Expanding the structure–activity relationships of alkynyl diphenylurea scaffold as promising antibacterial agents†
Hanzada T.
Nour El-Din‡
 a,
Mohamed M.
Elsebaie‡
a,
Mohamed M.
Elsebaie‡
 b,
Nader S.
Abutaleb
cd,
Ahmed M.
Kotb
b,
Ahmed S.
Attia
ae,
Mohamed N.
Seleem
b,
Nader S.
Abutaleb
cd,
Ahmed M.
Kotb
b,
Ahmed S.
Attia
ae,
Mohamed N.
Seleem
 *cf and
Abdelrahman S.
Mayhoub
*cf and
Abdelrahman S.
Mayhoub
 *bg
*bg
aDepartment of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo 11562, Egypt
bDepartment of Pharmaceutical Organic Chemistry, College of Pharmacy, Al-Azhar University, Cairo 11884, Egypt
cDepartment of Biomedical Sciences and Pathobiology, Virginia-Maryland College of Veterinary Medicine, Virginia Polytechnic Institute and State University, Blacksburg, Virginia 24061, USA
dDepartment of Microbiology and Immunology, Faculty of Pharmacy, Zagazig University, Zagazig 44519, Egypt
eDepartment of Microbiology and Immunology, School of Pharmacy, Newgiza University, Giza, Egypt
fCenter for One Health Research, Virginia Polytechnic Institute and State University, Blacksburg, Virginia 24061, USA
gNanoscience Program, University of Science and Technology, Zewail City of Science and Technology, Giza, Egypt. E-mail: amayhoub@zewailcity.edu.eg
First published on 30th November 2022
Abstract
With the continuous and alarming threat of exhausting the current antimicrobial arsenals, efforts are urgently needed to develop new effective ones. In this study, the antibacterial efficacy of a set of structurally related acetylenic-diphenylurea derivatives carrying the aminoguanidine moiety was tested against a panel of multidrug-resistant Gram-positive clinical isolates. Compound 18 was identified with a superior bacteriological profile than the lead compound I. Compound 18 demonstrated an excellent antibacterial profile in vitro: low MIC values, extended post-antibiotic effect, refractory ability to resistance development upon extended repeated exposure, and high tolerability towards mammalian cells. Finally, when assessed in a MRSA skin infection animal model, compound 18 showed considerable healing and less inflammation, decrease in the bacterial loads in skin lesions, and it surpassed fusidic acid in controlling the systemic dissemination of S. aureus. Collectively, compound 18 represents a promising lead anti-MRSA agent that merits further investigation for the development of new anti-staphylococcal therapeutics.
1. Introduction
The introduction of antibiotics into clinical use has had a huge impact as it elevated the possibility of treating life-threatening infectious diseases and controlling post-operative infections.1 Unfortunately, almost every new antibiotic introduced to the market was followed by the reported emergence of bacterial resistance against it. More seriously, pathogens have not only acquired resistance to single antibiotics but also to multiple antibiotics and to the majority of antibiotic classes discovered to date.2 Bacteria acquire resistance through multiple mechanisms, including the natural, acquired, and cross ones, leading to the emergence of multidrug-resistant (MDR) bacterial strains.3 It was reported that the frequency of life-threatening infectious diseases caused by MDR bacteria has reached alarming levels worldwide.4 It was estimated that by the year 2050, more than 10 million people would die annually as a result of drug-resistant bacterial infections. Therefore, if the situation remains as it is, infections that were previously treated with an antibiotic course in few days could become untreatable.5
Staphylococcus aureus is one of the leading causes of skin, wounds, and hospital acquired infections (HAIs). It is a leading cause of bacteremia, endocarditis, skin and soft tissue infections, and medical device-related infections.6 A successful treatment of infections caused by this resilient organism has become a daunting challenge with the emergence of clinical methicillin-resistant S. aureus (MRSA) isolates, which are currently a leading cause of both hospital-acquired and community-acquired infections worldwide.7,8 According to the Center for Disease Control and Prevention (CDC), MRSA infects about 323![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 patients yearly in the United States alone, resulting in 10
700 patients yearly in the United States alone, resulting in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 deaths and $1.7 billion healthcare costs.9 Linezolid and vancomycin are still the frontline drugs for the treatment of MRSA infections; however, resistance to both antibiotics is rising, highlighting the threat of upcoming non-treatable S. aureus strains.10,11 Consequently, there is a pressing need for the development of new effective anti-staphylococcal therapeutics, preferably with chemical characteristics that vastly differ from those of the existing ones to combat the rapid spread of drug resistance.12
600 deaths and $1.7 billion healthcare costs.9 Linezolid and vancomycin are still the frontline drugs for the treatment of MRSA infections; however, resistance to both antibiotics is rising, highlighting the threat of upcoming non-treatable S. aureus strains.10,11 Consequently, there is a pressing need for the development of new effective anti-staphylococcal therapeutics, preferably with chemical characteristics that vastly differ from those of the existing ones to combat the rapid spread of drug resistance.12
Accordingly, our work is directed towards the synthesis, optimization, and characterization of new anti-MRSA compounds in an effort to uncover and investigate potential new therapeutic candidates targeting this problematic organism.13–24 Previously, we screened a library of aminoguanidine-containing compounds for their anti-MRSA activity.25 The diphenylurea compound was one of the best leads obtained so far with a minimum inhibitory concentration (MIC) of 10 μg mL−1 against the MRSA USA300 strain (Fig. 1). Herein, we investigated the structure–activity relationship of this unique class of antibacterial drugs by replacing the iodine atom with a large library of alicyclic, aromatic, and open-chain acetylenic moieties. The antibacterial profile of the newly synthesized compounds was assessed, and the in vivo efficacy of the most promising compound was investigated in a MRSA skin infection mouse model.
2. Results and discussion
A. Synthetic procedure
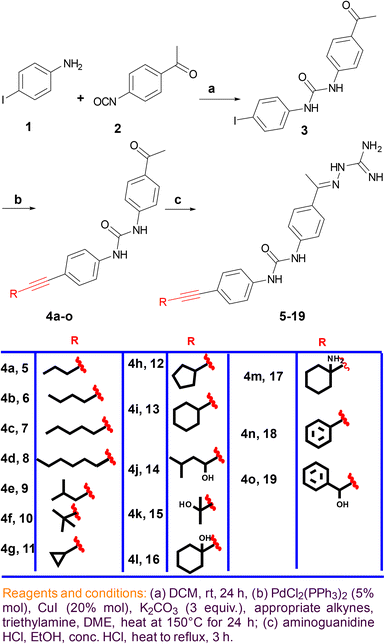
Reaction between 4-iodoaniline (1) and isocyanate 2 afforded the key starting material iododiphenylurea 3. The alkyne side chain was tethered using typical Sonogashira cross-coupling conditions, as outlined in Scheme 1. Finally, the desired aminoguanidine derivatives 5–19 were accessed through condensation reactions among 4a–o with aminoguanidine in the presence of a catalytic amount of hydrochloric acid (Scheme 1).B. Testing the synthesized compounds for their antibacterial activity
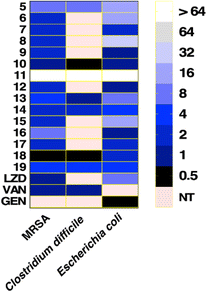
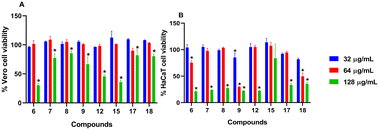
Compounds with open chain and branched substituents (5–10) showed potent anti-MRSA activity, with MIC values ranging from 0.5 μg mL−1 to 8 μg mL−1. Moreover, introducing polar groups, such as a hydroxyl group, had no significant effect on the MIC values. On the other hand, derivatives with aromatic substituents (compound 18) exhibited better anti-staphylococcal activity as the MIC value was found to be as low as 0.5 μg mL−1. Introducing a polar group, like hydroxyl group, resulted in an increase in the MIC value from 0.5 to 4 μg mL−1 (Fig. 2).
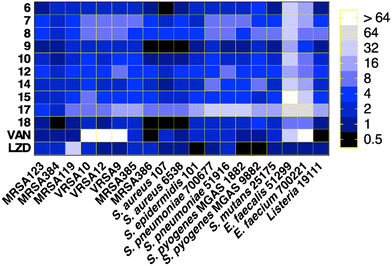
All of the tested compounds suppressed the growth of the tested strains (MSSA, MRSA, and VRSA) at concentrations as low as 0.5 μg mL−1 (Fig. 3). Compound 18 was the most potent antibacterial compound in this series with MIC values ranging from 0.5 to 2 μg mL−1. Similarly, compounds 6 and 9 inhibited the strains tested with MIC values ranging from 0.5 μg mL−1 to 2 μg mL−1. Interestingly, the antibacterial effect of the diphenylurea scaffold was extended to include linezolid-resistant and vancomycin-resistant staphylococcal strains, suggesting that they do not share the same resistance mechanism as linezolid or vancomycin.
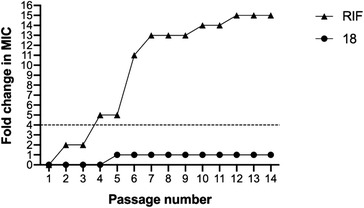
The MIC values of compound 18 remained unchanged for 4 passages and then showed only a one-fold increase in the MIC for the rest of the passages (Fig. 6). In contrast, the MIC value for rifampicin against MRSA USA300 increased rapidly and surpassed the four-fold change after only four passages. The MIC continued increasing until it reached around 15-fold increase by the end of the experiment (Fig. 6). The results demonstrated the low chances of resistance development to our tested compound in comparison to the control rifampicin antibiotic.
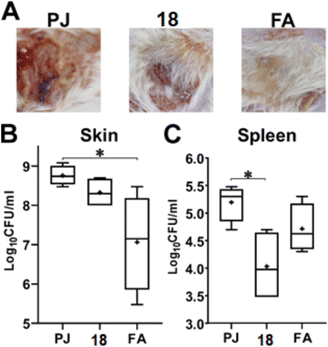
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10. Yet, this reduction was not significant as compared to the FA treatment, which generated around 1.5
log10. Yet, this reduction was not significant as compared to the FA treatment, which generated around 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 as compared to the vehicle-treated mice (Fig. 8B). On the other hand, compound 18 demonstrated superior activity compared to FA in controlling the systemic dissemination of the S. aureus, as evidenced by the higher reduction (>1
log10 as compared to the vehicle-treated mice (Fig. 8B). On the other hand, compound 18 demonstrated superior activity compared to FA in controlling the systemic dissemination of the S. aureus, as evidenced by the higher reduction (>1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10) in the bacterial burden detected in the spleens of the infected mice as compared to the vehicle-treated group (Fig. 8C).
log10) in the bacterial burden detected in the spleens of the infected mice as compared to the vehicle-treated group (Fig. 8C).
3. Experimental
3.1. Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford the desired compound (3).
1) to afford the desired compound (3).
1-(4-Acetylphenyl)-3-(4-iodophenyl)urea (3). Buff solid: mp 192–193 °C. as reported.32
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). Yields, physical properties, and spectral data of isolated purified products are listed below:
4). Yields, physical properties, and spectral data of isolated purified products are listed below:
1-(4-Acetylphenyl)-3-(4-(pent-1-yn-1-yl)phenyl)urea 4a. White solid (120 mg, 71.2%): mp 195–193 °C. 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 8.99 (brs, 1H), 7.93 (d, J = 8.7 Hz, 2H), 7.60 (d, J = 8.7 Hz, 2H), 7.49 (d, J = 8.7 Hz, 2H), 7.33 (d, J = 8.7 Hz, 2H), 2.46 (t, J = 7.5 Hz, 2H), 2.25 (s, 3H), 1.64–1.55 (m, 2H), 1.05 (t, J = 7.1 Hz, 3H); 13C NMR (DMSO-d6); δ 196.97, 152.46, 144.59, 139.76, 132.41, 131.03, 130.12, 118.72, 117.76, 116.46, 95.35, 80.92, 25.31, 22.14, 21.15, 17.47; MS (m/z); 320.39 (M+, 78.61%).
1-(4-Acetylphenyl)-3-(4-(hex-1-yn-1-yl)phenyl)urea 4b. Yellow oil (139 mg, 79%): 1H NMR (DMSO-d6); δ 9.84 (brs, 1H), 9.61 (brs, 1H), 7.93 (d, J = 8.8 Hz, 2H), 7.60 (d, J = 8.8 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 2.46 (t, J = 7.9 Hz, 2H), 2.25 (s, 3H), 1.64–1.57 (m, 4H), 1.05 (t, J = 6.8 Hz, 3H); 13C NMR (DMSO-d6); δ 196.25, 152.43, 148.42, 138.43, 138.20, 137.09, 135.70, 132.02, 126.84, 117.59, 92.52, 81.00, 28.35, 25.31, 22.13, 19.17, 17.48; MS (m/z); 334.41 (M+, 62.41%).
1-(4-Acetylphenyl)-3-(4-(hept-1-yn-1-yl)phenyl)urea 4c. Brown oil (200 mg, 76.9%): 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 8.99 (brs, 1H), 7.93 (d, J = 8.1 Hz, 2H), 7.60 (d, J = 8.5 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 2.80 (t, J = 7.9 Hz, 2H), 2.23 (s, 3H), 1.59–141 (m, 6H), 0.96 (t, J = 6.5 Hz, 3H); 13C NMR (DMSO-d6); δ 198.51, 155.68, 148.42, 138.20, 137.09, 135.70, 132.05, 126.84, 124.05, 117.59, 92.52, 81.00, 31.04, 28.35, 25.31, 22.13, 17.48; MS (m/z); 348.44 (M+, 69.41%).
1-(4-Acetylphenyl)-3-(4-(oct-1-yn-1-yl)phenyl)urea 4d. Brown oil (220 mg, 72%): 1H NMR (DMSO-d6); δ 9.19 (brs, 1H), 8.98 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 2.48 (t, J = 7.2 Hz, 2H), 2.23 (s, 3H), 1.58–1.43 (m, 8H), 0.96 (t, J = 6.1 Hz, 3H); 13C NMR (DMSO-d6); δ 198.51, 155.68, 152.43, 148.42, 138.20, 137.09, 135.70, 132.05, 126.84, 124.05, 117.59, 92.52, 81.00, 31.04, 28.35, 25.31, 22.13, 19.17, 17.48, 14.36; MS (m/z); 348.44 (M+, 69.41%).
1-(4-Acetylphenyl)-3-(4-(4-methylpent-1-yn-1-yl)phenyl)urea 4e. Yellowish white solid (230 mg, 87%); mp 198–199 °C. 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 9.00 (brs, 1H), 8.29 (d, J = 8.5 Hz, 2H), 7.94 (d, J = 8.5 Hz, 2H), 7.70 (d, J = 8.6 Hz, 2H), 7.51 (d, J = 8.7 Hz, 2H), 2.38 (d, J = 7.2 Hz, 2H), 2.23 (s, 3H), 1.92–1.85 (m, 1H), 1.04 (d, J = 7.2 Hz, 6H); 13C NMR (DMSO-d6); δ 200.65, 157.55, 139.24, 137.19, 132.21, 129.39, 127.46, 125.16, 123.99, 117.81, 91.98, 81.75, 29.87, 28.27, 25.24, 22.27; MS (m/z); 334.41 (M+, 92.31%).
1-(4-Acetylphenyl)-3-(4-(3,3-dimethylbut-1-yn-1-yl)phenyl)urea 4f. Yellow oil (195 mg, 74%); 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 9.00 (brs, 1H), 8.29 (d, J = 8.4 Hz, 2H), 7.94 (d, J = 8.4 Hz, 2H), 7.70 (d, J = 8.5 Hz, 2H), 7.51 (d, J = 8.5 Hz, 2H), 2.23 (s, 3H), 1.32 (s, 9H); 13C NMR (DMSO-d6); δ 200.24, 152.34, 148.35, 138.14, 137.09, 135.73, 132.04, 126.76, 123.90, 117.49, 100.27, 79.41, 31.22, 28.14, 25.31; MS (m/z); 334.41 (M+, 92.31%).
1-(4-Acetylphenyl)-3-(4-(cyclopropylethynyl)phenyl)urea 4g. Yellow oil (198 mg, 78.8%); 1H NMR (DMSO-d6); δ 9.16 (brs, 1H), 8.93 (brs, 1H), 7.94 (d, J = 8.5 Hz, 2H), 7.61 (d, J = 8.5 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 2.23 (s, 3H), 1.61–1.56 (m, 1H), 0.93–0.92 (m, 2H), 0.79–0.78 (m, 2H); 13C NMR (DMSO-d6); δ 202.85, 151.53, 147.51, 137.27, 136.23, 134.86, 131.23, 125.96, 123.10, 116.73, 94.81, 75.23, 24.47, 8.11, 0.50; MS (m/z); 318.37 (M+, 78.61%).
1-(4-Acetylphenyl)-3-(4-(cyclopentylethynyl)phenyl)urea 4h. Brown oil (200 mg, 73%); 1H NMR (DMSO-d6); δ 9.46 (brs, 1H), 9.17 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 2.91–2.87 (m, 1H), 2.23 (s, 3H), 2.03–1.60 (m, 8H); 13C NMR (DMSO-d6); δ 200.69, 157.55, 139.29, 137.19, 132.21, 129.39, 127.41, 125.19, 123.99, 117.75, 97.07, 80.36, 33.91, 30.60, 29.87, 25.11; MS (m/z); 346.43 (M+, 62.61%).
1-(4-Acetylphenyl)-3-(4-(cyclohexylethynyl)phenyl)urea 4i. Light brown oil (210 mg, 73.8%); 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 8.99 (brs, 1H), 7.93 (d, J = 8.7 Hz, 2H), 7.60 (d, J = 8.7 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 2.24 (s, 3H), 1.87–1.69 (m, 1H), 1.54–1.31 (m, 10H); 13C NMR (DMSO-d6); δ 196.97, 152.46, 144.59, 139.76, 132.41, 131.03, 130.12, 118.72, 117.76, 116.46, 95.35, 80.92, 29.23, 25.75, 25.19, 22.30, 21.47; MS (m/z); 360.46 (M+, 69.61%).
1-(4-Acetylphenyl)-3-(4-(3-hydroxy-5-methylhex-1-yn-1-yl)phenyl) urea 4j. White solid (220 mg, 76.5%); mp 201–203 °C. 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 8.99 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.42 (brs, 1H), 2.60 (s, 3H), 2.36 1.31–1.01 (m, 11H); 13C NMR (DMSO-d6); δ 200.65, 157.55, 139.24, 137.19, 132.21, 131.54, 129.39, 127.46, 125.16, 117.81, 91.98, 81.75, 47.82, 29.87, 28.27, 28.08, 25.24; MS (m/z); 364.44 (M+, 88.31%).
1-(4-Acetylphenyl)-3-(4-(3-hydroxy-3-methylbut-1-yn-1-yl)phenyl) urea 4k. Yellow oil (184 mg, 69.3%); 1H NMR (DMSO-d6); δ 9.27 (brs, 1H), 9.15 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.50 (d, J = 8.5 Hz, 2H), 7.34 (d, J = 8.5 Hz, 2H), 5.52 (brs, 1H), 2.23 (s, 3H), 1.51 (s, 6H); 13C NMR (DMSO-d6); δ 204.00, 160.36, 155.73, 152.21, 148.32, 137.12, 135.88, 132.21, 126.90, 117.71, 89.88, 70.29, 59.29, 29.15, 25.32; MS (m/z); 336.39 (M+, 72.45%).
1-(4-Acetylphenyl)-3-(4-((1-hydroxycyclohexyl)ethynyl)phenyl) urea 4l. Brown oil (205 mg, 69%); 1H NMR (DMSO-d6); δ 9.46 (brs, 1H), 9.17 (brs, 1H), 7.93 (d, J = 8.2 Hz, 2H), 7.60 (d, J = 8.2 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 6.23 (brs, 1H), 2.32 (s, 3H), 2.18–1.15 (m, 10H); 13C NMR (DMSO-d6); δ 196.97, 152.46, 144.59, 139.76, 132.41, 131.03, 130.12, 118.72, 117.76, 116.46, 95.35, 80.92, 58.38, 29.23, 25.75, 25.19, 22.30, 21.47; MS (m/z); 376.45 (M+, 61.61%).
1-(4-Acetylphenyl)-3-(4-((1-aminocyclohexyl)ethynyl)phenyl)urea 4m. Dark brown oil (196 mg, 66%); 1H NMR (DMSO-d6); δ 9.07 (brs, 1H), 8.89 (brs, 1H), 8.29 (d, J = 8.4 Hz, 2H), 8.13 (d, J = 8.4 Hz, 2H), 7.91 (brs, 2H), 7.70 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 2.67 (s, 3H), 1.72–1.56 (m, 10H); 13C NMR (DMSO-d6); δ 197.00, 153.02, 144.35, 135.03, 134.22, 132.62, 130.64, 122.16, 117,91, 91.01, 85.21, 49.41, 40.21, 27.71, 25.62, 21.90; MS (m/z); 376.45 (M+, 61.61%).
1-(4-Acetylphenyl)-3-(4-(phenylethynyl)phenyl)urea 4n. Brown oil (215 mg, 76.8%); 1H NMR (DMSO-d6); δ 9.85 (brs, 1H), 9.61 (brs, 1H), 8.17 (d, J = 8.4 Hz, 2H), 7.82–7.75 (m, 5H), 7.72 (d, J = 8.4 Hz, 2H), 7.62 (d, J = 8.4 Hz, 2H), 7.47 (d, J = 8.4 Hz, 2H), 2.25 (s, 3H); 13C NMR (DMSO-d6); δ 201.29, 155.79, 152.25, 148.44, 139.02, 137,13, 135.85, 132.24, 129.27, 127,01, 122.92, 122.72, 117.75, 91.01, 89.84, 25.34; MS (m/z); 354.40 (M+, 48.51%).
1-(4-Acetylphenyl)-3-(4-(3-hydroxy-3-phenylprop-1-yn-1-yl)phenyl)urea 4o. Brown oil (187 mg, 61.6%); 1H NMR (DMSO-d6); δ 9.37 (brs, 1H), 9.19 (brs, 1H), 8.62 (d, J = 8.4 Hz, 2H), 8.52 (d, J = 8.4 Hz, 2H), 8.38 (d, J = 8.4 Hz, 2H), 8.08 (d, J = 8.4 Hz, 2H), 7.44–6.99 (m, 5H), 4.92 (brs, 1H), 2.73 (s, 3H); 13C NMR (DMSO-d6); δ 200.69, 155.77, 152.22, 148.43, 139,45, 139.36, 137.14, 132.32, 128,97, 128.77, 127.74, 126.96, 122.42, 117.78, 89.72, 86.93, 63.90, 25.31; MS (m/z); 384.44 (M+, 38.31%).
2-(1-(4-(3-(4-(Pent-1-yn-1-yl)phenyl)ureido)phenyl)ethylidene) hydrazine-1-carboximidamide 5. White solid (90 mg, 76.6%): mp 210–211 °C; 1H NMR (DMSO-d6); δ 9.47 (brs, 1H), 9.17 (brs, 1H), 8.10 (d, J = 8.4 Hz, 2H), 7.82 (d, J = 8.4 Hz, 2H), 7.79 (d, J = 8.2 Hz, 2H), 7.53 (d, J = 8.2 Hz, 2H), 5.72 (brs, 2H), 5.52 (brs, 2H), 2.46 (t, J = 7.1 Hz, 2H), 2.25 (s, 3H), 1.62–1.55 (m, 2H), 1.05–1.01 (t, J = 7.6 Hz, 3H); 13C NMR (DMSO-d6); δ 160.33, 155.68, 152.41, 148.38, 138.22, 137.09, 135.73, 132.04, 126.85, 124.03, 117.59, 92.34, 81.16, 25.31, 22.14, 21.15, 17.47; HRMS (EI) m/z 376.2018 M+, calc. for C21H24N6O 376.2012; anal. calc. for: C21H24N6O (376.46): C, 67.00; H, 6.43; N, 22.32%; found: C, 67.91; H, 6.98; N, 20.66%.
2-(1-(4-(3-(4-(Hex-1-yn-1-yl)phenyl)ureido)phenyl)ethylidene) hydrazine-1-carboximidamide 6. Off-white solid (88 mg, 75%): mp 215–217 °C; 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 8.99 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.74 (brs, 2H), 5.55 (brs, 2H), 2.45 (t, J = 7.8 Hz, 2H), 2.24 (s, 3H), 1.61–130 (m, 4H), 0.93 (t, J = 7.2 Hz, 3H); 13C NMR (DMSO-d6); δ 160.29, 155.68, 152.43, 148.42, 138.20, 137.09, 135.70, 132.05, 126.84, 124.05, 117.59, 92.52, 81.00, 31.04, 28.35, 25.31, 22.13, 17.48; HRMS (EI) m/z 390.2164 M+, calc. for C22H26N6O 390.2168; anal. calc. for: C22H26N6O (390.46): C, 67.67; H, 6.71; N, 21.52%; found: C, 67.91; H, 7.02; N, 19.99%.
2-(1-(4-(3-(4-(Hept-1-yn-1-yl)phenyl)ureido)phenyl)ethylidene) hydrazine-1-carboximidamide 7. Light-brown solid (89 mg, 73.5%): mp 208–210 °C; 1H NMR (DMSO-d6); 9.30 (brs, 1H), 9.17 (brs, 1H), 8.10 (d, J = 8.8 Hz, 2H), 7.79 (d, J = 8.8 Hz, 2H), 7.49 (d, J = 8.6 Hz, 2H), 7.25 (d, J = 8.6 Hz, 2H), 5.71 (brs, 2H), 5.50 (brs, 2H), 2.47 (t, J = 7.2 Hz, 2H), 2.23 (s, 3H), 1.59–143 (m, 6H), 0.96 (t, J = 7.8 Hz, 3H); 13C NMR (DMSO-d6); δ 160.34, 155.68, 152.41, 148.39, 138.20, 137.73, 135.73, 132.03, 126.84, 124.04, 117.54, 92.47, 81.00, 30.74, 25,30, 21.93, 18.88, 17.96, 13.96; HRMS (EI) m/z 404.2336 M+, calc. for C23H28N6O 404.2325; anal. calc. for: C23H28N6O (404.51): C, 68.29; H, 6.98; N, 20.78%; found: C, 67.91; H, 7.38; N, 20.03%.
2-(1-(4-(3-(4-(Oct-1-yn-1-yl)phenyl)ureido)phenyl)ethylidene) hydrazine-1-carboximidamide 8. Brown solid (80 mg, 69%): mp 211–213 °C; 1H NMR (DMSO-d6); δ 9.19 (brs, 1H), 8.98 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 7.8 Hz, 2H), 7.33 (d, J = 7.8 Hz, 2H), 5.72 (brs, 2H), 5.42 (brs, 2H), 2.48 (t, J = 6.8 Hz, 2H), 2.23 (s, 3H), 1.58–143 (m, 8H), 0.96 (t, J = 7.2 Hz, 3H); 13C NMR (DMSO-d6); δ 160.35, 155.67, 152.40, 148.35, 138.21, 137.08, 135.73, 132.01, 126.85, 124.04, 117.59, 92.52, 81.01, 31.23, 28,60, 28.47, 25.32, 22.50, 17.46, 14.39; HRMS (EI) m/z 418.2495 M+, calc. for C24H30N6O 418.2481; anal. calc. for: C24H30N6O (418.58): C, 68.87; H, 7.23; N, 20.08%; found: C, 67.91; H, 7.38; N, 20.03%.
2-(1-(4-(3-(4-(4-Methylpent-1-yn-1-yl)phenyl)ureido)phenyl) ethylidene)hydrazine-1-carboximidamide 9. Yellowish white solid (93 mg, 79.6%); mp 198–199 °C, 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 9.00 (brs, 1H), 8.29 (d, J = 8.6 Hz, 2H), 7.94 (d, J = 8.6 Hz, 2H), 7.70 (d, J = 8.7 Hz, 2H), 7.51 (d, J = 8.7 Hz, 2H), 5.71 (brs, 2H), 5.51 (brs, 2H), 2.38 (d, J = 7.6 Hz, 2H), 2.23 (s, 3H), 1.92–1.85 (m, 1H), 1.04 (d, J = 7.6 Hz, 6H); 13C NMR (DMSO-d6); δ 160.33, 155.68, 152.41, 148.37, 138.22, 137.10, 135.74, 132.04, 126.85, 124.04, 117.60, 91.36, 81.90, 28.11, 25.32, 22.31, 17.47; HRMS (EI) m/z 390.2149 M+, calc. for C22H26N6O 390.2168; anal. calc. for: C22H26N6O (390.49): C, 67.67; H, 6.71; N, 21.52%; found: C, 67.91; H, 7.55; N, 20.89%.
2-(1-(4-(3-(4-(3,3-Dimethylbut-1-yn-1-yl)phenyl)ureido)phenyl) ethylidene) hydrazine-1-carboximidamide 10. Beige solid (96 mg, 82%); mp 201–202 °C, 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 9.00 (brs, 1H), 8.29 (d, J = 8.4 Hz, 2H), 7.94 (d, J = 8.4 Hz, 2H), 7.70 (d, J = 8.5 Hz, 2H), 7.51 (d, J = 8.5 Hz, 2H), 5.71 (brs, 2H), 5.51 (brs, 2H), 2.23 (s, 3H), 1.32 (s, 9H); 13C NMR (DMSO-d6); δ 160.36, 155.67, 152.34, 148.35, 138.14, 137.09, 135.73, 132.02, 126.79, 123.90, 117.56, 100.27, 79.41, 31.22, 28.14, 17.47; HRMS (EI) m/z 390.2170 M+, calc. for C22H26N6O 390.2168; anal. calc. for: C22H26N6O (390.49): C, 67.67; H, 6.71; N, 21.52%; found: C, 67.91; H, 7.55; N, 20.89%.
2-(1-(4-(3-(4-(Cyclopropylethynyl)phenyl)ureido)phenyl) ethylidene)hydrazine-1-carboximidamide 11. Yellow solid (97 mg, 82%); mp 205–207 °C, 1H NMR (DMSO-d6); δ 9.27 (brs, 1H), 9.15 (brs, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 8.4 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.72 (brs, 2H), 5.53 (brs, 2H), 2.23 (s, 3H), 1.61–1.56 (m, 1H), 0.93–0.92 (m, 2H), 0.79–0.78 (m, 2H); 13C NMR (DMSO-d6); δ 159.50, 154.82, 151.53, 147.51, 137.27, 136.23, 134.86, 131.23, 125.96, 123.10, 116.73, 94.81, 75.23, 24.47, 16.62, 0.50; HRMS (EI) m/z 374.1857 M+, calc. for C21H22N6O 374.1855; anal. calc. for: C21H22N6O (374.44): C, 67.36; H, 5.92; N, 22.44%; found: C, 67.81; H, 6.41; N, 21.89%.
2-(1-(4-(3-(4-(Cyclopentylethynyl)phenyl)ureido)phenyl) ethylidene)hydrazine-1-carboximidamide 12. Yellow solid (95 mg, 81.7%); mp 215–217 °C, 1H NMR (DMSO-d6); δ 9.46 (brs, 1H), 9.17 (brs, 1H), 7.93 (d, J = 8.6 Hz, 2H), 7.60 (d, J = 8.6 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.74 (brs, 1H), 5.55 (brs, 1H), 5.25 (brs, 2H), 2.91–2.87 (m, 1H), 2.23 (s, 3H), 2.03–1.60 (m, 8H); 13C NMR (DMSO-d6); δ 160.33, 155.67, 152.40, 148.39, 138.12, 137.08, 135.69, 132.00, 126.80, 124.07, 117.56, 96.45, 80.50, 33.95, 30.59, 25.10, 17.47; HRMS (EI) m/z 402.2160 M+, calc. for C23H26N6O 402.2168; anal. calc. for: C23H26N6O (402.50): C, 68.63; H, 6.51; N, 20.88%; found: C, 68.46; H, 7.11; N, 20.42%.
2-(1-(4-(3-(4-(Cyclohexylethynyl)phenyl)ureido)phenyl)ethylidene) hydrazine-1-carboximidamide 13. Brown solid (84 mg, 72.6%); mp 216–218 °C. 1H NMR (DMSO-d6); δ 9.25 (brs, 1H), 9.17 (brs, 1H), 7.93 (d, J = 8.2 Hz, 2H), 7.60 (d, J = 8.2 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.77 (brs, 2H), 5.61 (brs, 2H), 2.24 (s, 3H), 1.87–1.31 (m, 11H); 13C NMR (DMSO-d6); δ 160.27, 155.68, 152.43, 148.47, 138.16, 137.09, 135.67, 132.04, 126.82, 124.02, 117.57, 96.20, 80.97, 32.66, 29.37, 25.83, 25.29, 24.79; HRMS (EI) m/z 416.2333 M+, calc. for C24H28N6O 416.2325; anal. calc. for: C24H28N6O (416.52): C, 69.21; H, 6.78; N, 20.18%; found: C, 68.91; H, 7.67; N, 19.89%.
2-(1-(4-(3-(4-(3-Hydroxy-5-methylhex-1-yn-1-yl)phenyl)ureido) phenyl)ethylidene)hydrazine-1-carboximidamide 14. Yellow solid (89 mg, 77.2%); mp 222–224 °C. 1H NMR (DMSO-d6); δ 9.13 (brs, 1H), 8.95 (brs, 1H), 7.93 (d, J = 8.2 Hz, 2H), 7.60 (d, J = 8.2 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.71 (brs, 1H), 5.51 (brs, 2H), 5.20 (brs, 2H), 2.38 (t, J = 6.4 Hz, 1H), 2.23 (s, 3H), 1.93–1.83 (m, 3H), 1.04 (d, J = 7.4 Hz, 6H); 13C NMR (DMSO-d6); δ 160.33, 155.68, 152.41, 148.37, 138.22, 137.10, 135.74, 132.04, 126.85, 124.04, 117.60, 91.36, 81.90, 51.12, 28.11, 25.32, 22.31, 17.47; HRMS (EI) m/z 420.2290 M+, calc. for C23H28N6O2 420.2274; anal. calc. for: C23H28N6O2 (420.51): C, 65.69; H, 6.71; N, 19.99%; found: C, 65.91; H, 6.77; N, 19.86%.
2-(1-(4-(3-(4-(3-Hydroxy-3-methylbut-1-yn-1-yl)phenyl)ureido) phenyl)ethylidene)hydrazine-1-carboximidamide 15. Yellow solid (100 mg, 85.7%); mp 219–221 °C. 1H NMR (DMSO-d6); δ 9.17 (brs, 1H), 8.99 (brs, 1H), 7.93 (d, J = 8.6 Hz, 2H), 7.6 (d, J = 8.6 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 5.72 (brs, 2H), 5.52 (brs, 2H), 3.63 (brs, 1H), 2.23 (s, 3H), 1.51 (s, 6H); 13C NMR (DMSO-d6); δ 160.36, 155.73, 152.21, 148.32, 138.12, 135.88, 132.22, 126.89, 126.89, 124.04, 117.76, 83.60, 70.29, 59.29, 29.15, 25.32; HRMS (EI) m/z 392.1957 M+, calc. for C21H24N6O2 392.1961; anal. calc. for: C21H24N6O2 (392.46): C, 64.27; H, 6.16; N, 21.41%; found: C, 64.51; H, 6.37; N, 20.22%.
2-(1-(4-(3-(4-((1-Hydroxycyclohexyl)ethynyl)phenyl)ureido)phenyl) ethylidene)hydrazine-1-carboximidamide 16. Brown solid (85 mg, 73.9%); mp 223–224 °C. 1H NMR (DMSO-d6); δ 9.46 (brs, 1H), 9.17 (brs, 1H), 7.93 (d, J = 8.6 Hz, 2H), 7.60 (d, J = 8.7 Hz, 2H), 7.49 (d, J = 8.4 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 6.23 (brs, 1H), 6.01 (brs, 4H), 2.26 (s, 3H), 2.18–1.15 (m, 10H); 13C NMR (DMSO-d6); δ 159.80, 155.74, 152.58, 149.00, 137.20, 136.12, 132.23, 131.93, 126.93, 117.66, 93.05, 87.29, 58.38, 40.40, 29.23 25.75, 21.47, 17.68; HRMS (EI) m/z 432.2261 M+, calc. for C24H28N6O2 432.2274; anal. calc. for: C24H28N6O2 (416.52): C, 66.65; H, 6.53; N, 19.43%; found: C, 67.71; H, 6.67; N, 19.56%.
2-(1-(4-(3-(4-((1-Aminocyclohexyl)ethynyl)phenyl)ureido)phenyl) ethylidene)hydrazine-1-carboximidamide 17. Dark brown solid (77 mg, 67%); mp 208–210 °C. 1H NMR (DMSO-d6); δ 9.47 (brs, 1H), 9.12 (brs, 1H), 8.14 (d, J = 8.2 Hz, 2H), 7.83 (d, J = 8.2 Hz, 2H), 7.55 (d, J = 8.4 Hz, 2H), 7.29 (d, J = 8.4 Hz, 2H), 6.23 (brs, 2H), 6.03 (brs, 2H), 2.26 (s, 3H), 2.18–1.14 (m, 10H); 13C NMR (DMSO-d6); δ 159.80, 155.74, 152.58, 149.00, 137.20, 136.12, 132.23, 131.93, 126.93, 123.96, 117.66, 93.05, 87.29, 58.38, 40.40, 29.23, 25.75, 24.73, 21.47, 17.68; HRMS (EI) m/z 431.2428 M+, calc. for C24H29N7O 431.2434; anal. calc. for: C24H29N7O (431.54): C, 66.80; H, 6.77; N, 22.72%; found: C, 66.71; H, 6.68; N, 22.57%.
2-(1-(4-(3-(4-(Phenylethynyl)phenyl)ureido)phenyl)ethylidene) hydrazine-1-carboximidamide 18. Light brown solid (95 mg, 82%); mp 228–230 °C. 1H NMR (DMSO-d6); δ 9.11 (brs, 1H), 9.00 (brs, 1H), 8.21 (d, J = 7.6 Hz, 2H), 7.83–7.78 (m, 5H), 7.73 (d, J = 8.2 Hz, 2H), 7.53 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 8.2 Hz, 2H), 5.78 (brs, 2H), 5.60 (brs, 2H), 2.26 (s, 3H); 13C NMR (DMSO-d6); δ 160.33, 155.79, 152.25, 148.44, 139.02, 137.13, 135.85, 132.22, 131.88, 129.27, 127.01, 122.92, 117.77, 91.01, 89.84, 25.34; HRMS (EI) m/z 410.1865 M+, calc. for C24H22N6O 410.1855; anal. calc. for: C24H22N6O (410.48): C, 70.23; H, 5.40; N, 20.47%; found: C, 69.91; H, 5.89; N, 20.27%.
2-(1-(4-(3-(4-(3-Hydroxy-3-phenylprop-1-yn-1-yl)phenyl)ureido) phenyl)ethylidene)hydrazine-1-carboximidamide 19. Dark brown solid (65 mg, 56.7%); mp 165–167 °C; 1H NMR (DMSO-d6); δ 9.18 (brs, 1H), 8.97 (brs, 1H), 8.57 (d, J = 8.1 Hz, 2H), 8.41 (d, J = 8.2 Hz, 2H), 8.08 (d, J = 8.4 Hz, 2H), 7.98 (d, J = 8.2 Hz, 2H), 7.43–6.97 (m, 5H), 5.78 (brs, 2H), 5.67 (brs, 2H), 5.41 (s,1H), 2.61 (s, 3H); 13C NMR (DMSO-d6); δ 160.31, 155.77, 152.22, 148.43, 139.45, 137.14, 137.14, 135.88, 132.32, 128.97, 127.74, 126.96, 122.42, 117.78, 89.73, 86.93, 63.90, 25.31; HRMS (EI) m/z 440.1968 M+, calc. for C25H24N6O2 400.1961; anal. calc. for: C25H24N6O2 (440.50): C, 68.17; H, 5.49; N, 19.08%; found: C, 68.91; H, 5.56; N, 18.87%.
3.2. Biology
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
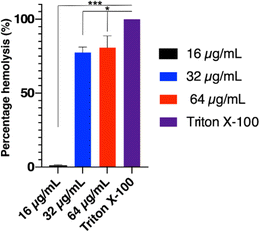
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a 1 % triton X-100 solution in order to induce complete hemolysis. The erythrocyte suspension was adjusted to an absorbance of 0.8 arbitrary units at 540 nm. Three tested concentrations (16, 32 and 64 μg mL−1) of compound 18 were prepared and mixed with equal volume of the freshly prepared erythrocyte suspension to reach a final concentration of (16, 32 and 64 μg mL−1) and incubated at 37 °C for 30 min. Negative control was PBS mixed with equal volume of the erythrocyte blood suspension. Finally, the mixtures were centrifuged at 13
1 with a 1 % triton X-100 solution in order to induce complete hemolysis. The erythrocyte suspension was adjusted to an absorbance of 0.8 arbitrary units at 540 nm. Three tested concentrations (16, 32 and 64 μg mL−1) of compound 18 were prepared and mixed with equal volume of the freshly prepared erythrocyte suspension to reach a final concentration of (16, 32 and 64 μg mL−1) and incubated at 37 °C for 30 min. Negative control was PBS mixed with equal volume of the erythrocyte blood suspension. Finally, the mixtures were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min and the absorbance of the supernatant was measured at 540 nm.36
000 × g for 5 min and the absorbance of the supernatant was measured at 540 nm.36
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
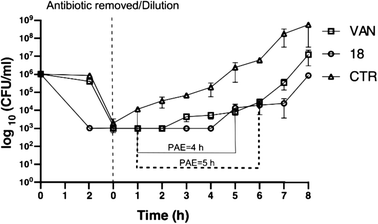
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in fresh TSB media, and viable counts were determined for all cultures at this time (time 0). The antibiotic-free bacterial suspensions were incubated at 37 °C with agitation at 180 RPM, and samples were collected hourly for 8 hours. Viable count was done at each time point on mannitol salt agar (MSA) plates, and the results were read after incubation for 18–24 h at 37 °C. The PAE was defined as follows: PAE = T − C, where T is the time (h) required for the viable count in the test culture to increase one log10 cycle above the count observed immediately after dilution, and C is the corresponding time for the control.43
1000 in fresh TSB media, and viable counts were determined for all cultures at this time (time 0). The antibiotic-free bacterial suspensions were incubated at 37 °C with agitation at 180 RPM, and samples were collected hourly for 8 hours. Viable count was done at each time point on mannitol salt agar (MSA) plates, and the results were read after incubation for 18–24 h at 37 °C. The PAE was defined as follows: PAE = T − C, where T is the time (h) required for the viable count in the test culture to increase one log10 cycle above the count observed immediately after dilution, and C is the corresponding time for the control.43
3.2.7.1. Ethics statement. Animal procedures were approved by the Research Ethics Committee of the Faculty of Pharmacy, Cairo University [approval no. (MI 2868)] following the Guide for the Care and Use of Laboratory Animals published by the Institute of Laboratory Animal Research (USA).
3.2.7.2. Murine skin infection model. The skin infection was performed as previously described.44,45 Briefly, three groups (n = 5) of six to eight-week-old female Balb/c mice, 18–20 g each, were included in the experiment. On the day prior to the infection, their backs were shaven using an electric hair clipper. On the day of the infection, the mice were anesthetized using 2,2,2-tribromoethanol (25 μg mL−1) and injected subcutaneously with 100 μL containing 2 × 109 CFU MRSA USA300, suspended in 0.5% hydroxypropyl methylcellulose (HPMC) in sterile pyrogen-free saline. The mice were kept in their cages and given food and water ad libitum. Seventy-two hours post-infection (day 3), the infection site was treated with either i) petroleum jelly (PG), ii) petroleum jelly containing 2% compound 18, or iii) commercially available topical ointment containing 2% fusidic acid (FA). The three treatments were applied twice daily for another three consecutive days (days 4 to 6). Twenty-four hours after the last dose, mice were euthanized with an overdose of the anesthesia, and the lesions were photographed using a digital camera. A skin patch equivalent to ∼1.5 cm2, surrounding the lesion site, was excised from the back of each mouse. In addition, the mice were dissected, and their spleens were excised to assess the systemic dissemination of the infection. The skin patch was shredded with a sterile surgical blade and then homogenized in 1 mL pyrogen-free saline. While the spleen was homogenized in only 0.5 mL saline. The homogenates were then serially diluted and plated on MSA. The plates were incubated at 37 °C overnight and the colonies were counted. A statistical analysis was performed using GraphPad Prism (version 9.0) (GraphPad Software, Inc., USA), applying ordinary one-way ANOVA, followed by the Tukey's multiple comparison test.
Conclusions
The SAR of alkynyldiphenylurea as new antibacterial agents was further explored against a set of clinically important Gram-positive cells. The phenylacetylene-containing derivative 18 showed the most promising antibacterial profile. Compound 18 was 20-times better than the lead compound I in terms of MIC values. In addition, compound 18 was highly tolerable to mammalian cells, and more importantly, it was comparable with fusidic acid in lowering the bacterial counts in infected mice wounds and even better in limiting systemic dissemination.Author contributions
H. T. N. and N. S. A. conducted the microbiological tests; M. M. E. and A. M. K. synthesized all compounds; M. M. E. was responsible for all chemical characterization and writing the experimental section. Supervision was the responsibility of A. S. A., M. N. S. and A. S. M.; all authors contributed in writing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This paper is based upon work supported by Science, Technology & Innovation Funding Authority (STDF) under grant number: 43883.Notes and references
- M. I. Hutchings, A. W. Truman and B. Wilkinson, Curr. Opin. Microbiol., 2019, 51, 72–80 CrossRef CAS PubMed.
- M. A. Cooper and D. Shlaes, Nature, 2011, 472, 32–32 CrossRef CAS PubMed.
- D. Ghernaout and N. Elboughdiri, Appl. Eng., 2020, 4, 1–6 Search PubMed.
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 109, 309–318 CrossRef PubMed.
- C. Antimicrobial Resistance, Lancet, 2022, 399, 629–655 CrossRef PubMed.
- S. Y. Tong, J. S. Davis, E. Eichenberger, T. L. Holland and V. G. Fowler, Jr., Clin. Microbiol. Rev., 2015, 28, 603–661 CrossRef CAS PubMed.
- S. Santajit and N. Indrawattana, BioMed Res. Int., 2016, 2016, 2475067 Search PubMed.
- E. F. Kong, J. K. Johnson and M. A. Jabra-Rizk, PLoS Pathog., 2016, 12, e1005837 CrossRef PubMed.
- Centers for Disease Control and Prevention, Antibiotic resistance threats in the United States, 2019, US Department of Health and Human Services, Centres for Disease Control and Prevention, 2019 Search PubMed.
- H. T. Nour El-Din, A. S. Yassin, Y. M. Ragab and A. M. Hashem, Infect. Drug Resist., 2021, 14, 1557–1571 CrossRef PubMed.
- A. Azhar, S. Rasool, A. Haque, S. Shan, M. Saeed, B. Ehsan and A. Haque, J. Med. Microbiol., 2017, 66, 1328–1331 CrossRef CAS PubMed.
- N. Desai, A. Maheta, K. Rajpara, V. Joshi, H. Vaghani and H. Satodiya, J. Saudi Chem. Soc., 2014, 18, 963–971 CrossRef.
- H. Mohammad, A. S. Mayhoub, A. Ghafoor, M. Soofi, R. A. Alajlouni, M. Cushman and M. N. Seleem, J. Med. Chem., 2014, 57, 1609–1615 CrossRef CAS PubMed.
- I. Eid, M. M. Elsebaei, H. Mohammad, M. Hagras, C. E. Peters, Y. A. Hegazy, B. Cooper, J. Pogliano, K. Pogliano, H. S. Abulkhair, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2017, 139, 665–673 CrossRef CAS PubMed.
- M. M. Elsebaei, H. Mohammad, M. Abouf, N. S. Abutaleb, Y. A. Hegazy, A. Ghiaty, L. Chen, J. Zhang, S. R. Malwal, E. Oldfield, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2018, 148, 195–209 CrossRef CAS PubMed.
- M. M. Elsebaei, H. Mohammad, A. Samir, N. S. Abutaleb, A. B. Norvil, A. R. Michie, M. M. Moustafa, H. Samy, H. Gowher, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2019, 175, 49–62 CrossRef CAS PubMed.
- M. Hagras, N. S. Abutaleb, A. O. Ali, J. A. Abdel-Aleem, M. M. Elsebaei, M. N. Seleem and A. S. Mayhoub, ACS Infect. Dis., 2018, 4, 1679–1691 CrossRef CAS PubMed.
- M. Hagras, Y. A. Hegazy, A. H. Elkabbany, H. Mohammad, A. Ghiaty, T. M. Abdelghany, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2018, 143, 1448–1456 CrossRef CAS PubMed.
- M. Hagras, H. Mohammad, M. S. Mandour, Y. A. Hegazy, A. Ghiaty, M. N. Seleem and A. S. Mayhoub, J. Med. Chem., 2017, 60, 4074–4085 CrossRef CAS PubMed.
- A. Hammad, N. S. Abutaleb, M. M. Elsebaei, A. B. Norvil, M. Alswah, A. O. Ali, J. A. Abdel-Aleem, A. Alattar, S. A. Bayoumi, H. Gowher, M. Seleem and A. S. Mayhoub, J. Med. Chem., 2019, 62, 7998–8010 CrossRef CAS PubMed.
- Y. Hosny, N. S. Abutaleb, M. Omara, M. Alhashimi, M. M. Elsebaei, H. S. Elzahabi, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2020, 185, 111830 CrossRef CAS PubMed.
- M. M. Elsebaei, N. S. Abutaleb, A. A. Mahgoub, D. Li, M. Hagras, H. Mohammad, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2019, 182, 111593 CrossRef PubMed.
- A. Mancy, N. S. Abutaleb, M. M. Elsebaei, A. Y. Saad, A. Kotb, A. O. Ali, J. A. Abdel-Aleem, H. Mohammad, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2019, 6, 80–90 Search PubMed.
- M. Hagras, N. S. Abutaleb, N. M. Elhosseiny, T. M. Abdelghany, M. Omara, M. M. Elsebaei, M. Alhashimi, A. B. Norvil, M. I. Gutay, H. Gowher, M. Seleem and A. S. Mayhoub, ACS Infect. Dis., 2020, 6, 2887–2900 CrossRef CAS PubMed.
- I. H. Eissa, H. Mohammad, O. A. Qassem, W. Younis, T. M. Abdelghany, A. Elshafeey, M. M. Abd Rabo Moustafa, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2017, 130, 73–85 CrossRef CAS PubMed.
- K. O. Olawale, S. O. Fadiora and S. S. Taiwo, Afr. J. Infect. Dis., 2011, 5, 40–46 Search PubMed.
- S. M. Jabbari Shiadeh, A. Pormohammad, A. Hashemi and P. Lak, Infect. Drug Resist., 2019, 12, 2713–2725 CrossRef PubMed.
- E. S. Shenoy, M. L. Paras, F. Noubary, R. P. Walensky and D. C. Hooper, BMC Infect. Dis., 2014, 14, 177 CrossRef PubMed.
- M. Otto, Nat. Rev. Microbiol., 2009, 7, 555–567 CrossRef CAS PubMed.
- O. Disson, A. Moura and M. Lecuit, Trends Microbiol., 2021, 29, 811–822 CrossRef CAS PubMed.
- H. Mohammad, W. Younis, H. G. Ezzat, C. E. Peters, A. AbdelKhalek, B. Cooper, K. Pogliano, J. Pogliano, A. S. Mayhoub and M. N. Seleem, PLoS One, 2017, 12, e0182821 CrossRef PubMed.
- CLSI, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard, 2012 Search PubMed.
- I. G. Shahin, N. S. Abutaleb, M. Alhashimi, A. E. Kassab, K. O. Mohamed, A. T. Taher, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2020, 202, 112497 CrossRef CAS PubMed.
- Y. Hosny, N. S. Abutaleb, M. Omara, M. Alhashimi, M. M. Elsebaei, H. S. Elzahabi, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2020, 185, 111830 CrossRef CAS PubMed.
- N. H. Mahdally, R. F. George, M. T. Kashef, M. Al-Ghobashy, F. E. Murad and A. S. Attia, Front. Microbiol., 2021, 12, 700494 CrossRef PubMed.
- A. W. Bernheimer, Methods Enzymol., 1988, 165, 213–217 CAS.
- M. A. Tag ElDein, A. S. Yassin, O. El-Tayeb and M. T. Kashef, Eur. J. Clin. Microbiol. Infect. Dis., 2021, 40, 2349–2361 CrossRef CAS PubMed.
- H. Mohammad, W. Younis, H. G. Ezzat, C. E. Peters, A. AbdelKhalek, B. Cooper, K. Pogliano, J. Pogliano, A. S. Mayhoub and M. N. Seleem, PLoS One, 2017, 12, e0182821 CrossRef PubMed.
- D. J. Farrell, M. Robbins, W. Rhys-Williams and W. G. Love, Antimicrob. Agents Chemother., 2011, 55, 1177–1181 CrossRef CAS PubMed.
- G. G. Zhanel, D. J. Hoban and G. K. Harding, DICP, Ann. Pharmacother., 1991, 25, 153–163 CAS.
- P. Gaibani, D. Lombardo, R. E. Lewis, M. Mercuri, S. Bonora, M. P. Landini and S. Ambretti, J. Antimicrob. Chemother., 2014, 69, 1856–1865 CrossRef CAS PubMed.
- M. M. Elsebaie, H. T. Nour El-Din, N. S. Abutaleb, A. A. Abuelkhir, H.-W. Liang, A. S. Attia, M. N. Seleem and A. S. Mayhoub, Eur. J. Med. Chem., 2022, 114204, DOI:10.1016/j.ejmech.2022.114204.
- J. P. Lavigne, R. Bonnet, S. Michaux-Charachon, J. Jourdan, J. Caillon and A. Sotto, J. Antimicrob. Chemother., 2004, 53, 616–619 CrossRef CAS PubMed.
- M. Hagras, N. S. Abutaleb, N. M. Elhosseiny, T. M. Abdelghany, M. Omara, M. M. Elsebaei, M. Alhashimi, A. B. Norvil, M. I. Gutay, H. Gowher, M. Seleem and A. S. Mayhoub, ACS Infect. Dis., 2020, 6, 2887–2900 CrossRef CAS PubMed.
- C. W. Tseng, M. Sanchez-Martinez, A. Arruda and G. Y. Liu, J. Visualized Exp., 2011, e2528 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2md00351a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |