, HA does not lose the ability to bind rCD44.
3.3. Uptake of HA-OVA conjugates into cancer cells
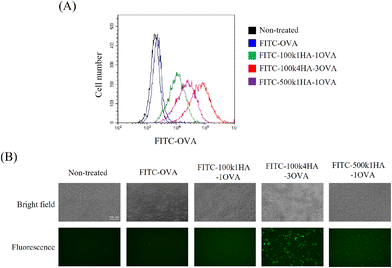
To evaluate the uptake of the conjugates into cells, FITC-labelled OVA or HA-OVA was added to CT26 cells. CT26 cells express a high level of CD44 on their surface.21 The cells treated with HA-OVA conjugates showed a high fluorescence intensity, while those treated with OVA did not (Fig. 4A). Among the HA-OVA conjugates, the cells treated with 100k4HA-3OVA showed the highest fluorescence intensity. Fluorescence microscopy images also indicated that a large number of 100k4HA-3OVA conjugates were incorporated into all of the cells, in contrast to the findings with the other conjugates (Fig. 4B). These results largely correlate with the frequency changes found in QCM analysis. We have demonstrated that conjugates consisting of multiple HA and BSA molecules and showing the same molecular size as HA were internalized into CT26 cells more than HA, and the cellular uptake was inhibited by the addition of an excess of HA.21 These findings suggest that 100k4HA-3OVA conjugates are actively internalized into cells through interaction between HA and CD44.3.4. The immune responses between CT26 cells treated with HA-OVA conjugates and splenocytes containing OVA-specific CTLs
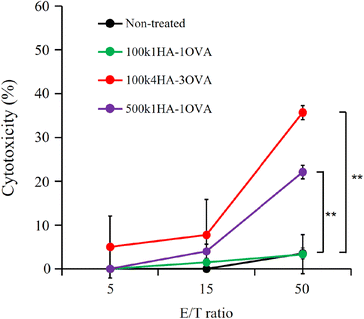
Because OVA protein is a foreign antigen for CT 26 cells, the fragments of OVA can bind to MHC class I and II molecules and be presented on the cell surface. If OVA-specific CTLs recognize the presented antigens, they can show immune responses. One of the most important immune responses for effective immunotherapy is the induction of the IFN-γ+ T-cell population.34 We obtained splenocytes from mice immunized twice with a mixture of OVA and CpG-DNA (Fig. 5A). The addition of OVA to the splenocytes led to them secreting IFN-γ, while that of BSA did not (Fig. S4†). This indicates that splenocytes included OVA-specific CTLs. To examine the immune responses between OVA antigens presented on CT26 cells and the splenocytes, the splenocytes were added to CT26 cells that had been pre-treated with OVA or HA-OVA conjugates for 24 h. The mixture of the CT26 cells treated with 100k4HA-3OVA and the splenocytes induced a large amount of IFN-γ (Fig. 5B). Although the treatment with 100k1HA-1OVA or 500k1HA-1OVA also led to the secretion of a certain amount of IFN-γ, the treatment with 100k4HA-3OVA produced far more secretion than these two treatments. The addition of an excess of HA in the reaction mixture significantly decreased the secretion of IFN-γ. The treatment with a mixture of free HA and OVA did not induce any immune responses. These results indicate that the internalization of OVA into CT26 cells is accelerated and mediated by HA in the conjugates. Furthermore, the level of the immune response is roughly proportional to the affinity with rCD44 and the uptake level into CT26 cells. We can conclude that, after internalization into the cells, OVA in the 100k4HA-3OVA conjugate is promptly degraded without being interfered with by HA, and the OVA fragments are presented sufficiently to be recognized by OVA-specific CTLs.3.5. Cytotoxicity of splenocytes including OVA-specific CTLs against CT 26 cells treated with HA-OVA conjugates
OVA-specific CTLs can exhibit cytotoxicity against cells presenting OVA fragments on MHC class I molecules. After incubation of the splenocytes including OVA-specific CTLs (effector cells) together with the CT26 cells treated with 100k4HA-3OVA (target cells) for 48 h, we observed a high level of LDH leakage in a manner dependent on the number of effector cells (Fig. 6). Half of the CT26 cells showed intense injury at an effector cell/target cell (E/T) ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. From microscopic observation, we confirmed that half of the CT26 cells had died at 48 h, while the remaining cells also showed morphological changes (Fig. S5†). Meanwhile, LDH leakage was not observed in the wells containing the splenocytes and untreated CT cells. These results indicate that the CT26 cells treated with the conjugate express OVA fragments on MHC class I molecules and become a target for the splenocytes. The splenocytes did not exhibit strong cytotoxicity against the CT 26 cells treated with 100k1HA-1OVA and 500k1HA-1OVA after 24 h, correlating with the findings on cellular uptake and IFN-γ production. However, after 48 h, LDH leakage from the CT26 cells treated with 500k1HA-1OVA was observed, at a level close to that for 100k4HA-3OVA at 24 h (Fig. S6†). This suggests that 500k1HA-1OVA (high-molecular-weight HA) is slowly degraded in the cells over time, and it takes a long time for the antigenic peptides to be presented and recognized by OVA-specific CTLs.
1. From microscopic observation, we confirmed that half of the CT26 cells had died at 48 h, while the remaining cells also showed morphological changes (Fig. S5†). Meanwhile, LDH leakage was not observed in the wells containing the splenocytes and untreated CT cells. These results indicate that the CT26 cells treated with the conjugate express OVA fragments on MHC class I molecules and become a target for the splenocytes. The splenocytes did not exhibit strong cytotoxicity against the CT 26 cells treated with 100k1HA-1OVA and 500k1HA-1OVA after 24 h, correlating with the findings on cellular uptake and IFN-γ production. However, after 48 h, LDH leakage from the CT26 cells treated with 500k1HA-1OVA was observed, at a level close to that for 100k4HA-3OVA at 24 h (Fig. S6†). This suggests that 500k1HA-1OVA (high-molecular-weight HA) is slowly degraded in the cells over time, and it takes a long time for the antigenic peptides to be presented and recognized by OVA-specific CTLs.
4. Discussion
Despite high expectations regarding the value of immunotherapy for treating cancer, there has been little progress in the response rate in the clinical phase. Immunity induced by the administration of TAAs and adjuvants is strongly dependent on the antigenicity. However, TAAs and even neoantigens can be considered to be self-antigens, so their antigenicity is not so high, compared with that of antigens derived from viruses and microbes. Therefore, if the TAAs on class MHC I molecules are replaced by antigens with high antigenicity, the CTLs induced by immunization with these antigens can strongly recognize and eradicate cancer cells. However, to achieve such an approach, it is necessary not only to prepare materials to deliver the antigens into cancer cells, but also to precisely characterize the interactions between the materials and the receptors and comprehensively understand the machinery involved in these interactions, as well as clarify the induction of immune responses by treatment with these materials. The present study demonstrates the relationship between the morphology of the prepared HA-OVA conjugates and the interaction with rCD44 and the cells expressing the CD44 molecule, as well as the subsequent antigen presentation.We succeeded in acquiring various conjugates by changing the molecular weight of HA, the mixing ratio, and the concentration of condensation reagents. A high concentration of condensation reagents led to the abundance of activated carboxylic groups of HA, facilitating reactions with amino groups. As a result, we obtained a conjugate consisting of multiple HA and OVA molecules (100k4HA-3OVA). A high modification rate of carboxylic groups is suspected of affecting the recognition of CD44 and subsequent cellular uptake. Determination of the substitution rate of amino groups on OVA can simultaneously reveal the substitution rate of carboxylic groups on HA (Table 2). Although many carboxylic groups in 100k4HA-3OVA are clearly used for the conjugations compared with the numbers for other conjugates, more than 96% of the carboxylic groups still remain, indicating that carboxylic groups are modified in every 10 disaccharide units. Oligo-HA of more than 6–8 disaccharide units in length is considered to be recognized by CD44.35 For every conjugate prepared in this study, because the substitution rate of carboxylic groups on HA guarantees many consecutive segments of eight disaccharide units each, the substitution level cannot strongly influence the recognition by CD44.
Not only the modification rate but also the morphology can influence the recognition of CD44. In our previous study, a conjugate containing multiple HA (80kHA) and BSA molecules and adopting a highly condensed structure was recognized by the rCD44 molecule more than 80kHA and was internalized into CT 26 cells with high efficiency.21 In this study, 100k4HA-3OVA was well recognized by the rCD44 molecule and the cells expressing CD44, compared with the recognition levels for 100kHA and 100k1HA-1OVA. These results suggest that CD44 prefers HA with a condensed structure. HA in the extracellular matrix shows rapid turnover with a half-life of ∼1.0–1.5 days.36 In mammals, liver sinusoidal endothelial cells express a recycling receptor that removes high-molecular-weight HA (>Mw = 1.0 × 106) from the circulation by endocytosis via the clathrin-coated pit pathway.37–39 CD44 can bind not only high-molecular-weight HA but also low-molecular-weight HA in the extracellular matrix degraded by hyaluronidases.40 One of the major cells expressing CD44 is T cells, which use HA as a material for breaking at inflammatory sites.41,42 Rolling via the interaction between CD44 on T cells and HA on endothelial cells activated by inflammatory cytokines is an early step leading to extravasation from the circulation into inflamed tissues. In contrast to HA adopting a random coil conformation in circulation, HA on cells aggregates with other matrix components such as proteoglycans. This HA is considered to exist as an entangled polymer and to be more condensed than that in circulation. Therefore, CD44 might show higher affinity to 100k4HA-3OVA than to HA and 100k1HA-1OVA.
In general, after the internalization of antigenic proteins into APCs, peptides produced by proteasomal degradation are further trimmed by enzymes in the endoplasmic reticulum to eight or nine amino acids in length, which is the optimal length for forming a stable complex with MHC class I molecules.43,44 In this study, after internalization of the HA-OVA conjugates into CT26 cells, HA would be promptly degraded by hyaluronidases, resulting in the exposure of OVA protein in the cytosol. Although some lysine residues of OVA protein are used for the conjugation with HA, we do not know whether the linkages between a carboxylic group on HA and a lysine residue on OVA are cleaved in the cytosol. In particular, 100k4HA-3OVA has the highest substitution level of amino groups in OVA, at 61% (Table 2). However, the IFN-γ response (Fig. 5) and LDH assay (Fig. 6) revealed that antigenic peptides are adequately presented with high efficiency. These findings suggest that HA attaches to the peptides that are not suitable as antigens or the epitopes showing high antigenicity for MHC class I H-2Kd contain no lysine residues. As a merit of antigenic protein delivery, we do not care about the functionality, compared with the case of the conjugation with anticancer proteins. If we deliver functional proteins such as anticancer proteins into cancer cells, we have to control and care about the conformational change and degradation. While, in contrast, the antigenic proteins can be actively subjected to a degradation process (antigen presentation process) whether they are being released from the carrier molecules or not. Anyway, the conjugates even 100k4HA-3OVA remain sufficient disaccharide units in length to be recognized by CD44 and peptide sequences to be presented and recognized by CTLs. However, the identification of the binding sites for lysine residues is still important. To identify the binding site and control the intracellular behaviors, peptide antigens can be preferred. One of the well-known OVA fragments presented on the MHC class I molecule (H-2Kb) is the following peptide: SIINFEKL. When the peptide was added to the cancer cells, it is possible to directly observe the presentation of the peptide by staining with the anti-mouse OVA257–264 (SIINFEKL) peptide bound to the H-2Kb antibody (data not shown). Now we are studying the conjugates by changing the OVA protein to the OVA peptide. After demonstrating and confirming the presentation of OVA fragments on the surface and interaction with OVA-specific CTLs, we will present these data in the next paper.
Although the levels of uptake of 500k1HA-1OVA into the CT26 cells and IFN responses were lower than those for 100k4HA-3OVA (Fig. 4 and 5), the cytotoxicity after adding splenocytes at 48 h approached that upon treatment with 100k4HA-3OVA. In Fig. 5 and 6, before adding the splenocytes, the amounts of uptake of each conjugate are considered to correspond to those shown in Fig. 4A. Meanwhile, after adding splenocytes, there was a strong correlation between the IFN response and the cytotoxicity at 24 h (Fig. 5 and 6). In particular, the cytotoxicity of the CT26 cells treated with 500k1HA-1OVA increased between 24 and 48 h (Fig. S6†). These findings suggest that high-molecular-weight HA took a long time to be degraded in cells, resulting in slow exposure of OVA proteins. Therefore, it required 48 h for the CT26 cells treated with 500k1HA-1OVA to be recognized and eradicated by CTLs. Indeed, the amounts of uptake of the conjugates for 500k1HA-1OVA and 100k4HA-3OVA (Fig. 4) are related to the cytotoxicity at 48 h (Fig. 6). This also implies that we can control the timing of antigen presentation by changing the molecular weight of HA.
In this study, we successfully modified the antigenicity of CT26 cells by delivering the highly antigenic protein OVA. It is easy for cells to undergo replacement of the presented low-antigenicity self-antigens with highly antigenic antigens. Other antigenic proteins with high antigenicity such as non-infected virus capsid protein can also be applied to this system. Furthermore, we can easily induce CTLs for highly antigenic proteins by immunization with effective adjuvants. Although cancer vaccine treatment has been impeded by major challenges, particularly the low antigenicity of cancer antigens, the use of this antigen modification and replacement system in combination with HA deliver technology could be a novel and effective strategy for treating cancers.
5. Conclusions
We prepared conjugates consisting of HA and the antigenic protein OVA with different molecular weights and sizes. A conjugate containing multiple HA and OVA molecules (100k4HA-3OVA) and showing a highly condensed structure was well recognized by the rCD44 molecule and incorporated into cancer cells expressing CD44. CT26 cells treated with 100k4HA-3OVA were recognized by OVA-specific CTLs, resulting in CLTs secreting a large amount of IFN-γ into the supernatant. By 48 h after mixing with the CTLs, almost all CT26 cells had died. These results indicate that, after the uptake of the 100k4HA-3OVA conjugate, CT26 cells present not only self-antigens, but also OVA fragments that are sufficiently antigenic to be recognized by OVA-specific CTLs. Therefore, this delivery of a foreign antigen using HA to cancer cells, followed by antigen replacement from self (intrinsic) to non-self, could be used as a novel strategy for treating cancers.Author contributions
Soichi Ogata: investigation, methodology, and writing – original draft. Reika Tsuji: investigation and methodology. Atsushi Moritaka: investigation and methodology. Shoya Ito: investigation and methodology. Shinichi Mochizuki: conceptualization, supervision, validation, writing – original draft, writing – review & editing, visualization, and funding acquisition.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (JP21H03825), the Grant-in-Aid for Challenging Pioneering Research (JP21K19925), and the Grant-in-Aid for Transformative Research Areas (A) (JP20H05873).References
- J. H. Russell and T. J. Ley, Annu. Rev. Immunol., 2002, 20, 323–370 CrossRef CAS PubMed.
- C. A. Klebanoff, L. Gattinoni and N. P. Restifo, Immunol. Rev., 2006, 211, 214–224 CrossRef CAS PubMed.
- W. Lee and M. Suresh, Front. Immunol., 2022, 13, 940047 CrossRef CAS PubMed.
- Y. Xie, L. Wang, A. Freywald, M. Qureshi, Y. Chen and J. Xiang, Cell. Mol. Immunol., 2013, 10, 72–77 CrossRef CAS PubMed.
- L. Novellino, C. Castelli and G. Parmiani, Cancer Immunol. Immunother., 2005, 54, 187–207 CrossRef CAS PubMed.
- K. Even-Desrumeaux, D. Baty and P. Chames, Cancers, 2011, 3, 2554–2596 CrossRef PubMed.
- A. N. Houghton, J. Exp. Med., 1994, 180, 1–4 CrossRef CAS PubMed.
- N. K. Nanda and E. E. Sercarz, Cell, 1995, 82, 13–17 CrossRef CAS PubMed.
- T. Kimura, S. Egawa and H. Uemura, Nat. Rev. Urol., 2017, 14, 501–510 CrossRef CAS PubMed.
- W. W. Overwijk, E. Wang, F. M. Marincola, H. G. Rammensee and N. P. Restifo, J. Immunother. Cancer, 2013, 1, 11 CrossRef PubMed.
- J. Rachmilewitz, Adv. Exp. Med. Biol., 2008, 640, 95–102 CrossRef CAS PubMed.
- Y. Yin, Y. Li and R. A. Mariuzza, Immunol. Rev., 2012, 250, 32–48 CrossRef PubMed.
- W. Reith, S. LeibundGut-Landmann and J. M. Waldburger, Nat. Rev. Immunol., 2005, 5, 793–806 CrossRef CAS PubMed.
- M. L. M. Jongsma, G. Guarda and R. M. Spaapen, Mol. Immunol., 2019, 113, 16–21 CrossRef CAS PubMed.
- T. C. Laurent and J. R. Fraser, FASEB J., 1992, 6, 2397–2404 CrossRef CAS PubMed.
- B. P. Toole, Nat. Rev. Cancer, 2004, 4, 528–539 CrossRef CAS PubMed.
- L. Bohaumilitzky, A. K. Huber, E. M. Stork, S. Wengert, F. Woelfl and H. Boehm, Front. Oncol., 2017, 7, 242 CrossRef PubMed.
- I. S. Bayer, Molecules, 2020, 25, 2649 CrossRef CAS PubMed.
- H. Knopf-Marques, M. Pravda, L. Wolfova, V. Velebny, P. Schaaf, N. E. Vrana and P. Lavalle, Adv. Healthcare Mater., 2016, 5, 2841–2855 CrossRef CAS PubMed.
- S. Asayama, M. Nogawa, Y. Takei, T. Akaike and A. Maruyama, Bioconjugate Chem., 1998, 9, 476–481 CrossRef CAS PubMed.
- R. Tsuji, S. Ogata and S. Mochizuki, Bioorg. Chem., 2022, 121, 105666 CrossRef CAS PubMed.
- C. B. Underhill, S. J. Green, P. M. Comoglio and G. Tarone, J. Biol. Chem., 1987, 262, 13142–13146 CrossRef CAS PubMed.
- I. Stamenkovic, M. Amiot, J. M. Pesando and B. Seed, Cell, 1989, 56, 1057–1062 CrossRef CAS PubMed.
- C. Hardwick, K. Hoare, R. Owens, H. P. Hohn, M. Hook, D. Moore, V. Cripps, L. Austen, D. M. Nance and E. A. Turley, J. Cell Biol., 1992, 117, 1343–1350 CrossRef CAS PubMed.
- A. Serafino, M. Zonfrillo, F. Andreola, R. Psaila, L. Mercuri, N. Moroni, D. Renier, M. Campisi, C. Secchieri and P. Pierimarchi, Curr. Cancer Drug Targets, 2011, 11, 572–585 CrossRef CAS PubMed.
- K. Tomihata and Y. Ikada, J. Biomed. Mater. Res., 1997, 37, 243–251 CrossRef CAS PubMed.
- Y. Deng, J. Ren, G. Chen, G. Li, X. Wu, G. Wang, G. Gu and J. Li, Sci. Rep., 2017, 7, 2699 CrossRef PubMed.
- S. Mochizuki, A. Kano, A. Yamayoshi and A. Maruyama, Sci. Technol. Adv. Mater., 2006, 7, 685–691 CrossRef CAS.
- Y. H. Lee, H. Y. Yoon, J. M. Shin, G. Saravanakumar, K. H. Noh, K. H. Song, J. H. Jeon, D. W. Kim, K. M. Lee, K. Kim, I. C. Kwon, J. H. Park and T. W. Kim, J. Controlled Release, 2015, 199, 98–105 CrossRef CAS PubMed.
- D. Elton, L. Medcalf, K. Bishop, D. Harrison and P. Digard, J. Virol., 1999, 73, 7357–7367 CrossRef CAS PubMed.
- A. Sajid, M. Castronovo and F. M. Goycoolea, Polymer, 2023, 15, 2115 CAS.
- L. Nilsson, Food Hydrocolloids, 2013, 30, 1–11 CrossRef CAS.
- S. Mochizuki, H. Morishita, Y. Adachi, Y. Yamaguchi and K. Sakurai, Carbohydr. Res., 2014, 391, 1–8 CrossRef CAS PubMed.
- C. Becker, H. Pohla, B. Frankenberger, T. Schuler, M. Assenmacher, D. J. Schendel and T. Blankenstein, Nat. Med., 2001, 7, 1159–1162 CrossRef CAS PubMed.
- T. Kaindl, H. Rieger, L. M. Kaschel, U. Engel, A. Schmaus, J. Sleeman and M. Tanaka, PLoS One, 2012, 7, e42991 CrossRef CAS PubMed.
- R. Tammi, A. M. Saamanen, H. I. Maibach and M. Tammi, J. Invest. Dermatol., 1991, 97, 126–130 CrossRef CAS PubMed.
- B. Zhou, J. A. Weigel, A. Saxena and P. H. Weigel, Mol. Biol. Cell, 2002, 13, 2853–2868 CrossRef CAS PubMed.
- B. Zhou, J. A. Weigel, L. Fauss and P. H. Weigel, J. Biol. Chem., 2000, 275, 37733–37741 CrossRef CAS PubMed.
- B. Zhou, J. A. Oka, A. Singh and P. H. Weigel, J. Biol. Chem., 1999, 274, 33831–33834 CrossRef CAS PubMed.
- V. Orian-Rousseau and J. Sleeman, Adv. Cancer Res., 2014, 123, 231–254 CrossRef PubMed.
- R. A. Clark, R. Alon and T. A. Springer, J. Cell Biol., 1996, 134, 1075–1087 CrossRef CAS PubMed.
- H. C. DeGrendele, P. Estess, L. J. Picker and M. H. Siegelman, J. Exp. Med., 1996, 183, 1119–1130 CrossRef CAS PubMed.
- T. Saric, S. C. Chang, A. Hattori, I. A. York, S. Markant, K. L. Rock, M. Tsujimoto and A. L. Goldberg, Nat. Immunol., 2002, 3, 1169–1176 CrossRef CAS PubMed.
- S. C. Chang, F. Momburg, N. Bhutani and A. L. Goldberg, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17107–17112 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm00439b |
| This journal is © The Royal Society of Chemistry 2023 |