Precise control of CNT-DNA assembled nanomotor using oppositely charged dual nanopores†
Chaofan
Ma
 ab,
Wei
Xu
ab,
Wei
Xu
 ab,
Wei
Liu
ab,
Changhui
Xu
ab,
Wei
Si
ab,
Wei
Liu
ab,
Changhui
Xu
ab,
Wei
Si
 *ab and
Jingjie
Sha
*ab
*ab and
Jingjie
Sha
*ab
aSchool of Mechanical Engineering, Southeast University, Nanjing, China. E-mail: major212@seu.edu.cn
bJiangsu Key Laboratory for Design and Manufacture of Micro-nano Biomedical Instruments, Southeast University, Nanjing, China
First published on 5th June 2023
Abstract
Inspired by nature, nanomotors have been developed that have great potential in microfluidics and biomedical applications. The development of the rotary nanomotor, which is an important type of nanomotor, is an essential step towards intelligent nanomachines and nanorobots. Carbon nanotubes (CNTs) are a crucial component of rotary nanomotors because of their excellent mechanical properties and adaptability to the human body. Herein, we introduce a convenient manipulation method for controlling the rotation of a nanomotor assembled from CNT-DNA, which uses the electroosmosis effect within oppositely charged dual nanopores. The central components of this nanomotor consist of a double-walled carbon nanotube (DWCNT) and a circular single-stranded DNA (ssDNA), which acts as the driving element for the nanomotor. Selective ion transport through charged nanopores can generate a robust electroosmotic flow (EOF), which serves as the primary power for the movement of circular ssDNA. The tangential force on the ssDNA is transmitted via electrostatic adsorption to the outer surface of the CNT, known as the rotor, resulting in the rotation of the nanomotor. By simply adjusting the electric field and surface charge density of each nanopore, rotational variables including speed, output power and torque can be readily regulated in this work. This proof-of-concept research provides a promising foundation for the future development of the precise control of nanomotors.
Introduction
Disease diagnosis and treatment are increasingly trending toward molecular or nanoscale operations for the precise excision of pathogenic structures and targeted drug delivery.1–3 In the past few decades, researchers have been dreaming of designing micro/nanodevices that can be placed and actuated in the human body to help diagnose and treat diseases.4,5 As modern science and technology is developing continuously, the technology of nanomotors is also developing rapidly in the field of disease prevention and treatment.6–9 The first successful preparation of molecular motors capable of sustained directional rotation in the presence of light was made by Feringa et al.10 in 1999 and was awarded the Nobel Prize in Chemistry in 2016. The development direction of nanomotors tends to be miniaturized and intelligent. They can carry out complex and accurate diagnosis and treatment operations on the premise of adapting to the human body. Therefore, promoting the design and manufacture of nanomotor systems has great application prospects for strengthening the diagnosis and treatment of various diseases.The drive method and motion control are critical issues in nanomotor design. For rotary nanomotors, chemical reactions, magnetism, ultrasound, light, heat and electricity can actuate and manipulate their rotation. With respect to chemically driven nanomotors, in which chemical energy is converted into rotational motion around single bonds, the concept is categorized into molecular switches, rotors and motors, where these comprise a rotating unit (rotor) and a stationary part (stator) that are interconnected by an axle. In some cases the rotational speed can also be regulated by external chemical stimuli.11 For magnetically driven nanomotors, which contain certain magnetic materials (such as Fe, Ni, etc.), the drive energy is the magnetic field gradient or magnetic field torque under the action of an external magnetic field.12 Nanomotors propelled by ultrasound,13 which are typically metallic rods that are a few micro-meters in length and a few hundred nanometers in diameter, exhibit several modes of motion. If photoactive materials are introduced into nanomotors, the absorption of light energy could lead to catalytic luminescence reaction, photoisomerization or photothermal conversion, which would generate an asymmetric field and realize the autonomous light-driven rotation of nanomotors.14 A carbon nanotube (CNT) nanomotor can be driven by temperature gradients.15–18 The CNT heats up when it is energized, and the electrodes at both ends can dissipate the heat so that the temperature gradient formed achieves the rotation of the nanomotor. The direction of rotation is independent of the current and is determined only by the temperature gradient. Alternatively, a CNT suspended in water is subjected to a rotating electric field of appropriate size and angular velocity, which can be rotated by means of the dipole direction of the water in MD simulations.19–21 In addition to the control methods mentioned above, theoretically, some atoms with inward radial deviation (IRD) are placed at the edge of the stator. The IRD atoms will repel the nanotubes in their thermal vibration-induced collisions and can drive the nanotubes to rotate when the non-zero moment around the axis of rotational symmetry of the ring is induced by the repulsion due to the IRD and the friction with the stator.22–25
It is undeniable that the driving methods of nanomotors described above have their unique characteristics and perform well in some specific tasks. In order to accelerate the development of micro/nanomotor technology, manipulation accuracy should be the focus. Researchers have made many improvements in the drive method. The rotational speed of the rotor can be specified by adjusting the temperature and the radial deviation of one of the end carbon atoms on the stator.26 For instance, a double-walled nanotube composite, obtained by placing black phosphate-based nanotubes (BPNTs) inside CNTs, functions with high structural stability when used as a rotator at temperatures of 250 K and above.27 A motor consists of thick carbon nanotubes horizontally sandwiched between two parallel gold substrates,28 subject to an antisymmetric thermal gradient, and the applied thermal field provides well-controlled speed and direction of motion of the motor. In order to measure the rotation of nanomotors, a probe test method has been proposed.29–31 Since the CNT probe is on the trajectory of the end tube that rotates with the rotor, it will collide with the end tube and sharp fluctuations that indicate the deflection of the probe tip can be observed and recorded. A rotational drive system made of co-axial multi-walled nanotubes has the function of regulating the input rotation of the nanomotor.32 A nanoscale rotating system consisting of a graphene substrate and defect-effect-driven triple-walled nanotubes performed well.33 Rotation of the nanotubes was found to be driven by the defects placed on the graphene with a stable rotation frequency. In another case, boron nitride carbon nanotubes were used as the brake tube.34 By increasing the depth of the brake tube inserted into the rotor, not only can the rotor speed be reduced, but also the rotor rotation direction can be reversed. Dipole symmetry of the induced charges on DWNTs is necessary for their application as nanomotors. Among the ways to obtain such systems is chemical or end-functionalization.35 For example, the symmetric charge distribution of a bare SWNT was observed to be disturbed after adding NH2 in the vicinity of the SWNT. A net positive and negative charge was observed to be induced on the opposite sides of the nanotube circumference, which is imperative for the application of nanomotors. It was found that the rotation speed could be adjusted by controlling the ratio of edge hydroxyl groups.36 The proportion of hydroxyl groups is positively correlated with the velocity, and the mechanism is that the strong hydrogen bonds formed between the interfaces increase the interfacial interactions. An increase of the hydroxyl group concentration causes more hydrogen bonding, which strengthens the interconnection and thus increases the rotation speed.
In addition to improving existing nanomotors to address these issues, another direct approach is to explore and innovate nanomotors with new structures and mechanisms. The rotary nanomotor mainly provides torque output or achieves curved displacement and can be used as an energy source for nanorobots or directly used for material delivery. Carbon nanotubes (CNTs) are important materials for rotating nanomotors because of their excellent mechanical properties and adaptability to organisms. DNA molecules, being flexible chain-like bodies, can act as chains or belts in macroscopic machinery to transmit power. It has already been reported that DNA has been directly used as a rotor for nanomotors. A 450 nm long six-helix DNA bundle is docked onto an ∼50 nm diameter nanopore, where an electro- or diffusiophoretic force locally bends and deforms the bundle, whereupon an osmotically induced water flow causes it to rotate.37,38 The DNA rotor arm is supported by a base made using DNA origami technology, and the rotation of the DNA rotor arm is driven by applying square wave AC currents on both sides to produce energy modulation of the fixed axis.39 In addition, through MD simulations, the electric field can directly drive the DNA backbone to rotate around the axis (phosphorus atoms of dsDNA).40 Nanopore electroosmotic flow can provide a powerful driving force and enable the regulation of speed and direction of molecular transport. Nanopore sensing technology developed based on the Coulter counter stands out among many single-molecule detection methods because of its rapid, low-cost and label-free advantages.41–49 A nanoscale film is used to separate the electrolyte solution, and a nanopore in the film connects the two sides of the solution. The Ag/AgCl electrode is connected to the electrolyte solution at both sides of the nanopore, and the ions electromigrate through the nanopore to form ionic currents under an applied bias voltage. When a biomolecule passes through a nanopore, it blocks the ion transport due to its volume effect inside the nanopore, generating a current blocking signal. Statistical analysis of parameters such as amplitude and duration of blocking signals can yield a wealth of biological information. Nanopore sensing has been widely used in biomolecule detection,50 DNA sequencing,51 metal ion detection,52etc. In the presence of an applied electric field, biomolecules enter the nanopore driven by electrophoretic53 or electroosmotic forces,54 and information about the biomolecules is obtained from the collected ionic current signal. In contrast to the directional determinism of the electroosmotic force, the direction of the electroosmotic flow is influenced by the surface charge property of nanopores,55 and the amplitude and direction of the electroosmotic flow can be controlled by adjusting the surface charge. Therefore, nanopore electroosmotic flow as a driving force can achieve precise control of the rotational behaviour of nanomotors.
Herein, having established that circular ssDNA can rotate directionally in a dual nanopore system, as shown in Fig. S1,† we designed a convenient manipulation method to precisely control the movement of a nanomotor assembled from CNT-DNA, which uses the electroosmosis effect within oppositely charged dual nanopores. By using the controlled strategies of applied electric field and surface charge density (σ) of each nanopore, the precise manipulations of nanomotors were then realized in this work.
Results and discussion
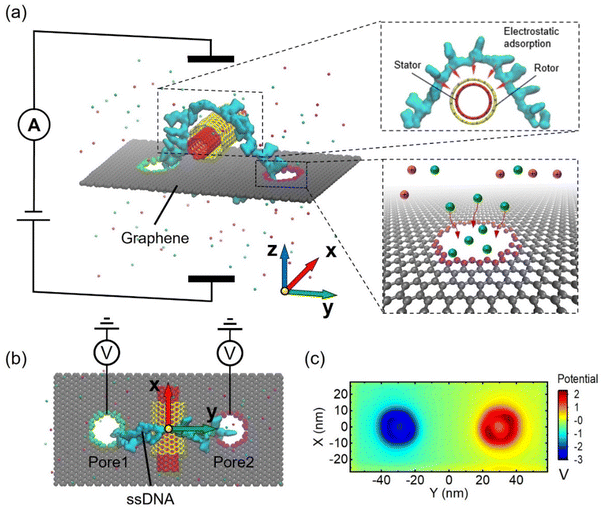
An example of the system setup for precise control of the nanomotor is illustrated in Fig. 1a and b, which show the side view and the top view of the system. The nanomotor is composed of two parts. One is a double-walled carbon nanotube, which is the power output component of the nanomotor, and the other is a circular single-stranded DNA, which is the power transfer component of the nanomotor. The inner and outer walls of the DWCNT serve as the stator and rotor of the nanomotor, respectively. The ssDNA is a circular shape by bonding the bases of the head and the tail of the strand, which is passed through pore1, then wrapped around the positively charged rotor from the outside, and then passed through pore2. The ssDNA is driven by the dual nanopore system and transmits rotational motion to the DWCNT, which can transform an irregular rotation into a regular circular rotation. The green and purple atoms of the nanopores were selected to tune their charges so as to manipulate the nanomotor. Selective ion transport through charged nanopores can induce cooperation and competition between electroosmosis and electrophoresis. The rotation of the circular ssDNA is driven by the electroosmotic effect in oppositely charged dual nanopores. There is an electrostatic adsorption between the ssDNA and the rotor, which forms a tangential force driving the nanomotor rotation.Specifically, as shown in Fig. 1a, nanopore 1 is negatively charged, while nanopore 2 is positively charged. An electric field is applied in the +Z direction, which generates a cation-dominated electroosmotic flow along the +Z direction in nanopore 1, and the friction between water molecules and ssDNA bases drives the bases near the nanopore along the +Z direction. Similarly, nanopore 2 generates an anion-dominated electroosmotic flow along the −Z direction, which drives the bases near the nanopore along the −Z direction. Thus, the two nanopores act together to drive the clockwise rotation of the circular ssDNA. Then tangential forces can transfer the clockwise rotation to the rotor. Fig. 1c shows the 2D electrostatic potential distribution of the graphene membrane. A significant potential enhancement and decrease is found around the positively and negatively charged nanopores, respectively.
Since the rotational behaviour of the CNT nanomotor is driven by the circular ssDNA in the two nanopores, the diameter of the nanopore is limited to 1.3 nm, which can accommodate only one ssDNA base at a time. However, it should be noted that techniques and strategies are needed to reproduce the fabrication of the 1.3 nm nanopore, as this currently remains a challenge. A pre-designed control strategy for the applied electric field and a tunable surface charge density for both nanopores was then used to achieve a rotational and precise control of the assembled CNT-DNA nanomotor, as discussed in detail below.
According to the system setup and the electrostatic potential distribution depicted in Fig. 1, we first investigated the possibility of the assembled CNT-DNA nanomotor being driven by the oppositely charged dual nanopores. The simulation details are described in the Experimental section. We set the surface charge density of both nanopores to 0.075 e Å−2, with the negatively charged pore 1 (green) and the positively charged pore 2 (purple). A voltage of 16 V was applied in the +Z direction.
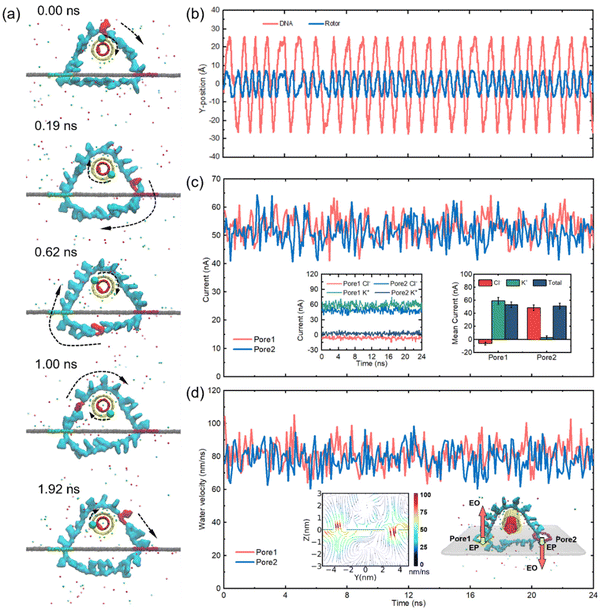
As shown in Fig. 2a, we have marked a base on the ssDNA and a carbon atom on the rotor in red and green, respectively, to clearly observe the rotational behaviour of the nanomotor at a certain time. The traces of the marked elements of the rotor and the ssDNA moving in the y-direction are plotted as shown in Fig. 2b. It is obvious that the displacement curve of the nanomotor is not that smooth. This phenomenon is a result of the competition among the electrophoretic force, electroosmotic force, van der Waals force, thermal force, etc. We found that a periodic rotation of the rotor and the ssDNA can be achieved in almost constant proportion, which proves that the electrostatic attraction between the rotor and the circular ssDNA enables relative slide-free transmission. During the operation of the nanomotor, the total ion current of both nanopores fluctuates as a function of time, as shown in Fig. 2c. This fluctuation is the result of a combination of irregular thermal motions of ions in solution and transport of the ssDNA through nanopores. In order to better compare the magnitude of the current, and the contribution of anions and cations to the electroosmotic flow, as shown in Fig. 2c, we take the +Z direction as the positive direction of the current in pore 1, and the −Z direction as the positive direction of the current in pore 2. The amplitude of the ion current in the two pores is almost the same, with an average value of about 52 pA. The insets in Fig. 2c show that the ionic currents generated by Cl− and K+ in the two nanopores depend on time (left panel) as well as on the distribution of Cl− and K+ contributions to the ionic currents (right panel). A K+-dominated ionic current is formed in pore 1 and Cl− moves in the opposite direction to K+; meanwhile, the ionic current in pore 2 is dominated by Cl− and K+ and Cl− move in the same direction. The Stokes friction between ions and water molecules leads to a robust EOF. From Fig. 2d and the inserted left-hand panel, it can be seen that the EOF velocities of pore 1 and pore 2 are basically the same, with the average value remaining at ∼80 nm ns−1. The EOF direction of pore 1 is positive along the Z-axis, the EOF direction of pore 2 is opposite to that of pore 1 and the maximum flux are both concentrated at the orifice. Since the ssDNA in this paper is in a circular shape, the total charge of dA20 is about −20e and the charge of each base is about −1e after being linked by a phosphodiester bond. Taking an electric field strength of 0.2 V Å−1 as shown in Fig. 2 as an example, the electrophoretic force on a single base is about 0.32 nN, which is much smaller than the driving force of electroosmotic flow of 1.24 nN (see the ESI Fig. S2† for the estimation method of the driving force of the electroosmotic flow). Thus, regardless of the direction of the electrophoretic force, there is no significant effect on the electroosmotic flow-dominated base transport. The embedded right panel qualitatively depicts the electroosmotic and electrophoretic forces exerted on the ssDNA in the two nanopores. ssDNA bases are negatively charged, so the electrophoretic force is in the −Z direction, resulting in a resistance force to the nanomotor in pore 1 and a driving force in pore 2.
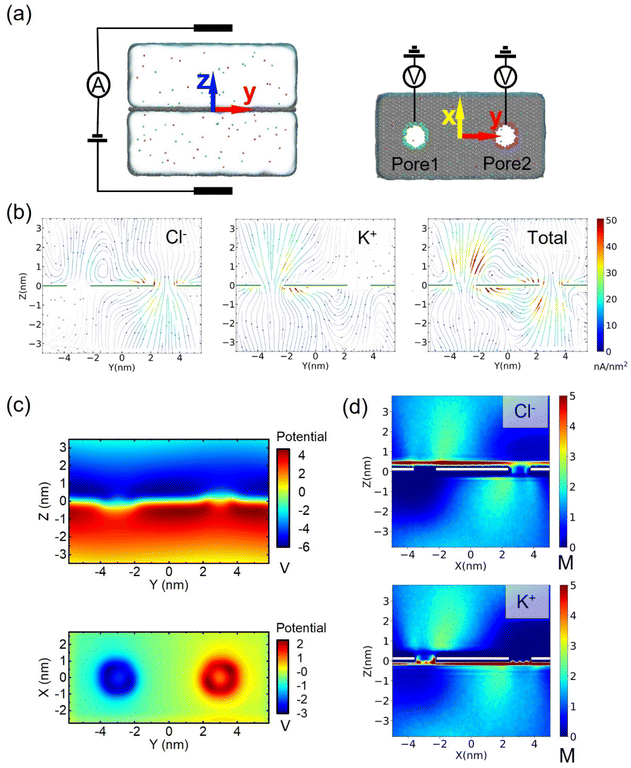
We found that water flows in the same direction as counterions in nanopores, suggesting that the main driving force of the nanomotor may be the EOF. To consolidate this judgment, we further simulated the selective transport of ions in the pristine charged nanopore, as shown in Fig. 3a, and established a bare dual nanopore system without an assembled CNT-DNA nanomotor to investigate the factors, including ionic current flux distribution (Fig. 3b), 2D potential distribution (Fig. 3c) and density maps of ion concentrations (Fig. 3d). When an external electrical field with an amplitude of 16 V was applied to the system, the ions started to move. More chloride ions were observed to translocate through the positively charged nanopores with σ (surface charge density) of 0.075 e Å−2, while few chloride ions could enter the negatively charged nanopores with σ of −0.075 e Å−2; the steady-state local density of the ionic current induced by Cl− is shown in Fig. 3b. The case of K+ is opposite to that of Cl−, i.e., a robust flux of counter ions in the nanopore compared to positive ions, which is practically due to selective ion transport in the charged nanopore. These charged nanopores will change the potential distribution in the system, as shown in Fig. 3c, so that the positively charged nanopores attract anions and negatively charged nanopores attract cations. Therefore, the surface charge of the nanopore also redistributes the ions around the nanopore, and despite the enrichment of ions on both sides of the graphene membrane due to the high electric field, the ion concentration of counter ions within the nanopore is significantly higher than that of positive ions, as shown in Fig. 3d. Thus, the mystery of the mechanism of strong electroosmosis generation is unraveled. This is because the high selectivity of ions passing through charged nanopores leads to a redistribution of electric potential, ion concentration and flux. Further simulations are shown in Fig. S13–S19, and the ESI† also confirms the above interesting findings.
In order to realize precise control of the assembled CNT- DNA nanomotor, a series of simulations were presented from the perspectives of applied electric field, tunable nanopore surface charge density and rotor radius. The primary driving force of the nanomotor manipulation method proposed in this paper is the EOF, so the electric field and the nanopore surface charge density are the chief parameters for precisely controlling the nanomotor speed. We have studied the voltages of 14 V, 15 V, 16 V, 17 V and 18 V, when the nanopore surface charge density is 0.075 e Å−2 and the corresponding electric field strengths are 0.175 V Å−1, 0.1875 V Å−1, 0.2 V Å−1, 0.2125 V Å−1 and 0.225 V Å−1, as shown in Fig. 4, where the case of 16 V is shown in Fig. 2, and more information is presented in Fig. S3–S6.†Fig. 4a shows the variation of the y-direction position of the marked elements of the ssDNA and the nanomotor rotor under different electric fields as a function of time. As the voltage rises, both the ssDNA and the rotor rotational frequency increase, and a constant ratio drive is almost maintained. Fig. 4b shows the plots of the ion current fluctuations with time for the two nanopores corresponding to Fig. 4a. The ion currents of pore 1 and pore 2 are essentially equal, and the ion current increases linearly with increasing voltage. Fig. 4c–e summarize the operating parameters of the nanomotors for different electric fields. Fig. 4c shows the rotational speed and transfer ratio of the ssDNA and the rotor. The rotational speed of ssDNA increased from 0.24 r per ns to 1.53 r per ns; the rotor rotational speed increased linearly from 0.37 r per ns to 2.37 r per ns. The transfer ratio is defined as the average speed ratio of the ssDNA and the nanomotor rotor. Due to the electrostatic adsorption force between the rotor and the ssDNA, the transfer ratio varies slightly at different voltages. Fig. 4c and d show the output power and torque of the nanomotor, respectively. The force analysis and calculation method of the nanomotor are illustrated in Fig. S2.† With rising voltage, the output power increases linearly from 2.6 nW to 42.6 nW, while the output torque increases almost linearly from 1.48 nN nm to 2.95 nN nm. According to the calculated output power and torque, the assembled CNT-DNA nanomotor can be applied to a number of mechanical parts that require energy input.
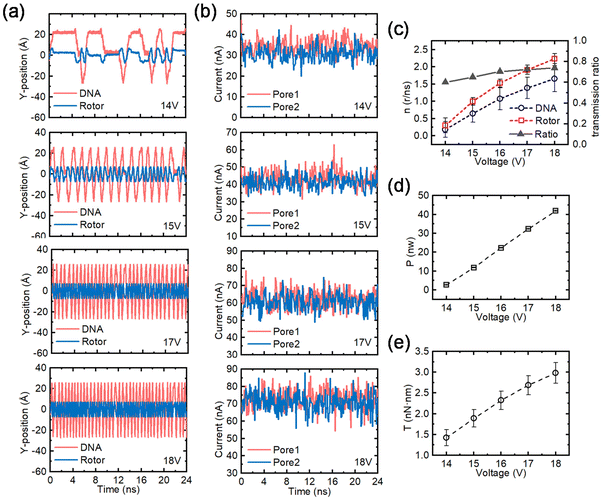 | ||
| Fig. 4 Effects of the applied electric field on the performance of the nanomotor. (a) The y-coordinates of the marked residue on the ssDNA and the C atom on the rotor under different electric fields as a function of time. From top to bottom the applied voltages are 14 V, 15 V, 17 V and 18 V. (b) The fluctuation of the currents in pore 1 and pore 2 as a function of time for the voltages corresponding to panel (a). (c) The effect of voltage on rotational speed and transmission ratio of the nanomotor. The red dashed lines and open squares, and the blue dashed lines and open circles represent the rotational speeds of the rotor and the ssDNA, respectively; the black solid line and solid triangles represent the transmission ratio. (d) and (e) The effects of the applied voltage on the output power (P) and the torque (T) of the nanomotor, respectively. The case of an applied voltage of 16 V can be obtained from Fig. 2. All error bars in the graph represent the standard deviation. | ||
In addition, the surface charge density of the nanopores can also be adjusted appropriately. We simulated four nanopore surface charge densities with amplitudes of 0.049 e Å−2, 0.075 e Å−2, 0.102 e Å−2 and 0.127 e Å−2 at a fixed voltage of 16 V, as shown in Fig. 5 (0.075 e Å−2 is shown in Fig. 2 and more data are shown in Fig. S7–S9†). Fig. 5a illustrates the variation of the y-directional position of the marked elements of the ssDNA and the nanomotor rotor with time for different nanopore surface charge densities. The rotational frequencies of both the ssDNA and the rotor increase and then decrease when the nanopore surface charge density increases, with a peak at 0.075 e Å−2. Fig. 5b shows the plots of ion current fluctuations with time for the two nanopores corresponding to Fig. 5a. The ionic current increases and then decreases as the surface charge density increases, which is positively correlated with the rotation frequency. However, it is interesting to note that simulations of the bare dual nanopore system at the same surface charge density show the phenomenon that nanopores with higher surface charge density produce higher ionic currents (see Fig. S17–S19).† The comparison reveals that the ionic currents of pore 1 and pore 2 are essentially equal when the nanopore surface charge density is, respectively, 0.049 e Å−2 and 0.075 e Å−2, while the ionic currents of pore 1 are larger than those of pore 2 at 0.102 e Å−2 and 0.127 e Å−2. This may be attributed to the higher density of positive surface charges generating a high surface potential, leading to electrostatic adsorption on ssDNA bases, creating a resistance that slows down the rotation of the ssDNA. Furthermore, the prolonged residence of negatively charged bases in pore 2 can repel Cl−, which leads to a weakening of the ionic current intensity in pore 2. Fig. 5c–e summarize the operating parameters of the nanomotors for different surface charge densities. Fig. 5c exhibits the rotational speed and transfer ratio of the ssDNA and the rotor. The maximum value of the ssDNA speed is 1.01 r per ns when the surface charge density is 0.075 e Å−2; the rotor also reaches a maximum value of 1.52 r per ns in this case. The transfer ratio has only a small variation and is basically stable at ∼0.67. Fig. 5c and d show the output power and the torque of the nanomotor, respectively. The rotational speed of the ssDNA and the rotor first increases and then decreases. The output power and the torque also increase first and then decrease. The maximum output power is 22.5 nW and the maximum output torque is 2.34 nN nm in this study. Therefore, a single increase in surface charge density does not always enhance the speed of the nanomotor, but should be set within a reasonable range. With the above-mentioned data reference on voltage and nanopore surface charge density, convenient and efficient operation, and precise control of the assembled CNT-DNA nanomotor can be achieved.
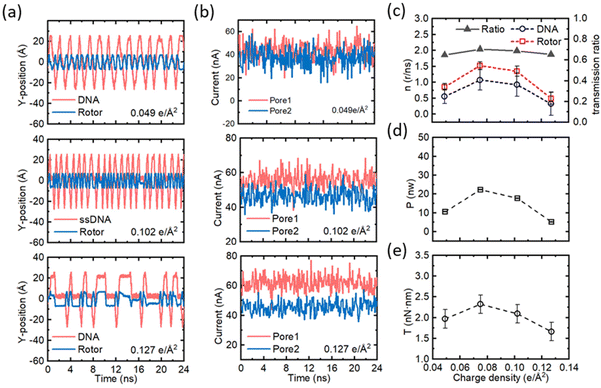 | ||
| Fig. 5 Effects of the surface charge density on the performance of the nanomotor. (a) The y-coordinates of the marked residue on the ssDNA and the C atom on the rotor under different surface charge densities as a function of time. From top to bottom the surface charge densities are 0.049 e Å−2, 0.102 e Å−2 and 0.127 e Å−2. (b) The fluctuation of the currents in pore 1 and pore 2 as a function of time for the surface charge densities corresponding to panel (a). (c) The effect of surface charge density on rotational speed and transmission ratio of the nanomotor. The red dashed lines and open squares, and the blue dashed lines and open circles represent the rotational speeds of the rotor and the ssDNA, respectively; the black solid line and solid triangles represent the transmission ratio. (d) and (e) The effects of surface charge density on the output power (P) and torque (T) of the nanomotor, respectively. The case of the surface charge density of 0.075 e Å−2 can be obtained from Fig. 2. All error bars in the graph represent the standard deviation. | ||
Furthermore, to explore the operating performance of the circular ssDNA drive, we investigated the manipulation of the nanomotor at different transfer ratios by altering the size of the CNT nanomotor. When the applied voltage of the system is 16 V and the surface charge density of both nanopores is 0.075 e Å−2, rotors with radii of 6.079 Å, 6.755 Å, 7.430 Å and 8.106 Å, respectively, are simulated. The stator radii corresponding to the rotor are 4.728 Å, 5.404 Å, 6.079 Å and 6.755 Å, respectively. (See Fig. 6, where 6.079 Å is shown in Fig. 2 and more data are shown in Fig. S10–S12†.) Fig. 6a shows the positional changes in the y-direction of the marked elements of the ssDNA and the rotor versus time for different rotor radii. The rotational frequency of both the ssDNA and the rotor decreases when the rotor radius increases. Fig. 6b shows the plots of the ion current distribution for the two nanopores corresponding to Fig. 6a. The ion current remains essentially unchanged with increasing rotor radius, which indicates that although the electroosmotic intensity does not change, the rotational speed of the ssDNA decreases, which may be due to the effect of the tension of the ssDNA on the rotational speed caused by the increase in rotor size. Similarly, the reduction of the rotational speed causes a slight reduction of the ionic current in pore 2. Fig. 6c–e summarize the operating conditions of the nanomotor at different rotor radii, where Fig. 6c shows the speed and transfer ratio of the ssDNA and the rotor. The speed of both the rotor and the ssDNA decreased nonlinearly with decreasing slope, from 1.04 r per ns to 0.35 r per ns for the ssDNA and from 1.58 r per ns to 0.45 r per ns for the rotor. The ssDNA-to-rotor transfer ratio increased linearly from 0.70 to 0.81, which was essentially consistent with the increase in rotor radius. Fig. 6c and d show the output power and torque of the nanomotor, respectively. As the voltage increases, the output power decreases nonlinearly from 22.5 nW to 6.1 nW, and the small rotor can provide a larger power output, while the torque output is basically stable at 2.35 nN nm, which is because the decrease in nanomotor speed is offset by the increase in rotor radius. The increased rotor radius will not keep the original constant ratio constant, and causes a relative sliding between the ssDNA and the rotor. Enhancing the positive electrical properties of the rotor could strengthen the electrostatic adsorption of ssDNA to the CNT nanomotor and, thus, may counteract the relative slipping.
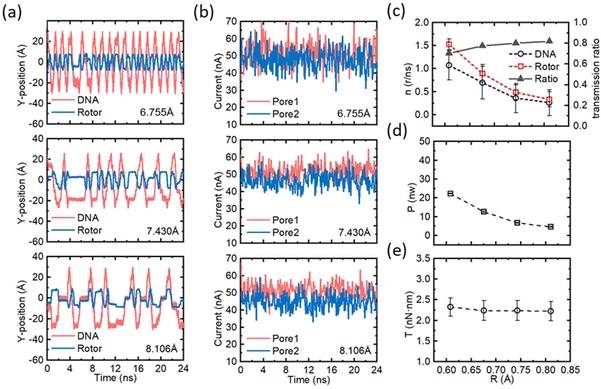 | ||
| Fig. 6 Effects of the rotor radius on the performance of the nanomotor. (a) The y-coordinates of the marked residue on the ssDNA and the C atom on the rotor under different rotor radii as a function of time. From top to bottom the rotor radii are 6.755 Å, 7.430 Å and 8.106 e Å−2. (b) The fluctuation of the currents in pore 1 and pore 2 as a function of time for the rotor radius corresponding to panel (a). (c) The effect of rotor radius on rotational speed and transmission ratio of the nanomotor. The red dashed lines and open squares, and the blue dashed lines and open circles represent the rotational speeds of the rotor and the ssDNA, respectively; the black solid line and solid triangles represent the transmission ratio. (d) and (e) The effects of rotor radius on the output power (P) and torque (T) of the nanomotor, respectively. The case of the rotor radius of 6.095 Å can be obtained from Fig. 2. All error bars in the graph represent the standard deviation. | ||
Conclusions
In this study, based on MD simulations, an assembled CNT-DNA nanomotor and its controlling method are proposed to achieve simplified operation and precise control. A DWCNT is the core of the nanomotor and a circular ssDNA is the driving component. The strong electroosmotic flow in dual nanopores created by selective ion transport is the main driving force of the nanomotor. The rotational speed, output power and torque of the nanomotor can be obtained by adjusting the electric field strength and the surface charge density to achieve effective kinetic input to the mechanical parts. The rotational behaviour of the nanomotor with different rotor radii was further investigated, and the relative sliding between the ssDNA and the rotor occurred as the rotor radius increased and a preliminary solution was proposed.The control method of the assembled CNT-DNA nanomotor using oppositely charged dual nanopores is a major innovation in this paper. Since the designed nanomotor is effective and controllable, it is expected to contribute to significant progress in the fields of disease detection, drug delivery, nano-surgery and surgical assistance. Based on the circular ssDNA drive, a series of nano-transfer machines can be formed, which have great prospects in nanomachinery. In conclusion, an experimental setup for manipulating assembled CNT-DNA nanomotors using a dual nanopore system is possible in the near future, although some challenges still need to be addressed, and this is a possible concern for future researchers. We hope that the results of this study will provide some insightful contributions to the design and driving of nanomotors for their wide application in the future.
Experimental section
General MD methods
In this paper, the program NAMD256 was used to perform all the simulations with periodic boundary conditions along x, y, and z directions. The time step is 1 fs in all simulations. The nanomotor was composed of a double-walled carbon nanotube (DWCNT) and a single-stranded DNA (ssDNA, dA20) molecule. The first nucleotide at the 3′-terminal of each ssDNA was bound to the end nucleotide at the 5′-terminal using a phosphodiester bond with a force constant of 10 kcal (mol Å2)−1 to form a circular shape. The ssDNA, graphene membrane, DWCNT, TIP3P water and ions were described by the CHARMM36 force field57 with CUFIX corrections.58 The entire system is solvated by water, and potassium ions and chloride ions were added to neutralize the system, where the ion concentrations for the simulation systems were all 2 M. VMD59 and Tcl scripts were used to view and analyze the trajectories of all the simulated systems.MD simulation of the rotation of circular ssDNA driven by two charged nanopores in a single-layer graphene membrane
The graphene membrane is ≈11.8 nm long and ≈5.6 nm wide. Two 1.3 nm nanopores are located on the graphene membrane separated by a distance of 5.6 nm. Using VMD's Solvate plugin, the system was solvated by adding water molecules to the system. Water molecules overlapping the graphene membrane were removed. Then, certain amounts of potassium ions and chloride ions were added into the system to bring the concentration to 2 M and also to make the system electrically neutral. The whole system consists of approximately 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms. Firstly, the conjugate gradient method was used to perform energy minimization for 3600 steps. Then, the system was equilibrated for ≈10 ns in the NPT ensemble with constant atom number N, pressure P and temperature T. During the simulation, the DNA and the graphene membrane were restrained at their initial positions with a spring constant of 0.1 kcal (mol Å2)−1 so as to avoid the interaction between them. A Langevin thermostat was applied to all atoms of the system to make sure that the temperature of the system would be stable at around 295 K. The production simulation under an electric field was performed in the NVT ensemble with a constant volume, while in the simulation ssDNA was driven by the two charged nanopores. The electric field with amplitude of ≈0.18 V Å−1 was set and the surface charge densities for pore 1 and pore 2 were −0.075 and 0.075 e Å−2 (see ESI Fig. 1).† Circular ssDNA can be driven to rotate at a steady speed by the EOF using oppositely charged dual nanopores. Ionic currents through the nanopores were calculated using the formulas below:
000 atoms. Firstly, the conjugate gradient method was used to perform energy minimization for 3600 steps. Then, the system was equilibrated for ≈10 ns in the NPT ensemble with constant atom number N, pressure P and temperature T. During the simulation, the DNA and the graphene membrane were restrained at their initial positions with a spring constant of 0.1 kcal (mol Å2)−1 so as to avoid the interaction between them. A Langevin thermostat was applied to all atoms of the system to make sure that the temperature of the system would be stable at around 295 K. The production simulation under an electric field was performed in the NVT ensemble with a constant volume, while in the simulation ssDNA was driven by the two charged nanopores. The electric field with amplitude of ≈0.18 V Å−1 was set and the surface charge densities for pore 1 and pore 2 were −0.075 and 0.075 e Å−2 (see ESI Fig. 1).† Circular ssDNA can be driven to rotate at a steady speed by the EOF using oppositely charged dual nanopores. Ionic currents through the nanopores were calculated using the formulas below: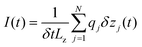 | (1) |
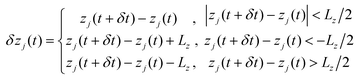 | (2) |
MD simulation of the rotation of an assembled CNT-DNA nanomotor driven by two charged nanopores in a single-layer graphene membrane
The DWCNT was located 18 nm above the center of the graphene membrane (see Fig. 3a). The inner tube of the DWCNT (stator) is 4 nm in length and is the stator of the motor. The outer tube (rotor) is 2.5 nm in length, and each atom has 2e positive charges to eliminate the relative slippage of the ssDNA and the rotor. A fixed constraint is applied to the stator and an SMD force is applied to the rotor to offset the displacement in the x-direction. Other simulation details about the system setup were similar to the MD simulation of the rotation of the circular ssDNA driven by two charged nanopores described above (see Fig. 3).MD simulation of selective ion transport through two bare charged nanopores without a nanomotor
Differing from the systems introduced above, the nanomotor was removed from the systems to investigate the selective ion transport dynamics through bare double nanopores. As shown in Fig. 3, the nanopores coloured in green and purple have surface charge densities of −0.075 e Å−2 and +0.075 e Å−2, respectively; after the minimization of 3600 steps and equilibration for ≈5 ns, the system was further simulated under an electric field of ≈0.18 V Å−1. The whole system was divided into 0.4 nm × 0.4 nm × 0.4 nm grids to calculate the ion concentration and ion flux distributions. The local current in each grid element was calculated using the formulas described in previous works. The Streamplot function in the Python matplotlib library was used to generate flux plots.Statistical analysis
All the data were sampled at 2.4 ps intervals, they were analysed from trajectories of MD simulations, and the ionic current was also block-averaged in 0.24 ns blocks. For each condition, one to five replica simulations were performed. The statistical analysis was based on VMD software and the corresponding Tcl scripts, and all data are expressed as mean ± standard error.Author contributions
Chaofan Ma carried out the MD simulation work, performed data analysis, conceptualized the research, prepared figures, and wrote and edited the manuscript. Chaofan Ma, Wei Xu, Wei Liu and Changhui Xu wrote and edited the manuscript. Wei Si and Jingjie Sha edited the manuscript.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The authors thank the National Natural Science Foundation of China (52075099 and 52035003) for financial support, and the Big Data Computing Center of Southeast University for computing resources.References
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. D. P. Rodriguez-Torres, L. S. Acosta-Torres, L. A. Diaz-Torres, R. Grillo, M. K. Swamy, S. Sharma, S. Habtemariam and H.-S. Shin, J. Nanobiotechnol., 2018, 16, 71 CrossRef PubMed.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Acc. Chem. Res., 2019, 52, 1750–1760 CrossRef CAS PubMed.
- J. Zhou, L. Rao, G. Yu, T. R. Cook, X. Chen and F. Huang, Chem. Soc. Rev., 2021, 50, 2839–2891 RSC.
- W. Si, M. Yu, G. Wu, C. Chen, J. Sha, Y. Zhang and Y. Chen, ACS Nano, 2020, 14, 15349–15360 CrossRef CAS PubMed.
- W. Si, Z. Zhu, G. Wu, Y. Zhang, Y. Chen and J. Sha, Small Methods, 2022, 6, 2200318 CrossRef CAS PubMed.
- I. Ortiz-Rivera, M. Mathesh and D. A. Wilson, Acc. Chem. Res., 2018, 51, 1891–1900 CrossRef CAS PubMed.
- W. Wang and C. Zhou, Adv. Healthcare Mater., 2021, 10, 2001236 CrossRef CAS PubMed.
- J. Dan, S. Shi, H. Sun, Z. Su, Y. Liang, J. Wang and W. Zhang, Crit. Rev. Food Sci. Nutr., 2022, 1–21 CrossRef PubMed.
- H. Choi, J. Yi, S. H. Cho and S. K. Hahn, Biomaterials, 2021, 279, 121201 CrossRef CAS PubMed.
- N. Koumura, R. W. J. Zijlstra, R. A. van Delden, N. Harada and B. L. Feringa, Nature, 1999, 401, 152–155 CrossRef CAS PubMed.
- A. Mondal, R. Toyoda, R. Costil and B. L. Feringa, Angew. Chem., Int. Ed. DOI:10.1002/anie.202206631.
- W. Xi, A. A. Solovev, A. N. Ananth, D. H. Gracias, S. Sanchez and O. G. Schmidt, Nanoscale, 2013, 5, 1294–1297 RSC.
- A. L. Balk, L. O. Mair, P. P. Mathai, P. N. Patrone, W. Wang, S. Ahmed, T. E. Mallouk, J. A. Liddle and S. M. Stavis, ACS Nano, 2014, 8, 8300–8309 CrossRef CAS PubMed.
- D. Wang, J. Jiang, B. Hao, M. Li, Z. Chen, H. Zhang, X. Wang and B. Dong, Appl. Mater. Today, 2022, 29, 101652 CrossRef.
- J. Chen, Y. Gao, C. Wang, R. Zhang, H. Zhao and H. Fang, J. Phys. Chem. C, 2015, 119, 17362–17368 CrossRef CAS.
- A. M. Fennimore, T. D. Yuzvinsky, W.-Q. Han, M. S. Fuhrer, J. Cumings and A. Zettl, Nature, 2003, 424, 408–410 CrossRef CAS PubMed.
- J. Shi, K. Cai and Q.-H. Qin, Nanotechnology, 2016, 27, 495704 CrossRef PubMed.
- G. Cuba-Supanta, H. N. Fernández-Escamilla, J. Guerrero-Sanchez, J. Rojas-Tapia and N. Takeuchi, Nanoscale, 2020, 12, 18313–18321 RSC.
- Z. Fu, D. Liang, S. Jiang, P. Zhao, K. Han and Z. Xu, AIP Adv., 2019, 9, 115008 CrossRef.
- M. M. Rahman, M. M. Chowdhury and M. K. Alam, Small, 2017, 13, 1603978 CrossRef PubMed.
- Z. Fu, D. Liang, S. Jiang, P. Zhao, K. Han and Z. Xu, J. Phys. Chem. C, 2019, 123, 30649–30656 CrossRef CAS.
- Z.-Y. Guo, Q.-W. Hou and B.-Y. Cao, J. Heat Transfer, 2012, 134, 051010 CrossRef.
- K. Cai, P. Wu, J. Shi, Z. Zhong and Y. Zhang, Int. J. Mech. Sci., 2022, 225, 107372 CrossRef.
- H. Duan, J. Shi, K. Cai and Q.-H. Qin, Int. J. Mol. Sci., 2018, 19, 3513 CrossRef PubMed.
- J. Shi, Z. Wang and Z. Chen, Comput. Mater. Sci., 2016, 120, 94–98 CrossRef CAS.
- K. Cai, J. Wan, Q. H. Qin and J. Shi, Nanotechnology, 2016, 27, 055706 CrossRef PubMed.
- J. Shi, H. Cai, K. Cai and Q.-H. Qin, J. Phys. D: Appl. Phys., 2017, 50, 025304 CrossRef.
- F. Zhu, Z. Guo and T. Chang, Appl. Mater. Today, 2020, 18, 100520 CrossRef.
- K. Cai, J. Yu, L. Liu, J. Shi and Q. H. Qin, Phys. Chem. Chem. Phys., 2016, 18, 22478–22486 RSC.
- K. Cai, J. Yu, J. Shi and Q. H. Qin, Sci. Rep., 2016, 6, 27338 CrossRef CAS PubMed.
- K. Cai, J. Yu, J. Wan, H. Yin, J. Shi and Q. H. Qin, Carbon, 2016, 101, 168–176 CrossRef CAS.
- X.-N. Zhang, K. Cai, J. Shi and Q.-H. Qin, Nanotechnology, 2018, 29, 045706 CrossRef PubMed.
- X. Lin and Q. Han, Mol. Simul., 2020, 46, 356–361 CrossRef CAS.
- H. Jin, H. Duan and J. Shi, Comput. Mater. Sci., 2019, 159, 327–332 CrossRef CAS.
- S. Negi, V. K. Bhartiya and S. Chaturvedi, Indian J. Phys., 2018, 92, 479–485 CrossRef CAS.
- B. Zhang, R. Li and Q. Peng, Nanomaterials, 2022, 12, 3363 CrossRef CAS PubMed.
- X. Shi, A.-K. Pumm, C. Maffeo, F. Kohler, W. Zhao, A. Aksimentiev, H. Dietz and C. Dekker, arXiv, 2022, DOI:10.48550/arXiv.2206.06612.
- X. Shi, A.-K. Pumm, J. Isensee, W. Zhao, D. Verschueren, A. Martin-Gonzalez, R. Golestanian, H. Dietz and C. Dekker, Nat. Phys., 2022, 18, 1105–1111 Search PubMed.
- A.-K. Pumm, W. Engelen, E. Kopperger, J. Isensee, M. Vogt, V. Kozina, M. Kube, M. N. Honemann, E. Bertosin, M. Langecker, R. Golestanian, F. C. Simmel and H. Dietz, Nature, 2022, 607, 492–498 CrossRef CAS PubMed.
- C. Maffeo, L. Quednau, J. Wilson and A. Aksimentiev, Nat. Nanotechnol., 2023, 18, 238–242 CrossRef CAS PubMed.
- S. Lu, X. Wu, M. Li, Y. Ying and Y. Long, View, 2020, 1, 20200006 CrossRef.
- Z. Hu, M. Huo, Y. Ying and Y. Long, Angew. Chem., Int. Ed., 2021, 60, 14738–14749 CrossRef CAS PubMed.
- Y.-L. Ying, Z.-L. Hu, S. Zhang, Y. Qing, A. Fragasso, G. Maglia, A. Meller, H. Bayley, C. Dekker and Y.-T. Long, Nat. Nanotechnol., 2022, 17, 1136–1146 CrossRef CAS PubMed.
- K. Zhan, Z. Li, J. Chen, Y. Hou, J. Zhang, R. Sun, Z. Bu, L. Wang, M. Wang, X. Chen and X. Hou, Nano Today, 2020, 33, 100868 CrossRef CAS.
- H. Lu, F. Giordano and Z. Ning, Genomics, Proteomics Bioinf., 2016, 14, 265–279 CrossRef PubMed.
- Y. Wang, Y. Zhao, A. Bollas, Y. Wang and K. F. Au, Nat. Biotechnol., 2021, 39, 1348–1365 CrossRef CAS PubMed.
- J. Sha, B. Xu, Y. Chen and Y. Yang, Acta Chim. Sin., 2017, 75, 1121 CrossRef CAS.
- F. Fu, Z. Zhang, Q. Sun, B. Xu and J. Sha, Acta Chim. Sin., 2019, 77, 287 CrossRef CAS.
- Y. Wang, X. Yu, Y. Liu, X. Xie, X. Cheng, S. Huang and Z. Wang, Acta Chim. Sin., 2014, 72, 378 CrossRef CAS.
- A. Nehra, S. Ahlawat and K. P. Singh, Sens. Actuators, B, 2019, 284, 595–622 CrossRef CAS.
- Y.-L. Ying, J. Zhang, R. Gao and Y.-T. Long, Angew. Chem., Int. Ed., 2013, 52, 13154–13161 CrossRef CAS PubMed.
- K. Zhan, Z. Li, J. Chen, Y. Hou, J. Zhang, R. Sun, Z. Bu, L. Wang, M. Wang, X. Chen and X. Hou, Nano Today, 2020, 33, 100868 CrossRef CAS.
- J. Saharia, Y. M. Nuwan, D. Y. Bandara, B. I. Karawdeniya, C. Hammond, G. Alexandrakis and M. J. Kim, RSC Adv., 2021, 11, 24398–24409 RSC.
- D. Pandey and S. Bhattacharyya, Appl. Math. Model., 2022, 111, 471–485 CrossRef.
- H. Niu, M.-Y. Li, Y.-L. Ying and Y.-T. Long, Chem. Sci., 2022, 13, 2456–2461 RSC.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov and A. D. Mackerell, J. Comput. Chem., 2009, 671–690 Search PubMed.
- J. Yoo and A. Aksimentiev, J. Phys. Chem. Lett., 2012, 3, 45 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr01912h |
| This journal is © The Royal Society of Chemistry 2023 |