Open Access Article
Open Access ArticleMolecular dynamics modelling of the interaction of a synthetic zinc-finger miniprotein with DNA†
Jessica
Rodriguez
 *ab,
Federica
Battistini
bc,
Soraya
Learte-Aymamí
*ab,
Federica
Battistini
bc,
Soraya
Learte-Aymamí
 a,
Modesto
Orozco
*bc and
José L.
Mascareñas
a,
Modesto
Orozco
*bc and
José L.
Mascareñas
 *a
*a
aCentro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CIQUS), and Departamento de Química Orgánica, Universidade de Santiago de Compostela Rúa Jenaro de la Fuente s/n, 15782, Santiago de Compostela, Spain. E-mail: jessicarodriguez.villar@usc.es; joseluis.mascarenas@usc.es
bInstitute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology (BIST), Baldiri Reixac 10-12, 08028 Barcelona, Spain
cDepartment of Biochemistry and Molecular Biology, University of Barcelona, 08028 Barcelona, Spain
First published on 19th May 2023
Abstract
We report the modelling of the DNA complex of an artificial miniprotein composed of two zinc finger modules and an AT-hook linking peptide. The computational study provides for the first time a structural view of these types of complexes, dissecting interactions that are key to modulate their stability. The relevance of these interactions was validated experimentally. These results confirm the potential of this type of computational approach for studying peptide–DNA complexes and suggest that they could be very useful for the rational design of non-natural, DNA binding miniproteins.
Introduction
The regulation of eukaryotic protein expression is mainly achieved at the level of transcription, and it is ultimately dependent on the interaction of specialized proteins called transcription factors (TFs) with specific DNA sequences.1 TFs are classified into families as a function of the structure of their DNA binding domain. Zinc fingers (ZFs) constitute the largest family of eukaryotic TFs, and play a key role in regulating the expression of numerous genes that are essential for different cellular processes.2 These proteins are composed of several repeats of zinc-containing modules, usually made of two β-sheets and one α-helix, that cooperate to bind specific DNA sequences.3 In the more classical Cys2–His2 ZF proteins, the zinc atom is coordinated by two cysteines in one chain and two histidines in the other, coordination that is key to stabilize the 3D folding. The binding to the DNA is mainly carried out by insertion of the α-helix into the major groove, where specific amino acids establish well-defined contacts with the edge of the bases. It is noteworthy that the DNA affinity of individual zinc-finger modules is low, and therefore the binding requires cooperative tandem repeats. The modular nature of these proteins has inspired the genetic engineering of a broad variety of non-natural polydactyl zinc-finger derivatives that bind the designed DNA sequences by programmed interactions through the major groove thread.4,5The zinc finger motif has also inspired the design of synthetic miniproteins capable of interacting with specific DNA sequences.6 In particular, our group has demonstrated that an appropriate conjugation of the zinc finger of the Drosophila transcription factor GAGA7 with minor groove binding units allows for high affinity DNA binding.8 Note that this zinc finger by itself (as an isolated module) fails to interact with its target site (GAGAG), something that Nature has solved in the GAGA factor by including two highly basic protein regions at the N-terminus, BR1 and BR2. Our designed conjugates interact with high affinity and selectivity to a DNA sequence bearing the peptide and the minor groove recognition regions in adjacent sites.8
Particularly appealing in our designs is the use of an AT-hook type of peptide as a minor groove anchor, because its peptidic nature facilitates the synthetic access to the conjugates.9–11 Specifically, we have reported the synthesis of three different non-natural DNA-binding miniproteins, i.e., Hk-gaga, gaga-Hk and gaga-Hk-gaga (Fig. 1),10 made by one AT-hook motif tethered to the ZF domain of the GAGA TF (Ser28 to Phe58 in the reference pdb structure).12 These newly designed miniproteins, with a fully peptide backbone, bind, with high affinity (two digit nanomolar) and excellent selectivity, composite DNA sequences of up to 14 base pairs.10
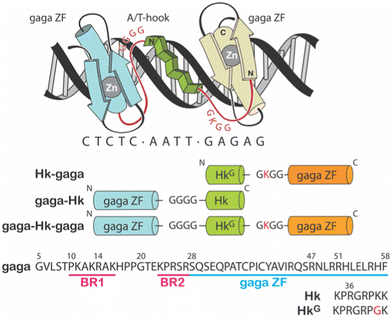 | ||
| Fig. 1 Top: Schematic representation of the major–minor–major groove interaction of the miniprotein gaga-Hk-gaga. The sequence of the peptidic linkers tethering the ZF domains of the GAGA TF and the AT-hook connector are highlighted in red. Bottom: Schematic illustration of the sequences of the designed miniproteins. Note that in the C-terminal GAGA fragment (in orange), the N-terminal Ser residue was removed.9 | ||
Whereas these results confirm the viability of making synthetic DNA binding agents, there is a lack of structural information on the DNA complexes, and all attempts to obtain crystallographic data have so far been unsuccessful. In this context, molecular modelling, and especially molecular dynamics (MD) simulations, may not only provide an overview of the interaction, but also unveil relevant information on the factors controlling the recognition.13
Molecular dynamics studies on protein–nucleic acid complexes,14 and particularly those entailing zinc finger modules,15 are scarce, and restricted to a few natural systems. For example, the group of Case used MDs in combination with NMR to study the hydration of the DNA complex of the transcription factor IIIA,16 while Gago performed a MD simulation on the DNA binding of TF Sp1. These studies helped to decode the bases of DNA-binding selectivity and gave results that were in consonance with previously reported experimental data.17 MDs of ZF-nucleic acid complexes have also been investigated for the TATA,18 CreA,19 WRKY,20 ZAP,21 NCp722 and GR23 proteins, among others. To our knowledge, MD simulations on DNA complexes involving non-natural protein binders have not been described.
Herein, we report a MD study of a complex between the synthetic miniprotein gaga-Hk-gaga and its target DNA site: CTCTC-AATT-GAGAG. The calculations revealed interactions in the DNA major groove that are key for the formation of a stable complex.24 The relevance of these interactions was experimentally confirmed using DNA-binding assays. Our results not only demonstrate the potential of the modelling to obtain a structural picture of the complex and dissect relevant contacts, but also pave the way for a future rational design of new miniproteins targeting selective DNA sequences.
Results and discussion
Structural model for the binding of gaga-Hk-gaga to its consensus DNA sequence
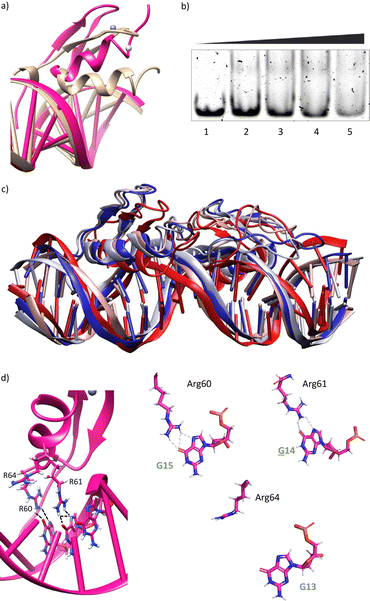
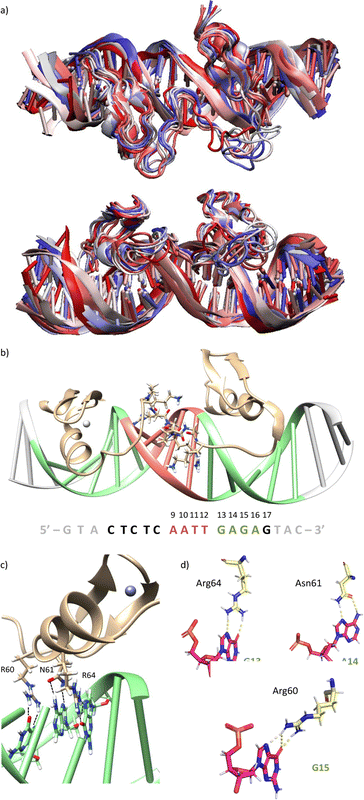
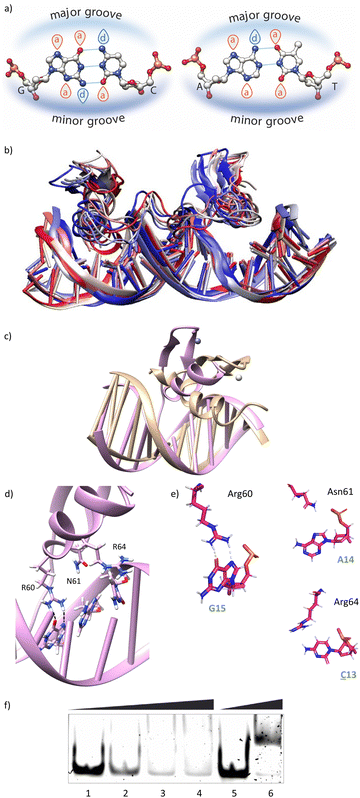
Using as starting point the structural data available for the DNA interaction of the GAGA ZF,12 and one AT-hook of HMG-I(Y)25 we assembled a hypothetical model for the DNA complex of gaga-Hk-gaga, with two ZF GAGA fragments bound to adjacent major grooves, and linked through an AT-hook anchor, inserted into the central minor groove. The Gly/Lys linkers were built and connected to the peptidic fragments using PyMOL. Once assembled, we carried out a MD simulation extending up to 500 ns using atomistic representation and explicit solvent (see the Experimental section). The resulting MD ensemble of structures shows that there is little structural variability over the simulation time (Fig. 2a). The final frame of the MD simulation, after ensuring that the structure has converged according to the RMSD values (Fig. S5, ESI†), was chosen as a representative snapshot of the trajectory. Inspection of this frame reveals that the ZF modules of gaga-Hk-gaga bind, as expected, the major grooves of the target DNA and recognize the first three GAG bases in a similar way to that observed in the solution structure of the native GAGA TF/DNA complex (Fig. 2b)12 whereas in the native GAGA protein the formation of a stable DNA complex requires the two basic regions in addition to the zinc finger module, in the designed conjugate gaga-Hk-gaga the additional contacts required for the binding are provided by the AT-hook, which inserts into the central minor groove facilitating the docking of both ZFs. We have previously shown that only sequences with a central A/T region are appropriately recognized by the miniproteins.10Of note, the polyglycine units linking the ZFs and the AT-hook do not exhibit interactions with the DNA,26 but provide the right connection to span the required distances between the binding modules. The design of the spacers is key to obtain efficient DNA-binding conjugates: the spacer must not only span the required distance between the DNA-binding domains, but also be flexible enough to allow the adaptation of the modules to their respective DNA binding sites.6g–n
The modelled structural information shows that the native interactions of the side chains of the ZF of GAGA with the DNA bases G13, A14 and G15, are conserved in both ZFs of the synthetic conjugate. In the simulation of the DNA complex of gaga-Hk-gaga, Arg64, Asn61 and Arg60 interact with the nucleobases G13, A14 and G15, respectively (Fig. 2c, d and Table S1, ESI†). The guanidium group of Arg64 recognizes the N7 and O6 atoms of G13 (hydrogen bonds observed in >38% of the simulation time), the carboxyamide of Asn61 interacts with the N7 and NH2 atoms of A14 (hydrogen bonds observed in >39% of the simulation time) and the guanidinium group of Arg60 with the O6 and N7 atoms of G15 (hydrogen bonds observed in 94% and 30% of the simulation time, respectively). Moreover, most of the supplementary electrostatic interactions of the ZF with the sugar phosphate backbone present in the DNA complex of the original GAGA TF are maintained.12 Importantly, we found 35 water molecules at the protein–DNA interface, which establish bridged hydrogen bonds with both the nucleobases and the amino acids (see the ESI†, Fig. S4), playing an important role in the stabilization of the protein–DNA complex.
The AT-hook module is essential for the formation of the complex, as it interacts with the DNA through its central Arg–Gly–Arg core deeply inserted into the minor groove and adopting an extended conformation, resembling that in the native AT-hook/DNA complex (Fig. 3).25 It establishes key interactions (Fig. 3b and Table S1, ESI†): the guanidium group of Arg78 interacts with the O2 atom of T12 (hydrogen bonds observed in 52% of the simulation time), the backbone amine group of Gly79 recognizes the O2 atom of T11 (hydrogen bonds observed in 27% of the simulation time) and the guanidium group of Arg80 forms a hydrogen bond with the N3 atom of A9 (hydrogen bonds observed in 17% of the simulation time).
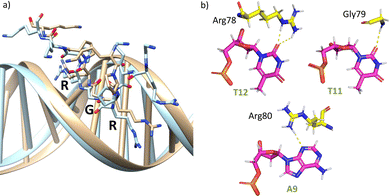 | ||
| Fig. 3 (a) Superposition of a snapshot for the minor groove interaction extracted from the final frame of the 1000 ns MD simulation of gaga-Hk-gaga (tan) with the structure of an AT-hook complex (PDB ID 2EZF, light blue). (b) Interactions of the AT-hook moiety of the miniprotein gaga-Hk-gaga with its target DNA. Key hydrogen bonds are shown as yellow dashed lines. | ||
Overall, the DNA recognition entails well balanced supramolecular contacts of the zinc finger modules with the GAG sequences, and of the AT-hook peptide with the edge of the bases in the minor groove. All these structural information obtained by modelling the ternary complex is also valid for the bivalent DNA binders gaga-Hk and Hk-gaga (confirmed from MD simulation of the binary complex Hk-gaga, see Fig. S3 in the ESI†).
Mutational studies
With the model at hand, we wondered whether it could be used to assess the relevance of individual interactions occurring in the major groove of the miniprotein/DNA complex. Thus, we carried out MD simulations with oligonucleotides mutated either at positions 13, 14 and 15 (see Fig. 2 for the numbering).G13 → C mutation
Having in mind the distribution of hydrogen bond donors and acceptors in the major groove of the DNA (Fig. 4a), we hypothesized that mutation of G13 by a cytosine, should prevent the formation of hydrogen bonds with the guanidium group of Arg64, and thus might have a considerable effect in the formation of the complex. Indeed, MD simulations showed that this mutation results in loss of the interaction with Arg64 (hydrogen bonds observed in <1% of the simulation time), and a subsequent displacement of the ZF helix out from the groove (Fig. 4b and c).27 This displacement also conveys the loss of the Asn61/A14 contact (hydrogen bonds observed in <1% of the simulation time, see also Fig. 4d and e). The only conserved contacts are with Arg60 (hydrogen bonds observed in >50% of the simulation time, see Table S2, ESI†).Overall, the model suggests a cancellation in the DNA binding of this zinc finger module. We therefore envisioned that this mutation should have a drastic effect in the DNA interaction of the bivalent peptide gaga-Hk (the trivalent gaga-Hk-gaga might still keep a substantial affinity due to interaction of the second ZF fragment).10 We therefore assessed the DNA binding of gaga-Hk to the mutated sequence using non-denaturing electrophoresis mobility shift assays (EMSA) in polyacrylamide gels.28 As can be deduced from Fig. 4f (lanes 1–4), we didn't observe retarded bands when using the double stranded (ds) oligonucleotide ![[C with combining low line]](https://www.rsc.org/images/entities/b_i_char_0043_0332.gif) AGAG containing a G to C mutation, which contrasts with the clear shifted band formed when using the dsDNA bearing the consensus target site (GAGAG, lane 6). It is interesting to note that the native transcription factor GAGA is quite tolerant to this and other mutations, likely because the presence of the complementary basic regions (BR1 and BR2), which make a significant contribution to the affinity.12 However, in our synthetic constructs, the presence of a low affinity minor groove binder cannot compensate the loss of binding upon mutation of the GAG region.
AGAG containing a G to C mutation, which contrasts with the clear shifted band formed when using the dsDNA bearing the consensus target site (GAGAG, lane 6). It is interesting to note that the native transcription factor GAGA is quite tolerant to this and other mutations, likely because the presence of the complementary basic regions (BR1 and BR2), which make a significant contribution to the affinity.12 However, in our synthetic constructs, the presence of a low affinity minor groove binder cannot compensate the loss of binding upon mutation of the GAG region.
Therefore, our synthetic bivalent miniproteins are fine-tuned to bind their consensus DNA sites, and exhibit a great sensitivity to single mutations, which is clearly beneficial in terms of selectivity.
A14 → C, A14 → G and G15 → A mutations
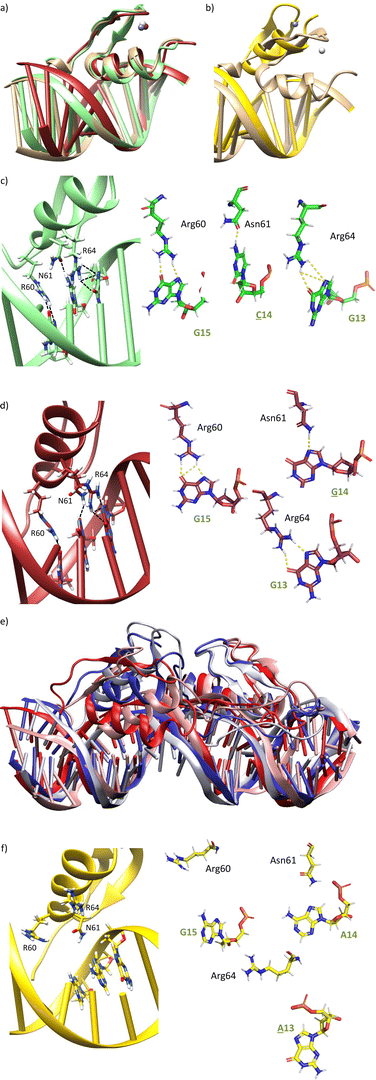
We next explored the impact of mutations at positions 14 and 15 of the target DNA. The mutation of A14 by any other nucleobase (C, T, G) may remove one of the hydrogen bonds between the carboxyamide of Asn61 and the DNA. Similarly, mutation of G15 by adenine (A) should abolish one of the hydrogen bonds with the guanidium group of Arg60.MD simulations of A14 → C or A14 → G mutations indeed display the loss of the bidentate DNA interaction with Asn61, but there is no displacement of the ZF helix from the major groove during the simulation time (Fig. 5a, c, d and Fig. S6a, b, ESI†). In Tables S3 and S4 (ESI†), it is shown that the frequency of hydrogen bonds with Asn61 is reduced to 17–19% of the simulation time, while it was present in >39% with the target DNA sequence. Moreover, it can be observed that the frequency of hydrogen bonds with Arg64 and Arg60 is also significantly reduced.
On the other hand, MD simulation of the complex with a G15 → A mutation showed loss of the interaction with Arg60, together with a huge displacement of the ZF helix out from the major groove (Fig. 5b, e and f), which triggered the loss of the Asn61/A14 and Arg64/G13 interactions (Fig. 5f). Indeed, no significant interaction was found in the hydrogen bond analysis of this simulation (Table S5, ESI†).
In agreement with MD simulations, EMSA analysis revealed that gaga-Hk does not elicit retarded bands when incubated with ds-oligonucleotides containing the mutated sequences G![[C with combining low line]](https://www.rsc.org/images/entities/b_i_char_0043_0332.gif) GAG, G
GAG, G![[G with combining low line]](https://www.rsc.org/images/entities/b_i_char_0047_0332.gif) GAG or GA
GAG or GA![[A with combining low line]](https://www.rsc.org/images/entities/b_i_char_0041_0332.gif) AG (Fig. 6). These results confirm that perturbing the interaction with either Asn61 or Arg60 results in a drastic effect on the DNA binding. Again, in the case of the native TF GAGA motif, this effect is less pronounced, and it exhibits a considerable interaction with DNAs featuring the mutations G
AG (Fig. 6). These results confirm that perturbing the interaction with either Asn61 or Arg60 results in a drastic effect on the DNA binding. Again, in the case of the native TF GAGA motif, this effect is less pronounced, and it exhibits a considerable interaction with DNAs featuring the mutations G![[C with combining low line]](https://www.rsc.org/images/entities/b_i_char_0043_0332.gif) GAG/G
GAG/G![[G with combining low line]](https://www.rsc.org/images/entities/b_i_char_0047_0332.gif) GAG in the major groove.12 This further highlights the advantage of our synthetic constructs in terms of selectivity.
GAG in the major groove.12 This further highlights the advantage of our synthetic constructs in terms of selectivity.
Taken together, these results demonstrate that single base DNA mutations can largely affect the DNA binding of our bivalent conjugates and inform about the relevance of specific hydrogen bonding interactions established by the zinc-finger unit.
Mutations in the synthetic miniprotein
After these studies, based on single base DNA mutations, we also made an initial assessment of mutations in the peptide. We questioned whether changing Asn61 (with a side chain exhibiting one hydrogen-bond donor and one hydrogen-bond acceptor) to Arg (with a side chain featuring a bidentate hydrogen-bond donor), the resulting peptides could bind a site with guanine instead of adenine (sequence: 5′-G![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) GAG-3′), as arginine might establish a bidentate interaction between its guanidium group and the N7 and O6 atoms of G. MD simulations with a mutated miniprotein gaga-Hk-gaga(N61R) bound to a ds-oligonucleotide containing the designed target sequence showed that while Arg61 is able to bind G14 (Fig. 7d and Table S6 (ESI†), hydrogen bonds observed in >22% of the simulation time) the higher length of Arg compared to Asn promotes a displacement of the helix, which abolishes the Arg64/G13 interaction (no significant interaction was found in the hydrogen bond analysis), and very likely the overall DNA binding (Fig. 7a, c and d). This effect was confirmed experimentally, with the mutated synthetic peptide gaga-Hk(N61R), which was not able to form stable complexes with the designed target ds-oligonucleotide, even at concentrations of 1 μM (Fig. 7b).
GAG-3′), as arginine might establish a bidentate interaction between its guanidium group and the N7 and O6 atoms of G. MD simulations with a mutated miniprotein gaga-Hk-gaga(N61R) bound to a ds-oligonucleotide containing the designed target sequence showed that while Arg61 is able to bind G14 (Fig. 7d and Table S6 (ESI†), hydrogen bonds observed in >22% of the simulation time) the higher length of Arg compared to Asn promotes a displacement of the helix, which abolishes the Arg64/G13 interaction (no significant interaction was found in the hydrogen bond analysis), and very likely the overall DNA binding (Fig. 7a, c and d). This effect was confirmed experimentally, with the mutated synthetic peptide gaga-Hk(N61R), which was not able to form stable complexes with the designed target ds-oligonucleotide, even at concentrations of 1 μM (Fig. 7b).
Conclusions
MD simulations allowed obtaining a detailed and realistic structural model for the DNA interaction of the miniprotein gaga-Hk-gaga, a synthetic, non-natural DNA binder made of two ZF domains linked to an AT-hook peptide. The high affinity and selective DNA binding of gaga-Hk-gaga, as well as of the corresponding binary analogues (gaga-Hk and Hk-gaga) derive from a cooperative major–minor groove recognition provided by the ZF and the AT-Hook moieties. The ZF has a strong preference for the sequence GAG, whereas the AT-hook inserts in the minor groove of the adjacent AATT sequence. The modelling allowed us to trace key recognition interactions, which were validated experimentally. Overall, MD simulations have proven very useful to dissect the relevance of single base-amino acid interactions in this type of protein–DNA complexes. Future work will seek to make use of MD simulations for the design of new miniproteins capable of interacting with different types of sequences.Experimental
Peptide synthesis and purification
Peptides were synthesized using a Liberty Blue Lite automatic microwave assisted peptide synthesizer from CEM Corporation, following the manufacturer's recommended procedures. Peptide synthesis was performed using the standard Fmoc solid-phase method on a PAL-PEG-PS resin (0.19 mmol g−1). Amino acids were coupled in a 5-fold excess using DIC (N,N′-diisopropylcarbodiimide) as the activator, Oxime as base, and DMF as solvent. Couplings were conducted for 4 min at 90 °C. Deprotection of the temporal Fmoc protecting group was performed by treating the resin with 20% piperidine in DMF for 1 min at 75 °C. The cleavage/deprotection step was performed by treatment of the resin-bound peptide for 1.5–2 h with the following cleavage cocktail: 940 μL TFA, 25 μL EDT, 25 μL H2O and 10 μL TIS (1 mL of cocktail/40 mg resin). The crude products were purified by RP-HPLC, 4 mL min−1, gradient 10 to 50% B over 40 min (A: H2O 0.1% TFA, B: CH3CN 0.1% TFA) and identified as the desired peptides.High-Performance Liquid Chromatography (HPLC) was performed using an Agilent 1100 series Liquid Chromatograph Mass Spectrometer system. Analytical HPLC was carried out using a Eclipse XDB-C18 analytical column (4.6 × 150 mm, 5 μm), 1 mL min−1, gradient 5 to 75% B over 30 min. Purification of the peptides was performed on a semipreparative Phenomenex Luna-C18 (250 × 10 mm) reverse-phase column.
EMSA experiments
EMSAs were performed using a BioRad Mini Protean gel system, powered by an electrophoresis power supplies Power Pac Basic model, maximum power 150 V, frequency 50–60 Hz at 140 V (constant V). Binding reactions were performed over 30 min in 18 mM Tris–HCl buffer (pH 7.5), 90 mM KCl, 1.8 mM MgCl2, 0.2 mM TCEP, 9% glycerol, 0.11 mg mL−1 BSA, 2.2% NP-40 and 0.02 mM of ZnCl2. In the experiments we used 75 nM of the ds-DNAs and a total incubation volume of 20 μL. After incubation for 30 min the products were resolved by PAGE using a 10% non-denaturing polyacrylamide gel and 0.5× TBE buffer for 40 min at 20 °C and analyzed by staining with SyBrGold (molecular probes: 5 μL in 50 mL of 1× TBE) for 10 min and visualized by fluorescence. 5× TBE buffer: 0.445 M Tris, 0.445 M boric acid.Molecular dynamics simulations
Starting structures were taken from the PDB database (1YUI, 2EZF), and the initial systems to be studied (gaga-Hk-gaga and Hk-gaga bound to their respective target DNAs) were assembled using PyMOL. All systems were hydrated by truncated octahedral box of TIP3P water molecules,29 with a minimum thickness of 10 Å around the solute. The system was neutralized with K+ cations and K+ and Cl− were added until a physiological concentration of 150 mM was reached. Systems were optimized, thermalized and equilibrated using standard procedures30 which involve energy minimizations of the solvent, slow thermalization and a final re-equilibration for 10 ns, prior to the 500 ns production runs. Trajectories were collected in the isothermal–isobaric ensemble (T = 298 K, P = 1 atm) using the parmbsc1 force field for DNA,31 Amber99SBildn force field for protein, Dang parameters for potassium and chlorine ions32 and ZAFF parameters for Zn.33 Simulations were performed using AMBER 18.34 All trajectories were processed using the cpptraj module of the AmberTools 18 package using default values (for hydrogen bonds a distance cutoff of 3.0 Å and an angle cutoff of 135°). Root mean square deviations (RMSDs) for each simulation were calculated using heavy atoms for both the protein and the DNA. DNA base pair parameters were derived using Curves+.35Author contributions
J. R. and F. B. performed the calculations and analyzed the data. S. L. performed the EMSA assays. J. R., M. O. and J. L. M. guided the research and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has received financial support from Spanish grants (IJC2019-040358-I funded by MCIN/AEI/10.13039/501100011033 to J. R., PID2019-108624RB-I00 funded by MCIN/AEI/10.13039/501100011033 to J. L. M. and RTI2018-096704-B-100 and PID2021-122478NB-I00 to M. O.), the Consellería de Cultura, Educación e Ordenación Universitaria (Grants 2015-CP082, ED431C-2017/19, ED431C-2021/ 25 and ED431G 2019/03: Centro Singular de Investigación de Galicia accreditation 2019–2022 to J. L. M.) and the European Union (European Regional Development Fund-ERDF corresponding to the multiannual financial framework 2014–2020 to J. L. M.). This work was also supported by the BioExcel-2. Centre of Excellence for Computational Biomolecular Research” (823830, M. O.) and the Instituto de Salud Carlos III–Instituto Nacional de Bioinformática (ISCIII PT 17/0009/0007 co-funded by the Fondo Europeo de Desarrollo Regional, M. O.). Funding was also provided by the MINECO Severo Ochoa Award of Excellence from the Government of Spain (awarded to IRB Barcelona; M.O. is an ICREA-Institució Catalana de Recerca i Estudis Avancats- academia researcher). The authors acknowledge RES-HPC (BCV-2022-3-0002 and BCV-2023-1-0004) for providing generous computational resources.References
- C. W. Garvie and C. Wolberger, Mol. Cell., 2001, 8, 937 CrossRef CAS PubMed.
- (a) C. O. Pabo, E. Peisach and R. A. Grant, Annu. Rev. Biochem., 2001, 70, 313 CrossRef CAS PubMed; (b) S. Iuchi and N. Kudell, Zinc finger proteins: from atomic contact to cellular function, Landes Biosciences, Georgetown, TX, 2004, ISBN: 0-306-48231-2 Search PubMed.
- (a) A. Klug, Annu. Rev. Biochem., 2010, 79, 213 CrossRef CAS PubMed; (b) J. Miller, A. D. McLachlan and A. Klug, EMBO J., 1985, 4, 1609 CrossRef CAS PubMed.
- (a) F. D. Urnov, E. J. Rebar, M. C. Holmes, H. S. Zhang and P. D. Gregory, Nat. Rev. Genet., 2010, 11, 636 CrossRef CAS PubMed; (b) S. A. Wolfe, L. Nekludova and C. O. Pabo, Annu. Rev. Biophys. Biomol. Struct., 2000, 29, 183 CrossRef CAS PubMed.
- G. A. Gersbach, T. Gaj and C. F. Barbas, Acc. Chem. Res., 2014, 47, 2309 CrossRef PubMed.
- (a) D. J. Segal and C. F. Barbas III, Curr. Opin. Biotechnol., 2001, 12, 632 CrossRef CAS PubMed; (b) D. Jantz, B. T. Amann, G. J. Gatto Jr. and J. M. Berg, Chem. Rev., 2004, 104, 789 CrossRef CAS PubMed . For other DNA-binding peptides, see: ; (c) Y. Ruiz García, Y. V. Pabon-Martinez, C. I. E. Smith and A. Madder, Chem. Commun., 2017, 53, 6653 RSC; (d) Y. Ruiz García, A. Iyer, D. Van Lysebetten, Y. Vladimir Pabon, B. Louage, M. Honcharenko, B. G. De Geest, C. I. Smith, R. Strömberg and A. Madder, Chem. Commun., 2015, 51, 17552 RSC; (e) Y. Ruiz García, J. Zelenka, Y. V. Pabon, A. Iyer, M. Buděšínský, T. Kraus, C. I. Smith and A. Madder, Org. Biomol. Chem., 2015, 13, 5273 RSC; (f) G. A. Woolley, A. S. I. Jaikaran, M. Berezovski, J. P. Calarco, S. N. Krylov, O. S. Smart and J. R. Kumita, Biochemistry, 2006, 45, 6075 CrossRef CAS PubMed; (g) J. Mosquera, J. Rodríguez, M. E. Vázquez and J. L. Mascareñas, ChemBioChem, 2014, 15, 1092 CrossRef CAS PubMed; (h) J. Rodríguez, C. Perez-Gonzalez, M. Martínez-Calvo, J. Mosquera and J. L. Mascareñas, RSC Adv., 2022, 12, 3500 RSC; (i) E. Oheix and A. F. A. Peacock, Chem. – Eur. J., 2014, 20, 2829 CrossRef CAS PubMed; (j) G. A. Bullen, J. H. Tucker and A. F. Peacock, Chem. Commun., 2015, 51, 8130 RSC; (k) J. Mosquera, A. Jiménez-Balsa, V. I. Dodero, M. E. Vázquez and J. L. Mascarenas, Nat. Commun., 2013, 4, 1874 CrossRef PubMed; (l) J. B. Blanco, M. E. Vázquez, J. Martinez-Costas, L. Castedo and J. L. Mascareñas, Chem. Biol., 2003, 10, 713 CrossRef CAS PubMed; (m) A. Jiménez-Balsa, E. Pazos, B. Martínez-Albardonedo, J. L. Mascareñas and M. E. Vázquez, Angew. Chem., Int. Ed., 2012, 51, 8825 CrossRef PubMed; (n) O. Vázquez, M. I. Sánchez, J. Martinez-Costas, M. E. Vázquez and J. L. Mascarenas, Org. Lett., 2010, 12, 216 CrossRef PubMed.
- P. V. Pedone, R. Ghirlando, G. M. Clore, A. M. Gronenborn, G. Felsenfeld and J. G. Omichinski, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 2822 CrossRef CAS PubMed.
- (a) O. Vázquez, M. E. Vázquez, J. B. Blanco, L. Castedo and J. L. Mascareñas, Angew. Chem., Int. Ed., 2007, 46, 6886 CrossRef PubMed; (b) J. Rodríguez, J. Mosquera, O. Vázquez, M. E. Vázquez and J. L. Mascareñas, Chem. Commun., 2014, 50, 2258 RSC.
- (a) J. Rodríguez, J. Mosquera, J. R. Couceiro, M. E. Vázquez and J. L. Mascareñas, Chem. Sci., 2015, 6, 4767 RSC; (b) J. Rodríguez, J. Mosquera, R. García-Fandiño, M. E. Vázquez and J. L. Mascareñas, Chem. Sci., 2016, 7, 3298 RSC.
- J. Rodríguez, S. Learte-Aymamí, J. Mosquera, G. Celaya, D. Rodríguez, M. E. Vázquez and J. L. Mascareñas, Chem. Sci., 2018, 9, 4118 RSC.
- (a) J. Rodríguez, J. Mosquera, M. E. Vázquez and J. L. Mascareñas, Chem. – Eur. J., 2016, 22, 13474 CrossRef PubMed; (b) S. Learte-Aymamí, J. Rodríguez, M. E. Vázquez and J. L. Mascareñas, Chem. – Eur. J., 2020, 26, 8875 CrossRef PubMed; (c) J. Rodríguez, J. Mosquera, S. Learte-Aymamí, M. E. Vázquez and J. L. Mascareñas, Acc. Chem. Res., 2020, 53, 2286 CrossRef PubMed.
- J. G. Omichinski, P. V. Pedone, G. Felsenfeld, A. M. Gronenborn and G. M. Clore, Nat. Struct. Biol., 1997, 4, 122 (PDB ID: 1YUI) Search PubMed.
- For docking studies of Ru(II)-based DNA binding agents, see: (a) D. Bouzada, I. Salvado, G. Barka, G. Rama, J. Martinez-Costas, R. Lorca, A. Somoza, M. Melle-Franco, M. E. Vazquez and M. Vazquez Lopez, Chem. Commun., 2018, 54, 658 RSC; (b) M. I. Sanchez, G. Rama, R. Calo-Lapido, K. Ucar, P. Lincoln, M. Vazquez-Lopez, M. Melle-Franco, J. L. Mascareñas and M. E. Vazquez, Chem. Sci., 2019, 10, 8668 RSC . For MD simulations of metallopeptides, see: ; (c) S. Learte, P. Martin-Malpartida, L. Roldán-Martín, G. Sciortino, J. R. Couceiro, J.-D. Maréchal, M. J. Macias, J. L. Mascareñas and M. E. Vázquez, Commun. Chem., 2022, 5, 75 CrossRef PubMed.
- (a) D. L. Beveridge, S. B. Dixit, B. L. Kormos, A. M. Baranger and B. Jayaram, Molecular Dynamics Simulations and Free Energy Calculations on Protein-Nucleic Acid Complexes, in Computational Studies of RNA and DNA, ed. J. Šponer and F. Lankaš, Springer, Dordrecht, 2006, vol. 2 Search PubMed; (b) S. Khalid and P. M. Rodger, Prog. React. Kinet. Mech., 2004, 29, 167 CrossRef CAS; (c) Z. Pirkhezranian, M. Tahmoorespur, X. Daura, H. Monhemi and M. H. Sekhavati, BMC Genomics, 2020, 21, 60 CrossRef CAS PubMed; (d) M. Garton and C. Laughton, J. Mol. Biol., 2013, 425, 2910 CrossRef CAS PubMed; (e) L. Etheve, J. Martin and R. Lavery, Nucleic Acids Res., 2016, 44, 1440 CrossRef CAS PubMed.
- (a) G. Roxstrom, I. Velazquez, M. Paulino and O. Tapia, J. Phys. Chem. B, 1998, 102, 1828 CrossRef; (b) B. R. Morgan and F. Massi, Prot. Sci., 2010, 19, 1222 CrossRef CAS PubMed; (c) M. Y. Hamed, J. Comput. Aided Mol. Des., 2018, 32, 657 CrossRef CAS PubMed.
- (a) V. Tsui, I. Radhakrishnan, P. E. Wright and D. A. Case, J. Mol. Biol., 2000, 302, 1101 CrossRef CAS PubMed . See also: ; (b) A. Dreab and C. A. Bayse, J. Chem. Inf. Model., 2022, 62, 903 CrossRef CAS PubMed.
- E. Marco, R. Garcia-Nieto and F. Gago, J. Mol. Biol., 2003, 328, 9 CrossRef CAS PubMed.
- B. Yang, Y. Zhu, Y. Wang and G. Chen, J. Comput. Chem., 2011, 32, 416 CrossRef CAS PubMed.
- M. Paulino, P. Esperón, M. Vega, C. Scazzocchio and O. Tapia, THEOCHEM, 2002, 580, 225 CrossRef CAS.
- B. Pandey, A. Grover and P. Sharma, BMC Genomics, 2018, 19, 132 CrossRef PubMed.
- S. Pal, A. Kumar and H. Vashisth, J. Chem. Inf. Model., 2023, 63, 1002 CrossRef CAS PubMed.
- W. Ren, D. Ji and X. Xu, PLoS One, 2018, 13, e0196662 CrossRef PubMed.
- Y. Wang, N. Ma, Y. Wang and G. Chen, PLoS One, 2012, 7, e35159 CrossRef CAS PubMed.
- “Key interactions” are defined as the hydrogen bond interactions which allow selective DNA binding of the miniprotein.
- (a) J. R. Huth, C. A. Bewley, M. S. Nissen, J. N. Evans, R. Reeves, A. M. Gronenborn and G. M. Clore, Nat. Struct. Biol., 1997, 4, 657 CrossRef CAS PubMed (PDB ID: 2EZF); (b) E. Fonfría-Subirós, F. Acosta-Reyes, N. Saperas, J. Pous, J. A. Subirana and J. L. Campos, PLoS One, 2012, 7, e37120 CrossRef PubMed (PDB ID: 3UXW).
- The lysine residue introduced in the linker connecting the C-terminal side of the AT-hook peptide and the N-terminus of the GAGA ZF doesn't establish electrostatic contacts with the phosphate backbone, as initially hypothesized (see ref. 10).
- Comparison of the roll parameter for the different MD simulations carried out in this study suggests that mutations barely affect the DNA morphology (see Fig. S7 in the ESI†).
- (a) L. M. Hellman and M. G. Fried, Nat. Protoc., 2007, 2, 1849 CrossRef CAS PubMed; (b) D. Lane, P. Prentki and M. Chandler, Microbiol. Rev., 1992, 56, 509 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- (a) A. Pérez, F. J. Luque and M. Orozco, J. Am. Chem. Soc., 2007, 129, 14739–14745 CrossRef PubMed; (b) P. D. Dans, L. Danilāne, I. Ivani, T. Dršata, F. Lankaš, A. Hospital, J. Walther, R. I. Pujagut, F. Battistini, J. L. Gelpí, R. Lavery and M. Orozco, Nucleic Acids Res., 2016, 44, 4052–4066 CrossRef CAS PubMed; (c) P. D. Dans, J. Walther and H. Gómez, Curr. Opin. Struct. Biol., 2016, 37, 29–45 CrossRef CAS PubMed.
- I. Ivani, P. D. Dans, A. Noy, A. Pérez, I. Faustino, A. Hospital, J. Walther, P. Andrio, R. Goñi, A. Balaceanu, G. Portella, F. Battistini, J. L. Gelpí, C. González, M. Vendruscolo, C. A. Laughton, S. A. Harris, D. A. Case and M. Orozco, Nat. Methods, 2015, 13, 55–58 CrossRef PubMed.
- (a) L. X. Dang, J. Am. Chem. Soc., 1995, 117, 6954–6960 CrossRef CAS; (b) L. X. Dang and P. A. Kollman, J. Am. Chem. Soc., 1990, 112, 5716–5720 CrossRef CAS.
- M. B. Peters, Y. Yang, B. Wang, L. Füsti-Molnár, M. N. Weaver and K. M. Merz, Jr., J. Chem. Theory Comput., 2010, 6, 2935–2947 CrossRef CAS PubMed.
- D. A. Case, I. Y. Ben-Shalom, S. R. Brozell, D. S. Cerutti, T. E. Cheatham III, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, D. Ghoreishi, M. K. Gilson, H. Gohlke, A. W. Goetz, D. Greene, R. Harris, N. Homeyer, Y. Huang, S. Izadi, A. Kovalenko, T. Kurtzman, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, D. J. Mermelstein, K. M. Merz, Y. Miao, G. Monard, C. Nguyen, H. Nguyen, I. Omelyan, A. Onufriev, F. Pan, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. L. Simmerling, J. Smith, R. Salomon-Ferrer, J. Swails, R. C. Walker, J. Wang, H. Wei, R. M. Wolf, X. Wu, L. Xiao, D. M. York and P. A. Kollman, AMBER 2018, University of California, San Francisco, 2018 Search PubMed.
- R. Lavery, M. Moakher, J. H. Maddocks, D. Petkeviciute and K. Zakrzewska, Nucleic Acids Res., 2009, 37, 5917–5929 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cb00053b |
| This journal is © The Royal Society of Chemistry 2023 |

![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/b_char_0043_0332.gif) GAG
GAG![[G with combining low line]](https://www.rsc.org/images/entities/b_char_0047_0332.gif) GAG
GAG![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) AG
AG