Recent methods of droplet microfluidics and their applications in spheroids and organoids
Yihe
Wang
 a,
Mengqi
Liu
a,
Mengqi
Liu
 a,
Yu
Zhang
a,
Yu
Zhang
 a,
Hong
Liu
a,
Hong
Liu
 *b and
Lin
Han
*b and
Lin
Han
 *ac
*ac
aInstitute of Marine Science and Technology, Shandong University, Qingdao, 266237 P. R. China. E-mail: hanlin@sdu.edu.cn
bState Key Laboratory of Crystal Materials, Shandong University, Jinan, 250100 P. R. China. E-mail: hongliu@sdu.edu.cn
cShandong Engineering Research Center of Biomarker and Artificial Intelligence Application, Jinan, 250100 P. R. China
First published on 11th January 2023
Abstract
Droplet microfluidic techniques have long been known as a high-throughput approach for cell manipulation. The capacity to compartmentalize cells into picolitre droplets in microfluidic devices has opened up a range of new ways to extract information from cells. Spheroids and organoids are crucial in vitro three-dimensional cell culture models that physiologically mimic natural tissues and organs. With the aid of developments in cell biology and materials science, droplet microfluidics has been applied to construct spheroids and organoids in numerous formats. In this article, we divide droplet microfluidic approaches for managing spheroids and organoids into three categories based on the droplet module format: liquid droplet, microparticle, and microcapsule. We discuss current advances in the use of droplet microfluidics for the generation of tumour spheroids, stem cell spheroids, and organoids, as well as the downstream applications of these methods in high-throughput screening and tissue engineering.
1. Introduction
In vitro cell culture models are created to mimic the natural environment of cells, tissues, and organs while allowing them to grow, proliferate, and differentiate in the laboratory. Animal models can most faithfully recapitulate the physiological features of the human body; however, they still have limitations in terms of imaging accessibility, throughput of analysis, and human–animal differences.1 For example, many experimental animals have compromised immune systems, lack the same stroma–tumour interaction as humans, and do not allow the precise spatial and temporal control of genetic alterations, which makes it difficult to effectively apply the latest studies to clinical settings for drug efficacy assessment and high-throughput screening (HTS).2,3 While a two-dimensional (2D) culture of human cells can provide insight into human biology, the complexity and cellular diversity of tissues in vivo are not sufficiently reflected in 2D cell culture. The cell–cell and cell–environment interactions responsible for cell fate in 2D cell culture are different from those in natural tissues.4 In addition, tumour cells cultured in 2D have shown lower resistance to chemotherapy than those cultured in three-dimensional (3D) systems, which may present misleading results in drug screening.5–7 Three-dimensional culture systems can more faithfully recapitulate cell–cell interactions, matrix deposition, and cell microenvironments, along with physiological flow conditions, oxygen, and nutrient gradients.8–10 As a promising method to bridge the gap between 2D and animal models, 3D cell culture models have attracted increased interest in many directions, from the fundamental research of cancer and stem cell biology to applications for drug toxicity tests and HTS.11,12 These 3D models provide a promising tool to advance personalized medicine and next-generation drug screening and to limit the need for animal experimentation.Among various in vitro 3D cell models, spheroids and organoids are the two most common: they have distinct but overlapping purposes and differ in terms of cell sources, protocol for culture, and time for establishment.13 In brief, spheroids are 3D cell clusters from a broad range of cell sources, including cell lines, primary cells, or tissue fragments.14,15 Organoids are also 3D multicellular structures but must be established from sources containing stem cells: sourced from tissue-resident adult stem cells (ASCs), directly from biopsy samples, or from pluripotent stem cells (PSCs), such as embryonic stem cells (ESCs) or induced PSCs (iPSCs), which are obtained by reprogramming differentiated cell types.16 The formation of spheroids involves cell association and adherence, and some spheroid models involve self-organization.17 The formation of organoids, on the other hand, often relies on the self-organization and differentiation of stem cells.18 In comparison with spheroids that transiently resemble 3D cell organizations, organoids have more complex structures and functions and can partially recapitulate the physiology of organs.16
To date, many techniques have been employed to create spheroids and organoids, as summarized in previous reviews.16–20 Briefly, the methods for the formation of spheroids can be classified into two main categories, i.e., scaffold-based methods and scaffold-free methods.21,22 In scaffold-based methods, cells are embedded into 3D matrices resembling the extracellular matrix (ECM) of cells for their growth and organization into spheroids.23 Hydrogels made from naturally derived materials (e.g., collagen24 or alginate25), synthetic polymers (e.g., poly(ethylene glycol) (PEG)),26 or composite materials (e.g., hybrid hydrogels of collagen and alginate)27 are commonly used as scaffolds to support and provide physical or chemical cues to the cells. In scaffold-free methods, spheroids are normally formed by the self-aggregation and mutual adhesion of cells into cell assemblies in the absence of scaffolds (e.g., in suspension, in a spinner flask, in hanging drops, or on nonadherent surfaces).28 For example, hanging drops have been used to produce cell spheroids from the sedimentation and aggregation of cells in a multiwell plate format29–31 and in a more recent microfluidic format.32–35 Organoids can be established by the self-organization and differentiation of cells imbedded in a 3D hydrogel matrix, such as Matrigel,36–38 or created by a scaffold-free method, such as the “air–liquid-interface”.39–41
Microfluidics offer superior flexibility and throughput during the spatial and temporal control of cell organizations in the 3D format.42,43 As a branch of microfluidics, droplet microfluidics (DMFs), which generate and manipulate discrete droplets from the interactions of multiphase flows inside microchannels, possess several remarkable advantages in 3D cell applications. First, DMFs enable the high-throughput isolation of cells into compartments on the nanolitre scale for further processing into 3D assemblies. Second, DMFs offer a versatile platform for the production of various droplet templates (e.g., microparticles and microcapsules) when incorporating materials such as hydrogels and thus provide increased flexibility and heterogeneity for the control of cells and their local environments. Finally, in addition to droplet generation, DMFs can also be used for the multiplexed analysis of 3D cell assemblies in droplets by the integration of various manipulation processes into a single system.44–46 These superior capabilities have facilitated the development of DMFs with a broad range of applications for 3D cell culture.47 Nevertheless, there remains much room to improve DMFs towards broader practical applications for 3D cell models, probably with contributions from multiple disciplines, such as novel fabrication technologies, advanced functional materials, and fundamental biology.47
Recent progress in DMFs for cell culture has been reviewed elsewhere.22,48 In the present paper, we will focus on the emerging methodologies and applications of DMFs specifically in two types of 3D cell models, i.e., spheroids and organoids. The first part of the review is centred on DMF methods for the formation of spheroids and organoids, as described in several subcategories of droplet-based modules (i.e., liquid droplets, microparticles, and microcapsules). Then, several manipulation methods for droplet-based modules on a chip are summarized. The next section is focused on the applications of these modules in the study of tumour spheroids, stem cell spheroids, and organoids. Finally, we provide a critical discussion of the current state and future perspectives in this field.
2. Droplet microfluidics for the formation of spheroids and organoids
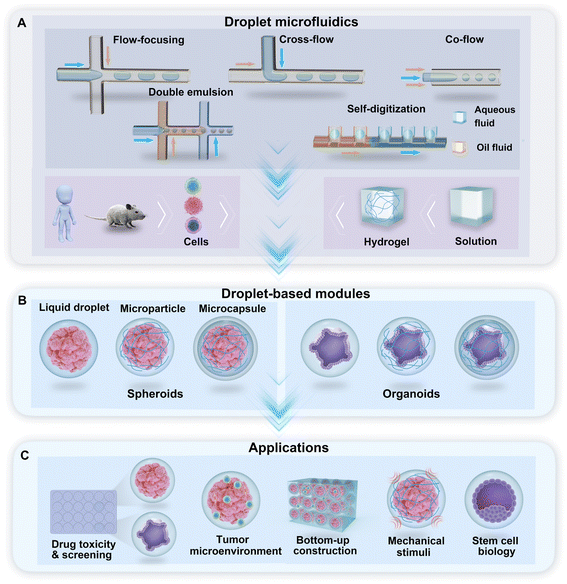
Droplet microfluidics is the technology used to generate a library of droplets by the manipulation of two or more immiscible fluids in microchannels. For the purpose of droplet generation and cell encapsulation, water-in-oil-based DMF technologies use a water-based solution as the dispersed phase and oil with a surfactant as the continuous phase. In the droplet generator, cross-flow, co-flow, and flow-focusing are the typical hydrodynamic junctions where two immiscible fluids meet and flow into a single channel and form droplets (Fig. 1A). Droplets with multiple phases, such as double emulsion (DE) droplets with water–oil–water phases, can be generated through sequential emulsions in multiple junctions (Fig. 1A). A hydrodynamic capillary method named “self-digitization” that relies on capillary force to trap droplets in “microwells” can be used to generate droplets and encapsulate cells (Fig. 1A).49–51 The incorporation of hydrogel materials in the formation of droplets creates diverse phases in the droplet template. According to the phase structure of the droplet, we divided the droplet-based modules for the generation of spheroids and organoids into three categories: liquid droplets, microparticles, and microcapsules (Fig. 1B). These modules provide a broad range of applications for research on drug toxicity and screening, the tumour environment, bottom-up construction, mechanical stimuli, and stem cell biology, among other topics (Fig. 1C).2.1 Liquid droplets
The formation of spheroids or organoids by DMFs often starts from the encapsulation of multiple cells in liquid droplets by the emulsification of a cell suspension solution in an oil fluid in certain geometries, such as flow-focusing junctions, T-junctions, coaxial channels, multijunctions, or side-well traps, as shown in Fig. 1A. The characteristics of the formation of droplets in these geometries have been reviewed in detail elsewhere.47 To maintain cell viability, proliferation, and phenotype in the droplets, a sufficient supply of cell-supporting components, such as nutrients, growth factors, amino acids, and oxygen, must be present in the aqueous solution (e.g., cell culture medium). In comparison with single cells in droplets, spheroids or organoids with a larger number of cells require larger droplet volumes containing more nutrients, growth factors, amino acids, and by-products to maintain their viability and functionality.48 The size of the spheroids can be tuned by varying the initial cell concentration in the aqueous phase and the flow rates of the aqueous phase and continuous phase.52Similar to hanging drops, which rely on the sedimentation and aggregation of cells on the liquid–air interface, DMFs compartmentalize cells and allow cells to settle and aggregate in the droplets. In particular, DMFs can enhance the spheroid formation process by generating droplets in a high-throughput manner and providing size control. For example, DMFs have been used to generate spheroids by the encapsulation, sedimentation and aggregation of cells within droplets (Fig. 2A).53 Upon the formation of tight cell–cell junctions and the secretion of ECM components, the cell aggregates grow into cohesive structures, namely, spheroids.
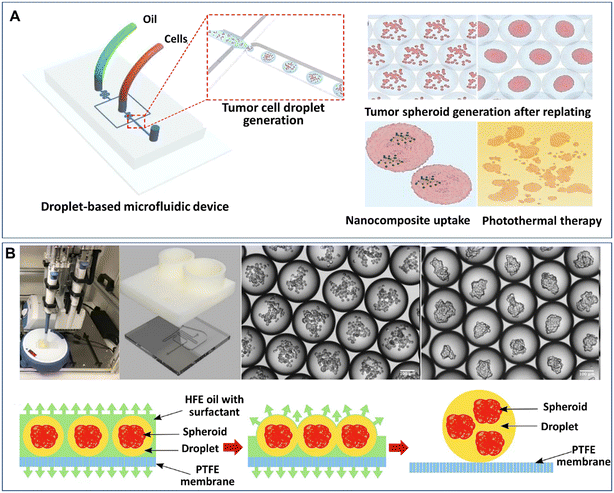 | ||
| Fig. 2 Formation of spheroids in liquid droplets by droplet microfluidics. (A) Spheroids formed in liquid droplets and used for photothermal therapy testing. Reproduced from ref. 53 with permission from Springer Nature, copyright 2020. (B) Spheroids formed in an automated droplet generator and recovered on a hydrophobic membrane. Reproduced from ref. 54 with permission from the Society for Laboratory Automation and Screening, copyright 2020. | ||
Although the process of cell encapsulation in liquid droplets is simple and rapid, there are challenges for the encapsulated cells to replenish nutrients from the surrounding oil, and thus it's difficult for the cells to maintain long-term viability and functionality in liquid droplets. It is particularly difficult to develop organoids in liquid droplets, as the cells require an even longer period to achieve functionalities. One possible solution is to replace the surrounding oil with a gas or an aqueous phase. Langer et al. developed an automated DMF platform to produce cell spheroids in scaffold-free droplets and recovered the spheroids by a membrane-based sample recovery process (Fig. 2B).54
2.2 Microparticles
Droplet microfluidics provides a template for the formation of spheroids and organoids in the presence of hydrogel scaffolds, namely, microparticles. Hydrogels can induce more complex cell-matrix and cell–cell interactions in cellular environments when employed in a 3D cell model. Extracellular matrix-derived materials, such as collagen, Matrigel, and fibrin, have been used as 3D matrices to culture spheroids and organoids.55–57 These hydrogels can allow cell adherence by providing cell-adhesive moieties, such as arginylglycylaspartic acid (RGD) peptides, on which cell viability and functionality may rely. Other non-ECM natural hydrogels, such as agarose and alginate, can also be used for the culture of spheroids due to their stable physical properties.58,59 In contrast to natural ECM components with diverse cues, batch-to-batch variations, and uncontrolled biodegradability, synthetic hydrogels have more consistent and flexible compositions.60,61 However, synthetic materials normally lack natural cell-adherence moieties, biodegradability sites, or biophysical structures of ECM, and thus these synthetic materials must be functionalized with cell-binding or enzymatically degradable sites. Hybrid hydrogels made from natural ECM components and synthetic materials and other functionalized materials are good candidates to provide biophysical and biochemical cues for the growth of spheroids or organoids, as they can provide not only cell-adhesive sites but also the desired physical properties.62–64 To date, microparticles made from hydrogels (e.g., Matrigel,65–67 methacrylic gelatine,68 and agarose46,58,69,70) have been created by DMFs for the study of spheroids or organoids.The formation of spheroids or organoids in microparticles starts from the encapsulation of cells into droplets containing hydrogel precursors, followed by cell aggregation or precursor gelation. The liquid droplets can be solidified after gelation and thus enable the replacement of the oil phase with the aqueous phase for long-term cell culture. Fig. 3A shows an example of the formation of organoids in Matrigel microparticles created from a “T-junction”.65 In this work, an aqueous solution of Matrigel suspended with cells was introduced into a T-junction and emulsified into droplets by an oil fluid at 8 °C. Upon temperature elevation to 37 °C, the Matrigel crosslinked into gels, forming cell-laden microparticles. After 7 days of culture, cells in the microparticles proliferated and differentiated into organoids (Fig. 3A). By employing this method, organoids derived from healthy liver tissue and liver tumour tissue were created with a defined population of cells and microstructures that resemble their parental tissues.66 Cell-laden microparticles can also be generated by the self-digitization method, as shown in Fig. 3B: a cell suspension with resolved hydrogel precursors and oil are introduced into a capillary trap channel in sequence; upon droplet formation and precursor gelation, the oil is subsequently replaced with cell culture medium; by perfusing a medium flow, cells in the microparticles eventually grow into spheroids in a 2D array of microwells.71
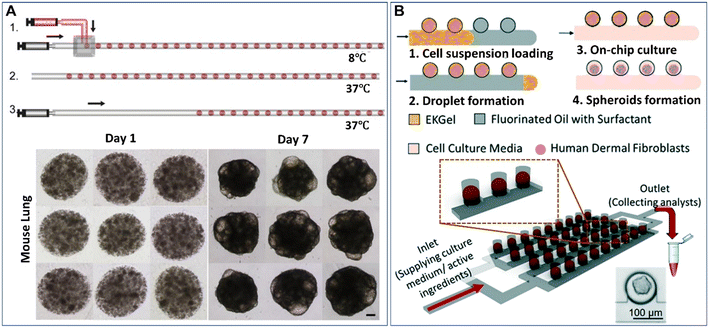 | ||
| Fig. 3 Formation of spheroids or organoids in microparticles by droplet microfluidics. (A) Formation of cell-laden Matrigel microparticles in a T-junction and growth of cells into organoids after 7 days of culture. The scale bar is 100 μm. Reproduced from ref. 65 with permission from Elsevier, copyright 2021. (B) A 2D array of spheroid-laden microparticles generated by self-digitization. Reproduced from ref. 71 with permission from the Royal Society of Chemistry, copyright 2021. | ||
2.3 Microcapsules
Core–shell particles (defined as microcapsules in the present paper) can be generated from multiphase droplet templates (e.g., DE droplet template) in a microfluidic configuration (e.g., flow-focusing junction). The methods for the manipulation and formation of cell-laden microcapsules and their applications have been reviewed elsewhere.72 The present review focuses mainly on the recent emerging methods and applications of microcapsules for spheroids and organoids. According to the phase organization in each domain of the droplet, the multiphase droplet templates used for spheroid or organoid formation can be defined as W/O/W,73 or W/O/G,74 in which W, O, and G represent the water, oil and gas phases, respectively. In addition to the DE droplet template, microcapsules can also be generated from a W/O droplet template, where the inner aqueous phase is separated into two immiscible phases (e.g., a solid hydrogel shell and a liquid core) and is defined as a W/W/O template.68,75–81 Alternatively, some aqueous phase systems have been implemented to fabricate various cell-laden microcapsules in an oil-free manner.82–84 In these systems, the formation of microcapsules often relies on the incompatibility of the continuous phase and the disperse phase, as well as the interactions of polymers along the interfaces of the two phases. For instance, microcapsules made from the interfacial complexation of oppositely charged Na-alginate and chitosan in the shell have been generated from an aqueous two-phase system for organoid culture.84 In these DMF-generated microcapsules, the inner core is either an aqueous phase containing suspended cells or a solid phase formed from the solidification of a precursor solution of a hydrogel, such as collagen,77 Matrigel,76 and alginate.75,78 The outer shell of the microcapsule can also be generated from a precursor hydrogel solution (e.g., alginate74,79,85) or generated directly from an immiscible oil phase,73 where the microcapsules are formed from W/W/O (Fig. 4A)77 or W/O/W templates (Fig. 4B),86 respectively.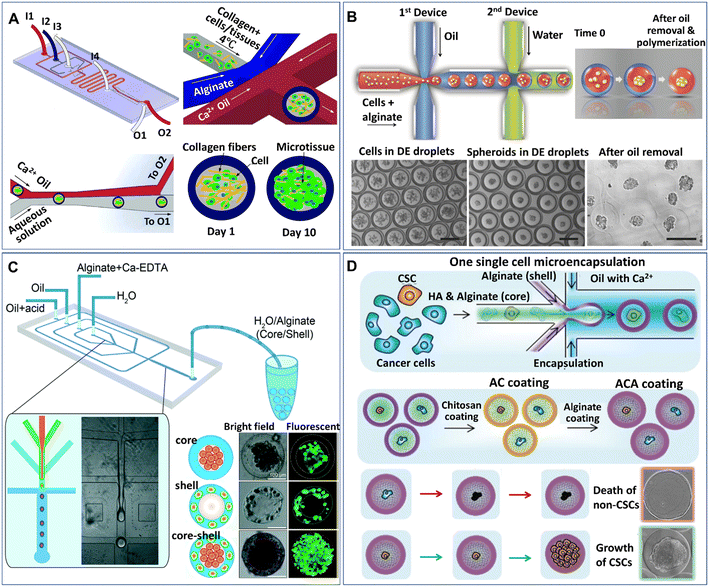 | ||
| Fig. 4 Formation of spheroids in core–shell microcapsules. (A) Spheroids formed in microcapsules with a collagen core and alginate shell in a one-step droplet generator. Reproduced from ref. 77 with permission from John Wiley & Sons Inc., copyright 2015. (B) Spheroids formed in microcapsules with alginate cores and oil shells and recovered after oil removal. Scale bars are 200 μm. Reproduced from ref. 86 with permission from John Wiley & Sons Inc., copyright 2016. (C) Formation of spheroids cocultured with fibroblasts in core–shell microcapsules. Reproduced from ref. 80 with permission from the Royal Society of Chemistry, copyright 2016. (D) Spheroids grown from single cells encapsulated in microcapsules coated with chitosan and alginate. Reproduced from ref. 90 with permission from John Wiley & Sons Inc., copyright 2020. | ||
Microcapsules formed from core–shell droplet templates offer extra flexibility in encapsulating different cells because of their unique structure. This kind of particle comprises an inner core and an outer shell, which enables the separation of different cells into different domains of the microcapsules. By introducing two types of cells, microcapsules for cell coculture have been produced.80,85Fig. 4C shows an example of the formation of core–shell spheroids with fibroblasts and hepatocytes cocultured in the core and shell domains of alginate microcapsules, respectively.80 Studies show that when used for stem cell culture, microcapsules composed of a liquid core and solid shell performed better in maintaining stemness than conventional 2D substrates or homogeneous hydrogel microparticles.79,87,88 The shell layer of the capsule can also serve as a protector to prevent cell detachment from the microgel; otherwise, the encapsulated cells on the interface of the microcapsules can easily escape.89 In addition, when permeable materials (e.g., hyaluronic acid) are incorporated into the core of the microcapsules, the outer surface can be coated with chitosan and alginate to prevent leakage of the material in the core, as shown in Fig. 4D.90
3. Manipulations of droplet-based modules on chips
Droplet microfluidics have made it possible to manipulate droplets on chips in various processes, such as trapping, sorting, merging, and splitting, which have been extensively reviewed.47,91,92 These manipulation processes have leveraged the uses of DMFs in more desired functions, such as cell identification, chemical reactions, and multiplex analysis. Particularly for 3D cell models, some manipulation processes in addition to droplet generation have been integrated into DMF platforms. In this section, we will review the recent developments in the manipulations of droplet-based modules on chips, i.e., trapping, merging, and arrangement in fibres.Despite the high-throughput production efficiency, DMFs used merely to generate mobile droplets have difficulty meeting the need to identify cell-laden droplets for subsequent analysis. One strategy to overcome this challenge is the integration of droplet trapping with a droplet generator on a chip.93–95 Sart et al. reported a microfluidic platform integrated from a droplet generator and a 2D array of microwells that are used to immobilize droplets (Fig. 5A).58 The formation of spheroids in the droplets starts from the sedimentation and aggregation of cells, followed by the gelation of agarose in the droplets and replacement of oil with the aqueous phase (Fig. 7B). The inherent organization and spatial heterogeneities of cells within the spheroid can be resolved during their self-organization (Fig. 5C).69 On the basis of this method, Tomasi et al. employed a “droplet pairing” strategy to bring a series of smaller droplets containing Matrigel, spheroids, or drugs to merge with spheroid-laden droplets (Fig. 5D).46 This approach also allows the modulation of the spheroids' physical or chemical environment and drug treatment over a large range of concentrations in a single parallelized experiment. In addition, the secretion of cytokines of individual MSC spheroids can be monitored from the fluorescence signals on magnetic beads decorated with antibodies in paired droplets, as shown in Fig. 5E.70 Recently, Wang et al. developed an all-in-water microfluidic platform for the formation of hydrogel fibres containing arranged cell-laden aqueous droplets (Fig. 5F).96 The formation and arrangement of the droplets relied on the actuation of the pump valve and the stable interface between the two immiscible liquids. By applying this method, calcium alginate fibre-containing islet organoids were generated through the encapsulation of pancreatic endocrine in the droplets and the subsequent gelation of alginate.
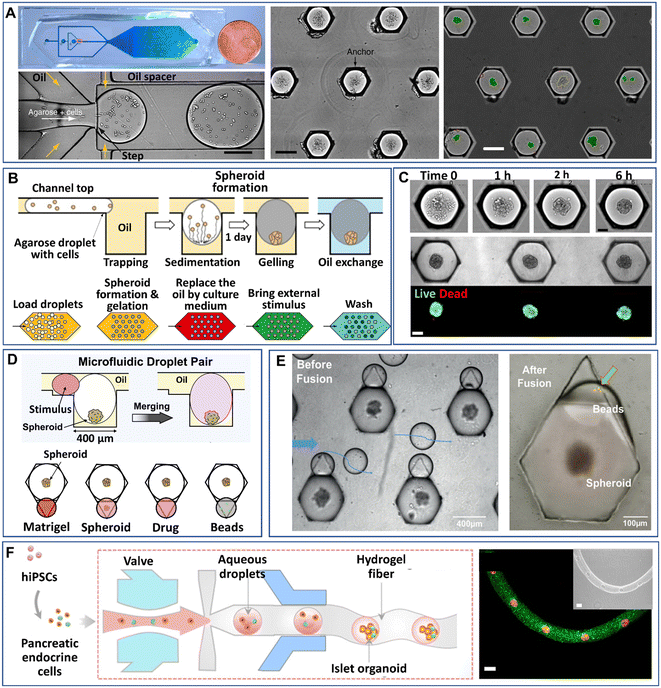 | ||
| Fig. 5 Manipulations of spheroids or organoids generated by droplet microfluidics on a chip. (A) Cells encapsulated into droplets by a flow-focusing droplet generator, anchored and allowed to form spheroids in a 2D array of microwells. Scale bars are 200 μm. Reproduced from ref. 58 with permission from Springer Nature, copyright 2017. (B) Process of spheroid generation and workflow of droplet treatment. Reproduced from ref. 58 with permission from Springer Nature, copyright 2017, and ref. 69 with permission from the American Association for the Advancement of Science, copyright 2020. (C) Formation of spheroids after cell aggregation. Reproduced from ref. 69 with permission from the American Association for the Advancement of Science, copyright 2020. Scale bars are 100 μm. (D) Manipulation of spheroids formed in anchored droplets by pairing droplets containing Matrigel, spheroids, drugs, or beads. Reproduced from ref. 46 with permission from Elsevier, copyright 2020. (E) Pairing of smaller droplets containing beads to detect cytokines secreted from individual spheroids in larger droplets. Reproduced from ref. 70 with permission from John Wiley & Sons Inc, copyright 2020. (F) An all-in-water droplet microfluidic platform for the formation of aqueous-droplet-filled hydrogel fibres as organoid carriers. Scale bars are 100 μm. Reproduced from ref. 96 with permission from the American Chemical Society, copyright 2021. | ||
4. Applications of droplet microfluidics in spheroids
4.1 Tumour spheroids
Tumour spheroids are in vitro tumour models that are developed to resemble the native structure of the tumour and stroma by the incorporation of stromal cells, ECMs, a gradient of nutrients and gas, and physical features such as stiffness.74 Tumour spheroids can be generated from a broad range of cell sources including cancer cell lines97,98 and primary tumour cells.99–101 Many techniques can be used to generate tumour spheroids, including liquid overlay,102,103 hanging drop,30,104 3D scaffold,26,55,105,106 and spinning flask methods.107,108 In recent decades, microfluidics have been implemented to culture tumour spheroids in the form of microwells,109,110 U-shaped microstructures,111,112 microrotational flow,113 acoustic tweezers,114 emulsions,45etc. Droplet microfluidics offers many great advantages over other microfluidic methods in the production and manipulation of tumour spheroids: 1) DMFs enable the high-throughput generation of tumour spheroids, which is particularly helpful for their applications in drug screening. For example, a DMF platform was used to produce spheroids from the encapsulation of cells into droplets at a rate of several tens of droplets per second, and the resultant spheroids were used for drug toxicity tests.115 2) The size and composition of spheroids can be easily tuned by DMFs, which is very useful to provide heterogeneous responses to target therapies. For example, tumour spheroids co-cultured with stroma cells were formed in Core–shell microcapsules and used for drug evaluation.85 3) The miniature tumours in hydrogel particles can be used as building blocks to create complex tissue/organ structures.116,117From the macroscopic view, tumour spheroids can be created by embedding individual cells into 3D hydrogel scaffolds. The hydrogels used in tumour spheroid formation can be derived from the natural ECMs of the tumour stroma (e.g., Matrigel,118 collagen,119 and hyaluronic acid120), natural materials (e.g., agarose58 and alginate59), or synthetic polymers (e.g., PEG).60 In the microfluidic device, the formation of tumour spheroids normally starts with the allocation of cells into individual hydrogel microparticles/microcapsules, followed by the growth of cells into spheroids. The hydrogels added to the droplets play important roles in the growth and functionality of tumour spheroids in many aspects. First, hydrogels can provide biophysical and biochemical cues to regulate tumour cell fate as the natural tumour stroma does to the tumour cells. For example, By testing cell invasion in collagen microcapsules with tunable mechanical properties, Nassoy et al. found that mechanical cues from the surrounding microenvironment may trigger cell invasion from a growing tumour.74 In addition, the hydrogels in the microparticles and microcapsules provide a porous scaffold for the exchange of nutrients for, and removal of waste from, the cells, while aqueous droplets in oil do not allow nutrient exchange, or waste removal. Natural materials, such as alginate,45,85,94,95,121 a mixture of alginate and Matrigel,122 or methacrylic gelatin,68 have been used to form hydrogel microparticles/microcapsules. Moreover, the use of hydrogels in droplet templates allows the assembly of cell-laden microparticles/microcapsules as building blocks for complex tumour tissue.123,124 Agarwal et al. developed a platform for the bottom-up construction of avascular tumour tissue from microcapsules encapsulated with tumour spheroids (Fig. 6) for the study of drug resistance.117
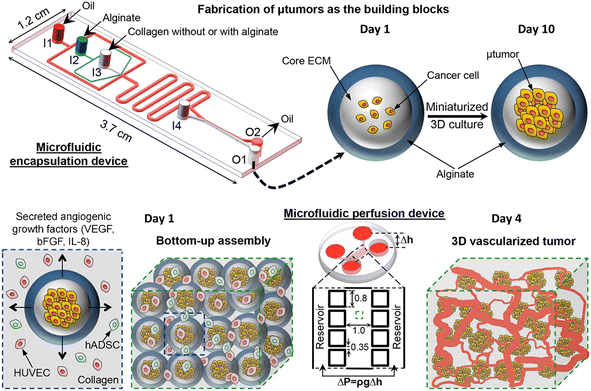 | ||
| Fig. 6 A droplet microfluidic device used for the formation of tumour-laden microcapsules and their assembly into vascularized tumour tissues. Reproduced from ref. 117 with permission from American Chemical Society, copyright 2017. | ||
Tumour spheroids generated by DMFs have been used to investigate cell–cell interactions,45,95 natural environmental effectors on cell behaviour,74,76 and the effects of chemical46,85,94,95,122 or photothermal treatments53 on tumours. In a natural tumour microenvironment, stromal cells, such as cancer-associated fibroblasts (CAFs), promote the survival, progression, metastasis and invasion of tumours by cytokine and chemokine secretion and downregulation of suppressor genes.125,126 The presence of CAFs can also impair the chemical treatment of tumours. The survival rate of tumour cells after chemical treatment was significantly increased when tumour cells and CAFs were coencapsulated into droplets to form tumour spheroids.95 Tumour spheroids can also be used to create a hypoxic tumour environment, in which hypoxic cells develop resistance and sensitivity to chemotherapeutic agents.127 Droplet microfluidics have been used to generate hypoxic microenvironments of tumours, in which the swelling–shrinkage behaviour of tumours and the response to chemical treatment of tumours were observed.76 Recently, Prince et al. developed a 2D array droplet microfluidic platform based on a self-digitalization method to form tumour spheroids (Fig. 7A).128 The tumour spheroids were generated in the presence of a biomimetic hydrogel derived from chemically modified cellulose nanocrystals and gelatine and cultured under perfusion conditions. Their responses to drugs at a gradient of concentrations were tested in a branched channel (Fig. 7B).
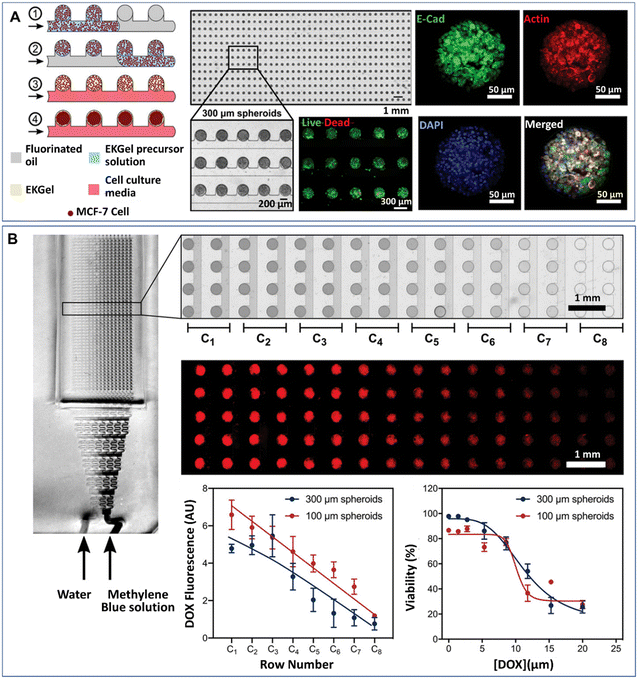 | ||
| Fig. 7 Formation of tumour spheroids and drug efficacy test in a 2D array DMF platform. (A) Tumour spheroids formed from the self-digitization of cells in a hydrogel suspension into fluorinated oil. (B) Cytotoxicity test of drugs in a concentration-gradient generator. Scale bars are 1 mm. (A) and (B) were reproduced from ref. 128 with permission from John Wiley & Sons Inc, copyright 2021. | ||
4.2 Stem cell spheroids
Aggregates of stem cells, including mesenchymal stem cells (MSCs) and pluripotent stem cells (PSCs), can grow into coherent spheroid structures named the mesenchymal body (MB) and embryonic body (EB), respectively. In comparison with the 2D format, the stem cells in 3D spheroids self-arrange in an organized spatial manner, such that the cellular functions linked with the 3D structure, such as anti-inflammatory129 and secretory activities,130–132 are accordingly improved. For example, human mesenchymal stem cells (hMSCs) in spheroids show higher expression of growth factors than those cultured in 2D.133Cell-laden microcapsules have been employed to investigate the effects of biochemical and biophysical cues on stem cell spheroids.134,135 A biomimetic microcapsule encapsulated with stem cell spheroids enables the control of cell–ECM interactions by tuning the composition and mechanical properties of the ECM, and the effects on the proliferation and pluripotency of stem cells were also investigated.77 A biomimetic ovarian microtissue was generated from an alginate/collagen microcapsule for the miniaturized 3D culture of early secondary preantral follicles.136 The crucial role of mechanical heterogeneity in the mammalian ovary in regulating follicle development and ovulation was revealed. Microcapsules were used to encapsulate a single cancer cell for the screening and culture of cancer stem cells (CSCs) (Fig. 4D).90 The screening of CSCs was based on the high survival rate of CSCs that later grow into aggregates/spheroids, while non-CSCs die of anoikis during culture. Microparticles made from degradable poly(vinyl alcohol) (PVA) were used to culture hMSC spheroids whose osteogenic differentiation was directed by the tailored microenvironment in the microparticles.137 The hMSC spheroids cultured in the biodegradable microparticles can be potentially used for in vivo tissue injection.
5. Applications of droplet microfluidics in organoids
Organoids are an organ form of differentiated cells derived from ASCs in tissue or PSCs.16,138 Compared with traditional 2D culture systems, 3D organoids better resemble the native organ in terms of gene and protein expression, metabolic function and microscale tissue architecture.18,139 In the typical formation of ASC-derived organoids, organoids are formed from ASCs imbedded into an ECM protein-rich matrix that resembles the stem cell niche. They can mimic the key features of native organs in terms of multicellular compositions, architectures, and functionalities. Over the past decade, multiple types of organoids have been successfully generated to delineate the physiological hallmarks of developing organs in humans, such as the brain,140–142 intestine,143–145 liver,146–148 and kidney.149–151Generally, the process of organoid formation relies on the spontaneous self-organization of stem cells in a 3D ECM. Animal-derived matrices (e.g., Matrigel) have been widely used as scaffolds for culturing organoids and promoting their further differentiation.152–155 Synthetic hydrogels, such as PEG, present great tunability in composition and mechanical properties and thus have been used to culture organoids.156–158 Microfluidics have also been implemented for the generation or manipulation of organoids in the form of organoids-on-a-chip with enhanced perfusion for nutrient exchange and controlled mechanical cues.159–163
Organoids derived from tissues (with tissue-resident ASCs) have been established in droplet templates. For example, an automated droplet microfluidic platform was developed to form organoids from healthy or tumour tissues in the presence of Matrigel, and the resultant organoids loaded into a multiwell plate were used for drug tests (Fig. 8).67 Mouse mammary tumour organoids were established by encapsulating tumour pieces in nonadhesive alginate microcapsules and subjected to phenotype analysis, drug uptake efficiency, and mechanical analysis, and showed potential usage for gene sequencing (Fig. 9).116
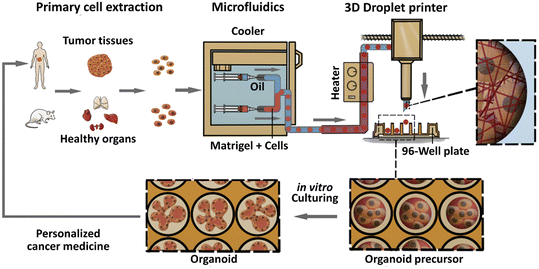 | ||
| Fig. 8 Formation of organoids in an automated droplet microfluidic system. Organoids were derived from patient tumour tissues or healthy organs in the presence of Matrigel for the development of personalized medicine. Reproduced from ref. 67 with permission from Elsevier, copyright 2020. | ||
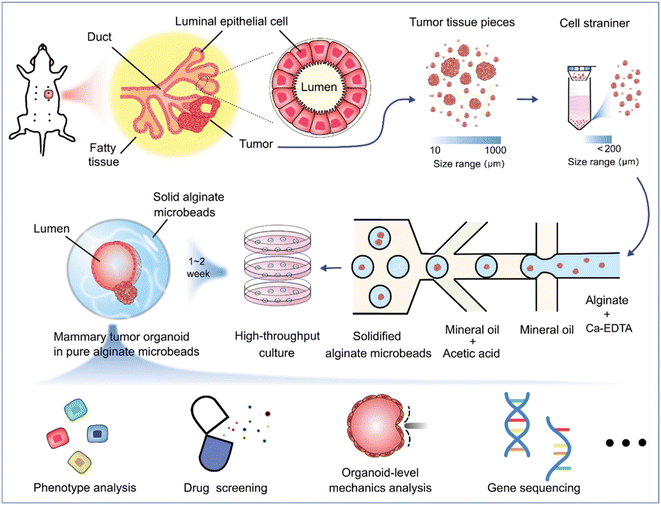 | ||
| Fig. 9 Formation and applications of organoids in microparticles in a DMF platform. Mouse mammary tumour organoids were established by encapsulating tumour pieces in nonadhesive alginate microcapsules with potential applications in phenotype analysis, drug screening, organoid-level mechanics analysis, and gene sequencing. Reproduced from ref. 116 with permission from John Wiley & Sons Inc, copyright 2021. | ||
Organoids developed from human-induced pluripotent stem cells (hiPSCs) are particularly useful for engineering organs from which cell retrieval is impractical. For example, hiPSCs have been used to form brain organoids that are difficult to be established from native tissues of the brain.142 Hydrogel microcapsules generated by DMFs have been used to form a large population of organoids derived from hiPSCs. A DMF system was developed for the controllable fabrication of hybrid hydrogel capsules, which allows massive 3D culture and the formation of functional and uniform islet organoids derived from hiPSCs (Fig. 10).84 In this work, a one-step DE process was adopted to generate core–shell microcapsules containing alginate and chitosan (Fig. 10A), and droplet generation was controlled by pneumatic single layer membrane (SLM) valves (Fig. 10B). The formation of microcapsules was based on the ionic crosslinking of chitosan and alginate diffused from the outer layer and inner core, respectively (Fig. 10C). By employing this strategy, islet organoids were formed in the microcapsules from the differentiated hiPSCs (Fig. 10D). Liver organoids derived from hiPSCs were generated by the same method with favourable cell viability and growth with a consistent size.83
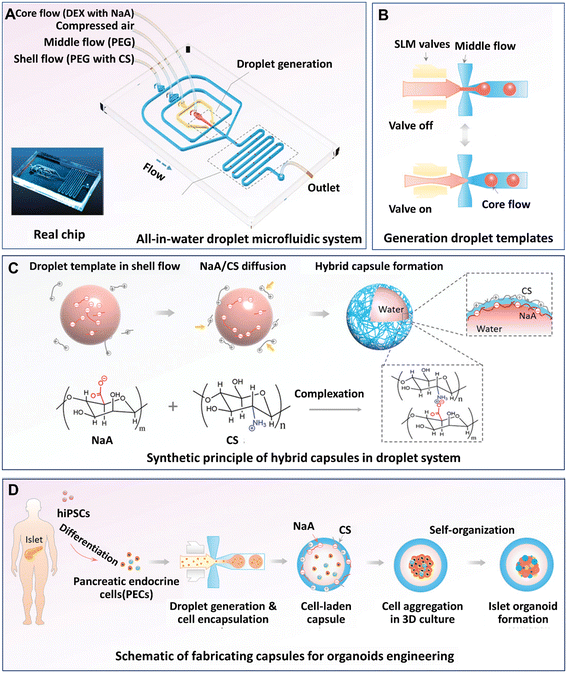 | ||
| Fig. 10 Formation and applications of organoids in core–shell microcapsules in a DMF platform. (A) An all-in-water droplet microfluidic system used for organoid generation. (B) Generation of droplet templates controlled by an SLM valve. (C) Core–shell microcapsules generated from a hybrid polymer from alginate and chitosan. (D) Schematic of the fabrication of capsules for organoid engineering. Reproduced from ref. 84 with permission from John Wiley & Sons Inc., copyright 2020. | ||
6. Discussion and perspectives
In the few recent decades, droplet microfluidics have been used for the generation and manipulation of spheroids and organoids as powerful tools in the fields of drug screening, cancer modelling, and stem cell biology. They offer great advantages in the modelling of spheroids and organoids in terms of efficiency, reproducibility, reduced sample consumption, and flexibility. Working in the interdisciplinary domains of fluid dynamics, cell biology and materials science, researchers have established many DMF platforms for the culture of spheroids and organoids that resemble tissues and organs in terms of the structure and functional properties. By incorporating functional materials such as hydrogels into droplets, researchers have been able to engineer droplets in the format of microparticles and microcapsules, expanding the range of their applications.Indeed, the majority of established DMF methods for cultivating and manipulating spheroids and organoids focus on addressing technique problems rather than bringing deeper biological insights. Nevertheless, an increasing depth in biological research based on these models has emerged thanks to several remarkable capacities of DMFs. First, the discrete droplets produced by DMFs can not only serve as vessels to cultivate spheroids and organoids, but also allow independent research on the biological properties of cells at the single-spheroid or single-organoid level, such as secretion properties of stem cell spheroids,70 local mechanical regulations on the stem cell fate,77 and the linkage between spatial heterogeneities and functionalities of stem cells in spheroids,69 which would be supressed or diluted in other methods that deal with a large population of spheroids or organoids. Moreover, DMFs offer opportunities to create unique physiological structures that are difficult to be produced by other methods. For example, iPSC spheroids were cultured in the core of microcapsules bearing a hydrogel shell that resembles the zona pellucida of natural blastomeres and showed significantly improved quality.164 Third, organoids formed in microparticles may have different biological properties from those formed under traditional culture conditions. For example, tissue-derived organoids formed in boundary-confined submicroliter microparticles presented different levels of gene or protein expression from organoids cultured in bulk Matrigel.66
Despite compelling developments, challenges remain to be resolved from the perspectives of fundamental research and practical applications. From the perspective of fundamental research, although DMFs provide many advantages for the generation of spheroids and organoids, the physiological similarity of these models to native tissue or organs is still limited. For example, a majority of these DMF-based spheroid and organoid models are established without vascular systems, the essential components of native tissues and organs, thus impeding the biological study of vascularized tissue and applications in high-fidelity drug discovery. Moreover, these models do not recapitulate the conditions of perfusion, in which soluble factors, such as nutrients and cytokines, are supplied in a dynamic manner. The integration of additional vasculature structures or perfusion conditions with these models may help to fill this gap. For example, tumour cells encapsulated in droplets formed by self-digitization have been cultured into tumour spheroids under perfusion conditions.128 Tumour spheroids formed in microcapsules were assembled with endothelial cells and other stromal cells to form 3D vascularized tumours in a microfluidic perfusion device for drug toxicity tests.117 Moreover, DMF systems are less well studied for organoid developments than for spheroid developments. The formation of organoids relies heavily on cellular self-association and presents high variability in different culture conditions, which may add additional challenges for the production of organoids by DMFs. Therefore, the development of DMF platforms with high reliability and reproducibility that can meet the variable requirements for forming organoids is particularly important.
From the perspective of practical applications, several daunting challenges need to be addressed before widespread industrial scaling and commercialization of DMF platforms for spheroids or organoids. First, the fabrication of DMF devices and manipulation of these systems need to be simple and robust, specifically for biologists with no expertise in microfluidics. The fabrication process may be facilitated by 3D printing, a high-throughput and automated technique for device fabrication, given that the materials of the device meet the requirements for droplet generation. The manipulation of these systems may be simplified with other fluid-driven processes, such as self-driven systems and capillary-driven flow. In addition, accurate, continuous, functional readouts are as important as optimization of the spheroids and organoid culture. Other than imaging methods, more analytical techniques for clinical applications may also need to be integrated with DMF platforms. Therefore, when transferring the proof-of-concept to the real world, much effort is still needed to overcome these challenges.
Author contributions
Conceptualization: Y. Wang, H. Liu, and L. Han; funding acquisition: Y. Wang, H. Liu, and L. Han; supervision: H. Liu and L. Han; visualization: Y. Wang, M. Liu, and Y. Zhang; writing – original draft: Y. Wang, M. Liu, and Y. Zhang; writing – review & editing: Y. Wang, H. Liu, and L. Han.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Major Scientific and Technological Innovation Project of Shandong Province (2021CXGC010603), the National Natural Science Foundation of China (Grant No. 32001018, 62174101), the Fundamental Research Funds of Shandong University (2020QNQT001), the Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong Province, and the Shandong University Foundation for Future Scholar Plan.References
- N. Shanks, R. Greek and J. Greek, Philos. Ethics Humanit. Med., 2009, 4, 2 CrossRef PubMed.
- M. Cekanova and K. Rathore, Drug Des., Dev. Ther., 2014, 8, 1911–1922 CrossRef PubMed.
- C. P. Day, G. Merlino and T. Van Dyke, Cell, 2015, 163, 39–53 CrossRef CAS PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed.
- J. C. Fontoura, C. Viezzer, F. G. dos Santos, R. A. Ligabue, R. Weinlich, R. D. Puga, D. Antonow, P. Severino and C. Bonorino, Mater. Sci. Eng., C, 2020, 107, 110264 CrossRef CAS PubMed.
- D. Lv, S. C. Yu, Y. F. Ping, H. Wu, X. Zhao, H. Zhang, Y. Cui, B. Chen, X. Zhang, J. Dai, X. W. Bian and X. H. Yao, Oncotarget, 2016, 7, 56904–56914 CrossRef PubMed.
- C. Bingel, E. Koeneke, J. Ridinger, A. Bittmann, M. Sill, H. Peterziel, J. K. Wrobel, I. Rettig, T. Milde, U. Fernekorn, F. Weise, A. Schober, O. Witt and I. Oehme, Cell Death Dis., 2017, 8, e3013 CrossRef CAS PubMed.
- M. Sun, A. Liu, X. Yang, J. Gong, M. Yu, X. Yao, H. Wang and Y. He, Adv. NanoBiomed Res., 2021, 1, 2000066 CrossRef.
- M. Ravi, V. Paramesh, S. R. Kaviya, E. Anuradha and F. D. Paul Solomon, J. Cell. Physiol., 2015, 230, 16–26 CrossRef CAS PubMed.
- N. Chaicharoenaudomrung, P. Kunhorm and P. Noisa, World J. Stem Cells, 2019, 11, 1065–1083 CrossRef PubMed.
- J. Xu, G. Qi, W. Wang and X. S. Sun, NPJ Sci. Food, 2021, 5, 14 CrossRef PubMed.
- Z. F. Bielecka, K. Maliszewska-Olejniczak, I. J. Safir, C. Szczylik and A. M. Czarnecka, Biol. Rev., 2017, 92, 1505–1520 CrossRef PubMed.
- S. Gunti, A. T. K. Hoke, K. P. Vu and N. R. London, Cancers, 2021, 13, 874 CrossRef CAS PubMed.
- L. B. Weiswald, D. Bellet and V. Dangles-Marie, Neoplasia, 2015, 17, 1–15 CrossRef PubMed.
- K. U. Kim, S. M. Wilson, K. S. Abayasiriwardana, R. Collins, L. Fjellbirkeland, Z. Xu, D. M. Jablons, S. L. Nishimura and V. C. Broaddus, Am. J. Respir. Cell Mol. Biol., 2005, 33, 541–548 CrossRef CAS PubMed.
- M. Hofer and M. P. Lutolf, Nat. Rev. Mater., 2021, 6, 402–420 CrossRef CAS PubMed.
- N. E. Ryu, S. H. Lee and H. Park, Cell, 2019, 8, 1620 CrossRef CAS PubMed.
- J. Kim, B. K. Koo and J. A. Knoblich, Nat. Rev. Mol. Cell Biol., 2020, 21, 571–584 CrossRef CAS PubMed.
- L. Shariati, Y. Esmaeili, S. Haghjooy Javanmard, E. Bidram and A. Amini, Stem Cells, 2021, 39, 1625–1649 CrossRef PubMed.
- A. Ashok, D. Choudhury, Y. Fang and W. Hunziker, Biotechnol. Adv., 2020, 39, 107460 CrossRef CAS PubMed.
- C. R. Thoma, M. Zimmermann, I. Agarkova, J. M. Kelm and W. Krek, Adv. Drug Delivery Rev., 2014, 69–70, 29–41 CrossRef CAS PubMed.
- M. Russo, C. M. Cejas and G. Pitingolo, Advances in microfluidic 3D cell culture for preclinical drug development, Elsevier Inc., 1st edn, 2022, vol. 187 Search PubMed.
- Y. Li and E. Kumacheva, Sci. Adv., 2018, 4, 1–11 Search PubMed.
- V. M. Le, M. D. Lang, W. Bin Shi and J. W. Liu, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 540–544 CrossRef CAS PubMed.
- M. Dvir-Ginzberg, T. Elkayam, E. D. Aflalo, R. Agbaria and S. Cohen, Tissue Eng., 2004, 10, 1806–1817 CrossRef CAS PubMed.
- D. Loessner, K. S. Stok, M. P. Lutolf, D. W. Hutmacher, J. A. Clements and S. C. Rizzi, Biomaterials, 2010, 31, 8494–8506 CrossRef CAS PubMed.
- C. Liu, D. Lewin Mejia, B. Chiang, K. E. Luker and G. D. Luker, Acta Biomater., 2018, 75, 213–225 CrossRef CAS PubMed.
- X. Cui, Y. Hartanto and H. Zhang, J. R. Soc., Interface, 2017, 14, 20160877 CrossRef PubMed.
- A. Y. Hsiao, Y. C. Tung, C. H. Kuo, B. Mosadegh, R. Bedenis, K. J. Pienta and S. Takayama, Biomed. Microdevices, 2012, 14, 313–323 CrossRef CAS PubMed.
- L. Zhao, J. Xiu, Y. Liu, T. Zhang, W. Pan, X. Zheng and X. Zhang, Sci. Rep., 2019, 9, 19717 CrossRef CAS PubMed.
- J. M. Kelm, E. Ehler, L. K. Nielsen, S. Schlatter, J. C. Perriard and M. Fussenegger, Tissue Eng., 2004, 10, 201–214 CrossRef CAS PubMed.
- D. Rodoplu, J. S. Matahum and C.-H. Hsu, Lab Chip, 2022, 22, 1275–1285 RSC.
- J. Park, H. Kim and J. K. Park, Analyst, 2020, 145, 6974–6980 RSC.
- H. W. Wu, Y. H. Hsiao, C. C. Chen, S. F. Yet and C. H. Hsu, Molecules, 2016, 21, 882 CrossRef PubMed.
- O. Frey, P. M. Misun, D. A. Fluri, J. G. Hengstler and A. Hierlemann, Nat. Commun., 2014, 5, 4250 CrossRef CAS PubMed.
- N. Barker, M. Huch, P. Kujala, M. van de Wetering, H. J. Snippert, J. H. van Es, T. Sato, D. E. Stange, H. Begthel, M. van den Born, E. Danenberg, S. van den Brink, J. Korving, A. Abo, P. J. Peters, N. Wright, R. Poulsom and H. Clevers, Cell Stem Cell, 2010, 6, 25–36 CrossRef CAS PubMed.
- L. Xin, R. U. Lukacs, D. A. Lawson, D. Cheng and O. N. Witte, Stem Cells, 2007, 25, 2760–2769 CrossRef CAS PubMed.
- F. Antonica, D. F. Kasprzyk, R. Opitz, M. Iacovino, X. H. Liao, A. M. Dumitrescu, S. Refetoff, K. Peremans, M. Manto, M. Kyba and S. Costagliola, Nature, 2012, 491, 66–71 CrossRef CAS PubMed.
- M. Takasato, P. X. Er, M. Becroft, J. M. Vanslambrouck, E. G. Stanley, A. G. Elefanty and M. H. Little, Nat. Cell Biol., 2014, 16, 118–126 CrossRef CAS PubMed.
- S. L. Giandomenico, S. B. Mierau, G. M. Gibbons, L. M. D. Wenger, L. Masullo, T. Sit, M. Sutcliffe, J. Boulanger, M. Tripodi, E. Derivery, O. Paulsen, A. Lakatos and M. A. Lancaster, Nat. Neurosci., 2019, 22, 669–679 CrossRef CAS PubMed.
- J. T. Neal, X. Li, J. Zhu, V. Giangarra, C. L. Grzeskowiak, J. Ju, I. H. Liu, S. H. Chiou, A. A. Salahudeen, A. R. Smith, B. C. Deutsch, L. Liao, A. J. Zemek, F. Zhao, K. Karlsson, L. M. Schultz, T. J. Metzner, L. D. Nadauld, Y. Y. Tseng, S. Alkhairy, C. Oh, P. Keskula, D. Mendoza-Villanueva, F. M. De La Vega, P. L. Kunz, J. C. Liao, J. T. Leppert, J. B. Sunwoo, C. Sabatti, J. S. Boehm, W. C. Hahn, G. X. Y. Zheng, M. M. Davis and C. J. Kuo, Cell, 2018, 175, 1972–1988.e16 CrossRef CAS PubMed.
- V. van Duinen, S. J. Trietsch, J. Joore, P. Vulto and T. Hankemeier, Curr. Opin. Biotechnol., 2015, 35, 118–126 CrossRef CAS PubMed.
- A. D. Castiaux, D. M. Spence and R. S. Martin, Anal. Methods, 2019, 11, 4220–4232 RSC.
- K. S. McMillan, M. Boyd and M. Zagnoni, Lab Chip, 2016, 16, 3548–3557 RSC.
- S. Yoon, J. A. Kim, S. H. Lee, M. Kim and T. H. Park, Lab Chip, 2013, 13, 1522–1528 RSC.
- R. F. X. Tomasi, S. Sart, T. Champetier and C. N. Baroud, Cell Rep., 2020, 31, 107670 CrossRef CAS PubMed.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed.
- S. Sart, G. Ronteix, S. Jain, G. Amselem and C. N. Baroud, Chem. Rev., 2022, 122, 7061–7096 CrossRef CAS PubMed.
- D. E. Cohen, T. Schneider, M. Wang and D. T. Chiu, Anal. Chem., 2010, 82, 5707–5717 CrossRef CAS PubMed.
- J. E. Kreutz, J. Wang, A. M. Sheen, A. M. Thompson, J. P. Staheli, M. R. Dyen, Q. Feng and D. T. Chiu, Lab Chip, 2019, 19, 1035–1040 RSC.
- X. Cui, L. Wu, Y. Wu, J. Zhang, Q. Zhao, F. Jing, L. Yi and G. Li, Anal. Chim. Acta, 2020, 1107, 127–134 CrossRef CAS PubMed.
- B. Kwak, Y. Lee, J. Lee, S. Lee and J. Lim, J. Controlled Release, 2018, 275, 201–207 CrossRef CAS PubMed.
- J. M. Lee, J. W. Choi, C. D. Ahrberg, H. W. Choi, J. H. Ha, S. G. Mun, S. J. Mo and B. G. Chung, Microsyst. Nanoeng., 2020, 6, 52 CrossRef CAS PubMed.
- K. Langer and H. N. Joensson, SLAS Technol., 2020, 25, 111–122 CrossRef CAS PubMed.
- J. Liu, Y. Tan, H. Zhang, Y. Zhang, P. Xu, J. Chen, Y. C. Poh, K. Tang, N. Wang and B. Huang, Nat. Mater., 2012, 11, 734–741 CrossRef CAS PubMed.
- K. M. Charoen, B. Fallica, Y. L. Colson, M. H. Zaman and M. W. Grinstaff, Biomaterials, 2014, 35, 2264–2271 CrossRef CAS PubMed.
- J. A. Bagley, D. Reumann, S. Bian, J. Lévi-Strauss and J. A. Knoblich, Nat. Methods, 2017, 14, 743–751 CrossRef CAS PubMed.
- S. Sart, R. F. X. Tomasi, G. Amselem and C. N. Baroud, Nat. Commun., 2017, 8, 469 CrossRef PubMed.
- C. Kim, BioChip J., 2015, 9, 105–113 CrossRef CAS.
- F. C. Chang, S. L. Levengood, N. Cho, L. Chen, E. Wang, J. S. Yu and M. Zhang, Adv. Ther., 2018, 1, 1–11 Search PubMed.
- B. J. Gill, D. L. Gibbons, L. C. Roudsari, J. E. Saik, Z. H. Rizvi, J. D. Roybal, J. M. Kurie and J. L. West, Cancer Res., 2012, 72, 6013–6023 CrossRef CAS PubMed.
- Y. Liang, J. Jeong, R. J. DeVolder, C. Cha, F. Wang, Y. W. Tong and H. Kong, Biomaterials, 2011, 32, 9308–9315 CrossRef CAS PubMed.
- L. J. Bray, M. Binner, A. Holzheu, J. Friedrichs, U. Freudenberg, D. W. Hutmacher and C. Werner, Biomaterials, 2015, 53, 609–620 CrossRef CAS PubMed.
- S. Pradhan, J. M. Clary, D. Seliktar and E. A. Lipke, Biomaterials, 2017, 115, 141–154 CrossRef CAS PubMed.
- W. Zhang, D. Li, S. Jiang, E. A. Galan, Z. Zhang, L. Huang and S. Ma, Chem. Eng. Sci., 2021, 238, 116632 CrossRef CAS.
- H. Zhao, Y. Cheng, J. Li, J. Zhou, H. Yang, F. Yu, F. Yu, D. Khutsishvili, Z. Wang, S. Jiang, K. Tan, Y. Kuang, X. Xing and S. Ma, Fundam. Res., 2022 DOI:10.1016/j.fmre.2022.05.018.
- S. Jiang, H. Zhao, W. Zhang, J. Wang, Y. Liu, Y. Cao, H. Zheng, Z. Hu, S. Wang, Y. Zhu, W. Wang, S. Cui, P. E. Lobie, L. Huang and S. Ma, Cell Rep. Med., 2020, 1, 100161 CrossRef CAS PubMed.
- D. Lee and C. Cha, Pharmaceutics, 2018, 10, 229 CrossRef CAS PubMed.
- S. Sart, R. F. X. Tomasi, A. Barizien, G. Amselem, A. Cumano and C. N. Baroud, Sci. Adv., 2020, 6, eaaw7853 CrossRef CAS PubMed.
- A. Saint-Sardos, S. Sart, K. Lippera, E. Brient-Litzler, S. Michelin, G. Amselem and C. N. Baroud, Small, 2020, 16, 2002303 CrossRef CAS PubMed.
- Z. Chen, S. Kheiri, A. Gevorkian, E. W. K. Young, V. Andre, T. Deisenroth and E. Kumacheva, Lab Chip, 2021, 21, 3952–3962 RSC.
- H. Huang, Y. Yu, Y. Hu, X. He, O. Berk Usta and M. L. Yarmush, Lab Chip, 2017, 17, 1913–1932 RSC.
- H. F. Chan, Y. Zhang, Y. P. Ho, Y. L. Chiu, Y. Jung and K. W. Leong, Sci. Rep., 2013, 3, 3462 CrossRef PubMed.
- K. Alessandri, B. R. Sarangi, V. V. Gurchenkov, B. Sinha, T. R. Kießling, L. Fetler, F. Rico, S. Scheuring, C. Lamaze, A. Simon, S. Geraldo, D. Vignjević, H. Doméjean, L. Rolland, A. Funfak, J. Bibette, N. Bremond and P. Nassoy, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14843–14848 CrossRef CAS PubMed.
- G. Zhao, X. Liu, K. Zhu and X. He, Adv. Healthcare Mater., 2017, 6, 1700988 CrossRef PubMed.
- S. M. Grist, S. S. Nasseri, L. Laplatine, J. C. Schmok, D. Yao, J. Hua, L. Chrostowski and K. C. Cheung, Sci. Rep., 2019, 9, 17782 CrossRef PubMed.
- P. Agarwal, J. K. Choi, H. Huang, S. Zhao, J. Dumbleton, J. Li and X. He, Part. Part. Syst. Charact., 2015, 32, 809–816 CrossRef CAS PubMed.
- L. Yu, C. Ni, S. M. Grist, C. Bayly and K. C. Cheung, Biomed. Microdevices, 2015, 17, 33 CrossRef PubMed.
- P. Agarwal, S. Zhao, P. Bielecki, W. Rao, J. K. Choi, Y. Zhao, J. Yu, W. Zhang and X. He, Lab Chip, 2013, 13, 4525–4533 RSC.
- Q. Chen, S. Utech, D. Chen, R. Prodanovic, J.-M. Lin and D. A. Weitz, Lab Chip, 2016, 16, 1346–1349 RSC.
- D. Lee, K. Lee and C. Cha, Adv. Biosyst., 2018, 2, 1800236 CrossRef.
- K. Zhu, Y. Yu, Y. Cheng, C. Tian, G. Zhao and Y. Zhao, ACS Appl. Mater. Interfaces, 2019, 11, 4826–4832 CrossRef CAS PubMed.
- Y. Wang, H. Liu, M. Zhang, H. Wang, W. Chen and J. Qin, Biomater. Sci., 2020, 8, 5476–5488 RSC.
- H. Liu, Y. Wang, H. Wang, M. Zhao, T. Tao, X. Zhang and J. Qin, Adv. Sci., 2020, 7, 1903739 CrossRef CAS PubMed.
- Q. Sun, S. H. Tan, Q. Chen, R. Ran, Y. Hui, D. Chen and C. X. Zhao, ACS Biomater. Sci. Eng., 2018, 4, 4425–4433 CrossRef CAS PubMed.
- H. F. Chan, Y. Zhang and K. W. Leong, Small, 2016, 12, 2720–2730 CrossRef CAS PubMed.
- S. Zhao, P. Agarwal, W. Rao, H. Huang, R. Zhang, Z. Liu, J. Yu, N. Weisleder, W. Zhang and X. He, Integr. Biol., 2014, 6, 874–884 CrossRef CAS PubMed.
- W. Rao, S. Zhao, J. Yu, X. Lu, D. L. Zynger and X. He, Biomaterials, 2014, 35, 7762–7773 CrossRef CAS PubMed.
- D. K. Nguyen, Y. M. Son and N. E. Lee, Adv. Healthcare Mater., 2015, 4, 1537–1544 CrossRef CAS PubMed.
- H. Wang, P. Agarwal, B. Jiang, S. Stewart, X. Liu, Y. Liang, B. Hancioglu, A. Webb, J. P. Fisher, Z. Liu, X. Lu, K. H. R. Tkaczuk and X. He, Adv. Sci., 2020, 7, 2000259 CrossRef CAS PubMed.
- R. Seemann, M. Brinkmann, T. Pfohl and S. Herminghaus, Rep. Prog. Phys., 2012, 75, 016601 CrossRef PubMed.
- X. Casadevall i Solvas and A. De Mello, Chem. Commun., 2011, 47, 1936–1942 RSC.
- F. Qu, S. Zhao, G. Cheng, H. Rahman, Q. Xiao, R. W. Y. Chan and Y. P. Ho, Microsyst. Nanoeng., 2021, 7, 38 CrossRef CAS PubMed.
- L. Yu, M. C. W. Chen and K. C. Cheung, Lab Chip, 2010, 10, 2424–2432 RSC.
- P. Sabhachandani, V. Motwani, N. Cohen, S. Sarkar, V. Torchilin and T. Konry, Lab Chip, 2016, 16, 497–505 RSC.
- H. Wang, H. Liu, X. Zhang, Y. Wang, M. Zhao, W. Chen and J. Qin, ACS Appl. Mater. Interfaces, 2021, 13, 3199–3208 CrossRef CAS PubMed.
- K. H. Lee and T. H. Kim, Biosensors, 2021, 11, 445 CrossRef CAS PubMed.
- S. J. Han, S. Kwon and K. S. Kim, Cancer Cell Int., 2021, 21, 152 CrossRef PubMed.
- G. Palazzolo, H. Mollica, V. Lusi, M. Rutigliani, M. Di Francesco, R. C. Pereira, M. Filauro, L. Paleari, A. DeCensi and P. Decuzzi, Transl. Oncol., 2020, 13, 100760 CrossRef PubMed.
- H. Shoval, A. Karsch-Bluman, Y. Brill-Karniely, T. Stern, G. Zamir, A. Hubert and O. Benny, Sci. Rep., 2017, 7, 10428 CrossRef PubMed.
- Z. Wu, Z. Gong, Z. Ao, J. Xu, H. Cai, M. Muhsen, S. Heaps, M. Bondesson, S. Guo and F. Guo, ACS Appl. Bio Mater., 2020, 3, 6273–6283 CrossRef CAS PubMed.
- W. Metzger, D. Sossong, A. Bächle, N. Pütz, G. Wennemuth, T. Pohlemann and M. Oberringer, Cytotherapy, 2011, 13, 1000–1012 CrossRef CAS PubMed.
- E. C. Costa, V. M. Gaspar, P. Coutinho and I. J. Correia, Biotechnol. Bioeng., 2014, 111, 1672–1685 CrossRef CAS PubMed.
- J. M. Kelm, N. E. Timmins, C. J. Brown, M. Fussenegger and L. K. Nielsen, Biotechnol. Bioeng., 2003, 83, 173–180 CrossRef CAS PubMed.
- A. V. Taubenberger, S. Girardo, N. Träber, E. Fischer-Friedrich, M. Kräter, K. Wagner, T. Kurth, I. Richter, B. Haller, M. Binner, D. Hahn, U. Freudenberg, C. Werner and J. Guck, Adv. Biosyst., 2019, 3, 1900128 CrossRef PubMed.
- Y. Li, N. Khuu, A. Gevorkian, S. Sarjinsky, H. Therien-Aubin, Y. Wang, S. Cho and E. Kumacheva, Angew. Chem., Int. Ed., 2017, 56, 6083–6087 CrossRef CAS PubMed.
- H. Song, O. David, S. Clejan, C. L. Giordano, H. Pappas-lebeau, L. Xu and K. C. O'connor, Tissue Eng., 2004, 10, 1266–1276 CrossRef CAS PubMed.
- S. L. Nyberg, J. Hardin, B. Amiot, U. A. Argikar, R. P. Remmel and P. Rinaldo, Liver Transpl., 2005, 11, 901–910 CrossRef PubMed.
- J. M. Lee, D. Y. Park, L. Yang, E. J. Kim, C. D. Ahrberg, K. B. Lee and B. G. Chung, Sci. Rep., 2018, 8, 17145 CrossRef PubMed.
- F. Mirab, Y. J. Kang and S. Majd, PLoS One, 2019, 14, e0211078 CrossRef CAS PubMed.
- W. Liu, J. C. Wang and J. Wang, Lab Chip, 2015, 15, 1195–1204 RSC.
- W. Liu, J. Xu, T. Li, L. Zhao, C. Ma, S. Shen and J. Wang, Anal. Chem., 2015, 87, 9752–9760 CrossRef CAS PubMed.
- H. Ota, T. Kodama and N. Miki, Biomicrofluidics, 2011, 5, 034105 CrossRef PubMed.
- K. Chen, M. Wu, F. Guo, P. Li, C. Y. Chan, Z. Mao, S. Li, L. Ren, R. Zhang and T. J. Huang, Lab Chip, 2016, 16, 2636–2643 RSC.
- B. Kwak, Y. Lee, J. Lee, S. Lee and J. Lim, J. Controlled Release, 2018, 275, 201–207 CrossRef CAS PubMed.
- G. Fang, H. Lu, L. Rodriguez de la Fuente, A. M. K. Law, G. Lin, D. Jin and D. Gallego-Ortega, Adv. Sci., 2021, 8, 2102418 CrossRef CAS PubMed.
- P. Agarwal, H. Wang, M. Sun, J. Xu, S. Zhao, Z. Liu, K. J. Gooch, Y. Zhao, X. Lu and X. He, ACS Nano, 2017, 11, 6691–6702 CrossRef CAS PubMed.
- M. E. Dolega, F. Abeille, N. Picollet-D'hahan and X. Gidrol, Biomaterials, 2015, 52, 347–357 CrossRef CAS PubMed.
- I. R. Calori, S. R. Alves, H. Bi and A. C. Tedesco, ACS Appl. Bio Mater., 2022, 5, 723–733 CrossRef CAS PubMed.
- X. Xu, L. A. Gurski, C. Zhang, D. A. Harrington, M. C. Farach-Carson and X. Jia, Biomaterials, 2012, 33, 9049–9060 CrossRef CAS PubMed.
- C. Kim, J. Park and J. Y. Kang, Biomicrofluidics, 2014, 8, 066504 CrossRef PubMed.
- Y. Wang and J. Wang, Analyst, 2014, 139, 2449–2458 RSC.
- Y. T. Matsunaga, Y. Morimoto and S. Takeuchi, Adv. Mater., 2011, 23, H90–H94 CrossRef CAS PubMed.
- Y. Fang, Y. Guo, M. Ji, B. Li, Y. Guo, J. Zhu, T. Zhang and Z. Xiong, Adv. Funct. Mater., 2021, 32, 2109810 CrossRef.
- Y. Mao, E. T. Keller, D. H. Garfield, K. Shen and J. Wang, Cancer Metastasis Rev., 2013, 32, 303–315 CrossRef PubMed.
- W. Zhang and P. Huang, Cancer Biol. Ther., 2011, 11, 150–156 CrossRef CAS PubMed.
- W. R. Wilson and M. P. Hay, Nat. Rev. Cancer, 2011, 11, 393–410 CrossRef CAS PubMed.
- E. Prince, S. Kheiri, Y. Wang, F. Xu, J. Cruickshank, V. Topolskaia, H. Tao, E. W. K. Young, A. P. McGuigan, D. W. Cescon and E. Kumacheva, Adv. Healthcare Mater., 2022, 11, 2101085 CrossRef CAS PubMed.
- T. J. Bartosh, J. H. Ylöstalo, A. Mohammadipoor, N. Bazhanov, K. Coble, K. Claypool, R. H. Lee, H. Choi and D. J. Prockop, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13724–13729 CrossRef CAS PubMed.
- A. Imamura, H. Kajiya, S. Fujisaki, M. Maeshiba, T. Yanagi, H. Kojima and J. Ohno, Biochem. Biophys. Res. Commun., 2020, 523, 458–464 CrossRef CAS PubMed.
- M. Kim, H. W. Yun, D. Y. Park, B. H. Choi and B. H. Min, Tissue Eng. Regener. Med., 2018, 15, 427–436 CrossRef CAS PubMed.
- S. Sart, A. C. Tsai, Y. Li and T. Ma, Tissue Eng., Part B, 2014, 20, 365–380 CrossRef PubMed.
- K. C. Murphy, J. Whitehead, D. Zhou, S. S. Ho and J. K. Leach, Acta Biomater., 2017, 64, 176–186 CrossRef CAS PubMed.
- C. Siltanen, M. Yaghoobi, A. Haque, J. You, J. Lowen, M. Soleimani and A. Revzin, Acta Biomater., 2016, 34, 125–132 CrossRef CAS PubMed.
- C. Kim, S. Chung, Y. E. Kim, K. S. Lee, S. H. Lee, K. W. Oh and J. Y. Kang, Lab Chip, 2011, 11, 246–252 RSC.
- J. K. Choi, P. Agarwal, H. Huang, S. Zhao and X. He, Biomaterials, 2014, 35, 5122–5128 CrossRef CAS PubMed.
- Y. Hou, W. Xie, K. Achazi, J. L. Cuellar-Camacho, M. F. Melzig, W. Chen and R. Haag, Acta Biomater., 2018, 77, 28–37 CrossRef CAS PubMed.
- J. Drost and H. Clevers, Nat. Rev. Cancer, 2018, 18, 407–418 CrossRef CAS PubMed.
- Y. Zhang, S. Huang, W. Zhong, W. Chen, B. Yao and X. Wang, Acta Pharm. Sin. B, 2021, 11, 1697–1707 CrossRef CAS PubMed.
- M. A. Lancaster, N. S. Corsini, S. Wolfinger, E. H. Gustafson, A. W. Phillips, T. R. Burkard, T. Otani, F. J. Livesey and J. A. Knoblich, Nat. Biotechnol., 2017, 35, 659–666 CrossRef CAS PubMed.
- Y. Miura, M. Y. Li, F. Birey, K. Ikeda, O. Revah, M. V. Thete, J. Y. Park, A. Puno, S. H. Lee, M. H. Porteus and S. P. Paşca, Nat. Biotechnol., 2020, 38, 1421–1430 CrossRef CAS PubMed.
- M. A. Lancaster, M. Renner, C. A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Nature, 2013, 501, 373–379 CrossRef CAS PubMed.
- N. Gjorevski, N. Sachs, A. Manfrin, S. Giger, M. E. Bragina, P. Ordóñez-Morán, H. Clevers and M. P. Lutolf, Nature, 2016, 539, 560–564 CrossRef CAS PubMed.
- K. W. McCracken, J. C. Howell, J. M. Wells and J. R. Spence, Nat. Protoc., 2011, 6, 1920–1928 CrossRef CAS PubMed.
- I. Heo, D. Dutta, D. A. Schaefer, N. Iakobachvili, B. Artegiani, N. Sachs, K. E. Boonekamp, G. Bowden, A. P. A. Hendrickx, R. J. L. Willems, P. J. Peters, M. W. Riggs, R. O'Connor and H. Clevers, Nat. Microbiol., 2018, 3, 814–823 CrossRef CAS PubMed.
- L. Broutier, G. Mastrogiovanni, M. M. A. Verstegen, H. E. Francies, L. M. Gavarró, C. R. Bradshaw, G. E. Allen, R. Arnes-Benito, O. Sidorova, M. P. Gaspersz, N. Georgakopoulos, B. K. Koo, S. Dietmann, S. E. Davies, R. K. Praseedom, R. Lieshout, J. N. M. IJzermans, S. J. Wigmore, K. Saeb-Parsy, M. J. Garnett, L. J. W. Van Der Laan and M. Huch, Nat. Med., 2017, 23, 1424–1435 CrossRef CAS PubMed.
- M. Huch, C. Dorrell, S. F. Boj, J. H. Van Es, V. S. W. Li, M. Van De Wetering, T. Sato, K. Hamer, N. Sasaki, M. J. Finegold, A. Haft, R. G. Vries, M. Grompe and H. Clevers, Nature, 2013, 494, 247–250 CrossRef CAS PubMed.
- H. Hu, H. Gehart, B. Artegiani, C. LÖpez-Iglesias, F. Dekkers, O. Basak, J. van Es, S. M. Chuva de Sousa Lopes, H. Begthel, J. Korving, M. van den Born, C. Zou, C. Quirk, L. Chiriboga, C. M. Rice, S. Ma, A. Rios, P. J. Peters, Y. P. de Jong and H. Clevers, Cell, 2018, 175, 1591–1606 CrossRef CAS PubMed.
- L. J. Hale, S. E. Howden, B. Phipson, A. Lonsdale, P. X. Er, I. Ghobrial, S. Hosawi, S. Wilson, K. T. Lawlor, S. Khan, A. Oshlack, C. Quinlan, R. Lennon and M. H. Little, Nat. Commun., 2018, 9, 5167 CrossRef PubMed.
- M. Takasato, P. X. Er, H. S. Chiu, B. Maier, G. J. Baillie, C. Ferguson, R. G. Parton, E. J. Wolvetang, M. S. Roost, S. M. C. De Sousa Lopes and M. H. Little, Nature, 2015, 526, 564–568 CrossRef CAS PubMed.
- R. Morizane, A. Q. Lam, B. S. Freedman, S. Kishi, M. T. Valerius and J. V. Bonventre, Nat. Biotechnol., 2015, 33, 1193–1200 CrossRef CAS PubMed.
- X. Qian, H. N. Nguyen, M. M. Song, C. Hadiono, S. C. Ogden, C. Hammack, B. Yao, G. R. Hamersky, F. Jacob, C. Zhong, K. J. Yoon, W. Jeang, L. Lin, Y. Li, J. Thakor, D. A. Berg, C. Zhang, E. Kang, M. Chickering, D. Nauen, C. Y. Ho, Z. Wen, K. M. Christian, P. Y. Shi, B. J. Maher, H. Wu, P. Jin, H. Tang, H. Song and G. L. Ming, Cell, 2016, 165, 1238–1254 CrossRef CAS PubMed.
- M. Crespo, E. Vilar, S. Y. Tsai, K. Chang, S. Amin, T. Srinivasan, T. Zhang, N. H. Pipalia, H. J. Chen, M. Witherspoon, M. Gordillo, J. Z. Xiang, F. R. Maxfield, S. Lipkin, T. Evans and S. Chen, Nat. Med., 2017, 23, 878–884 CrossRef CAS PubMed.
- Y. Yamamoto, S. Gotoh, Y. Korogi, M. Seki, S. Konishi, S. Ikeo, N. Sone, T. Nagasaki, H. Matsumoto, S. Muro, I. Ito, T. Hirai, T. Kohno, Y. Suzuki and M. Mishima, Nat. Methods, 2017, 14, 1097–1106 CrossRef CAS PubMed.
- T. Takebe, K. Sekine, M. Enomura, H. Koike, M. Kimura, T. Ogaeri, R. R. Zhang, Y. Ueno, Y. W. Zheng, N. Koike, S. Aoyama, Y. Adachi and H. Taniguchi, Nature, 2013, 499, 481–484 CrossRef CAS PubMed.
- Z. Ma, J. Wang, P. Loskill, N. Huebsch, S. Koo, F. L. Svedlund, N. C. Marks, E. W. Hua, C. P. Grigoropoulos, B. R. Conklin and K. E. Healy, Nat. Commun., 2015, 6, 7413 CrossRef CAS PubMed.
- H. S. Shin, H. J. Hong, W. G. Koh and J. Y. Lim, ACS Biomater. Sci. Eng., 2018, 4, 4311–4320 CrossRef CAS PubMed.
- S. S. Ng, K. Saeb-Parsy, S. J. I. Blackford, J. M. Segal, M. P. Serra, M. Horcas-Lopez, D. Y. No, S. Mastoridis, W. Jassem, C. W. Frank, N. J. Cho, H. Nakauchi, J. S. Glenn and S. T. Rashid, Biomaterials, 2018, 182, 299–311 CrossRef CAS PubMed.
- M. Kasendra, A. Tovaglieri, A. Sontheimer-Phelps, S. Jalili-Firoozinezhad, A. Bein, A. Chalkiadaki, W. Scholl, C. Zhang, H. Rickner, C. A. Richmond, H. Li, D. T. Breault and D. E. Ingber, Sci. Rep., 2018, 8, 2871 CrossRef PubMed.
- Y. S. Zhang, J. Aleman, S. R. Shin, T. Kilic, D. Kim, S. A. M. Shaegh, S. Massa, R. Riahi, S. Chae, N. Hu, H. Avci, W. Zhang, A. Silvestri, A. S. Nezhad, A. Manbohi, F. De Ferrari, A. Polini, G. Calzone, N. Shaikh, P. Alerasool, E. Budina, J. Kang, N. Bhise, J. Ribas, A. Pourmand, A. Skardal, T. Shupe, C. E. Bishop, M. R. Dokmeci, A. Atala and A. Khademhosseini, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E2293–E2302 CAS.
- Y. Wang, L. Wang, Y. Zhu and J. Qin, Lab Chip, 2018, 18, 851–860 RSC.
- Y. Wang, L. Wang, Y. Guo, Y. Zhu and J. Qin, RSC Adv., 2018, 8, 1677–1685 RSC.
- E. Karzbrun, A. Kshirsagar, S. R. Cohen, J. H. Hanna and O. Reiner, Nat. Phys., 2018, 14, 515–522 Search PubMed.
- J. Xu, J. G. Shamul, N. A. Staten, A. M. White, B. Jiang and X. He, Small, 2021, 17, 1–12 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |