Open Access Article
Open Access ArticleA DNA-engineered metal–organic-framework nanocarrier as a general platform for activatable photodynamic cancer cell ablation†
Yahui
Ma‡
,
Renzeng
Chen‡
,
Xianheng
Chen
,
Yuqi
Sun
,
Yuanbo
Wang
* and
Bo
Wang
 *
*
Frontiers Science Centre for High Energy Material, Advanced Technology Research Institute (Jinan), Key Laboratory of Cluster Science (Ministry of Education), Beijing Key Laboratory of Photoelectronic/Electrophotonic Conversion Materials, Advanced Research Institute of Multidisciplinary Science, School of Medical Technology, School of Chemistry and Chemical Engineering, Beijing Institute of Technology, Beijing 100081, P. R. China. E-mail: ybwang@bit.edu.cn; bowang@bit.edu.cn
First published on 29th November 2022
Abstract
Activatable photodynamic cancer cell ablation constitutes a promising approach to performing highly effective photodynamic therapy (PDT) with mitigated phototoxicity. Regretfully, so far strategies to fabricate activatable PDT agents are only applicable to a limited number of photosensitizers (PSs). Herein, an activatable photodynamic cancer cell ablation platform that can be adopted for versatile PSs is presented. Thereinto, by engineering an iron(III) carboxylate-based metal–organic framework (MOF), MIL-101(Fe), with DNA grafted after PS loading, both hydrophilic and hydrophobic PSs can undergo negligible unspecific leakage and significant suppression of photosensitization during delivery. Following the reaction between MIL-101 and H2O2 whose level is greatly increased inside the tumor, MIL-101 is selectively degraded to release the loaded PDT agents and recover their photosensitization, controllably killing cancer cells upon H2O2 activation. Such a strategy assisted by a DNA-functionalized MOF significantly expands the varieties of PSs applicable for activatable PDT.
Introduction
Photodynamic therapy (PDT) has emerged as an attractive modality for oncotherapy owing to its spatiotemporal precision, non-invasive nature, and low side effects.1–10 It relies on photosensitizers (PSs) to generate reactive oxygen species (ROS), including peroxides, superoxides, singlet oxygen (1O2), etc., to kill cancerous cells.11–17 However, the majority of administrated PSs do not necessarily distribute in malignant tissues, and PSs localized on the skin and in healthy tissues tend to cause unspecific phototoxic damage in an off-target manner.18,19 Hence, researchers have recently developed activatable PSs which can be activated for ROS production in cancerous cells while remaining inert in normal cells.20,21 However, these strategies are typically limited to certain types of PSs, which require specific molecular modification on each PS molecule before they can be adopted for targeted photodynamic cell ablation.18,19,22 Therefore, a general strategy to render various PSs applicable for activatable photodynamic cancer cell ablation is highly desirable.Metal–organic frameworks (MOFs) with bioactive metal nodes and ligands have demonstrated their great potential to serve as nanocarriers for biomedical applications, owing to their high porosity for cargo transport and structural diversity for biofunctionalization.23–32 Most recently, activatable PS systems assisted by nanoscale hydrophobic MOFs with Fe(III) or Cu(II) nodes as exquisite PS carriers have been developed to realize selective activation of PSs inside tumor tissues rather than normal tissues.19,22,33 However, such strategies are only applicable to hydrophobic photosensitizers.19,22,33 It is because hydrophilic photosensitizers transported by hydrophobic MOFs are prone to leak out of the pores and cavities of the MOFs and then enter aqueous milieus through hydrophilic interaction with water, free from the MOF control for activatable sensitization. This limitation motivated us to expand the application range of strategies on activatable PDT to both hydrophobic and hydrophilic PSs. We hypothesized that by blocking the aperture of MOFs after PS loading, both the PSs would be less likely to undergo undesired leakage from the pores and cavities of MOFs in the delivery process, which would allow both the PSs to be under the control of MOFs for activatable photodynamic cancer cell ablation.
As a proof-of-concept, herein, PSs including 2-(4-(diphenylamino)phenyl)anthracene-9,10-dione (TPAAQ) with an aggregation-induced-emission feature22 and methylene blue (MB) with an aggregation-caused-quenching feature34 were adopted as hydrophobic and hydrophilic PS models, respectively. As indicated in Fig. 1A, MIL-101(Fe) with aperture window diameters of up to 16 Å (ref. 35) was modified with an azide group at its surface to yield MIL-101-N3, before the two PSs were respectively loaded inside the MOF to fabricate PS@MIL-101-N3. Thereafter, PS@MIL-101-N3 was conjugated with dibenzocyclooctyne-functionalized DNA (DBCO-DNA) through a click reaction at its surface to generate DNA-PS@MIL-101. As shown in Fig. 1B, the photosensitizing capability of PSs encapsulated by MIL-101 was found to be inhibited because of the isolation of O2 from PSs by the framework of MIL-101. However, upon incubation with H2O2 which is excessively secreted inside tumor regions, the framework of MIL-101 collapsed. The encapsulated PSs were thus released to come into contact with O2 enriched during the reaction between MIL-101 and H2O2, which regained the potent capability of ROS generation under light irradiation for activatable photodynamic cancer cell ablation.
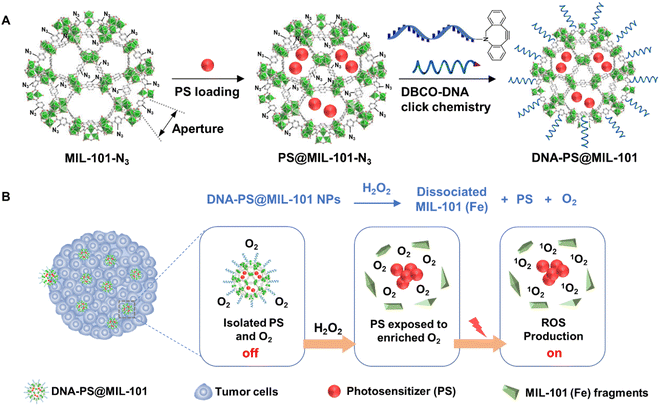 | ||
| Fig. 1 (A) Synthetic schemes of DNA-PS@MIL-101. (B) Schematic illustration of activatable photodynamic cancer cell ablation realized by DNA-PS@MIL-101. | ||
Results and discussion
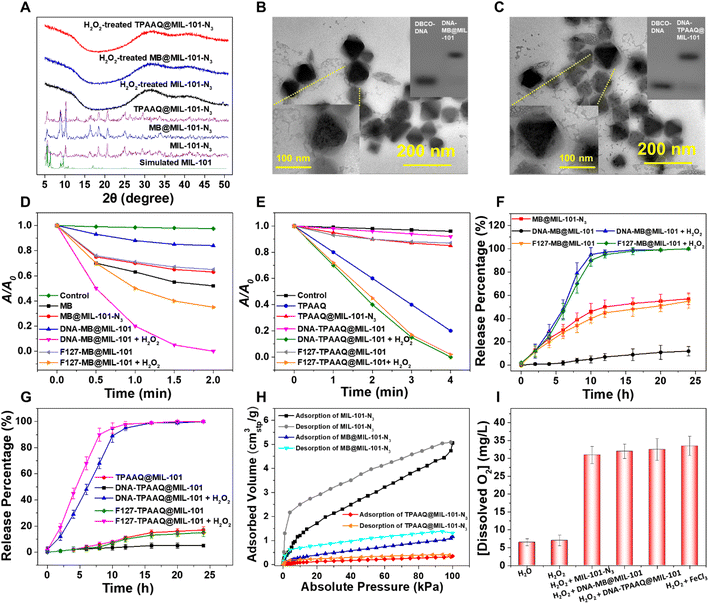
MIL-101 decorated with an amine group (MIL-101-NH2) was synthesized through a modified hydrothermal approach36 to produce MIL-101-N3 following tert-butyl nitrite (tBuONO) and azido-trimethyl silane (TMSN3) treatment. As shown in Fig. 2A, powder X-ray diffraction (PXRD) peaks of MIL-101-N3 matched well with those of MIL-101, suggesting that MIL-101-N3 inherits good crystallinity from MIL-101. Nuclear magnetic resonance (NMR) and Fourier transform infrared (FT-IR) spectra of MIL-101-NH2 and MIL-101-N3 post-digestion reveal that the amine group on MIL-101 was successfully converted to an azide group (Fig. S1–S3†). Under a transmission electron microscope (TEM), MIL-101-N3 exhibited a size of around 70 nm and similar crystalline morphology to nanoscale MIL-101(Fe) as reported (Fig. S4†). Such a size under 200 nm is favorable for cell uptake of nanoparticles.37,38MIL-101-N3 with a suitable size for cell uptake was subsequently used to respectively encapsulate PS models, i.e., MB and TPAAQ, of which 1H and 13C NMR spectra are presented in Fig. S5 and S6.† Through measurement of absorption spectra and thermogravimetric analysis (TGA) curves, the maximal loading weight percentages (wt%) of MB and TPAAQ inside MIL-101-N3 reached around 26% (Fig. S7†) within 16 h (Fig. S8†) and around 48% (Fig. S9†) within 8 h (Fig. S10†). After encapsulation of PSs, MIL-101-N3 was tested to maintain good crystallinity, as evidenced by PXRD analysis (Fig. 2A). To demonstrate that PSs were indeed loaded inside pores and cavities of MIL-101-N3 rather than being adsorbed on its surface, N2 adsorption and desorption isotherms and pore size distribution of MIL-101-N3, MB@MIL-101-N3, and TPAAQ@MIL-101-N3 were obtained. It was observed that after PS loading, the adsorbed volume of N2 by MIL-101-N3 significantly dropped, as did the pore volume of MIL-101-N3 (Fig. S11†), which verifies that in the process of PS loading, the PSs generally occupy the pores and cavities of MIL-101-N3. It is noteworthy that, after H2O2 exposure, all the MIL-101-N3, MB@MIL-101-N3, and TPAAQ@MIL-101-N3 lost their crystallinity and became amorphous. This result reveals the structural collapse of MIL-101-N3 in the presence of H2O2 following a Fenton-like reaction between Fe(III) in MIL-101-N3 and H2O2,39 which benefits the H2O2-triggered PS release from MB@MIL-101-N3 and TPAAQ@MIL-101-N3 for activatable ROS generation.
After PS loading, MIL-101-N3 was conjugated with single-stranded DBCO-DNA via a click reaction to yield DNA-encapsulated MOF particles. Through gel electrophoresis, it was detected that the mobility of DBCO-DNA greatly decreased after the click reaction (Fig. 2B and C), denoting that DNA was successfully grafted on MIL-101-N3 to respectively produce DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 post PS loading. As shown in Fig. S12,† with the emergence of the characteristic absorption peak of DNA at around 260 nm after the click reaction between MOF particles and DNA, DNA was proved to be successfully linked with both MB@MIL-101-N3 and TPAAQ@MIL-101-N3. Besides, the deceased zeta-potential values of MOF particles after DNA ligation indicate that the surfaces of both MB@MIL-101-N3 and TPAAQ@MIL-101-N3 nanoparticles were decorated with DNA. Images of DNA-MB@MIL-101 by energy dispersive X-ray spectroscopy (EDS) in scanning transmission electron microscopy (STEM) are shown in Fig. S13.† Since both MIL-101 and DNA contain C and O elements while the loaded MB exclusively contains the S element, the uniform distribution of the S element in the MOF particles and good overlap among C, O, and S elements denoted that MB was uniformly distributed in MIL-101 other than being adsorbed at the surface of MIL-101 particles. Both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 were imaged to be 80–120 nm in size, which generally agreed with the dynamic light scattering (DLS) studies (Fig. S14†), and some coronas were captured outside PS@MIL-101 under the TEM (Fig. 2B and C). To further verify the composition of the coronas, we treated both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 with deoxyribonuclease I (DNase-I) and observed that the as-mentioned coronas became indiscernible under the TEM (Fig. S4 and S15†). Given that DNase-I can specifically digest DNA after cleaving, the coronas outside PS@MIL-101 under the TEM were accordingly proven to be grafted with DNA onto the MOF surface. Notably, after H2O2 treatment, both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 lost their crystalline morphology and became fragments and debris (Fig. S4 and S15†), indicating H2O2-triggered degradation of DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101. Additionally, it can be observed that during the H2O2 incubation for up to 4 hours, both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 became more and more spherical in shape along with time (Fig. S16†), indicating the loss of crystallinity for MIL-101 nanoparticles upon H2O2 treatment. In particular, Pluronic F-127 was also used to encapsulate both MB@MIL-101-N3 and TPAAQ@MIL-101-N3 through hydrophobic interaction to produce F127-MB@MIL-101 and F127-TPAAQ@MIL-101 as two control PS agents. Both F127-MB@MIL-101 and F127-TPAAQ@MIL-101 possessed a similar size and morphology and similar H2O2-activated degradation to DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 (Fig. S17†), suggesting that the two control agents tend to demonstrate similar cell uptake and H2O2-responsive activatable behavior to DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101.
By adopting 9,10-anthracenediylbis(methylene)dimalonic acid (ABDA) as a 1O2 probe to measure the 1O2 generation efficiency of different PS agents, both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 exhibited greatly quenched 1O2 generation with a notably dropped 1O2 generation efficiency lower than 2 μg mL−1 min−1, as compared to free MB and TPAAQ, respectively (Fig. 2D and E). After H2O2 exposure, DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 regained the capability of 1O2 generation with a 1O2 generation efficiency of around 16 μg mL−1 min−1 and 5.5 μg mL−1 min−1, respectively. These indicate that both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 are H2O2-responsive regarding 1O2 generation. In contrast to DNA-MB@MIL-101, 1O2 generation of F127-MB@MIL-101 failed to be quenched, although that of its TPAAQ counterpart was quenched. The 1O2 generation efficiency of F127-MB@MIL-101 was similar to that of MB@MIL-101-N3, and it did not vary as strikingly as DNA-MB@MIL-101 did upon H2O2 exposure.
To figure out the reasons behind the aforementioned phenomena, the release processes of MB and TPAAQ in different agents along with time were monitored (Fig. 2F and G). It could be found that F127-MB@MIL-101 was prone to encounter MB leakage in water, similar to MB@MIL-101-N3 without any protection outside MIL-101 (Fig. 2F), although F127-TPAAQ@MIL-101 did not encounter a similar problem (Fig. 2G). Such a difference between hydrophilic MB and hydrophobic TPAAQ loaded inside MIL-101 with F127 encapsulation can be attributed to the fact that in water, hydrophilic MB adsorbed by MIL-101 with an aperture of up to 16 Å in diameter35 is much more likely to diffuse out of pores and cavities and enter the aqueous milieu outside, as compared with hydrophobic TPAAQ which tends to stay inside hydrophobic pores and cavities of MIL-101. It is noteworthy that DNA-MB@MIL-101 successfully trapped MB and avoided its leakage in water, different from its F127 counterpart (Fig. 2F). It benefits from the DNA encapsulation which effectively restricts the uncontrollable leakage of hydrophilic molecules from MIL-101 after DNA-MB@MIL-101 nanoparticles are brought into water. For both DNA-TPAAQ@MIL-101 and F127-TPAAQ@MIL-101, TPAAQ loaded in MIL-101 through hydrophobic interaction was not prone to leak out in water (Fig. 2G). Furthermore, O2 adsorption and desorption isotherms of MIL-101-N3, MB@MIL-101-N3, and TPAAQ@MIL-101-N3 suggest that the loaded PSs come into contact with scarce O2 inside the cavities of MIL-101, as compared to free or leaked PS molecules in water (Fig. 2H). It denotes that the trapped PS molecules in MIL-101 are relatively slow to sensitize 1O2 but free or leaked PS molecules in water remain to efficiently sensitize 1O2. These accordingly explain the phenomenon where a DNA–MIL-101 hybrid can uniquely trap both MB and TPAAQ for inhibition of 1O2 generation and degrade to release the PSs for activated 1O2 generation upon H2O2 exposure.
In addition, as shown in Fig. 2D and E, after H2O2-triggered activation of 1O2 generation, the generation efficiencies of both DNA-MB@MIL-101 and F127-MB@MIL-101 were higher than that of MB; similarly, those of both DNA-TPAAQ@MIL-101 and F127-TPAAQ@MIL-101 were also higher than that of TPAAQ. This can be ascribed to the fact that in the presence of H2O2 and MIL-101(Fe), dissolved O2 levels in water were greatly elevated after H2O2 decomposition under the catalysis of Fe(III) in MIL-101 (Fig. 2I and S18†). Besides, H2O2-treated DNA-MB@MIL-101 possessed a notably higher 1O2 generation efficiency than H2O2-treated F127-MB@MIL-101. It is because in the presence of DNA, released MB is apt to intercalate into DNA for intersystem crossing enhancement, which can greatly boost the 1O2 generation efficiency of MB.40 In this regard, the DNA–MIL-101 hybrid has proven to be a universal nanoplatform for PSs to realize both activable and enhanced ROS generation.
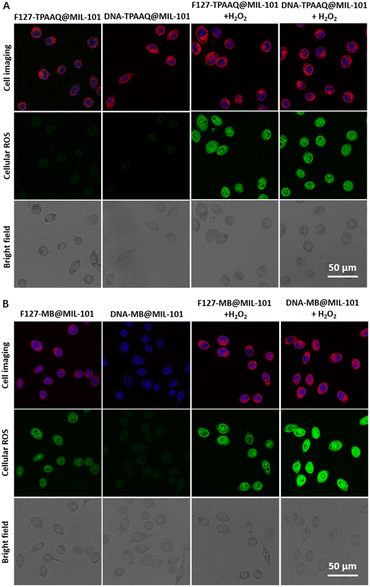
The promising activable and enhanced ROS generation of DNA-PS@MIL-101 motivated us to further explore its feasibility inside cancer cells. Prior to in vitro experiments, all DNA-MB@MIL-101, DNA-TPAAQ@MIL-101, F127-MB@MIL-101, and F127-TPAAQ@MIL-101 were immersed in water, DMEM, and FBS to study their stability in biological milieus. Both the hydrodynamic diameter and zeta-potential of the four nanoparticles showed negligible variation and the nanoparticles underwent modest degradation in biological media (Fig. S19 and S20†), verifying their good stability in biological milieus before reaction with H2O2. Cancerous HeLa cells were then incubated with different PS-involved agents for 24 h. Thereafter, the cells were labeled with a commercialized nuclear-specific dye, i.e., Hoechst with blue fluorescence and intracellular ROS probe, 2′,7′-dichlorofluorescein diacetate (DCFDA), before being visualized by confocal laser scanning microscopy (CLSM). DCFDA as a ROS detection agent becomes green fluorescent under oxidation of intracellular ROS. No intracellular PS and ROS signals were detected for HeLa cells without any incubation and the cells only incubated with H2O2 (Fig. S21†). As shown in Fig. 3A, both F127-TPAAQ@MIL-101 and DNA-TPAAQ@MIL-101 were endocytosed in similar quantities resulting from their similar sizes and surface charges. After H2O2 treatment, bright green fluorescence was observed in the cytoplasm of HeLa cells under light irradiation, in contrast to that of the cells free from H2O2 treatment (Fig. 3A and S22†), demonstrating that both F127-TPAAQ@MIL-101 and DNA-TPAAQ@MIL-101 are able to perform H2O2-triggered intracellular ROS generation.
Moreover, the MB-originated red fluorescence of cells incubated with DNA-MB@MIL-101 was much dimmer than that in cells treated with F127-MB@MIL-101. Still, after H2O2 treatment, the red fluorescence from MB in the cells incubated with DNA-MB@MIL-101 and F127-MB@MIL-101 was similar in intensity. It can be ascribed to the fact that, unlike MIL-101-loaded TPAAQ molecules, the fluorescence of MIL-101-loaded MB molecules without MB leakage was quenched as compared with free MB molecules in water (Fig. S23†), which is due to the aggregation-induced-quenching effect experienced by them aggregated in the pores and cavities of the MOF.22 When MB molecules were released or leaked out of MIL-101, their fluorescent intensity was found to recover, different from that of TPAAQ (Fig. S23†). After H2O2 treatment, the similar fluorescent intensity of MB in the cells incubated with DNA-MB@MIL-101 and F127-MB@MIL-101 (Fig. 3B) denotes that the endocytosis quantities of DNA-MB@MIL-101 and F127-MB@MIL-101 by HeLa cells are similar, owing to their similar sizes and surface charges. More importantly, discernible green fluorescence emitted from DCFDA could be observed in the cells treated with F127-MB@MIL-101 only, and the fluorescent intensity was comparable to the intensity of cells treated with both F127-MB@MIL-101 and H2O2 (Fig. 3B and S22†). This demonstrates that F127-MB@MIL-101 fails to perform H2O2-responsive intracellular ROS generation. In contrast to F127-MB@MIL-101, DNA-MB@MIL-101 was captured to quench ROS generation inside cells without H2O2 exposure whilst MB in this formulation regained its ROS generation after the incubated HeLa cells were treated with H2O2 (Fig. 3B and S22†). It verifies the unique controllable intracellular ROS activation endowed by the DNA–MIL-101 hybrid other than the F127–MIL-101 hybrid for hydrophilic MB. Besides, the green fluorescence of cells treated with DNA-MB@MIL-101 and H2O2 was much brighter than that of the cells treated with F127-MB@MIL-101 and H2O2, revealing that DNA can notably improve the ROS generation efficiency of MB inside cells, in accordance with the ROS titration results in the solution. In this regard, H2O2-activated and enhanced intracellular ROS generation of DNA-PS@MIL-101 is demonstrated.
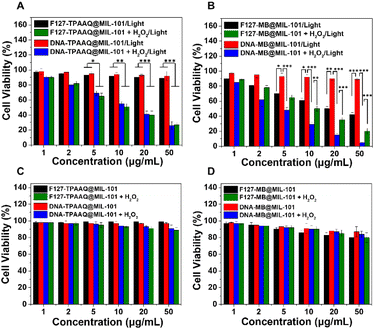
We next evaluated the activatable photodynamic cancer cell ablation effects of F127-TPAAQ@MIL-101, DNA-TPAAQ@MIL-101, F127-MB@MIL-101, and DNA-MB@MIL-101 through methylthiazolyldiphenyltetrazolium bromide assay. It was found that treating HeLa cells only with light or H2O2 cannot effectively kill them (Fig. S24†). As indicated in Fig. 4A, IC50 values of both F127-TPAAQ@MIL-101 and DNA-TPAAQ@MIL-101 under light toward HeLa cells are around 10 μg mL−1, much lower than the IC50 values of the two NPs without H2O2 activation. In contrast, F127-MB@MIL-101 did not show an inhibited ablation efficacy toward cancer cells whilst DNA-MB@MIL-101 was shown to successfully quench the photodynamic ablation (Fig. 4B). Furthermore, following the H2O2 trigger, DNA-MB@MIL-101 showed not only activated but also enhanced phototoxicity for cancer cells, as compared with F127-MB@MIL-101 post H2O2 activation. Meanwhile, neither F127-PS@MIL-101 nor DNA-PS@MIL-101 with/without H2O2 activation displayed toxicity toward HeLa cells in the dark. For 3T3 cells as a normal cell line, the DNA–MIL-101 hybrids loaded with PSs exhibited both low phototoxicity and negligible dark toxicity. Nevertheless, the F127–MIL-101 hybrids loaded with PSs could cause significant unspecific phototoxic damage to 3T3 cells although they were tested to be safe for the cells in the dark (Fig. S25†). These results demonstrate that as a universal nanoplatform, the DNA–MIL-101 hybrid can uniquely endow different PSs with the capability of H2O2-activated photodynamic cancer cell ablation and low toxicity toward normal cells.
The activatable photodynamic cancer cell ablation performed by the DNA–MIL-101 hybrids motivated us to further explore their antitumoral efficacies in a mouse model. The mouse model was established via subcutaneous inoculation of 4T1 cancer cells into BALB/c mice. The formed 4T1 breast tumor is a solid tumor, which provides a tumor microenvironment with upregulated H2O2 levels. Mice with 4T1 tumors were intravenously injected with PBS, DNA-MB@MIL-101, or DNA-TPAAQ@MIL-101. As indicated in Fig. S26A,† the tumor regions were gradually lit up by DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 and the fluorescence intensity in the tumor regions reached a peak at 8 h after intravenous injection, demonstrating the good imaging guiding capability of both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101. After administration of the two nanoparticles and white light irradiation at 8 h post injection, the nanoparticles were found to effectively inhibit the growth of mouse tumors, as compared to that of PBS-treated mice (Fig. S26B†). Moreover, the hematoxylin and eosin (H&E) staining results (Fig. S26C†) show that both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 caused obvious cell apoptosis in tumors, as compared with PBS, while they did not induce phototoxicity to normal skin cells, similar to PBS. This indicates that both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 can specifically ablate tumor tissues and cells in an activatable photodynamic manner with modest unspecific phototoxic damage toward normal tissues and cells.
To examine biocompatibility of the DNA–MIL-101 hybrids, H&E staining, blood chemistry analysis, and body weight measurement were also conducted on mice that received PBS, DNA-MB@MIL-101, or DNA-TPAAQ@MIL-101 treatment. As indicated in Fig. S26D, S27, and S28,† negligible impacts on normal organs, blood biochemistry, and body weights of tumor-bearing mice treated with the nanoparticles were respectively observed, which denote good biocompatibility of both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 toward mice. Additionally, ex vivo inductively coupled plasma mass spectrometry analysis of the Fe content in different organs after 1 and 7 days of intravenous injection was conducted (Fig. S29†), showing that both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 can be cleared from the mouse body with time. Along with the potent antitumoral effect of the two nanoparticles, both DNA-MB@MIL-101 and DNA-TPAAQ@MIL-101 are demonstrated to realize highly effective in vivo PDT with negligible side effects.
Conclusions
In conclusion, we have developed a general approach assisted by DNA-engineered MIL-101(Fe) for both hydrophilic and hydrophobic PSs to perform activable photodynamic cancer cell ablation. This approach loads PSs inside MIL-101 with minimal undesirable leakage via DNA encapsulation and quenches their photosensitization. Following exposure to H2O2 which is overexpressed inside tumor/cancer cells, MIL-101 structurally collapsed to release the loaded PSs and recover their ROS generation under light irradiation for triggered killing of cancer cells. With the advantage of DNA–MOF hybrids for activatable photodynamic cancer cell ablation, we foresee the great potential of the as-reported strategy for development of next-generation activatable PDT agents.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2020YFB1506300); National Natural Science Foundation of China (Grant No. 21625102, 21971017, 21922502, 22075018, 51991344, 52025025, and 52072400); Beijing Institute of Technology Research Fund Program for Young Scholars; Beijing Natural Science Foundation (Z190010); Chinese Postdoctoral Science Foundation.References
- X. Li, J. F. Lovell, J. Yoon and X. Chen, Nat. Rev. Clin. Oncol., 2020, 17, 657–674 CrossRef.
- Z. Meng, X. Zhou, J. Xu, X. Han, Z. Dong, H. Wang, Y. Zhang, J. She, L. Xu, C. Wang and Z. Liu, Adv. Mater., 2019, 31, 1900927 CrossRef PubMed.
- X. Li, N. Kwon, T. Guo, Z. Liu and J. Yoon, Angew. Chem., Int. Ed., 2018, 57, 11522–11531 CrossRef CAS.
- L. Zhang, S. Wang, Y. Zhou, C. Wang, X.-Z. Zhang and H. Deng, Angew. Chem., Int. Ed., 2019, 58, 14213–14218 CrossRef CAS PubMed.
- X.-Y. Lou, G. Zhang, N. Song and Y.-W. Yang, Biomaterials, 2022, 286, 121595 CrossRef CAS.
- D. Wang, H. Wu, S. Z. F. Phua, G. Yang, W. Qi Lim, L. Gu, C. Qian, H. Wang, Z. Guo, H. Chen and Y. Zhao, Nat. Commun., 2020, 11, 357 CrossRef CAS PubMed.
- H. Zhao, J. Xu, C. Feng, J. Ren, L. Bao, Y. Zhao, W. Tao, Y. Zhao and X. Yang, Adv. Mater., 2022, 34, 2106390 CrossRef CAS.
- D. Li, D. Hu, H. Xu, H. K. Patra, X. Liu, Z. Zhou, J. Tang, N. Slater and Y. Shen, Biomaterials, 2021, 264, 120410 CrossRef CAS.
- J. Wang, Y. Li and G. Nie, Nat. Rev. Mater., 2021, 6, 766–783 CrossRef CAS.
- N. Gong, X. Ma, X. Ye, Q. Zhou, X. Chen, X. Tan, S. Yao, S. Huo, T. Zhang, S. Chen, X. Teng, X. Hu, J. Yu, Y. Gan, H. Jiang, J. Li and X.-J. Liang, Nat. Nanotechnol., 2019, 14, 379–387 CrossRef CAS PubMed.
- B. Yang, Y. Chen and J. Shi, Chem. Rev., 2019, 119, 4881–4985 CrossRef CAS.
- K. Kenry, K. C. Chong and B. Liu, Acc. Chem. Res., 2019, 52, 3051–3063 CrossRef CAS.
- W. Wu, D. Mao, S. Xu, K. Kenry, F. Hu, X. Li, D. Kong and B. Liu, Chem, 2018, 4, 1937–1951 CAS.
- J. Sun, K. Du, J. Diao, X. Cai, F. Feng and S. Wang, Angew. Chem., Int. Ed., 2020, 59, 12122–12128 CrossRef CAS.
- T. Jiang, G. Xu, G. Chen, Y. Zheng, B. He and Z. Gu, Nano Res., 2020, 13, 1810–1824 CrossRef CAS.
- C. Liu, B. Liu, J. Zhao, Z. Di, D. Chen, Z. Gu, L. Li and Y. Zhao, Angew. Chem., Int. Ed., 2020, 59, 2634–2638 CrossRef CAS.
- Z. Cheng, Y. Cheng, Q. Chen, M. Li, J. Wang, H. Liu, M. Li, Y. Ning, Z. Yu, Y. Wang and H. Wang, Nano Today, 2020, 33, 100878 CrossRef CAS.
- J. F. Lovell, T. W. B. Liu, J. Chen and G. Zheng, Chem. Rev., 2010, 110, 2839–2857 CrossRef CAS.
- F. Hu, D. Mao, Y. Wang, W. Wu, D. Zhao, D. Kong and B. Liu, Adv. Funct. Mater., 2018, 28, 1707519 CrossRef.
- L. Shao, Y. Pan, B. Hua, S. Xu, G. Yu, M. Wang, B. Liu and F. Huang, Angew. Chem., Int. Ed., 2020, 59, 11779–11783 CrossRef CAS PubMed.
- Y. Cheng, J. Dai, C. Sun, R. Liu, T. Zhai, X. Lou and F. Xia, Angew. Chem., Int. Ed., 2018, 57, 3123–3127 CrossRef CAS.
- Y. Wang, W. Wu, J. Liu, P. N. Manghnani, F. Hu, D. Ma, C. Teh, B. Wang and B. Liu, ACS Nano, 2019, 13, 6879–6890 CrossRef CAS.
- K. Ni, G. Lan and W. Lin, ACS Cent. Sci., 2020, 6, 861–868 CrossRef CAS.
- G. Lan, K. Ni, S. S. Veroneau, X. Feng, G. T. Nash, T. Luo, Z. Xu and W. Lin, J. Am. Chem. Soc., 2019, 141, 4204–4208 CrossRef CAS PubMed.
- T. Simon-Yarza, A. Mielcarek, P. Couvreur and C. Serre, Adv. Mater., 2018, 30, 1707365 CrossRef.
- Y. Cheng, H. Zhang and X. Qu, Acc. Chem. Res., 2021, 2, 764–779 CAS.
- P. Gao, Y. Chen, W. Pan, N. Li, Z. Liu and B. Tang, Angew. Chem., Int. Ed., 2021, 60, 16763–16776 CrossRef CAS.
- J.-W. Wang, Q.-W. Chen, G.-F. Luo, Z.-Y. Han, W.-F. Song, J. Yang, W.-H. Chen and X.-Z. Zhang, ACS Nano, 2021, 15, 17870–17884 CrossRef CAS.
- Q.-X. Wang, Y.-F. Yang, X.-F. Yang, Y. Pan, L.-D. Sun, W.-Y. Zhang, Y. Shao, J. Shen, J. Lin, L. Li and C.-H. Yan, Nano Today, 2022, 43, 101439 CrossRef CAS.
- R. Xing, Q. Zou, C. Yuan, L. Zhao, R. Chang and X. Yan, Adv. Mater., 2019, 31, 1900822 CrossRef PubMed.
- S. Li, Q. Zou, Y. Li, C. Yuan, R. Xing and X. Yan, J. Am. Chem. Soc., 2018, 140, 10794–10802 CrossRef CAS PubMed.
- Y. Li, P. Sun, L. Zhao, X. Yan, D. K. P. Ng and P.-C. Lo, Angew. Chem., Int. Ed., 2020, 59, 23228–23238 CrossRef CAS PubMed.
- Y. Wang, S. Xu, L. Shi, C. Teh, G. Qi and B. Liu, Angew. Chem., Int. Ed., 2021, 60, 14945–14953 CrossRef CAS PubMed.
- S. Jockusch, N. J. Turro and D. A. Tomalia, Macromolecules, 1995, 28, 7416–7418 CrossRef CAS.
- G. Férey, C. Mellot-Draznieks, C. Serre, F. Millange, J. Dutour, S. Surblé and I. Margiolaki, Science, 2005, 309, 2040–2042 CrossRef.
- M. Savonnet, D. Bazer-Bachi, N. Bats, J. Perez-Pellitero, E. Jeanneau, V. Lecocq, C. Pinel and D. Farrusseng, J. Am. Chem. Soc., 2010, 132, 4518–4519 CrossRef CAS PubMed.
- K. Yin Win and S.-S. Feng, Biomaterials, 2005, 26, 2713–2722 CrossRef.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Mol. Pharm., 2008, 5, 505–515 CrossRef CAS.
- J.-X. Fan, M.-Y. Peng, H. Wang, H.-R. Zheng, Z.-L. Liu, C.-X. Li, X.-N. Wang, X.-H. Liu, S.-X. Cheng and X.-Z. Zhang, Adv. Mater., 2019, 31, 1808278 CrossRef.
- J. J. Nogueira, M. Oppel and L. González, Angew. Chem., Int. Ed., 2015, 54, 4375–4378 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2na00509c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |