Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The internal structure of gadolinium and perfluorocarbon-loaded polymer nanoparticles affects 19F MRI relaxation times†
Alvja
Mali
ab,
Margot
Verbeelen
c,
Paul B.
White
d,
Alexander H. J.
Staal
c,
N. Koen
van Riessen
ac,
Cyril
Cadiou
e,
Françoise
Chuburu
 e,
Olga
Koshkina
e,
Olga
Koshkina
 cf and
Mangala
Srinivas
cf and
Mangala
Srinivas
 *ag
*ag
aDepartment of Cell Biology and Immunology, Wageningen University and Research, Wageningen, The Netherlands. E-mail: mangala.srinivas@wur.nl
bDepartment of Radiology, Leiden University Medical Center, Leiden, The Netherlands
cDepartment of Tumor Immunology, Radboud Institute for Molecular Life Sciences, Radboud University Medical Center, Nijmegen, The Netherlands
dInstitute for Molecules and Materials, Radboud University, Nijmegen, The Netherlands
eUniversité de Reims Champagne Ardenne, CNRS, ICMR UMR 7312, Reims, France
fSustainable Polymer Chemistry Group, Department of Molecules and Materials, Mesa+ Institute for Nanotechnology, University of Twente, Enschede, The Netherlands
gCenya Imaging B.V., Amsterdam, The Netherlands
First published on 18th October 2023
Abstract
19F magnetic resonance imaging (19F MRI) is an emerging technique for quantitative imaging in novel therapies, such as cellular therapies and theranostic nanocarriers. Nanocarriers loaded with liquid perfluorocarbon (PFC) typically have a (single) core–shell structure with PFC in the core due to the poor miscibility of PFC with organic and inorganic solvents. Paramagnetic relaxation enhancement acts only at a distance of a few angstroms. Thus, efficient modulation of the 19F signal is possible only with fluorophilic PFC-soluble chelates. However, these chelates cannot interact with the surrounding environment and they might result in image artifacts. Conversely, chelates bound to the nanoparticle shell typically have a minimal effect on the 19F signal and a strong impact on the aqueous environment. We show that the confinement of PFC in biodegradable polymeric nanoparticles (NPs) with a multicore structure enables the modulation of longitudinal (T1) and transverse (T2) 19F relaxation, as well as proton (1H) signals, using non-fluorophilic paramagnetic chelates. We compared multicore NPs versus a conventional single core structure, where the PFC is encapsulated in the core(s) and the chelate in the surrounding polymeric matrix. This modulated relaxation also makes multicore NPs sensitive to various acidic pH environments, while preserving their stability. This effect was not observed with single core nanocapsules (NCs). Importantly, paramagnetic chelates affected both T1 and T219F relaxation in multicore NPs, but not in single core NCs. Both relaxation times of the 19F nucleus were enhanced with an increasing concentration of the paramagnetic chelate. Moreover, as the polymeric matrix remained water permeable, proton enhancement additionally was observed in MRI.
Introduction
19F Magnetic Resonance Imaging (19F MRI) is a promising noninvasive and quantitative technique which is in experimental clinical use.1–319F MRI is able to unambiguously localize and also quantify the imaging agent, without an endogenous background, combined with anatomic proton imaging.1,2,4,5 These features make 19F MRI very powerful in a broad range of applications, particularly in tracking of cellular therapies or therapeutic nanocarriers, and for the design of activatable probes.2,3,6–9 The combination of liquid perfluorocarbons (PFCs) and paramagnetic chelates in nanocarriers can provide multifunctional imaging probes. Paramagnetic chelates on the surface of nanocarriers can be used to sense the biological environment of the probe.10–12 Conversely, fluorophilic chelates that enhance the longitudinal 19F-relaxation rate increase the sensitivity of 19F MR signals.13–16 However, it remains difficult to develop nano-systems in which the paramagnetic chelate modulates both the longitudinal 19F-relaxation and the proton signal.Paramagnetic Relaxation Enhancement (PRE) acts over very short distances in a range of angstroms and decreases with the radius r6, as described by the Solomon–Bloembergen–Morgan equations.10,14,16,17 Hence, the paramagnetic chelate and the nucleus that should be modulated have to be very close to each other, in the range of angstroms.10,14,16 However, liquid PFC is both hydrophobic and lipophobic.18–20 As a result, common nanosized 19F MRI imaging agents are emulsions or single core capsules with a liquid PFC in the core that is stabilized either by a surfactant, polymer or a silica shell.1,2,4,5,21,22 The core sizes are usually at least several 10s of nanometers.1,2,4,5 As a result of the single core structure, currently, there are two strategies for the modification with paramagnetic chelates: (1) either covalent attachment or encapsulation of the hydrophilic or hydrophobic chelates in the organic shell of the agent,12,23,24 and (2) encapsulation of fluorophilic chelates in the fluorous core.13–15,25
When a paramagnetic chelate is encapsulated or covalently bound to the shell, it typically affects only a small fraction of 19F-nuclei that are close to the shell.23,24 Thus, only modest effects on 19F longitudinal relaxation rates can be achieved;23,24 moreover, the signal can become inhomogeneous due to different relaxation rates of 19F nuclei, depending on their distance from the chelate.25 On the other hand, this approach enables the modulation of protons in the surrounding medium and detection of other ions.12 Conversely, the fluorophilic chelates are soluble in the fluorous core; thus, the distance between 19F nuclei and paramagnetic ions decreases, altering the longitudinal relaxation properties of 19F nuclei.13–15,25 Depending on the ion, this strategy can be used to increase the imaging sensitivity of PFC-emulsions.13,14 However, in such emulsions the paramagnetic nuclei are isolated from the aqueous environment, making simultaneous modulation of the proton signal impossible.
Our group has introduced polymeric PFC-loaded NPs that display a structure different from conventional single core systems, namely the multicore structure (Fig. 1a).26,27 These NPs contain multiple small cores of perfluoro-15-crown-5 ether (PFCE) with a radius of 10–12 nm that are encapsulated in a matrix of biocompatible and biodegradable poly(lactic-co-glycolic acid) (PLGA) (Fig. 1a and b).26 This structure is advantageous for in vivo and clinical use, as it results in about 15-fold faster clearance of PFCE compared to the PFCE emulsions and it generates stable ultrasound.28–30 Conversely, emulsions and other single core systems can display organ accumulation times up to several months.28,29 These NPs can be produced at clinical grade and received approval for a clinical trial.26,30–33 Thus, combining these systems with paramagnetic probes could open up new opportunities in the development of probes for combined imaging, biosensing and delivery of therapeutics.
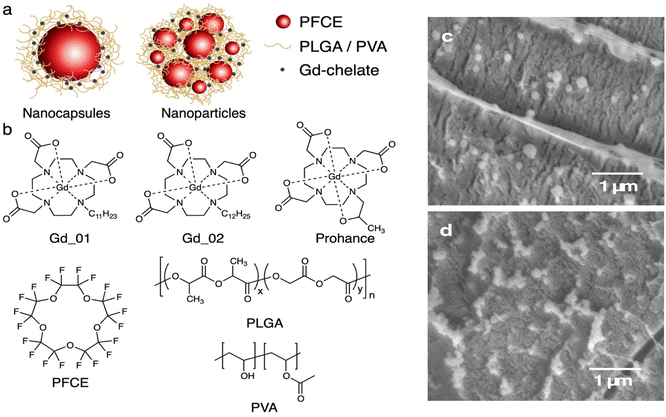 | ||
| Fig. 1 (a) Schematic of nanoparticles used in this study. Left: single core NCs, middle: multicore PFCE-PLGA NPs. (b) Chemical structures of components of NPs. Top row lipophilic chelates Gd_01 and Gd_02, and hydrophilic gadoteridol (Prohance). Bottom row: PFCE, PLGA and poly(vinyl alcohol) (PVA). (c and d) Cryogenic scanning electron microscopy (cryoSEM) micrographs of NPs (c) and NCs (d). See also Fig. S1 and S2† for larger images. Scale bar 1 μm. | ||
Here, we explored how the formation of a multicore structure and the resulting confinement of PFCE affect the PRE. Therefore, we used lipophilic gadolinium (Gd) chelates which were encapsulated in the polymer matrix of the multicore NPs. We compared their relaxation properties with single core NCs. The changes of 19F-relaxation in core–shell systems were minor, as expected. Conversely, in multicore nanoparticles, we detected enhancement of both T1 and T2 relaxation times, depending on the concentration of encapsulated gadolinium. Furthermore, unlike conventional emulsions, the proton signal could be modulated as well. A further interesting feature shown by multicore NPs was their T1 and T2 relaxation variation when the NPs were solubilized in various acidic solutions when compared to neutral pH. This variation was not present in single core NCs. In conclusion, this approach can be adopted for the development of dual 1H/19F MR imaging agents co-loaded with PFC and paramagnetic compounds for different applications, particularly biosensing and enhanced relaxation probes.
Results and discussion
Lipophilic chelates can be efficiently encapsulated in both multicore NPs and single core NCs
To investigate how structural properties affect the MR properties of NPs co-encapsulating Gd chelates and PFCE, we compared multicore PFCE-PLGA NPs with single core NCs (Fig. 1a). As gadolinium-agents we used two different hydrophobic Gd chelates that differ only in the length of a hydrophobic linker (Fig. 1b).34 These chelates are hydrophobic but not fluorophilic, ensuring they remain within the hydrophobic PLGA matrix.Both multicore NPs and single core NCs can be produced by a miniemulsion formulation approach,26 adjusting the concentration of the lipophilic Gd chelate in the PLGA matrix. This approach varies between NCs and NPs, and this distinction has been thoroughly investigated in previous research.26 Notably, the internal structure resulting from these distinct approaches was extensively characterized and confirmed using Small-Angle Neutron Scattering (SANS).
Various amounts of the lipophilic chelates, ranging from 0 to 2.4 mg Gd, were employed. Additionally, we included a hydrophilic clinical contrast agent, gadoteridol (Fig. 1b), as a control (Prohance; 140 mg Gd). Post-synthesis, repetitive centrifugation purification steps were performed to remove any free chelate.
Co-encapsulation of the Gd chelates did not impact the NP size (Fig. 1c and d, Tables S1 and S2†). Cryogenic Scanning Electron Microscopy (cryoSEM) images (Fig. 1c and d and Fig. S1 and S2†) revealed spherical NPs with relatively smooth surfaces. Gadolinium-loaded multicore NPs displayed diameters of around 200 nm and a monomodal size distribution (PDI ∼0.1), as determined by Dynamic Light Scattering (DLS, Table S1†). Capsules were slightly smaller with diameters ranging from 150 to 180 nm (Table S2†). Both size and PDI are similar to a control without a Gd chelate, and the variation between different samples is within a typical batch-to-batch variation range.26
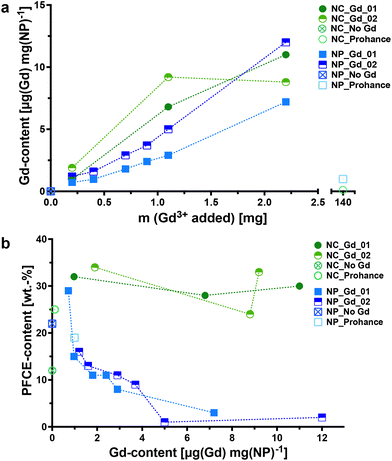
When lipophilic chelates were used for the synthesis, the concentration of encapsulated Gd increased in both multicore and single core particles with the added amount of the chelate, as shown by inductively coupled plasma-mass spectrometry (ICP-MS, Fig. 2a). The encapsulation of Gd_02, that has a longer hydrophobic chain, was slightly higher compared to that of Gd_01. In contrast, the encapsulation of hydrophilic gadoteridol was very low, despite using a nearly 50-fold higher Gd-concentration than the lipophilic chelates (Tables S3 and S4†). PLGA, being a hydrophobic polymer, is well-suited for encapsulating hydrophobic cargo like lipophilic chelates. In contrast, hydrophilic compounds like gadoteridol tend to remain in the aqueous phase during the miniemulsion process. Thus, it is possible to adjust the concentration of hydrophobic Gd chelates during the miniemulsion formulation.
Different trends were observed in the encapsulation of PFCE in multicore NPs and single-core NCs, as revealed by quantitative 19F NMR spectroscopy (Fig. 2b; indicated as NP_Gd_01, NP_Gd_02, NP_No Gd, and NP_Prohance). In multicore NPs, the PFCE content appeared to decrease with increasing Gd(III) concentrations, while in NCs, it was even higher compared to capsules without a Gd chelate. It is important to note that the encapsulation of paramagnetic chelates can have varying effects on the MR properties in both particle types, including line broadening, which can affect quantification (discussed further in the next section).
Finally, all NPs and NCs did not show any toxic effects, as demonstrated by a standard viability assay using the same dose as typically applied for labeling the cells for 19F MRI with multicore NPs (MTT-assay with RAW macrophages, Fig. S3†).27,30 The viability values were slightly higher compared to that of a live cell control. This behavior might originate from an increased phagocytic activity of the cells in the presence of NPs and was already observed with other cell types using PFCE-PLGA-NPs without Gd chelates.30 Hence, the encapsulation of Gd chelates did not affect the toxicity of NPs.
Confinement of PFC in multicore NPs enables the modulation of fluorine-19 and proton MR signals
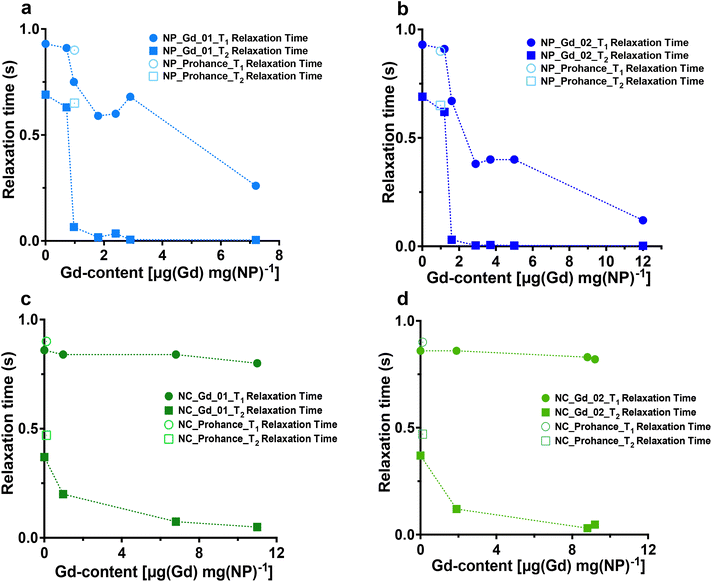
After synthesizing Gd-loaded NPs and NCs, we assessed their impact on 19F nucleus MR properties by measuring 19F relaxation times using NMR spectroscopy. The relaxation times without Gd chelates were consistent with prior studies.35 We then examined the effects of Gd encapsulation on longitudinal and transverse relaxation times (T1 and T2), with the results presented in Fig. 3 (refer to Tables S5–S8† for detailed data).NMR measurements revealed that the internal structure of NPs significantly influenced 19F nucleus relaxation properties. In multicore NPs (Fig. 3a and b), longitudinal (T1) relaxation times decreased by nearly 8-fold at the highest Gd-loading compared to Gd-free NPs. Additionally, transverse relaxation(T2) was notably affected, exhibiting approximately a 255-fold decrease. In single core NCs (Fig. 3c and d), there was a decrease in T2 relaxation time, albeit much less pronounced, approximately 2-fold, compared to multicore NPs. Furthermore, the encapsulation of Gd chelates induced a modest enhancement in T1 relaxation time in the single core capsules.
The stark decrease in transverse relaxation time observed in multicore NPs could explain the differences in PFCE quantification between multicore NPs and single core NCs. A shorter T2 leads to line broadening of the NMR signal (Fig. S4 and S5†), making it challenging to accurately integrate broad peaks within the baseline. As the concentration of encapsulated Gd increases, T2 relaxation time shortens further, broadening the PFCE peak and causing a decrease in detected PFCE concentration within multicore NPs. This effect did not occur in single core NCs.
What is the reason behind the altered relaxation properties? The effect of paramagnetic ions on the relaxation times of the 19F nucleus depends on the distance between the ion and the 19F nucleus.14,23,24 Previously, we reported the structural characterization of Gd chelate-free and gadoteridol-loaded multicore NPs and single core NCs by Small-Angle Neutron Scattering (SANS).26 Note that the radii obtained with SANS are typically smaller than the hydrodynamic sizes from DLS, as reported previously.26 In multicore NPs, each PFCE core typically displays a radius of 9–12 nm and a shell thickness of 4 nm, with some batch-to-batch variation.26 The single core capsules usually display a core radius of around 20 nm that is surrounded by a shell of a similar thickness.26
The distance between Gd and PFCE is shorter in multicore NPs compared to that in NCs, if we assume that Gd chelates are distributed homogenously within the PLGA matrix. Thus, the farthest possible distance between a PFCE molecule that is located in the center of the core and the Gd chelate in the outer part of the shell is approximately 15 nm. The distance between PFCE in the center or the core and the Gd chelate in the inner part of the shell is 9–12 nm. This distance decreases the closer a PFCE molecule is located to the shell. For the capsules, the farthest distance is 40 nm and the distance between PFCE in the center and the Gd molecule in the inner part of the shell is approximately 20 nm. Thus, this distance is almost twice as high compared to the distance in the multicore NPs. However, PRE typically decreases with the distance r6.14,16,17,10 Hence, shortening the distance alone would not be sufficient to modulate the PFCE molecules located in the center of the core. Thus, the diffusion of PFCE and its confinement in the multicore structure appear to affect the PRE.
Indeed, other reports also suggested that diffusion of PFCE can influence the PRE. Particularly, several reports showed that conjugation of Gd chelates to the surface of PFCE-loaded silica capsules results in the enhancement of transverse relaxation time.36–38 These studies did not report the PRE of the longitudinal relaxation time. Importantly, other groups did not observe PRE of longitudinal relaxation in 19F upon the incorporation of Gd in the surfactant layer of emulsions.12,23,24 Similarly, in this study we detected only a moderate effect on the longitudinal relaxation times in the control single core NCs, compared with multicore NPs. Thus, the formation of the confining multicore structure affects the enhancement of the longitudinal relaxation time. Typically, the diffusion speed of confined liquids is reduced compared to bulk.39 However, liquid PFCs display a very low intermolecular cohesiveness,40 which may invert these effects. Additionally, the distance that a PFCE molecule has to diffuse to relocate from the core center to the shell becomes lower due to the formation of the confining multicore structure. As a result, the PRE of both longitudinal and transverse relaxation times can be observed.
Furthermore, the concentration of Gd within the polymeric matrix influences the PRE. Higher Gd concentrations result in more Gd nuclei in the PLGA shell closer to the PFCE phase, leading to further decreases in both longitudinal and transverse relaxation times in multicore NPs.
Overall, our data show that the structural properties of NPs affect the paramagnetic relaxation of the 19F nucleus. The PRE effect enhances the PFC nano-emulsion imaging sensitivity and reduces the imaging time. It also enables the development of stimuli-responsive fluorinated nanoemulsions for on/off switching in response to various stimuli. This switch is achieved through the presence of paramagnetic metal ions, which significantly decrease the T2 relaxation time, leading to line broadening effects and modulation of the 19F MR signal for responsive imaging.
Finally, 1H/19F MRI demonstrated that the presence of paramagnetic chelates affected both the fluorine (Fig. 4a) and proton signals in multicore particles (Fig. 4b). As the concentration of Gd increased, the 19F signal intensity decreased due to the reduction in T2 relaxation time (Fig. 4a and c). Conversely, the proton signal increased with higher chelate encapsulation (Fig. 4c). The signal-to-noise-ratio of the proton signal increased from 17 to 49, when the Gd content increased from 0 to 2.4 μg per mg NPs. In multicore particles, Gd is encapsulated within the PLGA matrix, which retains hydration and permeability to water and small molecules.26 This property allows for the modulation of the proton signal.
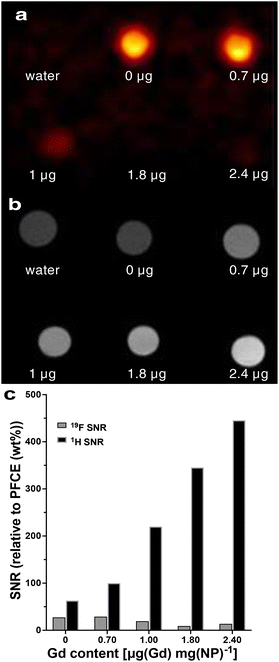 | ||
| Fig. 4 (a) 19F and (b) 1H MRI of multicore NPs that contain a different amount of Gd chelates. The Gd content in μg per mg of NPs is indicated under each sample. Water and NPs loaded only with PFCE were used as controls. (c) The signal-to-noise ratio of 19F and 1H MRI signals for different Gd contents. (See Table S9† for the signal-to-noise values). | ||
The encapsulation of paramagnetic chelates in multicore NPs allows for the modulation of both T1 and T2 relaxation times of the 19F nucleus, along with the proton signals from the aqueous environment of the NPs. In emulsions, it is typically possible to efficiently modulate either the proton or the fluorine relaxation, but not both simultaneously. When paramagnetic chelates are attached to the shell, they primarily alter 1H relaxation, with a minor effect on the PRE of 19F nuclei in the fluorous phase.23
In contrast, the fluorophilic chelates are dispersed in the PFC phase, shortening the 19F relaxation times, but remain inaccessible to water.14 Hence, only the PRE of the 19F-signal is obtained. In contrast to emulsions, co-encapsulation of PFC and Gd chelates in multicore NPs enables modulation of both 19F and 1H signals. Here we used gadolinium(III) chelates, which are known to cause a strong PRE of transverse relaxation. In future studies, these effects could be reduced by using other paramagnetic ions, similarly as Ahrens’ group reported for fluorophilic chelates.13,15
Acidic environment impacts the relaxation times in multicore NPs but not in single core NCs
As 19F paramagnetic relaxation enhancement (PRE) has been shown to be an efficient tool for the development of nano-systems with an on/off switching mechanism based on specific stimuli, we explored if multicore NPs can respond to a pH variation.36,38,41,42 Considering that the PRE-effect is concentration dependent, the Gd content for both single core and multicore NPs was kept constant for all measurements and samples were analyzed in environments with varied pH.Multicore NPs showed a more pronounced effect of pH compared with single core capsules (Fig. 5). Indeed, in addition to the negligible influence of Gd-loaded single core NCs on T2 at pH 7 when compared with the control NPs (Gd-free NCs), there was no impact on T1 and T2 relaxation times as a function of different pH used for both Gd_01 and Gd_02 (Fig. 5a; Tables S12 and S13†). While 19F relaxation properties of NCs remained almost constant, in multicore NPs, the relaxation times varied (Fig. 5b) from neutral to the most acidic environment. Thus, the transverse relaxation time increased gradually from pH 7 to pH 2 approximately 15-fold (Tables S10 and S11†) for both Gd_01 and Gd_02, indicating that the acidic solution can penetrate more easily into the multicore NP matrix as compared to single core NCs. This result is manifested by the sharp increase in the 19F's line-width as expected with an increase in T2 relaxation time (Fig. 5c). A minor change was observed in the T1 relaxation time of Gd_01 and Gd_02 encapsulated multicore NPs. This is attributed to the fact that Gd chelates are primarily 19F T2 relaxation agents.14
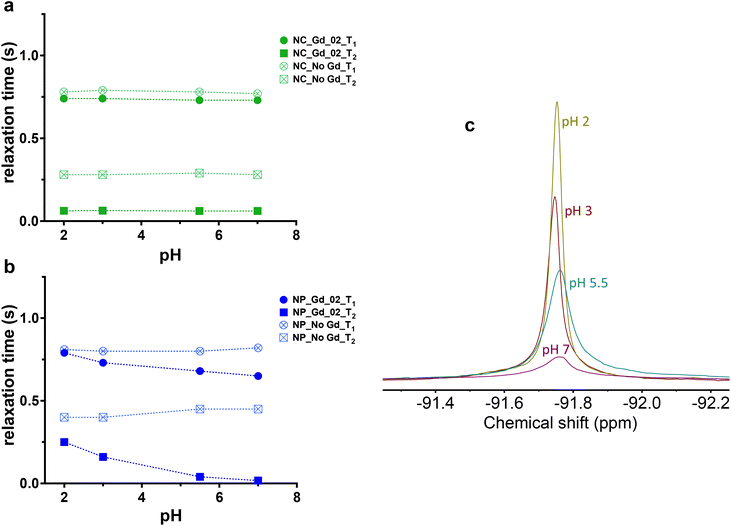 | ||
| Fig. 5 19F MR relaxation properties in the acidic environment of multicore NPs encapsulated wtih Gd-chelates compared to single core capsules. Longitudinal and transverse relaxation time of (a) Gd_02-loaded single core and (b) multicore NPs and related (c) 19F spectral line profile of multicore NPs under different pH environments, c = 10 mg mL−1, 500 MHz. When both type of NPs are loaded with Gd_01, the effect is the same (Table S10† for multicore nanoparticles loaded with Gd_01 and Table S12† for core-shell nanocapsules loaded with Gd_01, results not included in the graphs for clarity). | ||
It is well known from the literature43 that in strongly acidic solutions protons compete with Gd3+ to bind to a carboxylate ligand, thus increasing the dissociation rate of Gd3+ complexes. Therefore, it can be hypothesized that such a substantial change in T2 from pH variation is explained by the fact that a low pH environment might trigger the release of Gd from the chelate into the aqueous phase. Such release can be problematic, as the primary reason for toxicity caused by Gd-based contrast agents has been identified to be the dissociation of Gd from the chelated complex in vivo.44 As Gd3+ ions have a comparable ionic radius to Ca2+, they can interfere with calcium-mediated signaling pathways, thus making them exceedingly toxic.
To understand the mechanism of T2 variation of multicore NPs at different pH, we studied the transverse relaxation time of the residual solvent protons and fluorine-19 at pH 7 and 3 (Table 1). Three different samples were analyzed: (i) multicore NPs with no gadolinium (NPs_No Gd), (ii) NPs loaded with Gd chelates (NPs_ Gd), and (iii) NPs without Gd but supplemented with gadolinium chloride (NPs_ No Gd + GdCl3), used as a positive control to mimic Gd leakage from the NPs. Proton T2 relaxation time emerged to be equal in both NPs loaded with Gd and the ones supplied with salt, indicating that the PLGA matrix of the NPs, which additionally incorporates PVA,26 is permeable to water or at least to water protons. The exchange must be quite fast as the impact of the encapsulated Gd and the one supplemented with GdCl3 is negligible on T2 relaxation time. However, it is very unlikely that Gd3+ is permeable to the NPs as the 19F T2 does not change notably in the presence of the added Gd-based salt. Only encapsulated Gd has an enhanced impact (10-fold), which in turns further proves that encapsulation additionally provides a clue that Gd3+ does not dissociate under acidic conditions. If this was the case, then one would expect that 19F transverse relaxation time of NPs encapsulating Gd equals the one of NPs supplemented with gadolinium salt. Additionally, it is interesting to note that the 19F T2 value of NPs_ No Gd + GdCl3 at pH 3 is really close to that of NPs_No Gd than to NPs_ Gd demonstrating that in the acidic environment Gd3+ is not released from the chelate. These results suggest that the changes in the internal conformational structure change of NPs leads to a change in T2 relaxation time in the acidic environment.
| 1H T2(S) | 19F T2(S) | |||
|---|---|---|---|---|
| pH 7 | pH 3 | pH 7 | pH 3 | |
| D2O (Deuterium oxide) | 2.6 | 4.2 | n/a | n/a |
| NPs_No Gd | 1.1 | 2.7 | 0.45 | 0.49 |
| NPs_Gd | 0.18 | 0.36 | 0.02 | 0.21 |
| NPs_No Gd + GdCl3 | 012 | 0.36 | 0.12 | 0.50 |
A further possible mechanism which drives the variation of T2 values at different pH might be related to the stability of the NPs: at low pH the ester bonds in the polymer backbone typically hydrolyze. However, it can be distinctly observed that following dispersion of NPs, there is no difference in the 1H spectrum (Fig. 6a) between NPs in water (pH 7) compared to the ones in acidic solution (pH 3), even after 5 days, suggesting slow and negligible hydrolysis during this period. Moreover, the stability of the NPs was also confirmed by DLS. Indeed, size and PDI remain constant at neutral and acidic pH (Fig. 6b).
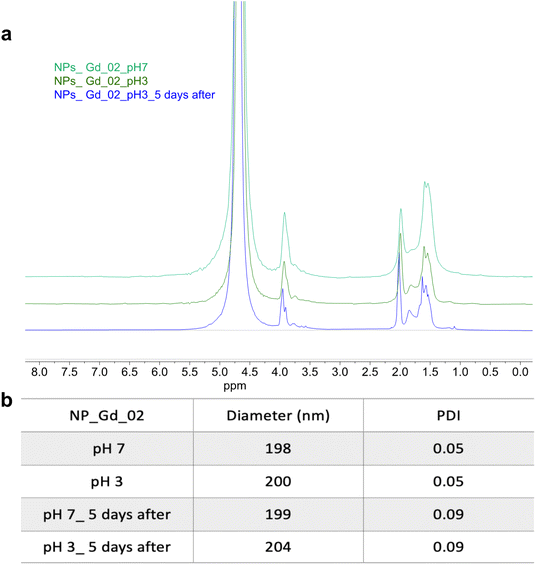 | ||
| Fig. 6 (a) 1H NMR spectrum of multicore NPs loaded with Gd_02 at pH 7 and 3, immediately after dispersing them in D2O (pH 7) and acidic solvent (pH 3), and five days after, 700 MHz, c = 10 mg mL−1. (b) The stability of the NPs is also demonstrated by the DLS values at pH 7 and 3. The stability was also confirmed for the multicore NPs loaded with Gd_01. (Results not included for clarity, Fig. S7†). | ||
These data might be explained by a morphology change inducing an increase in T2 relaxation time and signal intensity in acidic medium. Hence, it has been previously shown that in acidic pH PLGA particles have a different morphology than in alkaline and neutral pH.45,46 Both articles state that PLGA particles incubated in the acidic environment present fewer pores on the surface and inside compared to those under neutral pH. However, the particles at acidic pH keep having a smooth surface for a long period of time with a minimal loss in mass of PLGA particles. Therefore, we can hypothesize that this evident morphology alteration might push the Gd chelates farther from fluorine-19 atoms than the original morphology and hence increase the 19F T2 relaxation time.
The stability of the nanoprobe was also studied in terms of T2 values over time. For this purpose, the T1 and T2 variations of the system at pH 3 were measured right after and 3 hours after the solubilization (Fig. 7b). Fig. 7a shows the system stability in terms of signal intensity.
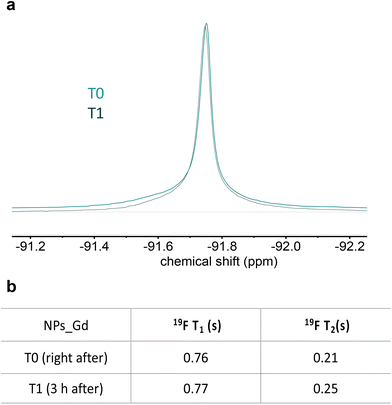 | ||
| Fig. 7 (a) 19F spectral line profile of multicore NPs at pH 3 in two timepoints and (b) related 19F MR relaxation properties. Multicore NPs: c = 10 mg mL−1, 500 MHz. | ||
When multicore NPs were dispersed in a basic environment (pH 14), the PLGA was hydrolyzed and in the presence of Gd3+, it resulted in a complete line broadening of the 19F NMR resonances (Fig. S6†). However, for NPs prepared without Gd3+, the high pH environment did not significantly impact the 19F line-shape (Fig. S6†), in agreement with our earlier results.35 Similarly, the DLS analysis showed an increase in size from 164 nm at pH 7 to 1300 nm at basic pH and in the PDI value from 0.05 to 0.4. As reported previously, NPs degrade at alkaline pH leading to the formation of agglomerates of the remaining surfactant and PFCE.35
Conclusion
A co-encapsulation of PFC and a paramagnetic chelate in NPs can lead to probes suitable for high-sensitive 19F MRI or sensing application. Yet, the modulation of both the 19F-signal of PFC inside the NPs and the proton signal from the aqueous environment of the probe remained not possible. We explored the impact of the nanoparticle structure on paramagnetic relaxation enhancement (PRE) and compared liquid PFC-loaded PLGA nanoparticles with multicore and single-core structures. Our findings highlight the crucial role of structural properties in 19F MR relaxation. In single-core capsules, the effect of the paramagnetic chelate was primarily observed as an increase in T2 relaxation time. Interestingly, multicore nanoparticles exhibited decreases in both 19F T1 and T2 relaxation time with increasing Gd chelate encapsulation, and they also allowed for the modulation of the proton signal. This approach holds promise for fine-tuning both 19F and 1H MR characteristics in agents containing liquid PFC and paramagnetic chelates, particularly for high-sensitivity probes and environmentally responsive agents. Finally, we showed that the multicore and single core structures of the NPs lead to different 19F relaxation times under different pH conditions. The 19F MR relaxation was not affected in single core NCs irrespective of the pH. However, an increase in T2 relaxation time has been observed in multicore NPs at the most acidic pH analyzed despite maintaining the physiochemical stability and integrity. In conclusion, multicore NPs can also be applied as a pH probe for acidic environment detection.Materials and methods
Materials
All chemicals were used as received: PLGA (Resomer 502H) from Evonik Industries, Germany; dichloromethane (DCM) from EMSURE, Merck Millipore, Darmstadt, Germany; polyvinyl alcohol (PVA), deuterium oxide and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich (St Louis, MO, USA); perfluoro-15-crown-5 ether (PFCE) from Exfluor, Texas, United States, and Prohance from Bracco, Amsterdam, The Netherlands. Lipophilic gadolinium chelates were synthesized as described previously.34 Water was purified by a Synergy water purification system from Merck.Methods
Synthesis of multicore nanoparticles. PVA (0.5 g) was dissolved in ultrapure water (25 g) under magnetic stirring. PLGA (0.1 g) was dissolved in dichloromethane (3 mL) and mixed with PFCE (0.9 mL) by pipetting it up and down with a glass pipette. In parallel, stock solutions of Gd-chelates in dichloromethane were prepared (c = 0.0166 mg μL−1). Different amounts of these chelates were added to the organic phase. For preparation of particles with gadoteridol, Prohance (1.78 mL, corresponds to 497 mg gadoteridol and 140 mg Gd) was added to the organic phase and sonicated for 15 s. Afterwards, the sonication of the aqueous surfactant solution was started, and the organic phase was added rapidly with a glass pipette (microtip Branson digital Sonifier s250, Missouri, United States). The miniemulsion was sonicated in an ice-water bath for 3 minutes at 40% amplitude. As PFCE is both hydrophobic and lipophobic, the organic phase has to be constantly premixed by pipetting during the addition to ensure that the PFCE and PLGA phases are added simultaneously. After sonication, DCM was evaporated overnight at room temperature under stirring to achieve solidification of the particles. The dispersions were transferred to 50 mL tubes and washed four times by centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g (4 °C, 35 min) and supernatant replacement. After the second washing step, the samples were sonicated in an ultrasonic bath (sonication bath, Diagenode Bioruptor, Seraign, Belgium) and the washing cycle was repeated. After washing, the particles were resuspended in water (5 mL), frozen with liquid nitrogen and freeze-dried. The resulting powder was stored at −20 °C.
000g (4 °C, 35 min) and supernatant replacement. After the second washing step, the samples were sonicated in an ultrasonic bath (sonication bath, Diagenode Bioruptor, Seraign, Belgium) and the washing cycle was repeated. After washing, the particles were resuspended in water (5 mL), frozen with liquid nitrogen and freeze-dried. The resulting powder was stored at −20 °C.
Synthesis of single core nanocapsules. For the synthesis of NCs, sodium cholate (0.375 g) that was used as a surfactant in the sonication step was dissolved in water (24.625 g, 1.5 wt% solution). PLGA (0.1 g) was dissolved in dichloromethane (2 mL) and mixed with PFCE (0.9 mL) by pipetting it up and down with a glass pipette. Different amounts of these chelates were added to the organic phase (corresponding to 1 mg, 5 mg and 10 mg of chelate). Some samples were prepared with the addition of Prohance (1.78 mL). The organic phase was then added with a glass pipette to the sodium cholate solution. As PFCE is both hydrophobic and lipophobic, the organic phase had to be premixed before sonication to ensure that PFCE and PLGA phases are added simultaneously. The miniemulsion was sonicated in an ice bath for 3 minutes at 40% amplitude (microtip Branson digital sonifier s250, Missouri, United States). After sonication, DCM was evaporated overnight at room temperature under stirring to achieve solidification of the particles. To exchange the surfactant, 10 g of PVA solution (1.96 wt%) was added to the suspension, followed by continuous stirring at 4 °C for 4 days. After 4 days, the capsules were washed 4 times with deionized water through centrifugation (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 098g, 35 minutes, 4 °C), resuspended in 5 mL of water, frozen (liquid N2) and freeze-dried. The NCs in a powder form were stored at −20 °C.
098g, 35 minutes, 4 °C), resuspended in 5 mL of water, frozen (liquid N2) and freeze-dried. The NCs in a powder form were stored at −20 °C.
Inductively coupled plasma-mass-spectrometry (ICP-MS). The Gd content was determined by ICP-MS using an X series I with a quadrupole mass spectrometer. Samples were prepared by adding 10 mg of the nanoparticles to 145 μL HNO3 (1%) and left overnight at room temperature in order to let the particles fully degrade. Afterwards, the samples were filled up to a total volume of 10 mL prior to the measurement.
Dynamic light scattering (DLS). The particle size and the polydispersity index (PDI) were measured by DLS on a Malvern Zetasizer ZS nano instrument. The particles were dispersed in ultrapure water at a concentration of 0.01 mg mL−1.
19 F NMR experiments. They were performed either with a Bruker Avance III HD 400 MHz nanobay spectrometer equipped with a 5 mm BBFO + probe at 298 K (Gd3+ loading relaxation experiments) or a JEOL ECZ-R 500 MHz spectrometer equipped with a RoyalHFX probe (pH effects on relaxation experiments).
Determination of PFCE content was done by quantitative 19F NMR. The NPs and NCs were resuspended in 400 μL of D2O and 100 μL of 0.1 vol% trifluoroacetic acid (TFA) as an internal reference. The interscan delay d1 was set to 25 (5 × T1).
For relaxation time measurements approximately 5 mg of particles were resuspended in D2O and transferred to an NMR tube. The 19F 90° pulse was calibrated prior to the measurement. The same method was used for the pH effect experiments except for the solvent which was a D2O solution of hydrogen chloride with the desired pH. For the latter experiment, the nanoparticles were resuspended prior to the measurement and T1 and T2 measured as described below.
T 1 measurements were done using the inversion recovery method of a 180° pulse followed by a variable delay period for transverse magnetization to relax longitudinally followed by a 90° detection pulse. The T1 was first estimated using a simple 1D sequence and then a full pseudo-2D acquisition was acquired with a list of delays with an interscan delay set to 5 × T1, the slowest. The recovery was fit to an exponential growth function which revealed the T1. The 19F T1 experiments were performed without 1H decoupling.
T 2 measurements were performed using the CPMG method of a 90° pulse followed by a train of tau-180-tau, where tau is a variable delay. For rapid T2-relaxing nuclei (T2 < 30 ms), a tau of 0.5 ms was used. For other, slower T2-relaxing nuclei, a tau of 1.2 ms was used. T2 was first estimated using a simple 1D sequence. Afterwards, a full pseudo-2D acquisition was acquired with a list of refocusing pulse repetitions ranging from 2 to 1750 and an interscan delay set to 5 × T1, the slowest. The recovery was fit to a mono-exponential decay function which revealed the T2.
The same setting was used to measure T1 and T2 relaxation times in an acidic environment. In this regard, roughly 5 mg of nanoparticles from the same sample were resuspended in 500 μL of several acidic solutions with pH 2, 3, 5.5 and 7. Gd-free PFCE nanoparticles were used as a control. T1 and T2 measurements were performed right after the resuspension.
Cryogenic scanning electron microscopy (CryoSEM). The images were obtained with a JEOL 6300F field emission scanning electron microscope. The samples (8 μL, 10 mg mL−1) were pipetted in 2 rivets that are placed together. Then the sample was frozen in liquid nitrogen slush and with a cryo transfer device placed in an Oxford Alto 2500 cryo-station. There the top rivet was broken and the sample was heated to −95 °C for 5 minutes. Then a coating of 60/40 Au/Pd was done and the sample was transferred to the cryoSEM.
1
H and
19
F magnetic resonance imaging. The imaging was carried out on a Bruker Biospec 11.7 T. The samples had Gd contents of 0, 0.7, 0.9, 1.8 and 2.5 μg mg−1. The concentration of nanoparticles was 10 mg mL−1, which is also often used for in vivo imaging. 1H MRI was acquired with the T1-weighted 2D fast low angle shot (FLASH) sequence: TR 104 ms, TE 2.25 ms, flip angle 40°, 2 averages, matrix 256 × 256, field of view (FOV) 32 × 32 mm, 12 slices with a slice thickness of 1 mm, scan time 53 s. 19F MRI was performed in a 3D RARE sequence with the following parameters: TR 1 s, TE 6.5 ms, turbo factor 8, averages, matrix 32 × 32×12, FOV 32 × 32×24 mm, scan time 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 min.
20 min.
Cell labelling and viability. A murine macrophage cell line (RAW cells) was cultured in RPMI medium (Gibco, Merelbeke, Belgium) enriched with 1% antimycotic-antibiotic (AA) (Gibco, Merelbeke, Belgium) + 10% foetal bovine serum (FBS). Cells were incubated at 37 °C for 24 hours, with nanoparticles at the concentration of 2 mg/1.0 × 106 cells. To investigate the influence of labelling on cell viability, an MTT assay was performed. For this purpose, the excess of label was removed, and cells were gently washed and resuspended in 100 μL of PBS. For the negative control, 100 μL of DMSO was added to cells instead of PBS and all the samples were placed in a 96-well plate. Next, 60 μL of complete medium and 10 μL of thiazolyl blue tetrazolium bromide (MTT) solution (4 mg MTT in 1 mL sterile water) (Sigma-Aldrich, Missouri, USA) was added to each well, followed by 1-hour incubation at 37 °C. After incubation, the plate was centrifuged for 2 minutes and 100 μL lysis buffer (9 mL isopropanol, 12.5 μL SDS, 200 μL of 2 N HCL, and 787.5 μL of deionized water) was added per well and the plates were incubated for 15 minutes in the dark at RT. The absorbance was then measured using an iMarkTM microplate absorbance reader (Bio-rad, California, USA) at 590 nm.
Author contributions
The experiments were designed by Alvja Mali, Margot Verbeelen, Paul B. White, Olga Koshkina and Mangala Srinivas. The experiments were conducted by Alvja Mali, Margot Verbeelen, Paul B. White, Alexander H.J. Staal, N. Koen van Riessen, and Cyril Cadiou. All the authors assisted with the data analysis and interpretation of the results. All authors contributed to the final draft. Alvja Mali and Margot Verbeelen contributed equally to this work.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This project has received funding from the Horizon 2020 Framework Programme Research and Innovation Programme under the Marie Sklodowska Curie grant agreement No NOVA-MRI (859908). The authors acknowledge the funding from ERC-2014-StG-336454-CoNQUeST, TTW-NWO open technology grant STW-14716, ERC-2015-PoC-713524-CONQUEST, ERC-2019-PoC-862989-CENYA, and ERA-CVD JTC2017-044. The authors kindly thank Bijal K. Bahuleyan (Univ. Rheims), Paul van der Ven (RU), Geert-Jan Janssen (RU) and the General Instruments Department (RU), Peter van Dijk (RU), Daniela Wilson (RU), Eric van Dinther (Radboudumc), Oya Tagit (Radboudumc), Massis Krekorian (Radboudumc), Edyta Swider-Cios (Radboudumc), and Kimberley R. G. Cortenbach (Radboudumc).References
- I. Tirotta, V. Dichiarante, C. Pigliacelli, G. Cavallo, G. Terraneo, F. B. Bombelli, P. Metrangolo and G. Resnati, F-19 Magnetic Resonance Imaging (MRI): From Design of Materials to Clinical Applications, Chem. Rev., 2015, 115, 1106–1129 CrossRef CAS PubMed.
- J. Ruiz-Cabello, B. P. Barnett, P. A. Bottomley and J. W. Bulte, Fluorine (19F) MRS and MRI in biomedicine, NMR Biomed., 2011, 24, 114–129 CrossRef CAS PubMed.
- F. Chapelin, C. M. Capitini and E. T. Ahrens, Fluorine-19 MRI for detection and quantification of immune cell therapy for cancer, J. Immunother. Cancer, 2018, 6, 11 CrossRef PubMed.
- J. M. Janjic and E. T. Ahrens, Fluorine-containing nanoemulsions for MRI cell tracking, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2009, 1, 492–501 CAS.
- E. Pisani, N. Tsapis, B. Galaz, M. Santin, R. Berti, N. Taulier, E. Kurtisovski, O. Lucidarme, M. Ourevitch, B. T. Doan, J. C. Beloeil, B. Gillet, W. Urbach, S. L. Bridal and E. Fattal, Perfluorooctyl Bromide Polymeric Capsules as Dual Contrast Agents for Ultrasonography and Magnetic Resonance Imaging, Adv. Funct. Mater., 2008, 18, 2963–2971 CrossRef CAS.
- C. Zhang, T. Liu, W. Wang, C. A. Bell, Y. Han, C. Fu, H. Peng, X. Tan, P. Král, K. Gaus, J. J. Gooding and A. K. Whittaker, Tuning of the Aggregation Behavior of Fluorinated Polymeric Nanoparticles for Improved Therapeutic Efficacy, ACS Nano, 2020, 14, 7425–7434 CrossRef CAS PubMed.
- T. Boissenot, E. Fattal, A. Bordat, S. Houvenagel, J. Valette, H. Chacun, C. Gueutin and N. Tsapis, Paclitaxel-loaded PEGylated nanocapsules of perfluorooctyl bromide as theranostic agents, Eur. J. Pharm. Biopharm., 2016, 108, 136–144 CrossRef CAS PubMed.
- H. Lin, X. Tang, A. Li and J. Gao, Activatable 19F MRI Nanoprobes for Visualization of Biological Targets in Living Subjects, Adv. Mater., 2021, 33, 2005657 CrossRef CAS PubMed.
- C. Wang, S. R. Adams and E. T. Ahrens, Emergent Fluorous Molecules and Their Uses in Molecular Imaging, Acc. Chem. Res., 2021, 54, 3060–3070 CrossRef CAS PubMed.
- D. Xie, M. Yu, R. T. Kadakia and E. L. Que, 19F Magnetic Resonance Activity-Based Sensing Using Paramagnetic Metals, Acc. Chem. Res., 2020, 53, 2–10 CrossRef CAS PubMed.
- G. Gambino, T. Gambino, R. Pohmann and G. Angelovski, A ratiometric F-19 MR-based method for the quantification of Ca2 + using responsive paramagnetic probes, Chem. Commun., 2020, 56, 3492–3495 RSC.
- G. Gambino, T. Gambino and G. Angelovski, Combination of Bioresponsive Chelates and Perfluorinated Lipid Nanoparticles Enables in vivo MRI Probe Quantification, Chem. Commun., 2020, 9433–9436, 10.1039/D0CC04416D.
- A. H. Jahromi, C. Wang, S. R. Adams, W. Zhu, K. Narsinh, H. Xu, D. L. Gray, R. Y. Tsien and E. T. Ahrens, Fluorous-Soluble Metal Chelate for Sensitive Fluorine-19 Magnetic Resonance Imaging Nanoemulsion Probes, ACS Nano, 2019, 13, 143–151 CrossRef CAS PubMed.
- A. A. Kislukhin, H. Y. Xu, S. R. Adams, K. H. Narsinh, R. Y. Tsien and E. T. Ahrens, Paramagnetic fluorinated nanoemulsions for sensitive cellular fluorine-19 magnetic resonance imaging, Nat. Mater., 2016, 15, 662–668 CrossRef CAS PubMed.
- J. Rho, E. Stares, S. R. Adams, D. Lister, B. Leach and E. T. Ahrens, Paramagnetic Fluorinated Nanoemulsions for in vivo F-19 MRI, Mol. Imaging Biol., 2020, 22, 665–674 CrossRef CAS PubMed.
- K. L. Peterson, K. Srivastava and V. C. Pierre, Fluorinated Paramagnetic Complexes: Sensitive and Responsive Probes for Magnetic Resonance Spectroscopy and Imaging, Front. Chem., 2018, 6, 160 CrossRef PubMed.
- G. M. Clore and J. Iwahara, Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and Their Complexes, Chem. Rev., 2009, 109, 4108–4139 CrossRef CAS PubMed.
- J. G. Riess, Fluorous micro-and nanophases with a biomedical perspective, Tetrahedron, 2002, 58, 4113–4131 CrossRef CAS.
- G. Cavallo, P. Metrangolo, R. Milani, T. Pilati, A. Priimagi, G. Resnati and G. Terraneo, The Halogen Bond, Chem. Rev., 2016, 116, 2478–2601 CrossRef CAS PubMed.
- J. G. Riess, Understanding the Fundamentals of Perfluorocarbons and Perfluorocarbon Emulsions Relevant to In Vivo Oxygen Delivery, Artif. Cells, Blood Substitutes, Biotechnol., 2005, 33, 47–63 CrossRef CAS PubMed.
- H. Matsushita, S. Mizukami, F. Sugihara, Y. Nakanishi, Y. Yoshioka and K. Kikuchi, Multifunctional Core–Shell Silica Nanoparticles for Highly Sensitive 19F Magnetic Resonance Imaging, Angew. Chem., Int. Ed., 2014, 53, 1008–1011 CrossRef CAS PubMed.
- A. L. Lee, C. T. Gee, B. P. Weegman, S. A. Einstein, A. R. Juelfs, H. L. Ring, K. R. Hurley, S. M. Egger, G. Swindlehurst, M. Garwood, W. C. K. Pomerantz and C. L. Haynes, Oxygen Sensing with Perfluorocarbon-Loaded Ultraporous Mesostructured Silica Nanoparticles, ACS Nano, 2017, 11, 5623–5632 CrossRef CAS PubMed.
- A. de Vries, R. Moonen, M. Yildirim, S. Langereis, R. Lamerichs, J. A. Pikkemaat, S. Baroni, E. Terreno, K. Nicolay, G. J. Strijkers and H. Grull, Relaxometric studies of gadolinium-functionalized perfluorocarbon nanoparticles for MR imaging, Contrast Media Mol. Imaging, 2014, 9, 83–91 CrossRef CAS PubMed.
- A. M. Neubauer, J. Myerson, S. D. Caruthers, F. D. Hockett, P. M. Winter, J. Chen, P. J. Gaffney, J. D. Robertson, G. M. Lanza and S. A. Wickline, Gadolinium-modulated 19F signals from perfluorocarbon nanoparticles as a new strategy for molecular imaging, Magn. Reson. Med., 2008, 60, 1066–1072 CrossRef CAS PubMed.
- Q. Peng, Y. Li, S. Bo, Y. Yuan, Z. Yang, S. Chen, X. Zhou and Z.-X. Jiang, Paramagnetic nanoemulsions with unified signals for sensitive 19F MRI cell tracking, Chem. Commun., 2018, 54, 6000–6003 RSC.
- O. Koshkina, G. Lajoinie, F. B. Bombelli, E. Swider, L. J. Cruz, P. White, R. Schweins, Y. Dolen, E. v. Dinther, N. K. v. Riessen, S. E. Rogers, R. Fokkink, I. K. Voets, E. R. H. v. Eck, A. Heerschap, M. Versluis, C. d. Korte, C. Figdor, I. J. M. d. Vrie and M. Srinivas, Multicore liquid perfluorocarbon-loaded multimodal nanoparticles for stable ultrasound and 19F MRI applied to in vivo cell tracking, Adv. Funct. Mater., 2019, 29, 1806485 CrossRef PubMed.
- M. Srinivas, L. J. Cruz, F. Bonetto, A. Heerschap, C. G. Figdor and I. J. M. De Vries, Customizable, multi-functional fluorocarbon nanoparticles for quantitative in vivo imaging using 19F MRI and optical imaging, Biomaterials, 2010, 31, 7070–7077 CrossRef CAS PubMed.
- C. Jacoby, S. Temme, F. Mayenfels, N. Benoit, M. P. Krafft, R. Schubert, J. Schrader and U. Flogel, Probing different perfluorocarbons for in vivo inflammation imaging by F-19 MRI: image reconstruction, biological half-lives and sensitivity, NMR Biomed., 2014, 27, 261–271 CrossRef CAS PubMed.
- A. H. J. Staal, K. Becker, O. Tagit, N. Koen van Riessen, O. Koshkina, A. Veltien, P. Bouvain, K. R. G. Cortenbach, T. Scheenen, U. Flögel, S. Temme and M. Srinivas, In vivo clearance of 19F MRI imaging nanocarriers is strongly influenced by nanoparticle ultrastructure, Biomaterials, 2020, 120307, DOI:10.1016/j.biomaterials.2020.120307.
- E. Hoogendijk, E. Swider, A. H. J. Staal, P. B. White, N. K. van Riessen, G. Glaßer, I. Lieberwirth, A. Musyanovych, C. A. Serra, M. Srinivas and O. Koshkina, Continuous-Flow Production of Perfluorocarbon-Loaded Polymeric Nanoparticles: From the Bench to Clinic, ACS Appl. Mater. Interfaces, 2020, 12, 49335–49345 CrossRef CAS PubMed.
- M. Krekorian, G. G. W. Sandker, K. R. G. Cortenbach, O. Tagit, N. K. van Riessen, R. Raavé, M. Srinivas, C. G. Figdor, S. Heskamp and E. H. J. G. Aarntzen, Characterization of Intrinsically Radiolabeled Poly(lactic-co-glycolic acid) Nanoparticles for ex Vivo Autologous Cell Labeling and in Vivo Tracking, Bioconjugate Chem., 2021, 32, 1802–1811 CrossRef CAS PubMed.
- E. Swider, K. Daoudi, A. H. Staal, O. Koshkina, N. K. van Riessen, E. van Dinther, I. J. M. de Vries, C. L. de Korte and M. Srinivas, Clinically-Applicable Perfluorocarbon-Loaded Nanoparticles For In vivo Photoacoustic, 19F Magnetic Resonance And Fluorescent Imaging, Nanotheranostics, 2018, 2, 258–268 CrossRef PubMed.
- E. Swider, O. Koshkina, J. Tel, L. J. Cruz, I. J. M. de Vries and M. Srinivas, Customizing poly(lactic-co-glycolic acid) particles for biomedical applications, Acta Biomater., 2018, 73, 38–51 CrossRef CAS PubMed.
- G. Rigaux, V. G. Roullin, C. Cadiou, C. Portefaix, L. Van Gulick, G. Bœuf, M. C. Andry, C. Hoeffel, L. Vander Elst, S. Laurent, R. Muller, M. Molinari and F. Chuburu, A new magnetic resonance imaging contrast agent loaded into poly(lacide-co-glycolide) nanoparticles for long-term detection of tumors, Nanotechnology, 2014, 25, 445103 CrossRef CAS PubMed.
- O. Koshkina, P. B. White, A. H. J. Staal, R. Schweins, E. Swider, I. Tirotta, P. Tinnemans, R. Fokkink, A. Veltien, N. K. van Riessen, E. R. H. van Eck, A. Heerschap, P. Metrangolo, F. Baldelli Bombelli and M. Srinivas, Nanoparticles for “two color” 19F magnetic resonance imaging: Towards combined imaging of biodistribution and degradation, J. Colloid Interface Sci., 2020, 565, 278–287 CrossRef CAS PubMed.
- K. Akazawa, F. Sugihara, T. Nakamura, S. Mizukami and K. Kikuchi, Highly Sensitive Detection of Caspase-3/7 Activity in Living Mice Using Enzyme-Responsive 19F MRI Nanoprobes, Bioconjugate Chem., 2018, 29, 1720–1728 CrossRef CAS PubMed.
- K. Akazawa, F. Sugihara, M. Minoshima, S. Mizukami and K. Kikuchi, Sensing caspase-1 activity using activatable 19F MRI nanoprobes with improved turn-on kinetics, Chem. Commun., 2018, 54, 11785–11788 RSC.
- T. Nakamura, H. Matsushita, F. Sugihara, Y. Yoshioka, S. Mizukami and K. Kikuchi, Activatable 19F MRI Nanoparticle Probes for the Detection of Reducing Environments, Angew. Chem., Int. Ed., 2015, 54, 1007–1010 CrossRef CAS PubMed.
- W. H. Thompson, Perspective: Dynamics of confined liquids, J. Chem. Phys., 2018, 149, 170901 CrossRef PubMed.
- J. G. Riess, Fluorous micro- and nanophases with a biomedical perspective, Tetrahedron, 2002, 58, 4113–4131 CrossRef CAS.
- M. B. Kok, A. De Vries, D. Abdurrachim, J. J. Prompers, H. Grüll, K. Nicolay and G. J. Strijkers, Quantitative 1H MRI, 19F MRI, and 19F MRS of cell-internalized perfluorocarbon paramagnetic nanoparticles, Contrast Media Mol. Imaging, 2011, 6, 19–27 CrossRef CAS PubMed.
- A. Mali, E. L. Kaijzel, H. J. Lamb and L. J. Cruz, 19F-nanoparticles: Platform for in vivo delivery of fluorinated biomaterials for 19F-MRI, J. Controlled Release, 2021, 338, 870–889 CrossRef CAS PubMed.
- T. J. Clough, L. Jiang, K.-L. Wong and N. J. Long, Ligand design strategies to increase stability of gadolinium-based magnetic resonance imaging contrast agents, Nat. Commun., 2019, 10, 1420 CrossRef PubMed.
- M. Rogosnitzky and S. Branch, Gadolinium-based contrast agent toxicity: a review of known and proposed mechanisms, BioMetals, 2016, 29, 365–376 CrossRef CAS PubMed.
- J. Li, G. Jiang and F. Ding, The effect of pH on the polymer degradation and drug release from PLGA-mPEG microparticles, J. Appl. Polym. Sci., 2008, 109, 475–482 CrossRef CAS.
- B. S. Zolnik and D. J. Burgess, Effect of acidic pH on PLGA microsphere degradation and release, J. Controlled Release, 2007, 122, 338–344 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr04577c |
| This journal is © The Royal Society of Chemistry 2023 |