Open Access Article
Open Access ArticleGreen synthesis of fluorescent carbon nanodots from sage leaves for selective anticancer activity on 2D liver cancer cells and 3D multicellular tumor spheroids
Shadi
Sawalha
 *a,
Samer
Abdallah
*a,
Samer
Abdallah
 b,
Amal
Barham
a,
Hala
Badawi
a,
Zeina
Barham
a,
Ahmad
Ghareeb
c,
Giuseppe
Misia
d,
Silvia
Collavini
e,
Alessandro
Silvestri
f,
Maurizio
Prato
b,
Amal
Barham
a,
Hala
Badawi
a,
Zeina
Barham
a,
Ahmad
Ghareeb
c,
Giuseppe
Misia
d,
Silvia
Collavini
e,
Alessandro
Silvestri
f,
Maurizio
Prato
 de and
Mohyeddin
Assali
de and
Mohyeddin
Assali
 *c
*c
aChemical Engineering Program, Faculty of Engineering and Information Technology, An-Najah National University, P.O. Box 7, Nablus, Palestine. E-mail: sh.sawalha@najah.edu
bDepartment of Biology and Biotechnology, Faculty of Science, An-Najah National University, P.O. Box 7, Nablus, Palestine
cDepartment of Pharmacy, Faculty of Medicine and Health Sciences, An-Najah National University, P.O. Box 7, Nablus, Palestine. E-mail: m.d.assali@najah.edu
dDepartment of Chemical and Pharmaceutical Sciences INSTM UdR Trieste, University of Trieste, via Licio Giorgieri 1, 34127 Trieste, Italy
eCenter for Cooperative Research in Biomaterials (CIC BiomaGUNE), Basque Research and Technology Alliance (BRTA), Donostia-San Sebastián 20014, Spain
fDepartment of Molecular Sciences and Nanosystems, Ca' Foscari University of Venice, Venezia 30170, Italy
First published on 28th September 2023
Abstract
Carbon nanodots, a family of carbon-based nanomaterials, have been synthesized through different methods from various resources, affecting the properties of the resulting product and their application. Herein, carbon nanodots (CNDs) were synthesized with a green and simple hydrothermal method from sage leaves at 200 °C for 6 hours. The obtained CNDs are well dispersed in water with a negative surface charge (ζ-potential = −11 mV) and an average particle size of 3.6 nm. The synthesized CNDs showed concentration-dependent anticancer activity toward liver cancer (Hep3B) cell lines and decreased the viability of the cancer cells to 23% at the highest used concentration (250 μg ml−1 of CNDs). More interestingly, the cytotoxicity of the CNDs was tested in normal liver cell lines (LX2) revealed that the CNDs at all tested concentrations didn't affect their viability including at the highest concentration showing a viability of 86.7%. The cellular uptake mechanisms of CNDs were investigated and they are thought to be through energy-dependent endocytosis and also through passive diffusion. The main mechanisms of endocytosis were lipid and caveolae-mediated endocytosis. In addition, the CNDs have hindered the formation of 3D spheroids from the Hep3B hepatocellular carcinoma cell line. Hence, it would be concluded that the synthesized CNDs from sage are more highly selective to liver cancer cells than normal ones. The CNDs' cancer-killing ability would be referred to as the production of reactive oxygen species.
1. Introduction
Cancer is one of the primary health issues affecting the global population and the second largest cause of death after cardiovascular disease. Despite recent substantial advancements in cancer treatment, aggressive tumors such as those of the lung, breast, pancreatic, and liver still cause significantly low patient survival rates.1In the treatment of cancer, the methods currently used, such as chemotherapy and radiation therapy, destroy healthy cells and harm the body's production of new cells at the same time as killing cancer cells, and they are drawn-out procedures.2 The chemotherapy currently in use lacks selectivity and affinities to particular malignancies. Recently, a few focused therapies have been created. However, they are pricey and only effective for specific cancers.3
Because of the damage caused by the current treatment used, researchers resorted to the use of targeted cancer therapy based on nanomaterials taking advantage of enhanced permeability and retention phenomena and the overexpression of specific receptors in cancer cells.4,5 One of these well-developed nanomaterials is carbon-based nanomaterials such as carbon nanotubes (CNTs),6,7 graphene,8 and carbon nanodots (CNDs).9
CNDs used in cancer theranostics have been documented over the past ten years.10 Numerous studies specifically examined the potential use of CNDs as targeted anticancer drug delivery systems.11 Furthermore, the eradication of cancer cells has been achieved by CNDs through either photothermal therapy (PTT) or photodynamic therapy (PDT).12
CNDs were discovered as a result of the purification of the single-walled carbon nanotube (SWNT) process in 2004 and became an important and interesting topic in nanomaterial and cancer therapy.13
“Top-down” and “bottom-up” synthesis are the two main categories of synthetic techniques used to create CNDs. The “top-down” approach is used to synthesize CNDs typically by breaking down carbon–carbon bonds of large molecules existing in bulk carbon materials such as charcoal, graphite, fullerene, etc.14 This can be achieved by different methods such as electrochemical exfoliation,15 laser ablation,16 and arc discharge.17 In “bottom-up”, methods such as microwave-assisted polymerization, pyrolysis,18–23 hydrothermal/solvothermal,24 and carbonization are used to convert small organic molecules contained in the precursor to carbon nanodots.25–27
CNDs have been synthesized from various resources, such as man-made28–31 and natural resources,32–36 and they have also been synthesized from various herbs and plants and have been used in different biomedical applications.37,38
One of the reasons that make CNDs good candidates for medical applications, especially as an anticancer agent, is their properties that caught and attracted the researcher's attention such as biocompatibility,39 water dispersibility,40 cytotoxicity,41 good conductivity,42 quantum yield and small size,43 and fluorescence emission.44
CNDs as anticancer agents have been studied by different researchers to treat a wide variety of cancer types such as breast cancer which has been treated by CNDs synthesized from bamboo.45 However, there have not been many studies on liver cancer. Chi-Lin Li et al. prepared CNDs from extracted ginger juice and used them to kill liver cancer cells (HepG2), where 50% of the cancer cells were killed at a concentration of 350 μg ml−1.46
In this research and for the first time (to the best of our knowledge) CNDs have been synthesized from sage leaves with a green hydrothermal method and characterized as being used as an anticancer agent for liver cancer cells (Hep3B) and performing cell viability tests for normal liver cells, to investigate the CND selectivity toward cancer cells. In addition, CNDs' effect on prohibiting 3D spheroid formation has been investigated. Finally, the possible mechanism of anticancer activity has been studied.
2. Materials and methods
Fresh sage leaves as the precursor for carbon nanodot synthesis were obtained from the local market. All used chemicals and reagents were purchased from Sigma-Aldrich and used without any further purification. Distilled water was the media used in the synthesis of CNDs.A Beckman Coulter DU 800 spectrophotometer collects the absorption of UV-Vis spectra between 200 and 800 nm. A PerkinElmer LS50B fluorescence spectrophotometer was used to measure the steady-state fluorescence in the emission range of 320 to 650 nm. Fluorescence spectra values were collected at excitation wavelengths ranging from 300 to 440 nm with a 20 nm increment. A Thermo Scientific Nicolet IS5 FTIR system equipped with an ATR sampling apparatus, 64 scans at 8 cm−1 resolution were used to record the Fourier transform infrared (FT-IR) spectra of synthesized CNDs in the range of 4000–650 cm−1. Atomic force microscopy (AFM) images and profiles were captured using a Nanosurf Core AFM microscope and the images were analyzed using Gwyddion software. The samples were prepared by drop-casting CND diluted solution on mica substrates and then vacuum drying at 120 °C. By utilizing Nano Brook Omni equipment using phase analysis light scattering (PALS), ζ-potential measurements were performed. Transmission electron microscope (TEM) images were obtained using a JEOL JEM 1400-Plus microscope with a Gatan US1000 CCD camera and 120 kV accelerating voltage; samples were prepared by drop-casting diluted CND solutions on lacey carbon film-covered copper TEM grids. X-ray photoelectron spectroscopy (XPS) measurements were conducted using a PHI 5000 VersaProbe III photoelectron spectrometer. For XPS samples, CND solutions were drop cast on a gold substrate and dried under vacuum for 24 h.
2.1. Synthesis of CNDs
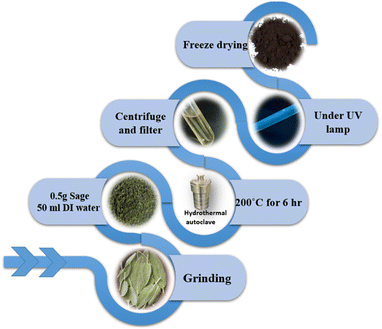
By a green hydrothermal method, CNDs were synthesized utilizing fresh sage leaves. First, sage leaves were washed well with water and then dried to remove excess water. 0.5 grams of sage leaves were cut into small pieces and then dispersed in 50 ml of distilled water. The mixture was inserted into a Teflon-lined stainless-steel autoclave and then heated to 200 °C for 6 h; subsequently, the autoclave was cooled down to room temperature, and the solution was centrifuged for 10 minutes at 3500 rpm. The resulting supernatant was filtered through a 0.45 μm microfilter. The formed CND solution was purified using a Sephadex 25-G size exclusion column. CND powder was obtained by freeze-drying the solution for 36 h and stored in the dark for further characterization and application.2.2. Cancer activity and cytotoxicity
We analyzed the cytotoxicity of prepared CNDs on human hepatocellular carcinoma cell line (Hep3B) cells purchased from ATCC (HB-8064) and normal liver cells (LX2) purchased from Sigma-Aldrich (SCC064).The first step in cell culture was culturing the cell line in T-175 cell culture flasks containing Roswell Park Memorial Institute (RPMI) basal medium supplemented with L-glutamine (1%), fetal bovine serum FBS (10%), and penicillin/streptomycin (1%). At 37 °C and 99% humidity, the cells were kept in a standard cell culture incubator at 5% CO2 equipped with a UV-Vis lamp. The medium was removed and washed with Ca2+-free phosphate buffer solution (PBS) before subculturing. Following that, cells were incubated in the incubator with 0.025% trypsin for up to 5 minutes until sufficient cells detached. Then, complete growth medium (CGM) was used to inactivate the trypsin, then the cell suspension was collected, and a trypan blue stain was used to determine the viable cell count before adjusting the cell concentration to 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 31
000 and 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for HeP3B and LX2 cells respectively. In a 96-well plate, the cells were seeded at 5000 cells per well. The cells were then allowed to adhere and accommodate overnight before tests were conducted. After 24 hours, we prepared different concentrations of CNDs which are 250 μg ml−1, 200 μg ml−1, 150 μg ml−1, 100 μg ml−1, and 50 μg ml−1 under 7.4 pH conditions. After that, we add 100 μl of each concentration per well. After 48 hours, 20 μl of MTS reagent was added to each well and incubated for 2 hours in the incubator. After that time, the absorbance was measured at 490 nm by using a plate reader.
000 for HeP3B and LX2 cells respectively. In a 96-well plate, the cells were seeded at 5000 cells per well. The cells were then allowed to adhere and accommodate overnight before tests were conducted. After 24 hours, we prepared different concentrations of CNDs which are 250 μg ml−1, 200 μg ml−1, 150 μg ml−1, 100 μg ml−1, and 50 μg ml−1 under 7.4 pH conditions. After that, we add 100 μl of each concentration per well. After 48 hours, 20 μl of MTS reagent was added to each well and incubated for 2 hours in the incubator. After that time, the absorbance was measured at 490 nm by using a plate reader.
2.3. Cellular uptake study of the synthesized carbon nanodots
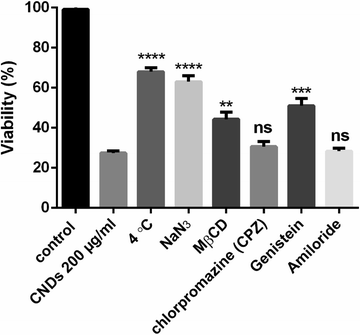
To investigate the cellular uptake mechanism of the CNDs, Hep3B cells were seeded in a 96 well plate at 5000 cells per well and maintained at 4 °C or incubated with one of the various endocytosis inhibitors (NaN3 10 mM, methyl-β-cyclodextrin 5 mg ml−1 (MβCD), chlorpromazine 10 μM (CPZ), genistein 200 μM, and amiloride 200 μM) for 1 h at 37 °C. The concentrations used are according to the literature.47,48 After that, 100 μl of 200 μg ml−1 CNDs was added to each well. After that, an MTS assay was performed.2.4. Capacity testing of 3D tumor spheroid formation
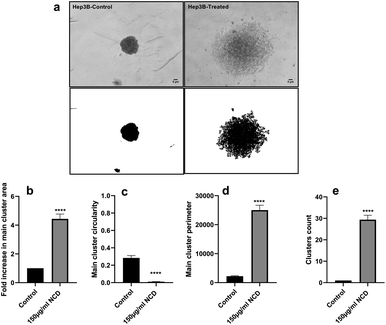
Human Hep3B liver cancer cells were cultured (4 × 103 cells) in ultra-low attachment 96-well plates in the presence of 150 μg ml−1 of carbon nanodots for 24 hours. Images of spheroids were captured 24 hours after incubation at a final magnification of 40× of an inverted microscope. ImageJ software was employed for analyzing images and their measurements.2.5. Glutathione deficiency test
A glutathione deficiency test was carried out to investigate the ability of synthesized CNDs to generate reactive oxygen species (ROS). 0.8 mM of glutathione (GSH) was prepared in 50 mM bicarbonate buffer with a pH of 8.6. The positive control was 0.8 mM of GSH solution with 1 mM H2O2, and the negative control was set using a GSH solution. 255 μl of synthesized CNDs was dispersed separately in the bicarbonate buffer with GSH. All samples were conducted in duplicate and wrapped with aluminum foil. The samples were agitated in a shaker for 2 h with a speed of 250 rpm. After that, 15 μl of 100 mM DNTB Ellman's reagent and 780 μl of prepared 0.05 M Tris-HCl were added to each sample and measured its absorbance at wavelength λ = 412 nm.3. Results and discussion
3.1. Synthesis and characterization
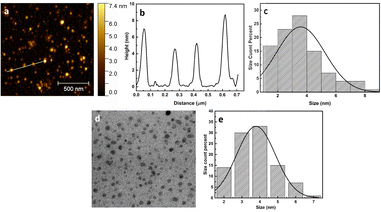
The bottom-up approach was followed up for the synthesis of CNDs from sage leaves, with the hydrothermal process at 200 °C for 6 hours. It is thought that the containing sage essential oils such as α-thujone, 1,8-cineole, and camphor49 would be polymerized during hydrothermal forming aromatic clusters, and when their concentration reaches a critical supersaturation point, burnt nucleation takes place, in addition to carbonization occurring for organic contents to form carbon dots.50 After the synthesis of the CNDs, we conducted various purification procedures to remove any intermediate or precursor materials. In this process, we performed centrifugation, filtration, and most importantly column chromatography separation using a Sephadex G-25 which confirms the purification of the carbon nanodots. The synthesized CNDs have been obtained with a 15% product yield. Fig. 1 illustrates the synthesis process.The morphology and size of synthesized carbon nanoparticles (CNDs) were characterized by using AFM and TEM microscopies. The CND height was measured to be in the range of 1–9 nm with an average of (3.6 ± 1.5 nm) as shown by the AFM results in Fig. 2a–c. On the other hand, the TEM micrograph (Fig. 2d & e) represents well-distributed particles with a size ranging from 2–7 nm with an average of 3.8 ± 1.6 nm. Both the AFM and TEM results would confirm that the formed CNDs are quasi-spherical in their shape and they are well-dispersed in the CND solution.
The chemical composition and surface functional groups were acquired by FTIR and XPS as shown in Fig. 3. The FTIR spectrum demonstrates different peaks at 3390, 3088, 2920, 2860, 1600, 1417, 1259, and 1084, which could be related to O–H, N–H, C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C
C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N, COO–, and C–O–C bands respectively as shown in Fig. 3a.51–54 The XPS measurement showed that the synthesized CNDs are composed of carbon, oxygen, and nitrogen elements with an atomic percent of 53.7, 44.1, and 2.2, respectively. The deconvolution of the high-resolution C 1s core (Fig. 3b) leads to three peaks at 284.8, 286.3, and 288.3 eV which can be related to C–C/C
O, C–N, COO–, and C–O–C bands respectively as shown in Fig. 3a.51–54 The XPS measurement showed that the synthesized CNDs are composed of carbon, oxygen, and nitrogen elements with an atomic percent of 53.7, 44.1, and 2.2, respectively. The deconvolution of the high-resolution C 1s core (Fig. 3b) leads to three peaks at 284.8, 286.3, and 288.3 eV which can be related to C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–O/C–N, and C
C, C–O/C–N, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional groups respectively.55,56 Besides that, the O 1s can be deconvoluted into two peaks of O
O functional groups respectively.55,56 Besides that, the O 1s can be deconvoluted into two peaks of O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C at 532.4 eV and O–C at 533.2 eV as shown in Fig. 3c.57 Finally, the nitrogen content is represented by the C–N functional group at 400.9 eV (Fig. 3d).58 The XPS results are well-matched with those of FT-IR emphasizing the existence of varied oxygen groups on the surface of the synthesized CNDs such as hydroxyl, carbonyl, and carboxylic groups. The existence of highly oxygenated groups would confirm the stability and water solubility of synthesized particles,59 in addition to the surface negative charge measured as PALS ζ-potential equal −11 ± 1.2 mV in phosphate buffer solution at (PBS @ PH ≈ 7.4).60 The obtained ζ-potential is nearly similar to those obtained by other researchers, for example, CNDs synthesized from ascorbic acid by the hydrothermal method have a negative charge of −23 mV.61 While CNDs from bovine gelatine gave a ζ-potential equal to −20 mV,62 on the other hand, carbon dots from bamboo showed a ζ-potential equal to −4.78 mV.63
C at 532.4 eV and O–C at 533.2 eV as shown in Fig. 3c.57 Finally, the nitrogen content is represented by the C–N functional group at 400.9 eV (Fig. 3d).58 The XPS results are well-matched with those of FT-IR emphasizing the existence of varied oxygen groups on the surface of the synthesized CNDs such as hydroxyl, carbonyl, and carboxylic groups. The existence of highly oxygenated groups would confirm the stability and water solubility of synthesized particles,59 in addition to the surface negative charge measured as PALS ζ-potential equal −11 ± 1.2 mV in phosphate buffer solution at (PBS @ PH ≈ 7.4).60 The obtained ζ-potential is nearly similar to those obtained by other researchers, for example, CNDs synthesized from ascorbic acid by the hydrothermal method have a negative charge of −23 mV.61 While CNDs from bovine gelatine gave a ζ-potential equal to −20 mV,62 on the other hand, carbon dots from bamboo showed a ζ-potential equal to −4.78 mV.63
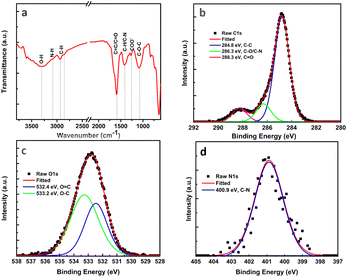 | ||
| Fig. 3 (a) FTIR spectrum of the synthesized CNDs, (b) high-resolution C 1s XPS spectra, (c) high-resolution O 1s XPS spectra, and (d) high-resolution N 1s XPS spectra. | ||
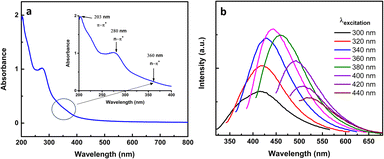
To investigate the optical characteristics of the synthesized CNDs, UV-Vis and photoluminescence spectral data were recorded. Under daylight, the synthesized CNDs are light yellow and tend to be transparent, but under UV light their color was blue. The UV-Vis spectrum exhibits strong absorption in the UV range with a tail extending to the visible region, as shown in Fig. 4a. The spectrum shows an absorption peak at 203 nm (shoulder) that is attributed to π–π* of the aromatic sp2 hybridized carbon core;64 besides that, two peaks at 280 nm and 360 nm are shown in the inset of Fig. 4a, which are ascribed to n–π* that is related to the existence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and N
O and N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bands.65
O bands.65
The fluorescence of synthesized CNDs was measured at different excitation wavelengths from 300–440 nm as shown in Fig. 4b. The CNDs gave a maximum emission of 443 nm when excited λex equals 360 nm. When the excitation increased above 360 nm the CNDs exhibited a red shift with a decay of the fluorescence. This behavior of CND fluorescence is a well-known phenomenon,66 and the photoluminescence of CNDs could be related to surface emission traps, surface defects, and surface functional groups.67
The quantum yield of CNDs was estimated @360 nm based on quinine sulfate (QY = 55%) as a reference using eqn (1).
 | (1) |
QY is the quantum yield, I is the estimated integrated emission intensity, A is the absorption of the sample, and n is the refractive index. R refers to the reference fluorophore (quinine sulfate dissolved in 0.1 M H2SO4) of known quantum yield. n is equal to 1.33 for water.68 The quantum yield was 1.95%.
Moreover, the stability of the prepared CNDs was examined for four months in the dark, and no precipitation or aggregations were formed. Furthermore, the fluorescence at 360 nm was examined weekly and the quantum yield (% QY) was estimated to be only a drop of about 6% (from 1.95 to 1.83%).
3.2. Cytotoxicity on Hep3B and LX2 cells
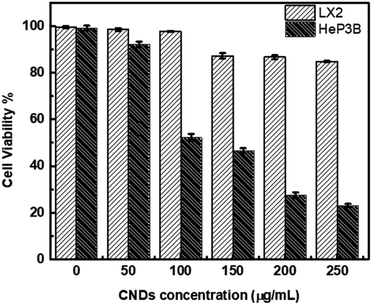
The cytotoxicity of the synthesized CNDs was tested against liver cancer cells (Hep3B) and normal liver cells (LX2) under exposure to light during the whole treatment process. The viability test was conducted to estimate the toxicity and efficiency of CNDs with selective targeting of the cancer cells.The results showed that the synthesized CNDs were active as an anticancer therapeutic agent in destroying cancer cells (Hep3B) at different concentrations, while the normal liver cells (LX2) remained unaffected with an availability of more than 86.7%. This means that these CNDs show selectivity to the liver cancer cells.
Human hepatocellular carcinoma cell line (Hep3B) and normal liver cells (LX2) were treated with CNDs at concentrations of 50–250 μg ml−1. As shown in Fig. 5 the Hep3B cell viability decreased with an increasing concentration to reach 23% at concentration 250 μg ml−1. At the same time the normal cell LX2 viability decreased slightly to reach 86.7% at the same highest concentration. The IC50 value of the CNDs on Hep3B cells was 100 μg ml−1 whereas at that concentration the LX2 cells have 97.8% cell viability. This means that our CNDs have a selective effect on the cancer cells without affecting the normal liver cells.
3.3. Cellular uptake study of the CNDs
The possible mechanism of the cellular uptake of the CNDs could be an energy-dependent or energy-independent pathway.69 The cytotoxicity of CNDs was determined after the incubation of the Hep3B cells at 4 °C or with sodium azide at 37 °C to examine the effect of temperature and ATP on the uptake process. As shown in Fig. 6, the cytotoxicity of the CNDs was reduced significantly in both cases which revealed an energy-dependent endocytosis. However, cytotoxicity is still observed which also indicates the existence of an energy-independent pathway such as passive diffusion. This behavior has also been reported previously by Zhou et al. with ultra-small carbon dots with a particle size of 1–5 nm.70 Furthermore, we have investigated the possible endocytosis process of the prepared carbon nanodots by using four different endocytosis inhibitors. Each one inhibits a specific process of the endocytosis.71 Methyl-β-cyclodextrin (MβCD) is considered an inhibitor of lipid raft-mediated endocytosis, chlorpromazine is an inhibitor of clathrin-mediated endocytosis, genistein inhibits caveolae-mediated endocytosis, and finally amiloride is an inhibitor of micropinocytosis. These compounds at the concentrations used don't have a cytotoxicity effect on the Hep3B cells. We have found that the cytotoxicity of CNDs decreased significantly in the case of MβCD and genistein as shown in Fig. 6 which revealed that the possible CND uptake is through caveolae and lipid-mediated endocytosis. According to the literature, small nanoparticles are internalized mainly through caveolae-mediated endocytosis.70,71 Moreover, Ji et al. have developed carbon nanodots from urea and citric acid and they observed that cellular uptake is energy-dependent through lipid endocytosis and micropinocytosis.47 In our study, the cellular uptake of our synthesized CNDs is energy-dependent and also by passive diffusion. Moreover, the main routes of endocytosis are lipid and caveolae-mediated endocytosis.3.4. CNDs blocked 3D MCTS spheroid formation
Over the last few years, three-dimensional (3D) cultures have become an innovative tool for cellular modelling, bridging the gap between in vitro research and animal model studies.72 The multicellular tumor spheroid model (MCTS) is currently coined as a 3D in vitro model that mimics the in vivo state of cancer tumors. The ability of cancer cells to group into spheroids in an in vitro setting indicates an enhanced tumorigenicity, resistance to chemotherapeutic drugs, and tumor-forming capacity in vivo.73,74 We studied the effect of the synthesized CNDs on Hep3B cells' capacity to form spheroids. Cells were allowed to group into spheroids under non-treated conditions (Control). Importantly, treated Hep3B cells with CNDs were not able to form spheroids. Cells remained spread in small groups/clusters over a large perimeter (Fig. 7). The occupied area and perimeter of the formed cluster under treated conditions were significantly increased (Fig. 7b and d) while its circularity decreased (Fig. 7c) indicating a disordered form. The number of small clusters was significantly increased compared to the control (Fig. 7e). Altogether, these results suggest a prohibiting effect of CNDs on Hep3B cancer spheroid formation. Previous researchers have proven the relevance of hindering spheroid formation in several types of cancer notably in liver cancer with the goal of decreasing tumorigenicity and resistance to chemotherapy.75–783.5. The possible mechanism of CND anticancer activity
The anticancer activity and selectivity could be referred to as the production of reactive oxygen species (ROS) by CNDs which have the responsibility to destroy many cell components such as proteins, DNA, and lipids resulting in cell death.79 Since the membrane of cancer cells is dilapidated and weaker than that of the normal cells,80 the CNDs select the cancer cells and kill them. The reactive oxygen species (ROS) term encompasses oxygen free radicals, such as the superoxide anion radical (O2−) and hydroxyl radical (OH−), and nonradical oxidants, such as hydrogen peroxide (H2O2) and singlet oxygen (O2−).81 Electrons (e−) and holes (h+) are produced upon excitation of the CNDs; therefore electrons are excited from the ground state (valence band) to the excitation state (conduction band). Several excited carriers are trapped on the CNDs because of the many surface flaws inhibiting e− and h+ recombination. And part of the h+ may combine with surface-adsorbed H2O to produce hydroxyl radicals, while the e− may be captured by oxygen dissolved in the solution, producing superoxide radicals.82To confirm the ability of the prepared CNDs to produce ROS, the percentage of glutathione oxidation was tested. Glutathione is a tripeptide that contains cysteine with a thiol group that can protect the cellular components from the damage that could be caused by ROS production.83 Due to its abundance in cancer cells it can be used as an indicator of the cellular oxidative stress generated by carbon nanomaterials.36,79 Herein, we utilized Ellman's assay to evaluate the percentage of glutathione activity loss (GSH%) upon incubation with the synthesized CNDs.84 As can be observed in Fig. 8, a great fraction of the glutathione is oxidized in the presence of the CNDs (88.2%) with comparable activity to the positive control (H2O2). This observation confirms the high ROS production of the oxygenated CNDs which could explain their potent anticancer activity in addition to the weakness of the cancer cell membrane that permits the higher penetration of the CNDs.
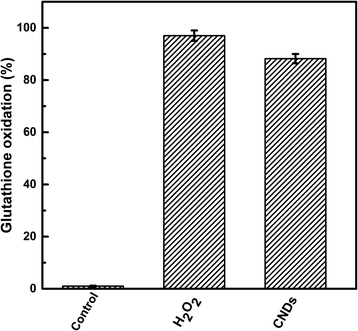 | ||
| Fig. 8 Percentage of glutathione oxidation mediated by prepared CNDs. H2O2 was used as a positive control. | ||
Interestingly, the CNDs prepared in this work show efficient inhibition toward Hep3B cancer cells at low inhibition concentrations compared to other research studies. To the best of our knowledge this is the first time that CNDs have been synthesized from sage leaves and utilized as anticancer agents. A comparison of CNDs from different resourses and in terms of treated cancer cell lines and inhibition concentrations is presented in Table 1.
| Resource | Method | Temperature (°C) | Time (h) | Size (nm) | Quantum yield (%) | Cancer cell | Inhibition concentration IC50 (μg ml−1) | Ref. |
|---|---|---|---|---|---|---|---|---|
| Walnut oil | Hydrothermal | 220 | 24 | 10–15 | 14.5 | MCF-7 | 1.25 | 85 |
| PC-3 | 5 | |||||||
| Cinnamon, black pepper, turmeric, and red chili | Hydrothermal | 200 | 12 | 3–5 | 43.6 | Kidney cells HK-2 | 2000 | 86 |
| Thumbai | Hydrothermal | 200 | 5 | 3.8 | — | Cervical cancer (HeLa) | 2000 | 87 |
| Lour | 2.9 | 2000 | ||||||
| Bamboo | Carbonization | 80 | 2 | 4 | 2.27 | Breast | 2000 | 45 |
| M. floribunda fruit | Hydrothermal | 200 | 12 | 3.5 | 18 | Human colon | 200 | 88 |
| Pineapple | Hydrothermal | 120 | 3 | 2.7 | 9.63 | Colon adenocarcinoma | 2000 | 45 |
| Ocimum sanctum | Hydrothermal | 180 | 4 | 5 | 9.3 | Breast | 1000 | 89 |
| Green tea | Carbonization | 300 | 2 | 2.5–4 | 4.3 | MCF-7 & MDA-MB-231 | 360 | 37 |
| Glucose | Acid-assisted ultrasonic | 40 | 4 | 10 | 6.8 | Lung cells (H157) | 5 | 90 |
| Ginger juice | Hydrothermal | 300 | 2 | 3.5–5 | 13.4 | Hepatocellular carcinoma cell | 350 | 68 |
| Waste onion | Hydrothermal | 120 | 2 | 7–25 | 28 | Hela cell | 1000 | 91 |
| Mint | Hydrothermal | 200 | 4 | — | 0.54 | Breast cells | 1200 | 38 |
| Sage leaves | Hydrothermal | 200 | 6 | 1–9 | 1.95 | HeP3B | 100 | This work |
4. Conclusions
In this work, CNDs have been successfully synthesized from fresh sage leaves by a simple green hydrothermal method. Simply put, CNDs have been synthesized under mild conditions of 200 °C for 6 hours, with no addition of any chemicals and external surface passivation agent and/or further modification. CNDs have an average size of 3.6 nm; they are highly water soluble, photoluminescent, and stable with a quantum yield equal to 1.95%. Our results clearly show that CNDs have an important role in inhibiting the growth of liver cancer cells (HeP3B). The synthesized CNDs showed impressive anticancer activity at various concentrations, and the cell activity was shown to decrease with increasing concentrations from 92% at 50 μg ml−1 to 23% at 250 μg ml−1. The normal cells (LX2) are not affected which means that the CNDs are selective for the cancer cells. The proposed anticancer mechanism is through the formation of reactive oxygen species. Interestingly, the synthesized CNDs were capable of inhibiting 3D spheroid formation indicating CNDs' potential in reducing tumorigenicity.Author contributions
Shadi Sawalha: conceptualization, investigation, validation, visualization, supervision, and writing – original draft. Mohyeddin Assali: conceptualization, investigation, formal analysis, and writing – review & editing. Samer Abdallah: methodology, investigation, and writing – review & editing. Amal Barham, Hala Badawi, and Zeina Barham: methodology, investigation, and writing – original draft. Ahmad Ghareeb: investigation and methodology. Giuseppe Misia, Silvia Collavini, Alessandro Silvestri, and Maurizio Prato: investigation, methodology, and visualization.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge Mr Ameed Amireh from the Department of Chemistry for help in FTIR and fluorescence analysis.Notes and references
- T. Boobalan, M. Sethupathi, N. Sengottuvelan, P. Kumar, P. Balaji, B. Gulyás, P. Padmanabhan, S. T. Selvan and A. Arun, ACS Appl. Nano Mater., 2020, 3, 5910–5919 CrossRef CAS.
- M. Arruebo, N. Vilaboa, B. Sáez-Gutierrez, J. Lambea, A. Tres, M. Valladares and Á. González-Fernández, Cancers, 2011, 3, 3279–3330 CrossRef CAS.
- D. T. Debela, S. G. Y. Muzazu, K. D. Heraro, M. T. Ndalama, B. W. Mesele, D. C. Haile, S. K. Kitui and T. Manyazewal, SAGE Open Med., 2021, 9 DOI:10.1177/20503121211034366.
- M. F. Attia, N. Anton, J. Wallyn, Z. Omran and T. F. Vandamme, J. Pharm. Pharmacol., 2019, 71, 1185–1198 CrossRef CAS.
- M. Assali, N. Kittana, S. A. Qasem, R. Adas, D. Saleh, A. Arar and O. Zohud, RSC Adv., 2019, 9, 1055–1061 RSC.
- M. Assali, A. N. Zaid, N. Kittana, D. Hamad and J. Amer, Nanotechnology, 2018, 29, 245101 CrossRef.
- M. Assali, N. Kittana, S. Dayyeh and N. Khiar, Nanotechnology, 2021, 32, 205101 CrossRef CAS.
- M. Assali, N. Kittana, I. Badran and S. Omari, RSC Adv., 2023, 13, 7000–7008 RSC.
- Q. Jia, Z. Zhao, K. Liang, F. Nan, Y. Li, J. Wang, J. Ge and P. Wang, Mater. Chem. Front., 2020, 4, 449–471 RSC.
- C.-L. Shen, H.-R. Liu, Q. Lou, F. Wang, K.-K. Liu, L. Dong and C.-X. Shan, Theranostics, 2022, 12, 2860–2893 CrossRef CAS.
- G. Nocito, G. Calabrese, S. Forte, S. Petralia, C. Puglisi, M. Campolo, E. Esposito and S. Conoci, Cancers, 2021, 13, 1991 CrossRef CAS.
- M.-H. Chan, B.-G. Chen, L. T. Ngo, W.-T. Huang, C.-H. Li, R.-S. Liu and M. Hsiao, Pharmaceutics, 2021, 13, 1874 CrossRef CAS.
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS.
- N. Papaioannou, A. Marinovic, N. Yoshizawa, A. E. Goode, M. Fay, A. Khlobystov, M.-M. Titirici and A. Sapelkin, Sci. Rep., 2018, 8, 6559 CrossRef.
- S. Anwar, H. Ding, M. Xu, X. Hu, Z. Li, J. Wang, L. Liu, L. Jiang, D. Wang, C. Dong, M. Yan, Q. Wang and H. Bi, ACS Appl. Bio Mater., 2019, 2, 2317–2338 CrossRef CAS.
- D. Tan, Y. Yamada, S. Zhou, Y. Shimotsuma, K. Miura and J. Qiu, Carbon, 2014, 69, 638–640 CrossRef.
- A. Szabó, C. Perri, A. Csató, G. Giordano, D. Vuono and J. B. Nagy, Materials, 2010, 3, 3092–3140 CrossRef.
- S. Sawalha, M. Assali, A. Nasasrah, M. Salman, M. Nasasrah, M. Jitan, H. S. Hilal and A. Zyuod, RSC Adv., 2022, 12, 4490–4500 RSC.
- S. Sawalha, R. Hamed and M. Assali, ChemistrySelect, 2023, 8, e202300522 CrossRef CAS.
- R. Hamed, S. Sawalha, M. Assali, R. A. Shqair, A. Al-Qadi, A. Hussein, R. Alkowni and S. Jodeh, Surf. Interfaces, 2023, 38, 102760 CrossRef CAS.
- Indriyati, M. M. Munir, M. Nasir and F. Iskandar, IOP Conf. Ser.: Earth Environ. Sci., 2022, 1017, 012009 CrossRef.
- M. Behi, L. Gholami, S. Naficy, S. Palomba and F. Dehghani, Nanoscale Adv., 2022, 4, 353–376 RSC.
- V. S. Sivasankarapillai, A. Vishnu Kirthi, M. Akksadha, S. Indu, U. Dhiviya Dharshini, J. Pushpamalar and L. Karthik, Nanoscale Adv., 2020, 2, 1760–1773 RSC.
- N. K. Quang, N. N. Hieu, V. V. Q. Bao, V. T. Phuoc, L. X. D. Ngoc, L. Q. Doc, N. M. Tri, L. V. T. Son, L. V. T. Son and C. T. C. Ha, New Carbon Mater., 2022, 37, 595–602 CrossRef CAS.
- A. V. Longo, A. Sciortino, M. Cannas and F. Messina, Phys. Chem. Chem. Phys., 2020, 22, 13398–13407 RSC.
- M. Aslan and H. Eskalen, Fullerenes, Nanotubes Carbon Nanostruct., 2021, 29, 1026–1033 CrossRef CAS.
- D. S. Chauhan, M. A. Quraishi and C. Verma, Carbon Lett., 2022, 32, 1603–1629 CrossRef.
- J. Lan, C. Liu, M. Gao and C. Huang, Talanta, 2015, 144, 93–97 CrossRef CAS.
- T. Ghosh, R. Ghosh, U. Basak, S. Majumdar, R. Ball, D. Mandal, A. K. Nandi and D. P. Chatterjee, J. Mater. Chem. A, 2018, 6, 6476–6492 RSC.
- J. Schneider, C. J. Reckmeier, Y. Xiong, M. von Seckendorff, A. S. Susha, P. Kasák and A. L. Rogach, J. Phys. Chem. C, 2017, 121, 2014–2022 CrossRef CAS.
- X. Dong, Y. Su, H. Geng, Z. Li, C. Yang, X. Li and Y. Zhang, J. Mater. Chem. C, 2014, 2, 7477–7481 RSC.
- B. De and N. Karak, RSC Adv., 2013, 3, 8286–8290 RSC.
- Y. Liu, Y. Zhao and Y. Zhang, Sens. Actuators, B, 2014, 196, 647–652 CrossRef CAS.
- M. Xue, Z. Zhan, M. Zou, L. Zhang and S. Zhao, New J. Chem., 2016, 40, 1698–1703 RSC.
- Isnaeni, I. Rahmawati, R. Intan and M. Zakaria, J. Phys.: Conf. Ser., 2018, 985, 012004 CrossRef.
- S. Sawalha, M. Assali, M. Raddad, T. Ghneem, T. Sawalhi, M. Almasri, A. Zarour, G. Misia, M. Prato and A. Silvestri, ACS Appl. Bio Mater., 2022, 5, 4860–4872 CrossRef CAS.
- P.-C. Hsu, P.-C. Chen, C.-M. Ou, H.-Y. Chang and H.-T. Chang, J. Mater. Chem. B, 2013, 1, 1774–1781 RSC.
- S. Shahid, S. Mohiyuddin and G. Packirisamy, J. Nanosci. Nanotechnol., 2020, 20, 6305–6316 CrossRef CAS.
- B. Chen, F. Li, S. Li, W. Weng, H. Guo, T. Guo, X. Zhang, Y. Chen, T. Huang, X. Hong, S. You, Y. Lin, K. Zeng and S. Chen, Nanoscale, 2013, 5, 1967–1971 RSC.
- L. Xiao and H. Sun, Nanoscale Horiz., 2018, 3, 565–597 RSC.
- X. Zhang, X. He, Y. Li, Z. Zhang, Y. Ma, F. Li and J. Liu, J. Nanosci. Nanotechnol., 2013, 13, 5254–5259 CrossRef CAS.
- P. Jayasekhar Babu, S. Saranya, Y. D. Singh, M. Venkataswamy, A. M. Raichur and M. Doble, Opt. Mater., 2021, 117, 111120 CrossRef CAS.
- A. A. Kokorina, E. S. Prikhozhdenko, N. V. Tarakina, A. V. Sapelkin, G. B. Sukhorukov and I. Y. Goryacheva, Carbon, 2018, 127, 541–547 CrossRef CAS.
- M. Righetto, F. Carraro, A. Privitera, G. Marafon, A. Moretto and C. Ferrante, J. Phys. Chem. C, 2020, 124, 22314–22320 CrossRef CAS.
- Y.-Y. Chen, W.-P. Jiang, H.-L. Chen, H.-C. Huang, G.-J. Huang, H.-M. Chiang, C.-C. Chang, C.-L. Huang and T.-Y. Juang, RSC Adv., 2021, 11, 16661–16674 RSC.
- C.-L. Li, C.-M. Ou, C.-C. Huang, W.-C. Wu, Y.-P. Chen, T.-E. Lin, L.-C. Ho, C.-W. Wang, C.-C. Shih, H.-C. Zhou, Y.-C. Lee, W.-F. Tzeng, T.-J. Chiou, S.-T. Chu, J. Cang and H.-T. Chang, J. Mater. Chem. B, 2014, 2, 4564–4571 RSC.
- Z. Ji, Z. Yin, Z. Jia and J. Wei, Langmuir, 2020, 36, 8632–8640 CrossRef CAS.
- D. Vercauteren, R. E. Vandenbroucke, A. T. Jones, J. Rejman, J. Demeester, S. C. De Smedt, N. N. Sanders and K. Braeckmans, Mol. Ther., 2010, 18, 561–569 CrossRef CAS.
- G. Privitera, T. Luca, S. Castorina, R. Passanisi, G. Ruberto and E. Napoli, Asian Pac. J. Trop. Biomed., 2019, 9, 24–28 CrossRef CAS.
- S. Sahu, B. Behera, T. K. Maiti and S. Mohapatra, Chem. Commun., 2012, 48, 8835–8837 RSC.
- B. R. Al-Hashimi, K. M. Omer, H. S. Rahman and H. H. Othman, Spectrochim. Acta, Part A, 2021, 244, 118835 CrossRef CAS.
- P. Das, M. Bose, S. Ganguly, S. Mondal, A. K. Das, S. Banerjee and N. C. Das, Nanotechnology, 2017, 28, 195501 CrossRef.
- K. Radhakrishnan, P. Panneerselvam and M. Marieeswaran, Anal. Methods, 2019, 11, 490–506 RSC.
- Y. Xu, C.-J. Tang, H. Huang, C.-Q. Sun, Y.-K. Zhang, Q.-F. Ye and A.-J. Wang, Chin. J. Anal. Chem., 2014, 42, 1252–1258 CAS.
- P. Roy, P.-C. Chen, A. P. Periasamy, Y.-N. Chen and H.-T. Chang, Mater. Today, 2015, 18, 447–458 CrossRef CAS.
- Y. Ding, F. Zhang, J. Xu, Y. Miao, Y. Yang, X. Liu and B. Xu, RSC Adv., 2017, 7, 28754–28762 RSC.
- H. Li, S. Han, B. Lyu, T. Hong, S. Zhi, L. Xu, F. Xue, L. Sai, J. Yang, X. Wang and B. He, Chin. Chem. Lett., 2021, 32, 2887–2892 CrossRef CAS.
- C. Dhanush and M. G. Sethuraman, Spectrochim. Acta, Part A, 2021, 260, 119920 CrossRef CAS.
- K. K. Karali, L. Sygellou and C. D. Stalikas, Talanta, 2018, 189, 480–488 CrossRef CAS.
- H. Sutanto, I. Alkian, N. Romanda, I. W. L. Lewa, I. Marhaendrajaya and P. Triadyaksa, AIP Adv., 2020, 10, 055008 CrossRef CAS.
- H. Shabbir, T. Tokarski, D. Ungor and M. Wojnicki, Materials, 2021, 14, 7604 CrossRef CAS.
- M. Amjad, M. Iqbal, A. Faisal, A. M. Junjua, I. Hussain, S. Z. Hussain, H. A. Ghramh, K. A. Khan and H. A. Janjua, Nanoscale Adv., 2019, 1, 2924–2936 RSC.
- Y. Liu, Y. Zhao and Y. Zhang, Sens. Actuators, B, 2014, 196, 647–652 CrossRef CAS.
- K. M. Omer, K. H. Hama Aziz and S. J. Mohammed, New J. Chem., 2019, 43, 12979–12986 RSC.
- T. G. Chatzimitakos, A. I. Kasouni, A. N. Troganis and C. D. Stalikas, ACS Appl. Mater. Interfaces, 2018, 10, 16024–16032 CrossRef CAS.
- A. Sharma, T. Gadly, A. Gupta, A. Ballal, S. K. Ghosh and M. Kumbhakar, J. Phys. Chem. Lett., 2016, 7, 3695–3702 CrossRef CAS.
- H. Ding, X.-H. Li, X.-B. Chen, J.-S. Wei, X.-B. Li and H.-M. Xiong, J. Appl. Phys., 2020, 127, 231101 CrossRef CAS.
- R.-J. Fan, Q. Sun, L. Zhang, Y. Zhang and A.-H. Lu, Carbon, 2014, 71, 87–93 CrossRef CAS.
- P. Foroozandeh and A. A. Aziz, Nanoscale Res. Lett., 2018, 13, 339 CrossRef.
- N. Zhou, S. Zhu, S. Maharjan, Z. Hao, Y. Song, X. Zhao, Y. Jiang, B. Yang and L. Lu, RSC Adv., 2014, 4, 62086–62095 CAS.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- A. G. Mitrakas, A. Tsolou, S. Didaskalou, L. Karkaletsou, C. Efstathiou, E. Eftalitsidis, K. Marmanis and M. Koffa, Int. J. Mol. Sci., 2023, 24, 6949 CrossRef CAS.
- R. B. Mokhtari, B. Qorri, M. Sambi, N. Baluch, S. Kumar, B. Das, M. R. Szewczuk, H. Yeger and H.-L. M. Cheng, Cancers, 2021, 13, 2784 CrossRef CAS.
- B. Pinto, A. C. Henriques, P. M. A. Silva and H. Bousbaa, Pharmaceutics, 2020, 12, 1186 CrossRef CAS.
- H. Lu, L. Noorani, Y. Jiang, A. W. Du and M. H. Stenzel, J. Mater. Chem. B, 2017, 5, 9591–9599 RSC.
- A. Wang, L. A. Madden and V. N. Paunov, J. Mater. Chem. B, 2020, 8, 10487–10501 RSC.
- D. Melnik, C. Sahana, N. Kopp, I. Wehland, D. Grimm and M. Krüger, Cells, 2020, 9, 367 CrossRef CAS.
- M. Hawash, M. T. Qaoud, N. Jaradat, S. Abdallah, S. Issa, N. Adnan, M. Hoshya, S. Sobuh and Z. Hawash, Biomimetics, 2022, 7, 247 CrossRef CAS.
- M. Assali, M. Almasri, N. Kittana and D. Alsouqi, ACS Biomater. Sci. Eng., 2019, 6, 112–121 CrossRef.
- C. Alibert, B. Goud and J.-B. Manneville, Biol. Cell, 2017, 109, 167–189 CrossRef PubMed.
- M. Schieber and N. S. Chandel, Curr. Biol., 2014, 24, R453–R462 CrossRef CAS PubMed.
- S. Prasad, P. Shanmugam, K. Bhuvaneswari, G. Palanisamy, T. Pazhanivel, T. Arunkumar, M. S. AlSalhi and M. J. Aljaafreh, Crystals, 2020, 10, 300 CrossRef CAS.
- A. Pompella, A. Visvikis, A. Paolicchi, V. D. Tata and A. F. Casini, Biochem. Pharmacol., 2003, 66, 1499–1503 CrossRef CAS PubMed.
- G. L. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–77 CrossRef CAS PubMed.
- E. Arkan, A. Barati, M. Rahmanpanah, L. Hosseinzadeh, S. Moradi and M. Hajialyani, Adv. Pharm. Bull., 2018, 8, 149–155 CrossRef CAS PubMed.
- N. Vasimalai, V. Vilas-Boas, J. Gallo, M. d. F. Cerqueira, M. Menéndez-Miranda, J. M. Costa-Fernández, L. Diéguez, B. Espiña and M. T. Fernández-Argüelles, Beilstein J. Nanotechnol., 2018, 9, 530–544 CrossRef CAS PubMed.
- P. T. Varsha Raveendran and N. K. Renuka, Mater. Chem. Phys., 2022, 288, 126236 CrossRef CAS.
- R. Atchudan, T. N. J. I. Edison, S. Perumal, N. Muthuchamy and Y. R. Lee, J. Photochem. Photobiol., A, 2020, 390, 112336 CrossRef CAS.
- A. Kumar, A. R. Chowdhuri, D. Laha, T. K. Mahto, P. Karmakar and S. K. Sahu, Sens. Actuators, B, 2017, 242, 679–686 CrossRef CAS.
- M. Ajmal, U. Yunus, A. Matin and N. U. Haq, J. Photochem. Photobiol., B, 2015, 153, 111–120 CrossRef CAS PubMed.
- R. Bandi, B. R. Gangapuram, R. Dadigala, R. Eslavath, S. S. Singh and V. Guttena, RSC Adv., 2016, 6, 28633–28639 RSC.
| This journal is © The Royal Society of Chemistry 2023 |