Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
14 examples of how LLMs can transform materials science and chemistry: a reflection on a large language model hackathon†
Kevin Maik
Jablonka
 *a,
Qianxiang
Ai‡
*a,
Qianxiang
Ai‡
 b,
Alexander
Al-Feghali‡
b,
Alexander
Al-Feghali‡
 c,
Shruti
Badhwar‡
c,
Shruti
Badhwar‡
 d,
Joshua D.
Bocarsly‡
d,
Joshua D.
Bocarsly‡
 e,
Andres M.
Bran‡
e,
Andres M.
Bran‡
 fg,
Stefan
Bringuier‡
fg,
Stefan
Bringuier‡
 h,
L. Catherine
Brinson‡
h,
L. Catherine
Brinson‡
 i,
Kamal
Choudhary‡
i,
Kamal
Choudhary‡
 j,
Defne
Circi‡
j,
Defne
Circi‡
 i,
Sam
Cox‡
i,
Sam
Cox‡
 k,
Wibe A.
de Jong‡
k,
Wibe A.
de Jong‡
 l,
Matthew L.
Evans‡
l,
Matthew L.
Evans‡
 mn,
Nicolas
Gastellu‡
mn,
Nicolas
Gastellu‡
 c,
Jerome
Genzling‡
c,
Jerome
Genzling‡
 c,
María Victoria
Gil‡
c,
María Victoria
Gil‡
 o,
Ankur K.
Gupta‡
o,
Ankur K.
Gupta‡
 l,
Zhi
Hong‡
l,
Zhi
Hong‡
 p,
Alishba
Imran‡
q,
Sabine
Kruschwitz‡
p,
Alishba
Imran‡
q,
Sabine
Kruschwitz‡
 r,
Anne
Labarre‡
r,
Anne
Labarre‡
 c,
Jakub
Lála‡
c,
Jakub
Lála‡
 s,
Tao
Liu‡
s,
Tao
Liu‡
 c,
Steven
Ma‡
c,
Steven
Ma‡
 c,
Sauradeep
Majumdar‡
c,
Sauradeep
Majumdar‡
 a,
Garrett W.
Merz‡
a,
Garrett W.
Merz‡
 t,
Nicolas
Moitessier‡
t,
Nicolas
Moitessier‡
 c,
Elias
Moubarak‡
c,
Elias
Moubarak‡
 a,
Beatriz
Mouriño‡
a,
Beatriz
Mouriño‡
 a,
Brenden
Pelkie‡
a,
Brenden
Pelkie‡
 u,
Michael
Pieler‡
u,
Michael
Pieler‡
 vw,
Mayk Caldas
Ramos‡
vw,
Mayk Caldas
Ramos‡
 k,
Bojana
Ranković‡
k,
Bojana
Ranković‡
 fg,
Samuel G.
Rodriques‡
fg,
Samuel G.
Rodriques‡
 s,
Jacob N.
Sanders‡
s,
Jacob N.
Sanders‡
 x,
Philippe
Schwaller‡
x,
Philippe
Schwaller‡
 fg,
Marcus
Schwarting‡
y,
Jiale
Shi‡
fg,
Marcus
Schwarting‡
y,
Jiale
Shi‡
 b,
Berend
Smit‡
b,
Berend
Smit‡
 a,
Ben E.
Smith‡
a,
Ben E.
Smith‡
 e,
Joren
Van Herck‡
e,
Joren
Van Herck‡
 a,
Christoph
Völker‡
a,
Christoph
Völker‡
 r,
Logan
Ward‡
r,
Logan
Ward‡
 z,
Sean
Warren‡
z,
Sean
Warren‡
 c,
Benjamin
Weiser‡
c,
Benjamin
Weiser‡
 c,
Sylvester
Zhang‡
c,
Sylvester
Zhang‡
 c,
Xiaoqi
Zhang‡
c,
Xiaoqi
Zhang‡
 a,
Ghezal Ahmad
Zia‡
a,
Ghezal Ahmad
Zia‡
 r,
Aristana
Scourtas
r,
Aristana
Scourtas
 aa,
K. J.
Schmidt
aa,
K. J.
Schmidt
 aa,
Ian
Foster
aa,
Ian
Foster
 ab,
Andrew D.
White
ab,
Andrew D.
White
 k and
Ben
Blaiszik
k and
Ben
Blaiszik
 *aa
*aa
aLaboratory of Molecular Simulation (LSMO), Institut des Sciences et Ingénierie Chimiques, Ecole Polytechnique Fédérale de Lausanne (EPFL), Sion, Valais, Switzerland. E-mail: mail@kjablonka.com
bDepartment of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA
cDepartment of Chemistry, McGill University, Montreal, Quebec, Canada
dReincarnate Inc., USA
eYusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge, CB2 1EW, UK
fLaboratory of Artificial Chemical Intelligence (LIAC), Institut des Sciences et Ingénierie Chimiques, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
gNational Centre of Competence in Research (NCCR) Catalysis, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
hIndependent Researcher, San Diego, CA, USA
iMechanical Engineering and Materials Science, Duke University, USA
jMaterial Measurement Laboratory, National Institute of Standards and Technology, Maryland 20899, USA
kDepartment of Chemical Engineering, University of Rochester, USA
lApplied Mathematics and Computational Research Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA
mInstitut de la Matière Condensée et des Nanosciences (IMCN), UCLouvain, Chemin des Étoiles 8, Louvain-la-Neuve, 1348, Belgium
nMatgenix SRL, 185 Rue Armand Bury, 6534 Gozée, Belgium
oInstituto de Ciencia y Tecnología del Carbono (INCAR), CSIC, Francisco Pintado Fe 26, 33011 Oviedo, Spain
pDepartment of Computer Science, University of Chicago, Chicago, Illinois 60637, USA
qComputer Science, University of California, Berkeley, CA 94704, USA
rBundesanstalt für Materialforschung und -prüfung, Unter den Eichen 87, 12205 Berlin, Germany
sFrancis Crick Institute, 1 Midland Rd, London NW1 1AT, UK
tAmerican Family Insurance Data Science Institute, University of Wisconsin–Madison, Madison, WI 53706, USA
uDepartment of Chemical Engineering, University of Washington, Seattle, WA 98105, USA
vOpenBioML.org, UK
wStability.AI, UK
xDepartment of Chemistry and Biochemistry, University of California, Los Angeles, CA 90095, USA
yDepartment of Computer Science, University of Chicago, Chicago, IL 60490, USA
zData Science and Learning Division, Argonne National Lab, USA
aaGlobus, University of Chicago, Data Science and Learning Division, Argonne National Lab, USA. E-mail: blaiszik@uchicago.edu
abDepartment of Computer Science, University of Chicago, Data Science and Learning Division, Argonne National Lab, USA
First published on 8th August 2023
Abstract
Large-language models (LLMs) such as GPT-4 caught the interest of many scientists. Recent studies suggested that these models could be useful in chemistry and materials science. To explore these possibilities, we organized a hackathon. This article chronicles the projects built as part of this hackathon. Participants employed LLMs for various applications, including predicting properties of molecules and materials, designing novel interfaces for tools, extracting knowledge from unstructured data, and developing new educational applications. The diverse topics and the fact that working prototypes could be generated in less than two days highlight that LLMs will profoundly impact the future of our fields. The rich collection of ideas and projects also indicates that the applications of LLMs are not limited to materials science and chemistry but offer potential benefits to a wide range of scientific disciplines.
1. Introduction
The intersection of machine learning (ML) with chemistry and materials science has witnessed remarkable advancements in recent years.1–9 Much progress has been made in using ML to, e.g., accelerate simulations10,11 or to directly predict properties or compounds for a given application.12 Thereby, developing custom, hand-crafted models for any given application is still common practice. Since science rewards doing novel things for the first time, we now face a deluge of tools and machine-learning models for various tasks. These tools commonly require input data in their own rigid, well-defined form (e.g., a table with specific columns or images from a specific microscope with specific dimensions). Further, they typically also report their outputs in non-standard and sometimes proprietary forms.This rigidity sharply contrasts the standard practice in the (experimental) molecular and materials sciences, which is intrinsically fuzzy and highly context-dependent.13 For instance, researchers have many ways to refer to a molecule (e.g., IUPAC name, conventional name, simplified molecular-input line-entry system (SMILES)14) and to report results and procedures. In particular, for the latter, it is known that small details such as the order of addition or the strength of stirring (e.g., “gently” vs. “strongly”) are crucial in determining the outcome of reactions. We do not have a natural way to deal with this fuzziness, and often a conversion into structured tabular form (the conventional input format for ML models) is impossible. Our current “solution” is to write conversion programs and chain many tools with plenty of application-specific “glue code” to enable scientific workflows. However, this fuzziness of chemistry and heterogeneity of tools have profound consequences: a never-ending stream of new file formats, interfaces, and interoperability tools exists, and users cannot keep up with learning.15 In addition, almost any transformation of highly context-dependent text (e.g., description of a reaction procedure) into structured, tabular form will lead to a loss of information.
One of the aims of this work is to demonstrate how large language models (LLMs) such as the generative pretrained transformer (GPT)-4,16–21 can be used to address these challenges. Foundation models such as GPTs are general-purpose technologies22 that can solve tasks they have not explicitly been trained on,23,24 use tools,25–27 and be grounded in knowledge bases.28,29 As we also show in this work, they provide new pathways of exploration, new opportunities for flexible interfaces, and may be used to effectively solve certain tasks themselves; e.g., we envision LLMs enabling non-experts to program (“malleable software”) using natural language as the “programming language”,30 extract structured information, and create digital assistants that make our tools interoperable—all based on unstructured, natural-language inputs.
Inspired by early reports on the use of these LLMs in chemical research,31–34 we organized a virtual hackathon event focused on understanding the applicability of LLMs to materials science and chemistry. The hackathon aimed to explore the multifaceted applications of LLMs in materials science and chemistry and encourage creative solutions to some of the pressing challenges in the field. This article showcases some of the projects (Table 1) developed during the hackathon.
| Name | Authors | Links |
|---|---|---|
| Predictive modeling | ||
| Accurate molecular energy predictions | Ankur K. Gupta, Garrett W. Merz, Alishba Imran, Wibe A. de Jong |

|
 https://doi.org/10.5281/zenodo.8104930
https://doi.org/10.5281/zenodo.8104930
|
||
| Text2Concrete | Sabine Kruschwitz, Christoph Völker, Ghezal Ahmad Zia |
 https://ghezalahmad/LLMs-for-the-Design-of-Sustainable-Concretes
https://ghezalahmad/LLMs-for-the-Design-of-Sustainable-Concretes
|
 https://doi.org/10.5281/zenodo.8091195
https://doi.org/10.5281/zenodo.8091195
|
||
| Molecule discovery by context | Zhi Hong, Logan Ward |
 https://globuslabs/ScholarBERT-XL
https://globuslabs/ScholarBERT-XL
|
 https://doi.org/10.5281/zenodo.8122087
https://doi.org/10.5281/zenodo.8122087
|
||
| Genetic algorithm without genes | Benjamin Weiser, Jerome Genzling, Nicolas Gastellu, Sylvester Zhang, Tao Liu, Alexander Al-Feghali, Nicolas Moitessier, Anne Labarre, Steven Ma |
 https://BenjaminWeiser/LLM-Guided-GA
https://BenjaminWeiser/LLM-Guided-GA
|
 https://doi.org/10.5281/zenodo.8125541
https://doi.org/10.5281/zenodo.8125541
|
||
| Text-template paraphrasing | Michael Pieler |
 https://micpie/text-template-paraphrasing-chemistry
https://micpie/text-template-paraphrasing-chemistry
|
 https://doi.org/10.5281/zenodo.8093615
https://doi.org/10.5281/zenodo.8093615
|
||
| Automation and novel interfaces | ||
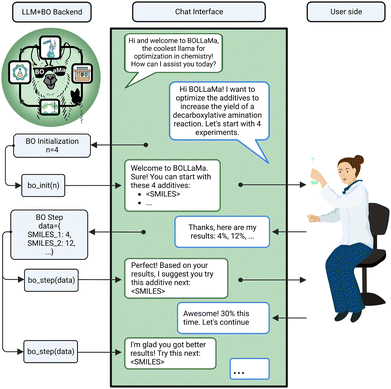
| BOLLaMa | Bojana Ranković, Andres M. Bran, Philippe Schwaller |
 https://doncamilom/BOLLaMa
https://doncamilom/BOLLaMa
|
 https://doi.org/10.5281/zenodo.8096827
https://doi.org/10.5281/zenodo.8096827
|
||
| sMolTalk | Jakub Lála, Sean Warren, Samuel G. Rodriques |
 https://jakublala/smoltalk-legacy
https://jakublala/smoltalk-legacy
|
 https://doi.org/10.5281/zenodo.8081749
https://doi.org/10.5281/zenodo.8081749
|
||
| MAPI-LLM | Mayk Caldas Ramos, Sam Cox, Andrew White |
 https://maykcaldas/MAPI_LLM
https://maykcaldas/MAPI_LLM
|
 https://maykcaldasMAPI_LLM
https://maykcaldasMAPI_LLM
|
||
 https://doi.org/10.5281/zenodo.8097336
https://doi.org/10.5281/zenodo.8097336
|
||
Conversational electronic lab notebook (ELN) interface ( ) ) |
Joshua D. Bocarsly, Matthew L. Evans and Ben E. Smith |
 https://the-grey-group/datalab
https://the-grey-group/datalab
|
 https://doi.org/10.5281/zenodo.8127782
https://doi.org/10.5281/zenodo.8127782
|
||
| Knowledge extraction | ||
| InsightGraph | Defne Circi, Shruti Badhwar |
 https://defnecirci/InsightGraph
https://defnecirci/InsightGraph
|
 https://doi.org/10.5281/zenodo.8092575
https://doi.org/10.5281/zenodo.8092575
|
||
| Extracting structured data from free-form organic synthesis text | Qianxiang Ai, Jacob N. Sanders, Jiale Shi, Stefan Bringuier, Brenden Pelkie, Marcus Schwarting |
 https://qai222LLM_organic_synthesis
https://qai222LLM_organic_synthesis
|
 https://doi.org/10.5281/zenodo.8091902
https://doi.org/10.5281/zenodo.8091902
|
||
| TableToJson: structured information from scientific data in tables | María Victoria Gil |
 https://vgvinter/TableToJson
https://vgvinter/TableToJson
|
 https://doi.org/10.5281/zenodo.8093731
https://doi.org/10.5281/zenodo.8093731
|
||
| AbstractToTitle & TitleToAbstract: text summarization and generation | Kamal Choudhary |
 https://usnistgov/chemnlp
https://usnistgov/chemnlp
|
 https://doi.org/10.5281/zenodo.8122419
https://doi.org/10.5281/zenodo.8122419
|
||
| Education | ||
| I-Digest | Beatriz Mouriño, Elias Moubarak, Joren Van Herck, Sauradeep Majumdar, Xiaoqi Zhang |
 https://XiaoqZhang/i-Digest
https://XiaoqZhang/i-Digest
|
 https://doi.org/10.5281/zenodo.8080962
https://doi.org/10.5281/zenodo.8080962
|
||
One of the conclusions of this work is that without these LLMs, such projects would take many months. The diversity of topics these projects address illustrates the broad applicability of LLMs; the projects touch many different aspects of materials science and chemistry, from the wet lab to the computational chemistry lab, software interfaces, and even the classroom. While the examples below are not yet polished products, the simple observation that such capabilities could be created in hours underlines that we need to start thinking about how LLMs will impact the future of materials science, chemistry, and beyond.35 The diverse applications show that LLMs are here to stay and are likely a foundational capability that will be integrated into most aspects of the research process. Even so, the pace of the developments highlights that we are only beginning to scratch the surface of what LLMs can do for chemistry and materials science.
Table 1 lists the different projects created in this collaborative effort across eight countries and 22 institutions (ESI Section V†). One might expect that 1.5 days of intense collaborations would, at best, allow a cursory exploration of a topic. However, the diversity of topics and the diversity in the participants' expertise, combined with the need to deliver a working prototype (within a short window of time) and the ease of prototyping with LLMs, generated not only many questions but also pragmatic prototypes. The projects were typically carried out in an exploratory way and without any evaluation of impact. In the remainder of this article, we focus on the insights we obtained from this collective effort. For the details of each project, we refer to the ESI.† While different challenges were explored during this hackathon, the results were preliminary. Digital Discovery did not peer review the soundness of each study. Instead, the peer review for this perspective was to scope the potential of LLMs in chemistry and materials science.
We have grouped the projects into four categories: (1) predictive modeling, (2) automation and novel interfaces, (3) knowledge extraction, and (4) education. The projects in the predictive modeling category use LLMs for classification and regression tasks—and also investigate ways to incorporate established concepts such as Δ-ML36 or novel concepts such as “fuzzy” context into the modeling. The automation and novel interfaces projects show that natural language might be the universal “glue” connecting our tools—perhaps in the future, we will need not to focus on new formats or standards but rather use natural language descriptions to connect across the existing diversity and different modalities.35
LLMs can also help make knowledge more accessible, as the projects in the “knowledge extraction” category show; they can extract structured information from unstructured text. In addition, as the project in the “education” category shows, LLMs can also offer new educational opportunities.
1.1 Predictive modeling
Predictive modeling is a common application of ML in chemistry. Based on the language-interfaced fine-tuning (LIFT) framework,37 Jablonka et al.32 have shown that LLMs can be employed to predict various chemical properties, such as solubility or HOMO–LUMO gaps based on line representations of molecules such as self-referencing embedded strings (SELFIES)38,39 and SMILES. Taking this idea even further, Ramos et al.34 used this framework (with in-context learning (ICL)) for Bayesian optimization—guiding experiments without even training models. These few-shot learning abilities have also been benchmarked by Guo et al.40The projects in the following build on top of those initial results and extend them in novel ways as well as by leveraging established techniques from quantum machine learning.
Given that these encouraging results could be achieved with and without fine-tuning (i.e., updates to the weights of the model) for the language-interfaced training on tabular datasets, we use the term LIFT also for ICL settings in which structured data is converted into text prompts for an LLM.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules in the QM9-G4MP2 dataset.
000 molecules in the QM9-G4MP2 dataset.
The Berkeley–Madison team (Ankur Gupta, Garrett Merz, Alishba Imran, and Wibe de Jong) used this dataset to fine-tune different LLMs using the LIFT framework. The team investigated if they could use an LLM to predict atomization energies with chemical accuracy. Jablonka et al.32 emphasized that these LLMs might be particularly useful in the low-data limit. Here, we have a relatively large dataset, so it is an ideal system to gather insights into the performance of these models for datasets much larger than those used by Jablonka et al.32
The Berkeley–Madison team showed that the LIFT framework based on simple line representations such as SMILES and SELFIES38,39 can yield good predictions (R2 > 0.95 on a holdout test set), that are, however, still inferior to dedicated models that have access to 3D information.44,45 An alternative approach to achieve chemical accuracy with LLMs tuned only on string representations is to leverage a Δ-ML scheme46 in which the LLM is tuned to predict the difference between G4(MP2) and B3LYP47 energies. Table 2 shows that good agreement could be achieved for the Δ-ML approach. This showcases how techniques established for conventional ML on molecules can also be applied with LLMs.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 232 molecules), using 10% (13
232 molecules), using 10% (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 026 molecules) as a holdout test set. GPTChem refers to the approach reported by Jablonka et al.,32 GPT-2-LoRA to PEFT of the GPT-2 model using LoRA. The results indicate that the LIFT framework can also be used to build predictive models for atomization energies, that can reach chemical accuracy using a Δ-ML scheme. Baseline performance (mean absolute error reported by Ward et al.45): 0.0223 eV for FCHL-based prediction of GP4(MP2) atomization energies and 0.0045 eV (SchNet) and 0.0052 eV (FCHL) for the Δ-ML scheme
026 molecules) as a holdout test set. GPTChem refers to the approach reported by Jablonka et al.,32 GPT-2-LoRA to PEFT of the GPT-2 model using LoRA. The results indicate that the LIFT framework can also be used to build predictive models for atomization energies, that can reach chemical accuracy using a Δ-ML scheme. Baseline performance (mean absolute error reported by Ward et al.45): 0.0223 eV for FCHL-based prediction of GP4(MP2) atomization energies and 0.0045 eV (SchNet) and 0.0052 eV (FCHL) for the Δ-ML scheme
| Mol. repr. & framework | G4(MP2) atomization energy | (G4(MP2)-B3LYP) atomization energy | ||
|---|---|---|---|---|
| R 2 | Median absolute deviation (MAD)/eV | R 2 | MAD/eV | |
| SMILES: GPTChem | 0.984 | 0.99 | 0.976 | 0.03 |
| SELFIES: GPTChem | 0.961 | 1.18 | 0.973 | 0.03 |
| SMILES: GPT2-LoRA | 0.931 | 2.03 | 0.910 | 0.06 |
| SELFIES: GPT2-LoRA | 0.959 | 1.93 | 0.915 | 0.06 |
Importantly, this approach is not limited to the OpenAI application programming interface (API). With parameter efficient fine-tuning (PEFT) with low-rank adaptors (LoRA)48 of the GPT-2 model,49 one can also obtain comparable results on consumer hardware. These results make the LIFT approach widely more accessible.
Interestingly, the largest LLMs can already give predictions without any fine-tuning. These models can “learn” from the few examples provided by the user in the prompt. Of course, such a few-shot approach (or ICL,20) does not allow for the same type of optimization as fine-tuning, and one can therefore expect it to be less accurate. However, Ramos et al.34 showed that this method could perform well—especially if only so few data points are available such that fine-tuning is not a suitable approach.
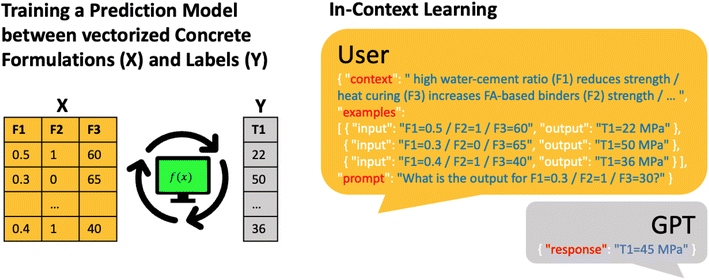
For their case study, the Text2Concrete team found a predictive accuracy comparable to a Gaussian process regression (GPR) model (but inferior to a random forest (RF) model). However, one significant advantage of LLMs is that one can easily incorporate context. The Text2Concrete team used this to include well-established design principles like the influence of the water-to-cement ratio on strength (Fig. 1) into the modeling by simply stating the relationship between the features in natural language (e.g., “high water/cement ratio reduces strength”). This additional context reduced the outliers and outperformed the RF model (R2 of 0.67 and 0.72, respectively).
The exciting aspect is that this is a typical example of domain knowledge that cannot be captured with a simple equation incorporable into conventional modeling workflows. Such “fuzzy” domain knowledge, which may sometimes exist only in the minds of researchers, is common in chemistry and materials science. With the incorporation of such “fuzzy” knowledge into LIFT-based predictions using LLMs, we now have a novel and very promising approach to leverage such domain expertise that we could not leverage before. Interestingly, this also may provide a way to test “fuzzy” hypotheses, e.g., a researcher could describe the hypothesis in natural language and see how it affects the model accuracy. While the Text2Concrete example has not exhaustively analyzed how “fuzzy” context alterations affect LLM performance, we recognize this as a key area for future research.
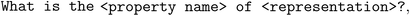 where the texts in chevrons are placeholders that are replaced with actual values such as “solubility” and “2-acetyloxybenzoic acid”. In the low-data regime, data points are “wasted” by the model needing to learn the syntax of the prompt templates. In the big-data regime, in contrast, one might worry that the model loses some of its general language modeling abilities by always dealing with the same template. This naturally raises the question if one can augment the dataset to mitigate these problems—thereby leveraging again, similar to Δ-ML, a technique that has found use in conventional ML previously. However, text-based data are challenging to augment due to their discrete nature and the fact that the augmented text still needs to be syntactically and semantically valid. Interestingly, as Michael Pieler (https://www.openbioml.org and Stability.AI) shows (and as has been explored by Dai et al.59), it turns out that LLMs can also be used to address this problem by simply prompting an LLM (e.g., GPT-4 or Anthropic's Claude) to paraphrase a prompt template (see ESI Section ID†).
where the texts in chevrons are placeholders that are replaced with actual values such as “solubility” and “2-acetyloxybenzoic acid”. In the low-data regime, data points are “wasted” by the model needing to learn the syntax of the prompt templates. In the big-data regime, in contrast, one might worry that the model loses some of its general language modeling abilities by always dealing with the same template. This naturally raises the question if one can augment the dataset to mitigate these problems—thereby leveraging again, similar to Δ-ML, a technique that has found use in conventional ML previously. However, text-based data are challenging to augment due to their discrete nature and the fact that the augmented text still needs to be syntactically and semantically valid. Interestingly, as Michael Pieler (https://www.openbioml.org and Stability.AI) shows (and as has been explored by Dai et al.59), it turns out that LLMs can also be used to address this problem by simply prompting an LLM (e.g., GPT-4 or Anthropic's Claude) to paraphrase a prompt template (see ESI Section ID†).
This approach will allow us to automatically create new paraphrased high-quality prompts for LIFT-based training very efficiently—to augment the dataset and reduce the risk of overfitting to a specific template. Latter might be particularly important if one still wants to retain general language abilities of the LLMs after finetuning on chemistry or material science data.
One might hypothesize that LLMs can make the evolution process more efficient, e.g., by using an LLM to handle the reproduction. One might expect that inductive biases in the LLM help create recombined molecules which are more chemically viable, maintaining the motifs of the two parent molecules better than a random operation.
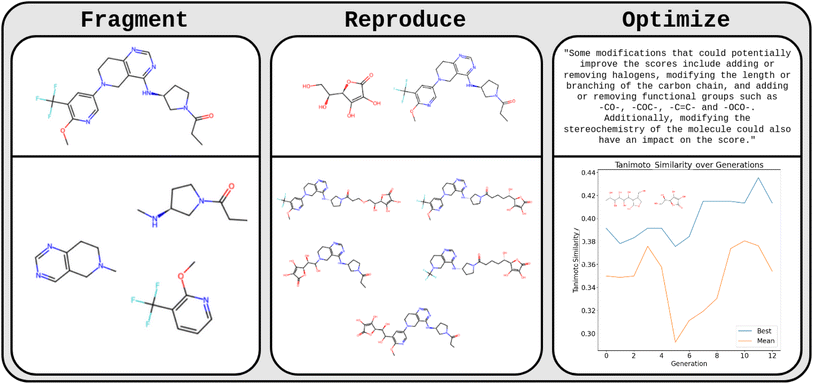
The team from McGill University (Benjamin Weiser, Jerome Genzling, Nicolas Gastellu, Sylvester Zhang, Tao Liu, Alexander Al-Feghali, Nicolas Moitessier) set out the first steps to test this hypothesis (Fig. 2). In initial experiments, they found that GPT-3.5, without any finetuning, can fragment molecules provided as SMILES at rotatable bonds with a success rate of 70%. This indicates that GPT-3.5 understands SMILES strings and aspects of their relation to the chemical structures they represent. Subsequently, they asked the LLMs to fragment and recombine two given molecules. The LLM frequently created new combined molecules with fragments of each species which were reasonable chemical structures more often than a random SMILES string combining operation (two independent organic chemists judged the LLM-GA-generated molecules to be chemically reasonable in 32/32 cases, but only in 21/32 cases for the random recombination operation).
Encouraged by these findings, they prompted an LLM with 30 parent molecules and their performance scores (Tanimoto similarity to vitamin C) with the task to come up with n new molecules that the LLM “believes” to improve the score. A preliminary visual inspection suggests that the LLM might produce chemically reasonable modifications. Future work will need to systematically investigate potential improvements compared to conventional GAs.
The importance of the results of the McGill team is that they indicate that these LLMs (when suitably conditioned) might not only reproduce known structures but generate new structures that make chemical sense.32,61
A current limitation of this approach is that most LLMs still struggle to output valid SMILES without explicit fine-tuning.33 We anticipate that this problem might be mitigated by building foundation models for chemistry (with more suitable tokenization62,63), as, for instance, the ChemNLP project of openbioml.org attempts to do (https://github.com/OpenBioML/chemnlp). In addition, the context length limits the number of parent molecules that can be provided as examples.
Overall, we see that the flexibility of the natural language input and the in-context learning abilities allows using LLMs in very different ways—to very efficiently build predictive models or to approach molecular and material design in entirely unprecedented ways, like by providing context—such as “fuzzy” design rules—or simply prompting the LLM to come up with new structures. However, we also find that some “old” ideas, such as Δ-ML and data augmentation, can also be applied in this new paradigm.
1.2 Automation and novel interfaces
Yao et al.64 and Schick et al.25 have shown that LLMs can be used as agents that can autonomously make use of external tools such as Web-APIs—a paradigm that some call MRKL (pronounced “miracle”) systems—modular reasoning, knowledge, and language systems.26 By giving LLMs access to tools and forcing them to think step-by-step,65 we can thereby convert LLMs from hyperconfident models that often hallucinate to systems that can reason based on observations made by querying robust tools. As the technical report for GPT-4 highlighted,66 giving LLMs access to tools can lead to emergent behavior, i.e., enabling the system to do things that none of its parts could do before. In addition, this approach can make external tools more accessible—since users no longer have to learn tool-specific APIs. It can also make tools more interoperable—by using natural language instead of “glue code” to connect tools.This paradigm has recently been used by Bran et al.67 to create digital assistants that can call and combine various tools such as Google search and the IBM RXN retrosynthesis tool when prompted with natural language. Boiko et al.68 used a similar approach and gave LLMs access to laboratories via cloud lab APIs. In their system, the LLM could use external tools to plan a synthesis, which it could execute using the cloud lab.
To answer these questions, state-of-the-art computational tools or existing databases can be used. However, their use often requires expert knowledge. To use existing databases, one must choose which database to use, how to query the database, and what representation of the compound is used (e.g., international chemical identifier (InChI), SMILES, etc.). Otherwise, if the data is not in a database, one must run calculations, which requires a deep understanding of technical details. LLMs can simplify this process. By typing in a question, we can prompt the LLM to translate this question into a workflow that leads to the answer.
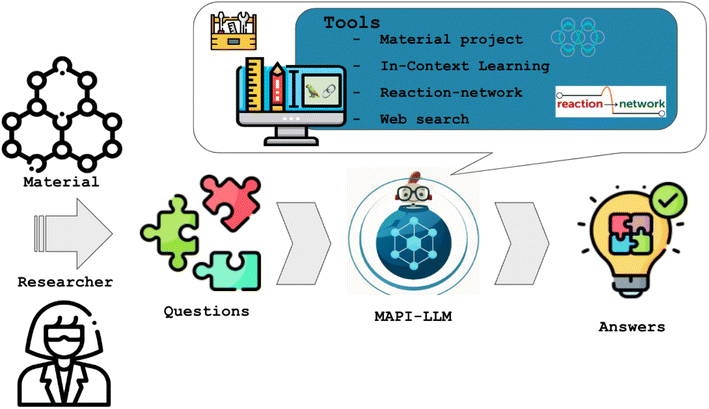
The MAPI-LLM team (Mayk Caldas Ramos, Sam Cox, Andrew White) made the first steps towards developing such a system (MAPI-LLM) and created a procedure to convert a text prompt into a query of the Materials Project API (MAPI) to answer questions such as “Is the material AnByCz stable?” In addition, MAPI-LLM is capable of handling classification queries, such as “Is Fe2O3 magnetic?”, as well as regression problems, such as “What is the band gap of Mg(Fe2O3)2?”.
Because an LLM is used to create the workflow, MAPI-LLM can process even more complex questions. For instance, the question “If Mn23FeO32 is not metallic, what is its band gap?” should create a two-step workflow first to check if the material is metallic and then obtain its band gap if it is not.
Moreover, MAPI-LLM applies ICL if the data for a material's property is unavailable via the MAPI. MAPI-LLM generates an ICL prompt, building context based on the data for similar materials available in Materials Project database. This context is then leveraged by an LLM to infer properties for the unknown material. This innovative use of ICL bridges data gaps and enhances MAPI-LLM's robustness and versatility (Fig. 3).
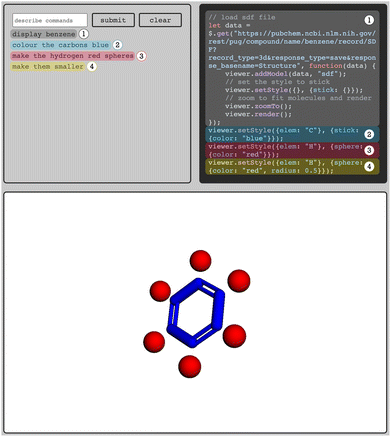
As the sMolTalk-team (Jakub Lála, Sean Warren, Samuel G. Rodriques) showed, one can use LLMs to write code for visualization tools such as  to address this inefficiency.70 Interestingly, few-shot prompting with several examples of user input with the expected JavaScript code that manipulates the
to address this inefficiency.70 Interestingly, few-shot prompting with several examples of user input with the expected JavaScript code that manipulates the  viewer is all that is needed to create a prototype of an interface that can retrieve protein structures from the protein data bank (PDB) and create custom visualization solutions, e.g., to color parts of a structure in a certain way (Fig. 4). The beauty of the language models is that the user can write the prompt in many different (“fuzzy”) ways: whether one writes “color” or “colour”, or terms like “light yellow” or “pale yellow” the LLM translates it into something the visualization software can interpret.
viewer is all that is needed to create a prototype of an interface that can retrieve protein structures from the protein data bank (PDB) and create custom visualization solutions, e.g., to color parts of a structure in a certain way (Fig. 4). The beauty of the language models is that the user can write the prompt in many different (“fuzzy”) ways: whether one writes “color” or “colour”, or terms like “light yellow” or “pale yellow” the LLM translates it into something the visualization software can interpret.
However, this application also highlights that further developments of these LLM-based tools are needed. For example, a challenge the sMolTalk tool faces is robustness. For instance, fragments from the prompt tend to leak into the output and must be handled with more involved mechanisms, such as retries (in which one gives the LLMs access to the error messages) or prompt engineering. Further improvement can also be expected if the application leverages a knowledge base such as the documentation of 
As the work of Hocky and White shows,71 an LLM-interface for software can also be used with other programs such as  ,72 and extended with speech-to-text models (such as Whisper73) to enable voice control of such programs. In particular, such an LLM-based agent approach might be implemented for the
,72 and extended with speech-to-text models (such as Whisper73) to enable voice control of such programs. In particular, such an LLM-based agent approach might be implemented for the  program, where various tools for protein engineering could be interfaced through a chat interface, lowering the barrier to entry for biologists to use recent advancements within in silico protein engineering (such as RosettaFold74 or RFDiffusion75).
program, where various tools for protein engineering could be interfaced through a chat interface, lowering the barrier to entry for biologists to use recent advancements within in silico protein engineering (such as RosettaFold74 or RFDiffusion75).
1.2.2.1 ELN interface:
 .
In addition to large, highly curated databases with well-defined data models76 (such as those addressed by the MAPI-LLM project), experimental materials and chemistry data is increasingly being captured using digital tools such as ELNs and laboratory information systems (LIMS). Importantly, these tools can be used to record both structured and unstructured lab data in a manner that is actionable by both humans and computers. However, one challenge in developing these systems is that it is difficult for a traditional user interface to have enough flexibility to capture the richness and diversity of real, interconnected, experimental data. Interestingly, LLMs can interpret and contextualize both structured and unstructured data and can therefore be used to create a novel type of flexible, conversational interface to such experimental data. The
.
In addition to large, highly curated databases with well-defined data models76 (such as those addressed by the MAPI-LLM project), experimental materials and chemistry data is increasingly being captured using digital tools such as ELNs and laboratory information systems (LIMS). Importantly, these tools can be used to record both structured and unstructured lab data in a manner that is actionable by both humans and computers. However, one challenge in developing these systems is that it is difficult for a traditional user interface to have enough flexibility to capture the richness and diversity of real, interconnected, experimental data. Interestingly, LLMs can interpret and contextualize both structured and unstructured data and can therefore be used to create a novel type of flexible, conversational interface to such experimental data. The  team (Joshua D. Bocarsly, Matthew L. Evans, and Ben E. Smith) embedded an LLM chat interface within
team (Joshua D. Bocarsly, Matthew L. Evans, and Ben E. Smith) embedded an LLM chat interface within  an open source materials chemistry data management system, where the virtual LLM-powered assistant can be “attached” to a given sample. The virtual assistant has access to responses from the JavaScript object notation (JSON) API of
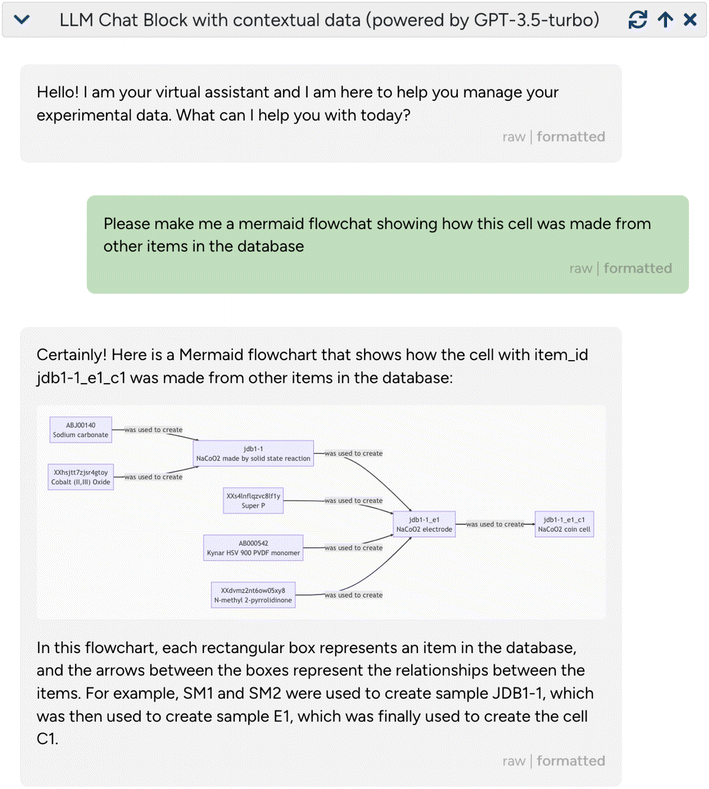
an open source materials chemistry data management system, where the virtual LLM-powered assistant can be “attached” to a given sample. The virtual assistant has access to responses from the JavaScript object notation (JSON) API of 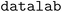 (containing both structured and unstructured/free text data) and can use them to perform several powerful tasks: first, it can contextualize existing data by explaining related experiments from linked responses, resolving acronyms/short-hand notations used by experimentalists, or creating concise textual summaries of complex and nested entries. Second, it can reformat or render the data, for instance, by creating (
(containing both structured and unstructured/free text data) and can use them to perform several powerful tasks: first, it can contextualize existing data by explaining related experiments from linked responses, resolving acronyms/short-hand notations used by experimentalists, or creating concise textual summaries of complex and nested entries. Second, it can reformat or render the data, for instance, by creating (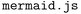 ) flowcharts or (Markdown) tables (Fig. 5). Third, it can use its generic reasoning abilities to suggest future experiments, for instance, related materials to study, synthesis protocols to try, or additional characterization techniques. This is shown in the examples given in ESI Section 2C,† where
) flowcharts or (Markdown) tables (Fig. 5). Third, it can use its generic reasoning abilities to suggest future experiments, for instance, related materials to study, synthesis protocols to try, or additional characterization techniques. This is shown in the examples given in ESI Section 2C,† where 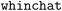 was able to provide hints about which NMR-active nuclei can be probed in the given sample.
was able to provide hints about which NMR-active nuclei can be probed in the given sample.
It is easy to envision that this tool could be even more helpful by fine-tuning or conditioning it on a research group's knowledge base (e.g., group Wiki or standard operating procedures) and communication history (e.g., a group's Slack history). An important limitation of the current implementation is that the small context window of available LLMs limits the amount of JSON data one can directly provide within the prompt, limiting each conversation to analyzing a relatively small number of samples. Therefore, one needs to either investigate the use of embeddings to determine which samples to include in the context or adopt an “agent” approach where the assistant is allowed to query the API of the ELN (interleaved with extraction and summarization calls).
As the examples in this section show, we find that LLMs have the potential to greatly enhance the efficiency of a diverse array of processes in chemistry and materials science by providing novel interfaces to tools or by completely automating their use. This can help streamline workflows, reduce human error, and increase productivity—often by replacing “glue code” with natural language or familiarising oneself with a software library by chatting with an LLM.
1.3 Knowledge extraction
Beyond proving novel interfaces for tools, LLMs can also serve as powerful tools for extracting knowledge from the vast amount of chemical literature. With LLMs, researchers can rapidly mine and analyze large volumes of data, enabling them to uncover novel insights and advance the frontiers of chemical knowledge. Tools such as paper-qa28 can help to dramatically cut down the time required for literature search by automatically retrieving, summarizing, and contextualizing relevant fragments from the entire corpus of the scientific literature—for example, answering questions (with suitable citations) based on a library of hundreds of documents.35 As the examples in the previous section indicated, this is particularly useful if the model is given access to search engines on the internet.However, for certain applications, one can construct powerful prototypes using only careful prompting. For instance, the InsightGraph team (Defne Circi, Shruti Badhwar) showed that GPT-3.5-turbo, when prompted with an example JSON containing a high-level schema and information on possible entities (e.g., materials) and pairwise relationships (e.g., properties of materials), can, as Fig. 7 illustrates, provide a knowledge graph representation of the entities and their relationships in a text describing the properties and composition of polymer nanocomposites. A further optimized version of this tool might offer a concise and visual means to understand and compare material types quickly and uses across sets of articles—a task that currently is very laborious. An advanced potential application is the creation of structured, materials-specific datasets for fact-based question-answering and downstream machine-learning tasks.
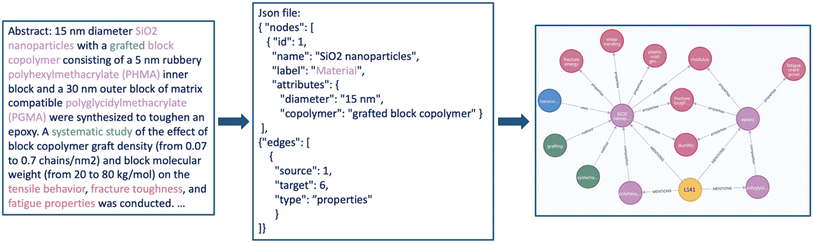 | ||
| Fig. 7 The InsightGraph interface. A suitably prompted LLM can create knowledge graph representations of scientific text that can be visualized using tools such as neo4j's visualization tools.83 | ||
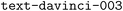 ) can, after finetuning on only 300 prompt–completion pairs, extract 93% of the components from the free-text reaction description into valid JSONs (Fig. 8). Such models might significantly increase the data available for training models on tasks such as predicting reaction conditions and yields. In contrast to previous approaches, such as the one of Guo et al.,85 the use of LLM does not require a specialized modeling setup but can be carried out with relatively little expertise. It is worth noting that all reaction data submitted to ORD are made available under the CC-BY-SA license, which makes ORD a suitable data source for fine-tuning or training an LLM to extract structured data from organic procedures. A recent study on gold nanorod growth procedures also demonstrated the ability of LLM in a similar task.82 In contrast to the LIFT-based prediction of atomization energies reported in the first section by the Berkeley–Madison team, parameter-efficient fine-tuning of the open-source Alpaca model86–88 using LoRA48 did not yield a model that can construct valid JSONs.
) can, after finetuning on only 300 prompt–completion pairs, extract 93% of the components from the free-text reaction description into valid JSONs (Fig. 8). Such models might significantly increase the data available for training models on tasks such as predicting reaction conditions and yields. In contrast to previous approaches, such as the one of Guo et al.,85 the use of LLM does not require a specialized modeling setup but can be carried out with relatively little expertise. It is worth noting that all reaction data submitted to ORD are made available under the CC-BY-SA license, which makes ORD a suitable data source for fine-tuning or training an LLM to extract structured data from organic procedures. A recent study on gold nanorod growth procedures also demonstrated the ability of LLM in a similar task.82 In contrast to the LIFT-based prediction of atomization energies reported in the first section by the Berkeley–Madison team, parameter-efficient fine-tuning of the open-source Alpaca model86–88 using LoRA48 did not yield a model that can construct valid JSONs.
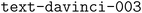 model, when prompted with a desired JSON schema and the HyperText Markup Language (HTML) of a table contained in a scientific paper, can generate structured JSON with the data in the table.
model, when prompted with a desired JSON schema and the HyperText Markup Language (HTML) of a table contained in a scientific paper, can generate structured JSON with the data in the table.
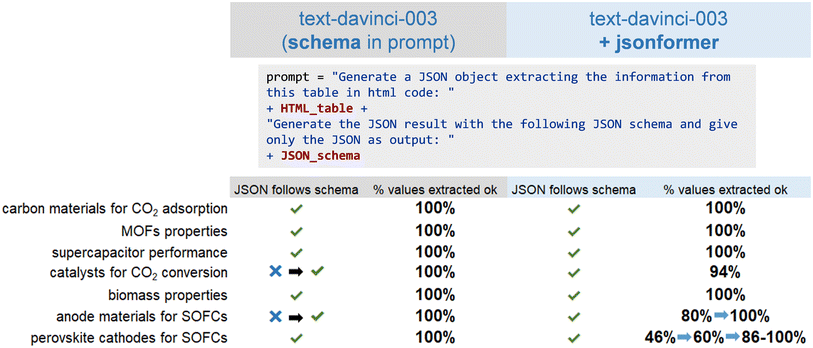
First, the OpenAI 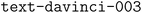 model was directly used to generate JSON objects from the table information. This approach was applied to several examples using tables collected from papers on different research topics within the field of chemistry.89–95 The accuracy for those different examples, calculated as the percentage of schema values generated correctly, is shown in Fig. 9. When the OpenAI model was prompted with the table and desired schema to generate a JSON object, it worked remarkably well in extracting the information from each table cell and inserting it at the expected place in the schema. As output, it provided a valid JSON object with a 100% success rate of error-free generated values in all the studied examples. However, in some examples, the model did not follow the schema.
model was directly used to generate JSON objects from the table information. This approach was applied to several examples using tables collected from papers on different research topics within the field of chemistry.89–95 The accuracy for those different examples, calculated as the percentage of schema values generated correctly, is shown in Fig. 9. When the OpenAI model was prompted with the table and desired schema to generate a JSON object, it worked remarkably well in extracting the information from each table cell and inserting it at the expected place in the schema. As output, it provided a valid JSON object with a 100% success rate of error-free generated values in all the studied examples. However, in some examples, the model did not follow the schema.
To potentially address this problem the team utilized the 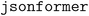 approach. This tool reads the keys from the JSON schema and only generates the value tokens, guaranteeing the generation of a syntactically valid JSON (corresponding to the desired schema) by the LLM.96,97 Using an LLM without such a decoding strategy cannot guarantee that valid JSON outputs are produced. With the
approach. This tool reads the keys from the JSON schema and only generates the value tokens, guaranteeing the generation of a syntactically valid JSON (corresponding to the desired schema) by the LLM.96,97 Using an LLM without such a decoding strategy cannot guarantee that valid JSON outputs are produced. With the 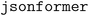 approach, in most cases, by using a simple descriptive prompt about the type of input text, structured data can be obtained with 100% correctness of the generated values. In one example, an accuracy of 80% was obtained due to errors in the generation of numbers in scientific notation. For a table with more complex content (long molecule names, hyphens, power numbers, subscripts, and superscripts,…) the team achieved an accuracy of only 46%. Most of these issues could be solved by adding a specific explanation in the prompt, increasing the accuracy to 100% in most cases.
approach, in most cases, by using a simple descriptive prompt about the type of input text, structured data can be obtained with 100% correctness of the generated values. In one example, an accuracy of 80% was obtained due to errors in the generation of numbers in scientific notation. For a table with more complex content (long molecule names, hyphens, power numbers, subscripts, and superscripts,…) the team achieved an accuracy of only 46%. Most of these issues could be solved by adding a specific explanation in the prompt, increasing the accuracy to 100% in most cases.
Overall, both approaches performed well in generating the JSON format. The OpenAI 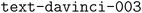 model could correctly extract structured information from tables and give a valid JSON output, but it cannot guarantee that the outputs will always follow the provided schema.
model could correctly extract structured information from tables and give a valid JSON output, but it cannot guarantee that the outputs will always follow the provided schema. 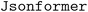 may present problems when special characters need to be generated, but most of these issues could be solved with careful prompting. These results show that LLMs can be a useful tool to help to extract scientific information in tables and convert it into a structured form with a fixed schema that can be stored in a database, which could encourage the creation of more topic-specific databases of research results.
may present problems when special characters need to be generated, but most of these issues could be solved with careful prompting. These results show that LLMs can be a useful tool to help to extract scientific information in tables and convert it into a structured form with a fixed schema that can be stored in a database, which could encourage the creation of more topic-specific databases of research results.
Large datasets of chemistry-related text are available from open-access platforms such as arXiv and PubChem. These articles contain titles, abstracts, and often complete manuscripts, which can be a testbed for evaluating LLMs as these titles and abstracts are usually written by expert researchers. Ideally, an LLM should be able to generate a title of an abstract close to the one developed by the expert, which can be considered a specialized text-summarization task. Similarly, given a title, an LLM should generate text close to the original abstract of the article, which can be considered a specialized text-generation task.
These tasks have been introduced by the AbstractToTitle & TitleToAbstract team (Kamal Choudhary) in the JARVIS-ChemNLP package.98 For text summarization, it uses a pre-trained Text-to-Text Transfer Transformer (T5) model developed by Google99 that is further fine-tuned to produce summaries of abstracts. On the arXiv condensed-matter physics (cond-mat) data, the team found that fine-tuning the model can help improve the performance (Recall-Oriented Understudy for Gisting Evaluation (ROUGE)-1 score of 39.0% which is better than an untrained model score of 30.8% for an 80/20 split).
For text generation, JARVIS-ChemNLP finetunes the pretrained GPT-2-medium49 model available in the HuggingFace library.100 After finetuning, the team found a ROUGE score of 31.7%, which is a good starting point for pre-suggestion text applications. Both tasks with well-defined train and test splits are now available in the JARVIS-Leaderboard platform for the AI community to compare other LLMs and systematically improve the performance.
In the future, such title to abstract capabilities can be extended to generating full-length drafts with appropriate tables, figures, and results as an initial start for the human researcher to help in the technical writing processes. Note that there have been recent developments in providing guidelines for using LLM-generated text in technical manuscripts,101 so such an LLM model should be considered as an assistant of writing and not the master/author of the manuscripts.
1.4 Education
Given all the opportunities LLM open for materials science and chemistry, there is an urgent need for education to adapt. Interestingly, LLMs also provide us with entirely novel educational opportunities,102 for example, by personalizing content or providing almost limitless varied examples.The I-Digest (Information-Digestor) hackathon team (Beatriz Mouriño, Elias Moubarak, Joren Van Herck, Sauradeep Majumdar, Xiaoqi Zhang) created a path toward such a new educational opportunity by providing students with a digital tutor based on course material such as lecture recordings. Using the Whisper model,73 videos of lecture recordings can be transcribed to text transcripts. The transcripts can then be fed into an LLM with the prompt to come up with questions about the content presented in the video (Fig. 10). In the future, these questions might be shown to students before a video starts, allowing them to skip parts they already know or after the video, guiding students to the relevant timestamps or additional material in case of an incorrect answer.
 | ||
| Fig. 10 The I-digest interface. (a) A video (e.g., of a lecture recording) can be described using the Whisper model. Based on the transcript, an LLM can generate questions (and answers). Those can assist students in their learning. (b) The LLM can also detect mentions of chemicals and link to further information about them (e.g., on PubChem103–105). | ||
Importantly, and in contrast to conventional educational materials, this approach can generate a practically infinite number of questions and could, in the future, continuously be improved by student feedback. In addition, it is easy to envision extending this approach to consider lecture notes or books to guide the students further or even recommend specific exercises.
2. Conclusion
The fact that the groups were able to present prototypes that could do quite complex tasks in such a short time illustrates the power of LLMs. Some of these prototypes would have taken many months of programming just a few months ago, but the fact that LLMs could reduce this time to a few hours is one of the primary reasons for the success of our hackathon. Combined with the time-constrained environment in teams (with practically zero cost of “failure”), we found more energy and motivation. The teams delivered more results than in most other hackathons we participated in.Through the LIFT framework, one can use LLMs to address problems that could already be addressed with conventional approaches—but in a much more accessible way (using the same approach for different problems), while also reusing established concepts such as Δ-ML. At the same time, however, we can use LLMs to model chemistry and materials science in novel ways; for example, by incorporating context information such as “fuzzy” design rules or directly operating on unstructured data. Overall, a common use case has been to use LLMs to deal with “fuzziness” in programming and tool development. We can already see tools like Copilot and ChatGPT being used to convert “fuzzy abstractions” or hard-to-define tasks into code. These advancements may soon allow everyone to write small apps or customize them to their needs (end-user programming). Additionally, we can observe an interesting trend in tool development: most of the logic in the showcased tools is written in English, not in Python or another programming language. The resulting code is shorter, easier to understand, and has fewer dependencies because LLMs are adept at handling fuzziness that is difficult to address with conventional code. This suggests that we may not need more formats or standards for interoperability; instead, we can simply describe existing solutions in natural language to make them interoperable. Exploring this avenue further is exciting, but it is equally important to recognize the limitations of LLMs, as they currently have limited interpretability and lack robustness.
It is interesting to note that none of the projects relied on the knowledge or understanding of chemistry by LLMs. Instead, they relied on general reasoning abilities and provided chemistry information through the context or fine-tuning. However, this also brings new and unique challenges. All projects used the models provided by OpenAI's API. While these models are powerful, we cannot examine how they were built or have any guarantee of continued reliable access to them.
Although there are open-source language models and techniques available, they are generally more difficult to use compared to simply using OpenAI's API. Furthermore, the performance of language models can be fragile, especially for zero- or few-shot applications. To further investigate this, new benchmarks are needed that go beyond the tabular datasets we have been using for ML for molecular and materials science—we simply have no frameworks to compare and evaluate predictive models that use context, unstructured data, or tools. Without automated tests, however, it is difficult to improve these systems systematically. On top of that, consistent benchmarking is hard because de-duplication is ill-defined even if the training data are known. To enable a scientific approach to the development and analysis of these systems, we will also need to revisit versioning frameworks to ensure reproducibility as systems that use external tools depend on the exact versions of training data, LLM, as well as of the external tools and prompting setup.
The diversity of the prototypes presented in this work shows that the potential applications are almost unlimited, and we can probably only see the tip of the iceberg—for instance, we didn't even touch modalities other than text thus far. In addition, we also want to note that the projects in the workshop mostly explored the use of LLMs as tools or oracles but not as muses.106 From techniques such as rubber duck debugging (describing the problem to a rubber duck),107 we know that even simple—non-intelligent—articulation or feedback mechanisms can help overcome roadblocks and create creative breakthroughs. Instead of explaining a problem to an inanimate rubber duck, we could instead have a conversation with an LLM, which could probe our thinking with questions or aid in brainstorming by generating diverse new ideas. Therefore, one should expect an LLM to be as good as a rubber duck—if not drastically more effective.
Given these new ways of working and thinking, combined with the rapid pace of developments in the field, we believe that we urgently need to rethink how we work and teach. We must discuss how we ensure safe use,108 standards for evaluating and sharing those models, and robust and reliable deployments. But we also need to discuss how we ensure that the next generation of chemists and materials scientists are proficient and critical users of these tools—that can use them to work more efficiently while critically reflecting on the outputs of the systems. This work showcased some potential applications of LLMs that will benefit from further investigation. We believe that to truly leverage the power of LLMs in the molecular and material sciences, however, we need a community effort—including not only chemists and computer scientists but also lawyers, philosophers, and ethicists: the possibilities and challenges are too broad and profound to tackle alone.
Data availability
The code and data for the case studies reported in this article can be found in the GitHub repositories linked in Table 1.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to specifically thank Jim Warren (NIST) for his contributions to discussions leading up to the hackathon and his participation as a judge during the event. We would also like to thank Anthony Costa and Christian Dallago (NVIDIA) for supporting the hackathon. B. B., I. T. F, and Z. H. acknowledge support from the the National Science Foundation awards #2226419 and #2209892. This work was performed under the following financial assistance award 70NANB19H005 from the U.S. Department of Commerce, National Institute of Standards and Technology as part of the Center for Hierarchical Materials Design (CHiMaD). K. J. S, A. S. acknowledge support from the the National Science Foundation award #1931306. K. M. J., S. M., J. v. H., X. Z., B. M., E. M., and B. S. were supported by the MARVEL National Centre for Competence in Research funded by the Swiss National Science Foundation (grant agreement ID 51NF40-182892) and the USorb-DAC Project, which is funded by a grant from The Grantham Foundation for the Protection of the Environment to RMI's climate tech accelerator program, Third Derivative. B. M. was further supported by the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 945363. M. C. R., S. C., and A. D. W. were supported by the National Science Foundation and the National Institute of General Medical Sciences under Grant No. 1764415 and award number R35GM137966, respectively. Q. A.'s contribution to this work was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number U18TR004149. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. M. V. G. acknowledges support from the Spanish National Research Council (CSIC) through the Programme for internationalization i-LINK 2021 (Project LINKA20412), and from the Spanish Agencia Estatal de Investigación (AEI) through the Grant TED2021-131693B-I00 funded by MCIN/AEI/10.13039/501100011033 and by the “European Union NextGenerationEU/PRTR” and through the Ramón y Cajal Grant RYC-2017-21937 funded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your future”. The project (M. L. E., B. E. S. and J. D. B.) has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement 957189 (DOI: 10.3030/957189), the Battery Interface Genome – Materials Acceleration Platform (BIG-MAP), as an external stakeholder project. M. L. E. additionally thanks the BEWARE scheme of the Wallonia-Brussels Federation for funding under the European Commission's Marie Curie-Skłodowska Action (COFUND 847587). B. E. S. acknowledges support from the UK's Engineering and Physical Sciences Research Council (ESPRC). B. P. acknowledges support from the National Science Foundation through NSF-CBET Grant No. 1917340. The authors thank Phung Cheng Fei, Hassan Harb, and Vinayak Bhat for their helpful comments on this project. D. C. and L. C. B. thank NSF DGE-2022040 for the aiM NRT funding support. K. C. thank the National Institute of Standards and Technology for funding, computational, and data-management resources. Please note certain equipment, instruments, software, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement of any product or service by NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose. A. K. G., G. W. M., A. I., and W. A. d. J. were supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, Materials Sciences and Engineering Division under Contract No. DE-AC02-05CH11231, FWP No. DAC-LBL-Long, and by the U.S. Department of Energy, Office of Science, Office of High Energy Physics under Award Number DE-FOA-0002705. M. B, B. R., and P. S. were supported by the NCCR Catalysis (grant number 180544), a National Centre of Competence in Research funded by the Swiss National Science Foundation. S. G. R. and J. L. acknowledge the generous support of Eric and Wendy Schmidt, and the core funding of the Francis Crick Institute, which receives its funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust.
project (M. L. E., B. E. S. and J. D. B.) has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement 957189 (DOI: 10.3030/957189), the Battery Interface Genome – Materials Acceleration Platform (BIG-MAP), as an external stakeholder project. M. L. E. additionally thanks the BEWARE scheme of the Wallonia-Brussels Federation for funding under the European Commission's Marie Curie-Skłodowska Action (COFUND 847587). B. E. S. acknowledges support from the UK's Engineering and Physical Sciences Research Council (ESPRC). B. P. acknowledges support from the National Science Foundation through NSF-CBET Grant No. 1917340. The authors thank Phung Cheng Fei, Hassan Harb, and Vinayak Bhat for their helpful comments on this project. D. C. and L. C. B. thank NSF DGE-2022040 for the aiM NRT funding support. K. C. thank the National Institute of Standards and Technology for funding, computational, and data-management resources. Please note certain equipment, instruments, software, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement of any product or service by NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose. A. K. G., G. W. M., A. I., and W. A. d. J. were supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, Materials Sciences and Engineering Division under Contract No. DE-AC02-05CH11231, FWP No. DAC-LBL-Long, and by the U.S. Department of Energy, Office of Science, Office of High Energy Physics under Award Number DE-FOA-0002705. M. B, B. R., and P. S. were supported by the NCCR Catalysis (grant number 180544), a National Centre of Competence in Research funded by the Swiss National Science Foundation. S. G. R. and J. L. acknowledge the generous support of Eric and Wendy Schmidt, and the core funding of the Francis Crick Institute, which receives its funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust.
References
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Machine learning for molecular and materials science, Nature, 2018, 559, 547–555 CrossRef CAS PubMed.
- S. M. Moosavi, K. M. Jablonka and B. Smit, The Role of Machine Learning in the Understanding and Design of Materials, J. Am. Chem. Soc., 2020, 142, 20273–20287 CrossRef CAS PubMed.
- D. Morgan and R. Jacobs, Opportunities and Challenges for Machine Learning in Materials Science, Annu. Rev. Mater. Res., 2020, 50, 71–103 CrossRef CAS.
- R. Ramprasad, R. Batra, G. Pilania, A. Mannodi-Kanakkithodi and C. Kim, Machine learning in materials informatics: recent applications and prospects, npj Comput. Mater., 2017, 3, 54 CrossRef.
- J. Schmidt, M. R. G. Marques, S. Botti and M. A. L. Marques, Recent advances and applications of machine learning in solid-state materials science, npj Comput. Mater., 2019, 5, 83 CrossRef.
- K. Choudhary, B. DeCost, C. Chen, A. Jain, F. Tavazza, R. Cohn, C. W. Park, A. Choudhary, A. Agrawal and S. J. Billinge, et al., Recent advances and applications of deep learning methods in materials science, npj Comput. Mater., 2022, 8, 59 CrossRef.
- K. M. Jablonka, D. Ongari, S. M. Moosavi and B. Smit, Big-Data Science in Porous Materials: Materials Genomics and Machine Learning, Chem. Rev., 2020, 120, 8066–8129 CrossRef CAS PubMed.
- J. Shi, M. J. Quevillon, P. H. Amorim Valença and J. K. Whitmer, Predicting Adhesive Free Energies of Polymer–Surface Interactions with Machine Learning, ACS Appl. Mater. Interfaces, 2022, 14, 37161–37169 CrossRef CAS PubMed.
- J. Shi, F. Albreiki, Y. J. Colón, S. Srivastava and J. K. Whitmer, Transfer Learning Facilitates the Prediction of Polymer–Surface Adhesion Strength, J. Chem. Theory Comput., 2023, 4631–4640 CrossRef CAS PubMed.
- F. Noé, A. Tkatchenko, K.-R. Müller and C. Clementi, Machine Learning for Molecular Simulation, Annu. Rev. Phys. Chem., 2020, 71, 361–390 CrossRef PubMed.
- S. Batzner, A. Musaelian, L. Sun, M. Geiger, J. P. Mailoa, M. Kornbluth, N. Molinari, T. E. Smidt and B. Kozinsky, E(3)equivariant graph neural networks for data efficient and accurate interatomic potentials, Nat. Commun., 2022, 13, 2453 CrossRef CAS PubMed.
- B. Sanchez-Lengeling and A. Aspuru-Guzik, Inverse molecular design using machine learning: generative models for matter engineering, Science, 2018, 361, 360–365 CrossRef CAS PubMed.
- J. F. Gonthier, S. N. Steinmann, M. D. Wodrich and C. Corminboeuf, Quantification of “fuzzy” chemical concepts: a computational perspective, Chem. Soc. Rev., 2012, 41, 4671 RSC.
- D. Weininger, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- K. M. Jablonka, L. Patiny and B. Smit, Making the collective knowledge of chemistry open and machine actionable, Nat. Chem., 2022, 14, 365–376 CrossRef CAS PubMed.
- R. Bommasani, et al., On the Opportunities and Risks of Foundation Models, CoRR 2021, abs/2108.07258 Search PubMed.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, Ł. Kaiser and I. Polosukhin, Attention is all you need, Adv. Neural Inf. Process. Syst., 2017, 30, 6000–6010 Search PubMed.
- A. Chowdhery, et al., PaLM: Scaling Language Modeling with Pathways, arXiv, 2022, preprint, arXiv:2204.02311, DOI:10.48550/arXiv.2204.02311.
- J. Hoffmann, et al., Training Compute-Optimal Large Language Models, arXiv, 2022, preprint, arXiv:2203.15556, DOI:10.48550/arXiv.2203.15556.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry and A. Askell, et al., Language models are few-shot learners, Adv. Neural Inf. Process. Syst., 2020, 33, 1877–1901 Search PubMed.
- C. N. Edwards, T. Lai, K. Ros, G. Honke and H. Ji, Translation between Molecules and Natural Language, Conference On Empirical Methods In Natural Language Processing, 2022 Search PubMed.
- T. Eloundou, S. Manning, P. Mishkin and D. Rock, GPTs are GPTs: An Early Look at the Labor Market Impact Potential of Large Language Models, arXiv, 2023, preprint, arXiv:2303.10130, DOI:10.48550/arXiv.2303.10130.
- A. Srivastava, et al., Beyond the Imitation Game: Quantifying and extrapolating the capabilities of language models, arXiv, 2022, preprint, arXiv:2206.04615, DOI:10.48550/arXiv.2206.04615.
- S. Bubeck, V. Chandrasekaran, R. Eldan, J. Gehrke, E. Horvitz, E. Kamar, P. Lee, Y. T. Lee, Y. Li, S. Lundberg, H. Nori, H. Palangi, M. T. Ribeiro and Y. Zhang, Sparks of Artificial General Intelligence: Early experiments with GPT-4, arXiv, 2023, preprint, arXiv:2303.12712, DOI:10.48550/arXiv.2303.12712.
- T. Schick, J. Dwivedi-Yu, R. Dessì, R. Raileanu, M. Lomeli, L. Zettlemoyer, N. Cancedda and T. Scialom, Toolformer: Language Models Can Teach Themselves to Use Tools, arXiv, 2023, preprint, arXiv:2302.04761, DOI:10.48550/arXiv.2302.04761.
- E. Karpas, et al., MRKL Systems: a modular, neuro-symbolic architecture that combines large language models, external knowledge sources and discrete reasoning, arXiv, 2022, preprint, arXiv:2205.00445, DOI:10.48550/arXiv.2205.00445.
- Y. Shen, K. Song, X. Tan, D. Li, W. Lu and Y. Zhuang, HuggingGPT: Solving AI Tasks with ChatGPT and its Friends in HuggingFace, arXiv, 2023, preprint, arXiv:2303.17580, DOI:10.48550/arXiv.2303.17580.
- A. White, paper-qa, 2022, https://github.com/whitead/paper-qa Search PubMed.
- J. Liu, LlamaIndex, 2022, https://github.com/jerryjliu/llama_index, last accessed 2023-05-30 Search PubMed.
- A. Karpathy, The Hottest New Programming Language Is English, 2023, https://twitter.com/karpathy/status/1617979122625712128, last accessed 2023-05-11 Search PubMed.
- G. M. Hocky and A. D. White, Natural language processing models that automate programming will transform chemistry research and teaching, Digit. Discov., 2022, 1, 79–83 RSC.
- K. M. Jablonka, P. Schwaller, A. Ortega-Guerrero and B. Smit, Is GPT-3 all you need for low-data discovery in chemistry?, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-fw8n4.
- A. D. White, G. M. Hocky, H. A. Gandhi, M. Ansari, S. Cox, G. P. Wellawatte, S. Sasmal, Z. Yang, K. Liu and Y. Singh, et al., Assessment of chemistry knowledge in large language models that generate code, Digit. Discov., 2023, 368–376 RSC.
- M. C. Ramos, S. S. Michtavy, M. D. Porosoff and A. D. White, Bayesian Optimization of Catalysts With In-context Learning, arXiv, 2023, preprint, arXiv:2304.05341, DOI:10.48550/arXiv.2304.05341.
- A. D. White, The future of chemistry is language, Nat. Rev. Chem., 2023, 7, 457–458 CrossRef CAS PubMed.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. Von Lilienfeld, Big data meets quantum chemistry approximations: the Δ-machine learning approach, J. Chem. Theory Comput., 2015, 11, 2087–2096 CrossRef CAS PubMed.
- T. Dinh, Y. Zeng, R. Zhang, Z. Lin, M. Gira, S. Rajput, J.-Y. Sohn, D. Papailiopoulos and K. Lee, Language-Interfaced Fine-Tuning for Non-Language Machine Learning Tasks, arXiv, 2022, preprint, arXiv:2206.06565, DOI:10.48550/arXiv.2206.06565.
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Self-referencing embedded strings (SELFIES): a 100% robust molecular string representation, Mach. Learn.: Sci. Technol., 2020, 1, 045024 Search PubMed.
- M. Krenn, Q. Ai, S. Barthel, N. Carson, A. Frei, N. C. Frey, P. Friederich, T. Gaudin, A. A. Gayle and K. M. Jablonka, et al., SELFIES and the future of molecular string representations, Patterns, 2022, 3, 100588 CrossRef CAS PubMed.
- T. Guo, K. Guo, B. Nan, Z. Liang, Z. Guo, N. V. Chawla, O. Wiest and X. Zhang, What indeed can GPT models do in chemistry? A comprehensive benchmark on eight tasks, arXiv, 2023, preprint, arXiv:2305.18365, DOI:10.48550/arXiv.2305.18365.
- L. A. Curtiss, P. C. Redfern and K. Raghavachari, Gaussian-4 theory using reduced order perturbation theory, J. Chem. Phys., 2007, 127, 124105 CrossRef PubMed.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. Von Lilienfeld, Quantum chemistry structures and properties of 134 kilo molecules, Sci. Data, 2014, 1, 1–7 Search PubMed.
- B. Narayanan, P. C. Redfern, R. S. Assary and L. A. Curtiss, Accurate quantum chemical energies for 133000 organic molecules, Chem. Sci., 2019, 10, 7449–7455 RSC.
- A. K. Gupta and K. Raghavachari, Three-Dimensional Convolutional Neural Networks Utilizing Molecular Topological Features for Accurate Atomization Energy Predictions, J. Chem. Theory Comput., 2022, 18, 2132–2143 CrossRef CAS PubMed.
- L. Ward, B. Blaiszik, I. Foster, R. S. Assary, B. Narayanan and L. Curtiss, Machine learning prediction of accurate atomization energies of organic molecules from low-fidelity quantum chemical calculations, MRS Commun., 2019, 9, 891–899 CrossRef CAS.
- R. Ramakrishnan, P. O. Dral, M. Rupp and O. A. von Lilienfeld, Big Data Meets Quantum Chemistry Approximations: The Δ-Machine Learning Approach, J. Chem. Theory Comput., 2015, 11, 2087–2096 CrossRef CAS PubMed.
- A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- E. J. Hu, Y. Shen, P. Wallis, Z. Allen-Zhu, Y. Li, S. Wang, L. Wang and W. Chen, Low-Rank Adaptation of Large Language Models, arXiv, 2021, preprint, arXiv:2106.09685, DOI:10.48550/arXiv.2106.09685.
- A. Radford, J. Wu, R. Child, D. Luan, D. Amodei and I. Sutskever, Language Models are Unsupervised Multitask Learners, 2019, https://d4mucfpksywv.cloudfront.net/better-language-models/language_models_are_unsupervised_multitask_learners.pdf Search PubMed.
- K. L. Scrivener, V. M. John and E. M. Gartner, Eco-efficient cements: Potential economically viable solutions for a low-CO2 cement-based materials industry, Cem. Concr. Res., 2018, 114, 2–26 CrossRef CAS.
- C. Völker, B. M. Torres, T. Rug, R. Firdous, G. Ahmad, J. Zia, S. Lüders, H. L. Scaffino, M. Höpler, F. Böhmer, M. Pfaff, D. Stephan and S. Kruschwitz, Green building materials: a new frontier in data-driven sustainable concrete design, 2023, DOI DOI:10.13140/RG.2.2.29079.85925.
- G. M. Rao and T. D. G. Rao, A quantitative method of approach in designing the mix proportions of fly ash and GGBS-based geopolymer concrete, Aust. J. Civ. Eng., 2018, 16, 53–63 CrossRef.
- V. Tshitoyan, J. Dagdelen, L. Weston, A. Dunn, Z. Rong, O. Kononova, K. A. Persson, G. Ceder and A. Jain, Unsupervised word embeddings capture latent knowledge from materials science literature, Nature, 2019, 571, 95–98 CrossRef CAS PubMed.
- T. Mikolov, K. Chen, G. Corrado and J. Dean, Efficient Estimation of Word Representations in Vector Space, International Conference On Learning Representations, 2013 Search PubMed.
- E. A. Olivetti, J. M. Cole, E. Kim, O. Kononova, G. Ceder, T. Y.-J. Han and A. M. Hiszpanski, Data-driven materials research enabled by natural language processing and information extraction, Appl. Phys. Rev., 2020, 7, 041317 CAS.
- S. Selva Birunda and R. Kanniga Devi, A review on word embedding techniques for text classification, Innovative Data Communication Technologies and Application: Proceedings of ICIDCA 2020, 2021, pp. 267–281 Search PubMed.
- Z. Hong, A. Ajith, G. Pauloski, E. Duede, C. Malamud, R. Magoulas, K. Chard and I. Foster, Bigger is Not Always Better, arXiv, 2022, preprint, arXiv:2205.11342, DOI:10.48550/arXiv.2205.11342.
- J. Li, Y. Liu, W. Fan, X.-Y. Wei, H. Liu, J. Tang and Q. Li, Empowering Molecule Discovery for Molecule-Caption Translation with Large Language Models: A ChatGPT Perspective, arXiv, 2023, preprint, arXiv: 2306.06615, DOI:10.48550/arXiv.2306.06615.
- H. Dai, et al., AugGPT: Leveraging ChatGPT for Text Data Augmentation, arXiv, 2023, preprint, arXiv:2302.13007, DOI:10.48550/arXiv.2302.13007.
- V. Venkatasubramanian, K. Chan and J. M. Caruthers, Computer-aided molecular design using genetic algorithms, Comput. Chem. Eng., 1994, 18, 833–844 CrossRef CAS.
- D. Flam-Shepherd and A. Aspuru-Guzik, Language models can generate molecules, materials, and protein binding sites directly in three dimensions as XYZ, CIF, and PDB files, arXiv, 2023, preprint, arXiv:2305.05708, DOI:10.48550/arXiv.2305.05708.
- R. Taylor, M. Kardas, G. Cucurull, T. Scialom, A. Hartshorn, E. Saravia, A. Poulton, V. Kerkez and R. Stojnic, Galactica: A Large Language Model for Science, arXiv, 2022, preprint, arXiv:2211.09085, DOI:10.48550/arXiv.2211.09085.
- P. Schwaller, T. Gaudin, D. Lányi, C. Bekas and T. Laino, “Found in Translation”: predicting outcomes of complex organic chemistry reactions using neural sequence-to-sequence models, Chem. Sci., 2018, 9, 6091–6098 RSC.
- S. Yao, J. Zhao, D. Yu, N. Du, I. Shafran, K. Narasimhan and Y. Cao, ReAct: Synergizing Reasoning and Acting in Language Models, arXiv, 2023, preprint, arXiv:2210.03629, DOI:10.48550/arXiv.2210.03629.
- J. Wei, X. Wang, D. Schuurmans, M. Bosma, E. Chi, F. Xia, Q. Le and D. Zhou, Chain-of-Thought Prompting Elicits Reasoning in Large Language Models, Neural. Inf. Process. Syst., 2022, 24824–24837 Search PubMed.
- OpenAI, GPT-4 Technical Report, arXiv, 2023, preprint, arXiv:2303.08774v3, DOI:10.48550/arXiv.2303.08774.
- A. M. Bran, S. Cox, A. D. White and P. Schwaller, ChemCrow: Augmenting large-language models with chemistry tools, arXiv, 2023, preprint, arXiv:2304.05376, DOI:10.48550/arXiv.2304.05376.
- D. A. Boiko, R. MacKnight and G. Gomes, Emergent autonomous scientific research capabilities of large language models, arXiv, 2023, preprint, arXiv:2304.05332, DOI:10.48550/arXiv.2304.05332.
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner, G. Ceder and K. A. Persson, Commentary: The Materials Project: A materials genome approach to accelerating materials innovation, APL Mater., 2013, 1, 011002 CrossRef.
- N. Rego and D. Koes, 3Dmol.js: molecular visualization with WebGL, Bioinformatics, 2014, 31, 1322–1324 CrossRef PubMed.
- A. White and G. Hocky, marvis – VMD Audio/Text control with natural language, 2022, https://github.com/whitead/marvis Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- A. Radford, J. W. Kim, T. Xu, G. Brockman, C. McLeavey and I. Sutskever, Robust speech recognition via large-scale weak supervision, arXiv, 2022, preprint, arXiv:2212.04356, DOI:10.48550/arXiv.2212.04356.
- M. Baek, et al., Accurate prediction of protein structures and interactions using a three-track neural network, Science, 2021, 373, 871–876 CrossRef CAS PubMed.
- J. L. Watson, et al., Broadly applicable and accurate protein design by integrating structure prediction networks and diffusion generative models, bioRxiv, 2022, preprint, DOI:10.1101/2022.12.09.519842.
- C. W. Andersen, et al., OPTIMADE, an API for exchanging materials data, Sci. Data, 2021, 8, 217 CrossRef PubMed.
- A. A. Volk, R. W. Epps, D. T. Yonemoto, B. S. Masters, F. N. Castellano, K. G. Reyes and M. Abolhasani, AlphaFlow: autonomous discovery and optimization of multi-step chemistry using a self-driven fluidic lab guided by reinforcement learning, Nat. Commun., 2023, 14, 1403 CrossRef CAS PubMed.
- B. J. Shields, J. Stevens, J. Li, M. Parasram, F. Damani, J. I. M. Alvarado, J. M. Janey, R. P. Adams and A. G. Doyle, Bayesian reaction optimization as a tool for chemical synthesis, Nature, 2021, 590, 89–96 CrossRef CAS PubMed.
- C. N. Prieto Kullmer, J. A. Kautzky, S. W. Krska, T. Nowak, S. D. Dreher and D. W. MacMillan, Accelerating reaction generality and mechanistic insight through additive mapping, Science, 2022, 376, 532–539 CrossRef PubMed.
- B. Ranković, R.-R. Griffiths, H. B. Moss and P. Schwaller, Bayesian optimisation for additive screening and yield improvements in chemical reactions – beyond one-hot encodings, ChemRxiv, 2022, preprint DOI:10.26434/chemrxiv-2022-nll2j.
- A. Dunn, J. Dagdelen, N. Walker, S. Lee, A. S. Rosen, G. Ceder, K. A. Persson and A. Jain, Structured information extraction from complex scientific text with fine-tuned large language models, arXiv, 2022, preprint, arXiv:2212.05238, DOI:10.48550/arXiv.2212.05238.
- N. Walker, J. Dagdelen, K. Cruse, S. Lee, S. Gleason, A. Dunn, G. Ceder, A. P. Alivisatos, K. A. Persson and A. Jain, Extracting Structured Seed-Mediated Gold Nanorod Growth Procedures from Literature with GPT-3, arXiv, 2023, preprint, arXiv:2304.13846, DOI:10.48550/arXiv.2304.13846.
- Neo4j, Neo4j – The World's Leading Graph Database, 2012, http://neo4j.org/ Search PubMed.
- S. M. Kearnes, M. R. Maser, M. Wleklinski, A. Kast, A. G. Doyle, S. D. Dreher, J. M. Hawkins, K. F. Jensen and C. W. Coley, The Open Reaction Database, J. Am. Chem. Soc., 2021, 143, 18820–18826 CrossRef PubMed.
- J. Guo, A. S. Ibanez-Lopez, H. Gao, V. Quach, C. W. Coley, K. F. Jensen and R. Barzilay, Automated Chemical Reaction Extraction from Scientific Literature, J. Chem. Inf. Model., 2021, 62, 2035–2045 CrossRef PubMed.
- R. Taori, I. Gulrajani, T. Zhang, Y. Dubois, X. Li, C. Guestrin, P. Liang and T. B. Hashimoto, Stanford Alpaca: An Instruction-following LLaMA model, 2023, https://github.com/tatsu-lab/stanford_alpaca Search PubMed.
- Alpaca-LoRA, https://github.com/tloen/alpaca-lora.
- H. Touvron, T. Lavril, G. Izacard, X. Martinet, M.-A. Lachaux, T. Lacroix, B. Rozière, N. Goyal, E. Hambro, F. Azhar, et al., Llama: open and efficient foundation language models, arXiv, 2023, preprint, arXiv:2302.13971, DOI:10.48550/arXiv.2302.13971.
- Z. G. Mamaghani, K. A. Hawboldt and S. MacQuarrie, Adsorption of CO2 using biochar – review of the impact of gas mixtures and water on adsorption, J. Environ. Chem. Eng., 2023, 11, 109643 CrossRef.
- Y. Peng, V. Krungleviciute, I. Eryazici, J. T. Hupp, O. K. Farha and T. Yildirim, Methane Storage in Metal–Organic Frameworks: Current Records, Surprise Findings, and Challenges, J. Am. Chem. Soc., 2013, 135, 11887–11894 CrossRef CAS PubMed.
- B. Sahoo, V. Pandey, A. Dogonchi, P. Mohapatra, D. Thatoi, N. Nayak and M. Nayak, A state-of-art review on 2D material-boosted metal oxide nanoparticle electrodes: Supercapacitor applications, J. Energy Storage, 2023, 65, 107335 CrossRef.
- D. D. Suppiah, W. M. A. W. Daud and M. R. Johan, Supported Metal Oxide Catalysts for CO2 Fischer–Tropsch Conversion to Liquid Fuels-A Review, Energy Fuels, 2021, 35, 17261–17278 CrossRef CAS.
- M. González-Vázquez, R. García, M. Gil, C. Pevida and F. Rubiera, Comparison of the gasification performance of multiple biomass types in a bubbling fluidized bed, Energy Convers. Manage., 2018, 176, 309–323 CrossRef.
- M. Mohsin, S. Farhan, N. Ahmad, A. H. Raza, Z. N. Kayani, S. H. M. Jafri and R. Raza, The electrochemical study of NixCe1−xO2−δ electrodes using natural gas as a fuel, New J. Chem., 2023, 47, 8679–8692 RSC.
- P. Kaur and K. Singh, Review of perovskite-structure related cathode materials for solid oxide fuel cells, Ceram. Int., 2020, 46, 5521–5535 CrossRef CAS.
- R. Sengottuvelu, jsonformer, 2018, https://github.com/1rgs/jsonformer Search PubMed.
- R. Sengottuvelu, jsonformer, 2018, https://github.com/martinezpl/jsonformer/tree/add-openai Search PubMed.
- K. Choudhary and M. L. Kelley, ChemNLP: A Natural Language Processing based Library for Materials Chemistry Text Data, arXiv, 2022, preprint, arXiv:2209.08203, DOI:10.48550/arXiv.2209.08203.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, Exploring the limits of transfer learning with a unified text-to-text transformer, J. Mach. Learn. Res., 2020, 21, 5485–5551 Search PubMed.
- T. Wolf, et al., Transformers: State-of-the-Art Natural Language Processing, Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, 2020 Search PubMed.
- N. Editorials, Tools such as ChatGPT threaten transparent science; here are our ground rules for their use, Nature, 2023, 613, 10–1038 Search PubMed.
- E. R. Mollick and L. Mollick, Using AI to Implement Effective Teaching Strategies in Classrooms: Five Strategies, Including Prompts, SSRN Electron. J., 2023 DOI:10.2139/ssrn.4391243.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, PubChem 2023 update, Nucleic Acids Res., 2022, 51, D1373–D1380 CrossRef PubMed.
- S. Kim, P. A. Thiessen, T. Cheng, B. Yu and E. E. Bolton, An update on PUG-REST: RESTful interface for programmatic access to PubChem, Nucleic Acids Res., 2018, 46, W563–W570 CrossRef CAS PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, PubChem 2019 update: improved access to chemical data, Nucleic Acids Res., 2018, 47, D1102–D1109 CrossRef PubMed.
- M. Krenn, R. Pollice, S. Y. Guo, M. Aldeghi, A. Cervera-Lierta, P. Friederich, G. dos Passos Gomes, F. Häse, A. Jinich, A. Nigam, Z. Yao and A. Aspuru-Guzik, On scientific understanding with artificial intelligence, Nat. Rev. Phys., 2022, 4, 761–769 CrossRef PubMed.
- A. Hunt and D. Thomas, The Pragmatic programmer : from journeyman to master, Addison-Wesley, Boston, 2000 Search PubMed.
- Q. Campbell, J. Herington and A. D. White, Censoring chemical data to mitigate dual use risk, arXiv, 2023, preprint, arXiv:2304.10510, DOI:10.48550/arXiv.2304.10510.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00113j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |