Open Access Article
Open Access ArticleExploring the binding effects and inhibiting mechanism of hyperoside to lipase using multi-spectroscopic approaches, isothermal titration calorimetry, inhibition kinetics and molecular dynamics†
Zhen Zeng ,
Di Wu
,
Di Wu *,
Lan Tang,
Xia Hu
*,
Lan Tang,
Xia Hu ,
Jing Zhang and
Fang Geng
,
Jing Zhang and
Fang Geng
Meat Processing Key Laboratory of Sichuan Province, School of Food and Biological Engineering, Chengdu University, Chengdu 610106, China. E-mail: diwulab@163.com
First published on 24th February 2023
Abstract
Hyperoside (HYP) is a flavonoid with various physiological activities. The present study examined the interaction mechanism between HYP and lipase using multi-spectrum and computer-aided techniques. Results demonstrated that the force type of HYP on lipase was mainly hydrogen bond, hydrophobic interaction force, and van der Waals force, and HYP had an excellent binding affinity with lipase at 1.576 × 105 M−1. HYP dose-dependently inhibited lipase in the inhibition experiment, and its IC50 value was 1.92 × 10−3 M. Moreover, the results suggested that HYP could inhibit the activity by binding to essential groups. Conformational studies indicated that the conformation and microenvironment of lipase were slightly changed after the addition of HYP. Computational simulations further confirmed the structural relationships of HYP to lipase. The interaction between HYP and lipase can provide ideas for the development of functional foods related to weight loss. The results of this study help comprehend the pathological significance of HYP in biological systems, as well as its mechanism.
1 Introduction
Obesity has developed into a global health problem. Its prevention is also the main goal of the “2013–2020 Global Action Plan for the Prevention and Control of Noncommunicable Diseases” formulated by the World Health Organization (WHO).1 Excessive accumulation of body lipids may lead to obesity, which causes various chronic diseases such as diabetes, cardiovascular disease, and cancer.2 The structure of pancreatic lipase, one of the digestive enzymes, is well known.3 It is a small single-chain spherical protein consisting of 449 amino acids with a relative molecular mass of 50–52 kDa. It has a cap-shaped catalytic center (active site) at the N-terminal end, including amino acids Ser153, Asp177 and His264. There is a surface ring covering the active site of lipase between residues Cys239 and Cys263 called the lid, which is usually in the closed state. The conformation is known as the closed state of lipase. In addition, the enzyme is stabilized by van der Waals forces within the lid and the N-terminal structural domain of the two rings β5 (residues 76–85) and β9 (residues 207–217). Colipase, a 9.3 kDa coenzyme with a β-sandwich structure, binds to the C-terminal structural domain of the protein, causing the lid to move from its closed state to a maximum displacement of 29 Å as the lipid–water interface is formed, which is referred to as the open state. In addition, the β5 loop bends toward the core of the protein, providing a free channel for the active site in this state. Lipase is in charge of the hydrolysis of 50–70% of total dietary lipids, and dietary lipid is hydrolyzed by pancreatic lipase into monoacylglycerols and free fatty acids, resynthesizing lipid.4,5 The level of pancreatic lipase activity is related to the effect of lipid absorption. The hydrolysis and absorption of excess dietary lipid can be reduced by inhibiting pancreatic lipase activity, which is an important method to prevent and control obesity.6 Presently, orlistat is the main commercially and clinically used weight loss drug but safety issues have arisen due to serious adverse effects.7,8 Therefore, there is a need to find more effective and safer pancreatic lipase inhibitors as alternative drugs for the prevention and treatment of obesity.Some studies have reported that various natural dietary phytochemicals with low toxicity, high efficiency and low side effects have shown potential for weight loss.9,10 Hyperoside (HYP, quercetin-3-O-β-D-galactoside) belongs to flavonoids and quercetin glycoside derivatives, which is widely found in many natural plants, such as Zanthoxylum bungeanum leaves, hawthorn, and Hypericum perforatum L.11–14 It has various positive physiological functions, such as anti-hyperglycemic, anti-oxidant, anti-depressant, anti-inflammatory, and central analgesic.15–17 HYP has been widely considered because of its positive benefits on human health.13 Interestingly, quercetin 3-rhamnoside also belongs to quercetin glycoside derivatives. Its structure is one less oxygen atom than HYP, and quercetin 3-rhamnoside is effective in lipase inhibition.18–20 HYP and quercetin 3-rhamnoside are quercetin glycoside derivatives and are usually used as bioactive compounds in functional foods and pharmaceutical preparations. Therefore, the possible inhibitory effect, medicine, and food homology of HYP have been examined to analyze HYP's interaction and inhibitory effect on lipase.
Given this background, this study aimed to investigate the interaction of HYP to lipase using multi-spectral techniques, isothermal titration calorimetry (ITC), inhibition kinetics, molecular simulation, and saturation transfer difference-nuclear magnetic resonance (STD-NMR) experiments. First, we determined the quenching behavior, binding constants, and the effect of HYP on lipase conformation by multi-spectral techniques and ITC. Then, HYP and orlistat were compared by enzyme kinetics to determine the inhibition type, and obtain the IC50 value for HYP. Finally, molecular docking and molecular dynamics were carried out to corroborate the experimental results. STD-NMR experiments confirmed the interaction of HYP with lipase. This work provides a theoretical basis for the development of natural pancreatic lipase inhibitors.
2 Materials and methods
2.1 Reagents and chemicals
The high-purity pancreatic lipase used in the experiment was purchased from Sigma Aldrich Co., Ltd. (Milwaukee, USA). Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China) provided HYP (CAS: 482-36-0, ≥97% purity) and orlistat (CAS: 96829-58-2, ≥98% purity). A stock solution in triaminomethane-HCl (Tris–HCl) buffer (5.0 × 10−2 M, pH 7.4) was prepared. The other materials and reagents were of analytical grade, and ultrapure water was utilized for all tests.2.2 Fluorescence titration measurements
A Cary Eclipse fluorescence spectrophotometer (Varian, California, USA) was used for the fluorescence titration. During fluorescence titration, the final lipase concentration was maintained at 6.0 × 10−6 M, and the final HYP concentration ranged from 0 M to 4.2 × 10−5 M. The samples were mixed before fluorescence measurement, and the fluorescence measurement was performed at three temperatures (277, 298, and 310 K). The excitation wavelength was 280 nm, the emission wavelength in the scope was 300–500 nm, and the slit widths for excitation and emission wavelengths were 5 nm. All fluorescence intensities were corrected based on a previously reported method to remove internal filtering effects.21,22 The quenching mechanism was determined using the Stern–Volmer eqn (1), as follows:23| F0/F = 1 + KSV[Q] | (1) |
log(F0 − F)/F = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka + n Ka + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q]
| (2) |
The microenvironment changes in tryptophan (Trp) and tyrosine (Tyr) residues in lipase were determined by synchronous fluorescence spectroscopy.25 At 298 K, the wavelength intervals (Δλ) were set at 15 and 60 nm.
Three-dimensional (3D) fluorescence was used to obtain fluorescence intensity information when the excitation and emission wavelengths or other variables were varied simultaneously. The final concentrations of free lipase and HYP were 6.0 × 10−6 M and 2.4 × 10−5 M, respectively. In this experiment, the excitation and emission wavelengths were set to 200–400 nm and 200–500 nm, respectively.
2.3 Time-resolved fluorescence measurements
Time-resolved fluorescence measurement was performed using the Horiba Jobin Yvon fluoromax-3 spectrofluorometer (Horiba, LesUlis, France). At a temperature of 298 K, the excitation and emission wavelengths were set to 280 and 345 nm, respectively. The tail fitting method and chi-square (χ2) value were used to study the fluorescence lifetime data and evaluate the fitting results. The average fluorescence lifetime (τ) data were calculated using the formula:26
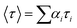 | (3) |
2.4 ITC analysis
The thermodynamic parameters of lipase and HYP were measured with an isothermal titration calorimeter CN-ITC200 (MicroCal, USA). All other solutions were degassed, except for the prepared protein solution containing 1% ethanol. Stored in a measuring cell, 1.0 × 10−4 M lipase solution was titrated with 1.0 × 10−3 M HYP solution (2.5 μL each injection, at 180 s intervals) under stirring at 250 rpm. The first injection was ignored in this experiment, and the change in enthalpy (ΔH) and entropy (ΔS), as well as the Gibbs free energy (ΔG) were obtained from eqn (4) and (5), where R is the gas constant (8.314 J mol−1 K−1):27,28
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka Ka
| (4) |
| ΔG = ΔH − TΔS | (5) |
2.5 Enzymatic assay of lipase activity
Typically, lipase activity and type of inhibition are assessed by the degree of hydrolysis of pNPC.29 The stock solution was made from the supernatant after the lipase solution was formed with Tris–HCl buffer and centrifuged for 5 min. After cooling to room temperature, 450 μL pNPC was added to start the reaction, the absorbance was measured using an enzyme marker (Berten Instrument Co. Ltd., Vermont, USA), and the inhibition was calculated, according to eqn (6).
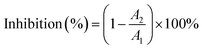 | (6) |
The types of inhibition of HYP and orlistat were measured using a UV-1901-PC spectrophotometer (Aucy, Shanghai, China) equipped with a 1.0 cm × 1.0 cm quartz cuvette within 0–200 s at 310 K and 405 nm. The final lipase concentration was 3.0 × 10−6 M, and the concentrations of HYP were 0, 1.5 × 10−5 M and 3.0 × 10−5 M. The concentrations of pNPC ranged within 2.0–3.6 × 10−3 M, and the absorbance curves were calculated for the initial velocity (V0) of the released product at 0–2 min. The inhibitory types of HYP and orlistat were determined using Lineweaver–Burk plots.30
2.6 Circular dichroism measurements
The samples were scanned using a circular dichroic spectrometer (Applied Photophysics, Surrey, UK) at 298 K with a cell path length of 1 mm. The scan wavelengths were from 200 nm to 260 nm. The average time was 0.5 s, and the step size was 1 nm. In all measurements, the molar ratio of lipase to HYP was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and the lipase concentration was fixed at 2.0 × 10−4 M.
4, and the lipase concentration was fixed at 2.0 × 10−4 M.
2.7 Fourier-transform infrared spectroscopy
A Nicolet-6700 Fourier-transform infrared (FT-IR) spectrometer with a smart OMNIC sampler adapter (Thermo Fisher Scientific, Sunnyvale, USA) was used to retrieve the FT-IR spectral data. At a temperature of 298 K, the sampler was used in this study to acquire infrared data in 128 runs with a resolution of 4 cm−1. After subtracting the background spectrum, individual protein (2.0 × 10−4 M), and different scales of HYP, the HYP samples were scanned at 1700–1500 cm−1 and analyzed using Peakfit v4.12 software (Beijing HuanZhongRuiChi Technology Co., Ltd., Beijing, China).312.8 Molecular docking and molecular dynamics methods
Molecular docking was performed using YASARA software (v18.3.23) based on the Autodock program.32 The desired crystal structure (PDB ID: 1ETH) (Triacylglycerol acyl-hydrolase Chain A and Colipase Chain B were used) was obtained from the RCSB Protein Data Bank. The ligand structure of HYP (CID: 5281643) was gained from PubChem with energy minimized.33 Lipase was defined as a receptor. All water molecules and non-polar hydrogen atoms in lipase were removed, and the polar and hydrogen atoms were added simultaneously. The charge distribution at pH 7.4 was changed and set to semi-flexible docking procedure during docking (lipase for rigid, HYP for flexible). A box of size 95 Å × 95 Å × 95 Å was formed for molecular docking, and it completely covered the protein. The Lamarckian Genetic Algorithm (LGA) method was applied to search for possible docking modes. The docking runs were set to 100, and the best model was selected based on the binding energy. The molecular docking results were visualized using Discovery Studio 4.5.21The optimal conformation of molecular docking was selected for MD simulation analysis. MD was conducted with the AMBER ff14SB force field and the YASARA program in the presence and absence of HYP. The AM1-BCC model was used to calculate the partial atomic charge of HYP, and the TIP3P water model was conducted. To completely cover the conformation, the box of MD was set as a cube to accommodate all molecules. Equilibrium ions were added to maintain electrical neutrality, and the energy of the system was minimized. The system used the NVT ensemble (298.15 K) and NPT ensemble (1.0 bar) to equilibrium. For the long-range Coulomb interactions, the cut-off of 8 Å and particle-mesh Ewald summation were used. The 1.25 fs time step was for the intramolecular forces, and the 2.50 fs time step was for the intermolecular forces. Data were recorded every 100 ps, and the total simulation running time was 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps. An initial simulation period was discarded as the equilibration time. The contribution of each interaction energy term to the binding process can be calculated by the following formula:34,35
000 ps. An initial simulation period was discarded as the equilibration time. The contribution of each interaction energy term to the binding process can be calculated by the following formula:34,35
| ΔE = ΔEvdw + ΔEelec + ΔEinter | (7) |
| ΔEinter = ΔEbond + ΔEangle + ΔEdihedral + ΔEplanarity | (8) |
2.9 STD-NMR experiment
An STD-NMR experiment was conducted at 298 K on a Varian Inova 400 MHz spectrometer with VNMRJ software. Lipase powder was dissolved in 1.0 × 10−2 M phosphate-buffered saline (PBS) (D2O) and centrifuged. The supernatant was taken to obtain 1.0 × 10−3 M lipase stock solution. HYP was dissolved in deuterated dimethyl sulfoxide to obtain 5.0 × 10−2 M HYP stock solution. The final lipase and HYP concentrations (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) were 2.0 × 10−5 M and 1.0 × 10−3 M, respectively. The sweep width was 256 transients and 8389.26 Hz to obtain the STD spectrum. At the same time, the sample was scanned 1024 times, and the saturation time was set to 2 s.
50) were 2.0 × 10−5 M and 1.0 × 10−3 M, respectively. The sweep width was 256 transients and 8389.26 Hz to obtain the STD spectrum. At the same time, the sample was scanned 1024 times, and the saturation time was set to 2 s.
2.10 Statistical analysis
In this study, data analysis and processing were achieved using Origin 2018 software (OriginLab Corporation, Northampton, USA). Data were analyzed by the one-way variance method (ANOVA), followed by the Least Significant Difference (LSD) test at 5% significance level using SPSS 20.0 software (Inc., Chicago, IL).3 Results and discussion
3.1 Fluorescence quenching of lipase
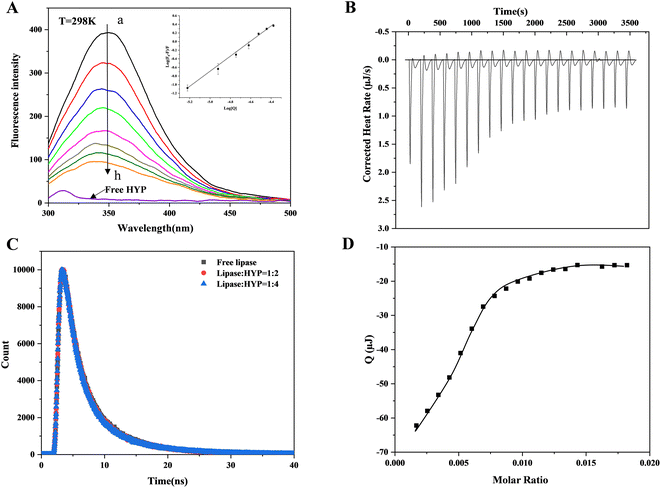
Dynamic and static quenching are the usual reasons for protein fluorescence quenching caused by small active molecules. Static quenching is the binding of small active molecules and proteins in the ground state to form a non-fluorescent complex; dynamic quenching is a process of electron or energy transfer between small active molecules and excited protein molecules.36 Whether in dynamic or static quenching, the action process conforms to the Stern–Volmer equation. Fig. 1A indicates that the lipase produced a maximum absorption peak at 345 nm at a temperature of 298 K, and the fluorescence intensity gradually decreased as the HYP concentration increased, showing that HYP and lipase formed a complex. However, changing the HYP concentration slightly varied the maximum wavelength of fluorescence (345 nm). Meanwhile, the addition of HYP slightly changed the shape of the lipase spectrum, indicating that HYP had a small effect on Trp's microenvironment.The relationship between log[Q] and log(F0 − F)/F at 298 K is displayed in Fig. 1A, which shows a good linear relationship. In accordance with eqn (1), KSV values were calculated and are displayed in Table 1. At 277, 298, and 310 K, the values of KSV were 7.113 ± 0.029 × 104 M−1, 6.826 ± 0.010 M−1, and 6.247 ± 0.060 × 104 M−1, respectively. These results show that the values of KSV decreased as the temperature increased, which is consistent with static quenching characteristics.37 The n values were calculated via double-logarithmic regression (eqn (2)), and at different temperatures, the n values were greater than 1, suggesting the presence of at least one binding site between HYP and lipase.
| System | T (K) | KSV × 104 (M−1) | Ka × 106 (M−1) | n | Rb | ΔG (kJ mol−1) | ΔH (kJ mol−1) | ΔS (J mol−1 K−1) |
|---|---|---|---|---|---|---|---|---|
| a Rb correlation coefficient for the Ka values. In each column, different superscript letters indicate significant difference (p < 0.05). | ||||||||
| HYP–lipase | 277 | 7.113 ± 0.029a | 3.535 ± 0.153a | 1.379 | 0.9969 | −34.81 | −25.46 | 33.77 |
| 298 | 6.826 ± 0.010b | 1.901 ± 0.074a,b | 1.337 | 0.9956 | −35.52 | |||
| 310 | 6.247 ± 0.060b | 1.048 ± 0.039c | 1.286 | 0.9962 | −35.93 | |||
Time-resolved fluorescence data can be further studied to explore dynamic or static quenching mechanisms. Fig. 1C shows the fluorescence lifetime curves with and without HYP. The chi-square (χ2) values were calculated through eqn (3) and are displayed in Table S1,† which indicates that the fitting values were acceptable. The lipase τ value was 1.989 ns, which is close to the τ value of lipase in the literature.38 When HYP was added at different concentrations, the τ values changed from 1.912 to 1.860 ns. Obviously, the τ value was almost unaffected. In general, the decay time of the uncomplexed fluorophore is virtually unaffected by the static quenching of the formation of the ground-state complex.39 The current results demonstrate that HYP's binding to lipase exhibited static quenching and did not affect the fluorescence life curve, which is consistent with the fluorescence titration results.
3.2 Thermodynamic parameters
ITC simply measures the heat absorbed or released by the interaction between the receptor and ligand, and provides the thermodynamic value of the interaction between them. Fig. 1B presents the corrected heat rate of HYP and lipase over time (μJ s−1). Fig. 1D shows the integral heat after dilution effect correction, which is a function of the ligand/protein mole ratio. A negative peak was observed when the HYP solution was titrated into the lipase solution, indicating that the interaction was exothermic (ΔH = −23.30 kJ mol−1). With the progress of titration, the peak decreased gradually because lipase was stably saturated by HYP. At the same time, during simultaneous binding, the ΔG and ΔS values were calculated as −29.67 kJ mol−1 and 21.36 J mol−1 K−1, respectively. These results suggest that the interaction was thermodynamically favorable, and the disorder degree of the system increased after the addition of HYP. The fluorescence thermodynamic parameters of lipase binding to HYP are displayed in Table 1. The ΔH, ΔG, and ΔS values calculated by the thermodynamics between HYP and lipase were −25.46 kJ mol−1, −35.52 kJ mol−1, and 33.77 J mol−1 K−1, respectively, which are in general agreement with the data calculated by ITC. The binding constant based on the ITC data was 1.576 × 105 M−1, indicating a medium-strength binding.Usually, four binding forces exist between HYP and lipase: van der Waals force, electrostatic force, hydrogen bond, and hydrophobic interaction force. The positive ΔS and negative ΔH values indicated that the hydrogen bond and hydrophobic interaction forces were characteristic.21 Furthermore, the good consistency between the fitting curve and the experimental data in Fig. 1(B and D) confirmed the rationality of the selected model. Therefore, hydrogen bonds and hydrophobic interaction force play an essential role in HYP–lipase complex formation in this experiment.
3.3 Synchronous and 3D fluorescence spectrum
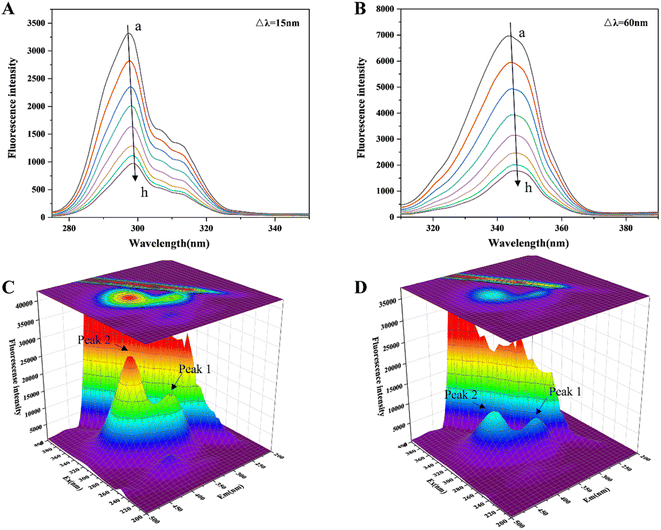
The changes in the microenvironment around the chromophore molecules containing Trp and Tyr residues can be seen by referring to the acquired synchronous fluorescence spectra.40 Fig. 2(A and B) indicates that at Δλ = 15 and 60 nm, the fluorescence intensity steadily decreased with increasing HYP in the system. Furthermore, when the concentration of HYP was 4.2 × 10−5 M, the Trp residue's fluorescence intensity decreased by 73.65%. However, the fluorescence intensity of the Tyr residue decreased by only 65.86%, indicating that the Trp residue was attributed to the fluorescence quenching of lipase, probably because the Trp residue was close to the binding site of HYP in lipase. The Tyr and Trp residues were slightly red-shifted, indicating that the lipase chromophore of hydrophobicity decreased and the polarity increased.3D fluorescence is often utilized to investigate the structural changes in proteins bound to small molecules (HYP), and it shows the changes in protein Trp and Tyr residues.41 Fig. 2(C and D) reveals that with the expansion of the HYP concentrations, the fluorescence intensities of Peak 1 and 2 decreased. Peak 1 was the protein main chain peak, and Peak 2 was the overlapping fluorescence peak of Trp and Tyr. At the same time, the fluorescence of the Rayleigh scattering peak decreased. This indicated that as the interaction with HYP destroyed the protective water layer on the lipase surface, the originally dispersed protein became increasingly dispersed, the protein particle size decreased, and the Rayleigh scattering intensity of the system decreased. The difference in fluorescence intensity between Peaks 1 and 2 suggests that the binding relationship allowed for the progression of the lipase microenvironment and conformation. This result is in line with the previous fluorescence analysis.
Lipase has seven Trp residues, and its fluorescence is mainly attributed to these Trp emissions.42 According to the results of the fluorescence data, there was a correlation between the maximum wavelength of fluorescence emission (345 nm) and HYP concentration, indicating that HYP had altered the microenvironment of Trp. We inferred that this might be associated with the exposure of Trp residues and the unfolding or loosening of the local structure of lipase.43,44
3.4 Inhibition activity and inhibition type of HYP
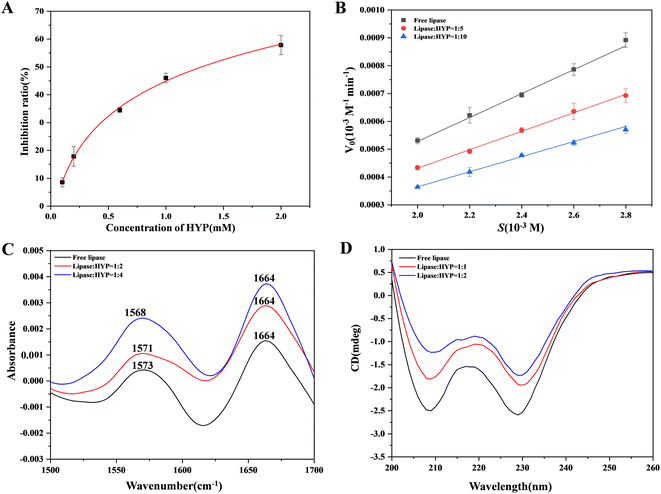
In the current study, the effect of various concentrations of HYP on lipase was evaluated in vitro. The inhibition of lipase activity by HYP is shown in Fig. 3A. The ratio of inhibition indicated that HYP inhibited lipase activity in a concentration-dependent manner. Moreover, the IC50 value of HYP on lipase was 1.92 × 10−3 M, and the IC50 value of orlistat on lipase was 6.0 × 10−5 M, indicating that HYP has inhibitory activity on lipase.45Generally, three types of interaction exist between inhibitors and enzymes: competitive, non-competitive, and mixed competitive inhibition. We can determine the suppression mode by calculating the Km and Vmax values.46 We compared the mechanisms of inhibition result of HYP and orlistat in the same manner. We found that the mechanisms of inhibition of HYP and orlistat were completely different. The inhibition of lipase with and without HYP was assessed by observing the hydrolysis of pNPC by lipase in Fig. 3B, and orlistat was regarded as a positive control in Fig. S2.† The results displayed in Fig. S1† indicated that the addition of HYP reduced the rate of pNPC hydrolysis, and its inhibition increased exponentially with the increase in HYP concentration (Fig. 3B). Our calculation also showed that the double reciprocal curves of the three groups of data roughly intersected the X-axis, implying that HYP is a non-competitive inhibitor to lipase. Meanwhile, according to the calculation results of the binding constant, HYP and lipase had medium binding constants (1.048 × 106 M−1 at 310 K), which may suggest that the binding force and inhibitory effect were related. Compared with the inhibition of HYP to lipase, orlistat had irreversible inhibition.47 With the increase in orlistat concentration, the double reciprocal curve of the three groups of data roughly intersected with the Y-axis, which is in line with the characteristics of competitive inhibition (Fig. S2†). Therefore, the inhibition type of this reaction was competitive inhibition. As a listed lipase inhibitor, orlistat has serious side effects on the human body. Therefore, considering the homologous safety of HYP and the binding properties between HYP and lipase, HYP can be a potential lipase inhibitor and needs to be verified by further in vivo experiments.
3.5 Conformational changes in lipase binding of HYP
FT-IR was utilized to investigate the changes in free lipase and after the expansion of various concentrations of HYP with lipase.48 As illustrated in Fig. 3C, the FT-IR spectra of free lipase and different concentrations of HYP binding with lipase showed two amide spectral bands. For the most part, the amide I band, which has been extensively examined, was sensitive to the progressions of protein. The stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O was represented by the amide I band, which was at 1700–1600 cm−1. The amide II band was at 1600–1500 cm−1, which could be viewed as a change in its structural content. With the increment in HYP concentrations, the peak intensity fluctuated. Moreover, in the amide I band (1664 cm−1), the shifts of all three groups of peaks were unchanged. Meanwhile, in the amide II band (1600–1500 cm−1), the peaks at the ratios of 1
O was represented by the amide I band, which was at 1700–1600 cm−1. The amide II band was at 1600–1500 cm−1, which could be viewed as a change in its structural content. With the increment in HYP concentrations, the peak intensity fluctuated. Moreover, in the amide I band (1664 cm−1), the shifts of all three groups of peaks were unchanged. Meanwhile, in the amide II band (1600–1500 cm−1), the peaks at the ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 were shifted by 2 and 5 cm−1, respectively. When the ratio of lipase to HYP was increased from 1
4 were shifted by 2 and 5 cm−1, respectively. When the ratio of lipase to HYP was increased from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 1
0 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, the α-helix in lipase decreased by 1.6%, and some changes occurred in the β-sheet, β-turn, and random coiling content. This result confirms that the addition of HYP slightly changed the conformation of lipase, which is consistent with the circular dichroism (CD) experimental results.
4, the α-helix in lipase decreased by 1.6%, and some changes occurred in the β-sheet, β-turn, and random coiling content. This result confirms that the addition of HYP slightly changed the conformation of lipase, which is consistent with the circular dichroism (CD) experimental results.
CD spectroscopy is a fast strategy to recognize conformational changes after adding a ligand.49 Fig. 3D shows that two characteristic negative peaks were presented at 208 and 228 nm, suggesting that lipase had a significant amount of α-helix. Even though the positions of the peaks were not significantly altered, the addition of HYP caused a change in all of the spectra. The presence of HYP still maintained the α-helix structure. The decrease in α-helix implied an increase in β-turn, β-sheet, and random coiling, but the typical secondary structure was affected. Thus, HYP was bound to some extent to the active site of lipase containing the α-helix, leading to structural instability.
3.6 Molecular docking
Molecular docking shows the binding energy and sites between proteins and small molecules.50 Fig. 4A shows that the binding energy for the best conformation of lipase and HYP binding after 100 operations was 35.06 kJ mol−1. HYP bound to Asp A:385, Asn A:295, Arg A:340, Asn A:329, and Asp A:388 to form hydrogen bonds, and the non-competitive components of HYP may be located at this site. Moreover, there was van der Waals force in the whole system (Fig. 4B), which was conducive to the conformational stability of the HYP–lipase complex. For example, HYP formed van der Waals interactions with amino acid residues Tyr A:373, Glu A:371, Ile A:372, Glu A:386, Thr A:293, Phe A:387, Leu B:41, and Pi–Pi T-shaped with amino acid residue Tyr A:370 with a bond length of 4.75 Å and 5.19 Å.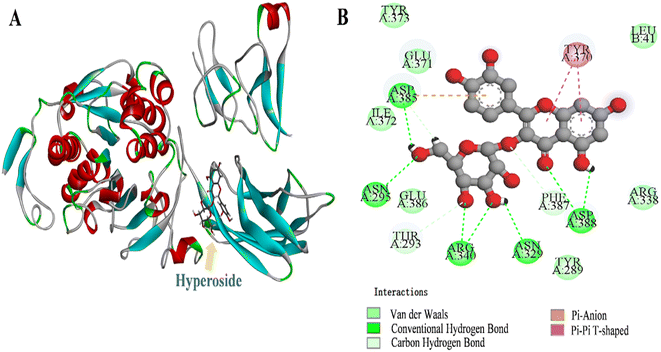 | ||
| Fig. 4 The 3D depiction of the HYP binding modalities with lipase. (B) Two–dimensional representation of the interaction maps between HYP and lipase. | ||
The active compound quercetin 3-rhamnoside has only one more oxygen atom than HYP.51 We found that the predicted types of interaction forces of molecular docking with lipase were van der Waals force and hydrogen bond, with the binding site of quercetin 3-rhamnoside being close to the active site of lipase. However, the binding site of HYP was far from the active site of lipase. The results of the inhibition mechanisms, together with the molecular docking results, speculated that HYP was bound to an essential group other than the active center, resulting in inhibited lipase activity.20 A previous work has found that the number of aromatic rings of small bioactive molecules and the ability of lipase to interact may also be related to the number of free hydroxyl groups in the structure.52
3.7 Molecular dynamics method
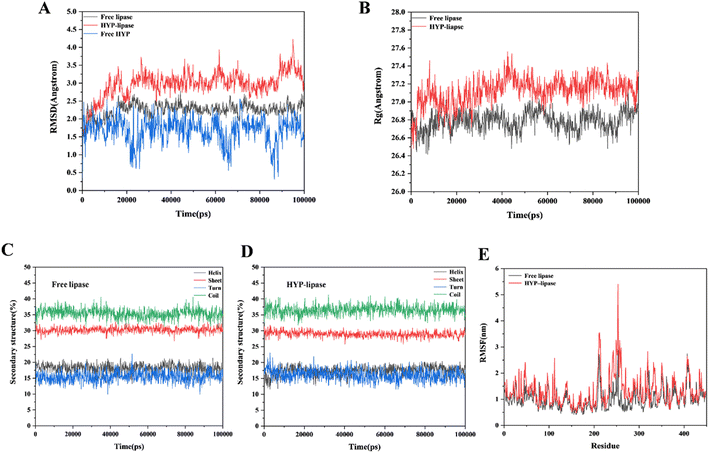
In accordance with the YASARA simulation platform, we evaluated the conformation and properties of the optimal conformational complex of HYP–lipase by using the change values of RMSD, Rg, secondary structure content and RMSF. RMSD values can be used to measure the stability of the system. From the RMSD analysis in Fig. 5A, it can be found that the system reaches equilibrium at about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps. Fig. 5A shows that within 20
000 ps. Fig. 5A shows that within 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 100
000 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps, the average value of RMSD was less than 6.0 Å in systems, indicating that the value was within an acceptable range. The average RMSD of the HYP–lipase complex was 3.08 Å, which is higher than that of free lipase (2.30 Å). The result suggests that the structure of free lipase may be more stable than that of the HYP–lipase complex. It can be seen that free lipase has been in a stable state after reaching equilibrium. Fig. 5E shows the fluctuations of each amino acid in both systems. The RMSF value of the corresponding amino acid in the HYP–lipase complex was higher than that of lipase, indicating that the HYP–lipase complex had higher mobility. This suggests that the structure of free lipase may be more stable than that of the HYP–lipase complex, corroborating with the RMSD value. Rg values can reflect the tightness of the protein structure to a certain extent, as exhibited in Fig. 5B. The average Rg value of free lipase was 26.80 Å, and that of the HYP–lipase complex was 27.16 Å. The binding radius slightly changed. Generally, the larger the Rg value of the system is, the more sparse the protein structure is.53 The system's Rg value increased as a result of the HYP binding, suggesting that the addition of HYP made the structure of lipase relatively unstable, resulting in a reduction of catalytic activity at the lipase's functional location.54 This finding corroborates the FT-IR and CD results.
000 ps, the average value of RMSD was less than 6.0 Å in systems, indicating that the value was within an acceptable range. The average RMSD of the HYP–lipase complex was 3.08 Å, which is higher than that of free lipase (2.30 Å). The result suggests that the structure of free lipase may be more stable than that of the HYP–lipase complex. It can be seen that free lipase has been in a stable state after reaching equilibrium. Fig. 5E shows the fluctuations of each amino acid in both systems. The RMSF value of the corresponding amino acid in the HYP–lipase complex was higher than that of lipase, indicating that the HYP–lipase complex had higher mobility. This suggests that the structure of free lipase may be more stable than that of the HYP–lipase complex, corroborating with the RMSD value. Rg values can reflect the tightness of the protein structure to a certain extent, as exhibited in Fig. 5B. The average Rg value of free lipase was 26.80 Å, and that of the HYP–lipase complex was 27.16 Å. The binding radius slightly changed. Generally, the larger the Rg value of the system is, the more sparse the protein structure is.53 The system's Rg value increased as a result of the HYP binding, suggesting that the addition of HYP made the structure of lipase relatively unstable, resulting in a reduction of catalytic activity at the lipase's functional location.54 This finding corroborates the FT-IR and CD results.
Table 2 summarizes the average values of the three types of energy terms in the HYP–lipase system within 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 100
000 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps of simulation. The numerical contribution of ΔEvdw and ΔEinter was relatively small in the whole system. However, ΔEelec had a high proportion in the total energy contribution, and its negative value, so it may contribute the most in the three kinds of energy terms. These observations indicated that the formed hydrogen bond as an electrostatic interaction may play a leading role in the interaction between HYP and lipase.55 The interaction between HYP and lipase and the subsequent changes in the conformation and stability of lipase may be attributed to the changes in interaction energy, including hydrogen bond, van der Waals and electrostatic interaction. Collectively, HYP mainly combined with lipase by hydrogen bond. These simulation results support the experimental results.
000 ps of simulation. The numerical contribution of ΔEvdw and ΔEinter was relatively small in the whole system. However, ΔEelec had a high proportion in the total energy contribution, and its negative value, so it may contribute the most in the three kinds of energy terms. These observations indicated that the formed hydrogen bond as an electrostatic interaction may play a leading role in the interaction between HYP and lipase.55 The interaction between HYP and lipase and the subsequent changes in the conformation and stability of lipase may be attributed to the changes in interaction energy, including hydrogen bond, van der Waals and electrostatic interaction. Collectively, HYP mainly combined with lipase by hydrogen bond. These simulation results support the experimental results.
| Complex | ΔEelec | ΔEvdw | ΔEinter | ΔE | |||
|---|---|---|---|---|---|---|---|
| ΔEbond | ΔEangle | ΔEdihedral | ΔEplanarity | ||||
| HYP–lipase | −16.124 | 1.863 | 0.985 | 0.606 | 1.231 | 0.010 | −11.429 |
| 2.832 | |||||||
We analyzed the secondary structure to further understand the conformational changes in free lipase and the HYP–lipase complex. Fig. 5(C and D) indicates that the β-turn content of the HYP–lipase complex increased within 0–2500 ps, tended to decrease, and finally reached equilibrium. On the contrary, the α-helix of the complex decreased in the range of 0–2500 ps and increased after 2500 ps until equilibrium. At the same time, the content of the coil and β-sheet in the complex presented a variation. In conclusion, the addition of HYP slightly interfered with the secondary structure of lipase to a certain extent.
All of the tests presented in this paper showed that the interaction between HYP and lipase occurred smoothly. However, upon the addition of HYP, the conformation of lipase was slightly changed. The strong signals at 208 and 228 nm (Fig. 3D) indicated the presence of the α-helix in lipase and a slight red shift, which may be indicative of the n → π* transition for the peptide bond of the α-helix.56 Based on the change in the secondary structure content within 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 100
000 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps in MD, the α-helix contents of free lipase and the HYP–lipase complex system were 18.10% and 17.39%, respectively, which decreased by 0.71% and showed an overall decreasing trend. This result is consistent with the FT-IR and CD results. The predominant conformation in lipase was the α-helix. According to the MD, FT-IR, and CD results, the α-helix content in lipase decreased upon the addition of HYP, and the decrease in the α-helix content could have had some effect on lipase activity and other functions. Studying the change in the relationship of the α-helix content in lipase is beneficial to providing clues for a deep understanding of its binding relationship with HYP and its effect on the biological function of lipase.
000 ps in MD, the α-helix contents of free lipase and the HYP–lipase complex system were 18.10% and 17.39%, respectively, which decreased by 0.71% and showed an overall decreasing trend. This result is consistent with the FT-IR and CD results. The predominant conformation in lipase was the α-helix. According to the MD, FT-IR, and CD results, the α-helix content in lipase decreased upon the addition of HYP, and the decrease in the α-helix content could have had some effect on lipase activity and other functions. Studying the change in the relationship of the α-helix content in lipase is beneficial to providing clues for a deep understanding of its binding relationship with HYP and its effect on the biological function of lipase.
3.8 STD-NMR experiment
STD-NMR technology is a powerful tool that can be used to screen small molecules to understand their interactions with macromolecules.57 In this study, we selected the protein-to-ligand concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 because it is appropriate.58 Moreover, the STD-NMR spectrum showed that the signal strength of the ligand protons was related to the tight binding of proteins. The 1H NMR spectra of HYP in the presence and absence of lipase are displayed in Fig. 6. Some hydrogen atoms present in the figure could not be seen in the spectrum due to the exchange of protons with D2O or because they did not reach the concentration of the ligand. Other hydrogen atoms were observed in the STD-NMR spectra, indicating that the hydrogen atoms were bound to lipase. An increased signal of HYP was observed in the presence of lipase, and slight changes occurred in the chemical shifts of all hydrogens due to the intermolecular interaction between HYP and lipase. According to the integrated peak areas, we infer that H-2, H-4, H-5, H-9, H-11, H-12, H-13, H-14, H-15, and H-16 had medium-intensity to strong-intensity interacting hydrogens with lipase, and H-6 had a weak affinity with lipase.
50 because it is appropriate.58 Moreover, the STD-NMR spectrum showed that the signal strength of the ligand protons was related to the tight binding of proteins. The 1H NMR spectra of HYP in the presence and absence of lipase are displayed in Fig. 6. Some hydrogen atoms present in the figure could not be seen in the spectrum due to the exchange of protons with D2O or because they did not reach the concentration of the ligand. Other hydrogen atoms were observed in the STD-NMR spectra, indicating that the hydrogen atoms were bound to lipase. An increased signal of HYP was observed in the presence of lipase, and slight changes occurred in the chemical shifts of all hydrogens due to the intermolecular interaction between HYP and lipase. According to the integrated peak areas, we infer that H-2, H-4, H-5, H-9, H-11, H-12, H-13, H-14, H-15, and H-16 had medium-intensity to strong-intensity interacting hydrogens with lipase, and H-6 had a weak affinity with lipase.
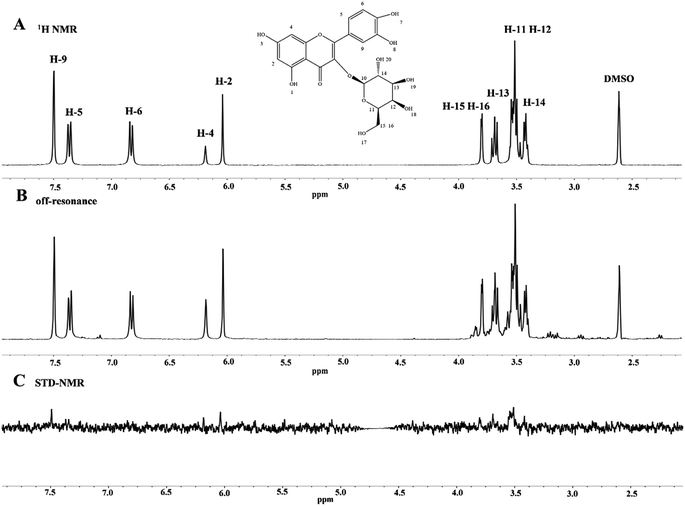 | ||
| Fig. 6 (A) 1H NMR spectrum of HYP alone (1.0 × 10−3 M). (B, C) The off-resonance spectra and STD-NMR of the HYP–lipase system (Clipase = 2.0 × 10−5 M, and CHYP = 1.0 × 10−3 M). | ||
4 Conclusions
In the present study, the interaction and inhibition mechanism of HYP to lipase were investigated using multi-spectral techniques, ITC, enzyme kinetics and MD methods. The complex formed by HYP and lipase resulted in static quenching. Meanwhile, the reaction between HYP and lipase became exothermic on its own. In the fluorescence spectrum, the addition of HYP generated a slight shift in the sample's maximum absorption wavelength, indicating a slight alteration in the microenvironment of the amino acid Trp residue and Tyr residue in lipase. Although HYP interacted with lipase, slight changes in its secondary structure occurred, which could be corroborated by the CD, FT-IR, and MD results. Furthermore, STD-NMR results revealed that HYP interacted with lipase. This study supplies a theoretical foundation for further development of HYP, a pancreatic lipase inhibitor of natural origin, as an anti-obesity supplement.Author contribution
Zhen Zeng: investigation, data curation; writing-original draft preparation. Di Wu: conceptualization, methodology, writing-reviewing and editing, project administration. Lan Tang: software, visualization. Xia Hu: data curation. Jing Zhang: supervision. Fang Geng: project administration, funding acquisition, resources.Conflicts of interest
All of the authors declare that there is no conflict of interests.Abbreviations
| HYP | Hyperoside |
| CD | Circular dichroism |
| FT-IR | Fourier-Transform Infrared |
| MD | Molecular dynamics |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| Rg | Radius of gyration |
| Trp | Tryptophan |
| Tyr | Tyrosine |
| STD-NMR | Saturation Transferred Differnce-Nuclear Magnetic Resonance |
| pNPC | p-Nitrophenol-cellobioside |
Acknowledgements
This work was supported by the fund of Science & Technology Department of Sichuan Provice. We express our sincere gratitude to all of the experts from the Analytical Testing Center of Sichuan University and the group of Prof. Li Hui from the School of Chemical Engineering for their help.References
- Y. H. Chang and H. Y. Hung, Eur. J. Med. Chem., 2022, 237, 114405 CrossRef CAS PubMed.
- J. Carretero Gómez, J. Ena, J. C. Arévalo Lorido, J. M. Seguí Ripoll, F. J. Carrasco-Sánchez, R. Gómez-Huelgas, M. I. Pérez Soto, J. Delgado Lista and P. Pérez Martínez, Rev. Clin. Esp., 2021, 221, 509–516 CrossRef PubMed.
- T. T. Liu, X. T. Liu, Q. X. Chen and Y. Shi, Biomed. Pharmacother., 2020, 128, 110314 CrossRef CAS PubMed.
- M. Ying, M. D. Meti, H. Xu, Y. Wang and J. Lin, Int. J. Biol. Macromol., 2018, 120, 1345–1353 CrossRef CAS PubMed.
- M. E. Lowe, Gastroenterology, 1994, 107, 1524–1536 CrossRef CAS PubMed.
- X. Huang, J. Zhu, L. Wang, H. Jing, C. Ma, X. Kou and H. Wang, Int. J. Biol. Macromol., 2020, 164, 1927–1938 CrossRef CAS PubMed.
- E. M. H. Mathus-liegen, M. L. V. I.-a. Leeuwen and A. Terpstra, Aliment. Pharmacol. Ther., 2015, 19, 601–611 CrossRef PubMed.
- A. K. Kakkar and N. Dahiya, Eur. J. Intern. Med., 2015, 26, 89–94 CrossRef CAS PubMed.
- M. González-Castejón and A. Rodriguez-Casado, Pharmacol. Res., 2011, 64, 438–455 CrossRef PubMed.
- M. Singh, T. Thrimawithana, R. Shukla and B. Adhikari, Future Foods, 2020, 1–2, 100002 CrossRef.
- Y. Zhang, X. Z. Zhang, Y. Wang and J. G. Liu, Zeitschrift für Naturforschung A, 2017, 72, 77–86 CrossRef CAS.
- X. Han, W. Li, D. Huang and X. Yang, Chem.-Biol. Interact., 2016, 257, 132–140 CrossRef CAS PubMed.
- X. Wang, F. Peng, F. Liu, Y. Xiao, F. Li, H. Lei, J. Wang, M. Li and H. Xu, Lwt, 2020, 133, 109869 CrossRef CAS.
- D. Wu, L. Tang, Z. Zeng, J. Zhang, X. Hu, Q. Pan, F. Geng and H. Li, Food Chem., 2022, 386, 132837 CrossRef CAS PubMed.
- Z. Li, J. Hu, Y. L. Li, F. Xue, L. Zhang, J. Q. Xie, Z. H. Liu, H. Li, D. H. Yi, J. C. Liu and S. W. Wang, Free Radical Biol. Med., 2013, 57, 132–140 CrossRef CAS PubMed.
- X. Hu, H. Li, L. Fu, F. Liu, H. Wang, M. Li, C. Jiang and B. Yin, Microb. Pathog., 2019, 127, 116–120 CrossRef CAS PubMed.
- N. Verma, G. Amresh, P. K. Sahu, N. Mishra, C. V. Rao and A. P. Singh, Indian J. Exp. Biol., 2013, 51, 65–72 CAS.
- D. Wu, R. Duan, L. Tang, X. Hu, F. Geng, Q. Sun, Y. Zhang and H. Li, Food Chem., 2021, 359, 129960 CrossRef CAS PubMed.
- R. Huang, Y. Zhang, S. Shen, Z. Zhi, H. Cheng, S. Chen and X. Ye, Food Chem., 2020, 326, 126785 CrossRef CAS PubMed.
- A. I. Martinez-Gonzalez, E. Alvarez-Parrilla, A. G. Diaz-Sanchez, L. A. de la Rosa, J. A. Nunez-Gastelum, A. A. Vazquez-Flores and G. A. Gonzalez-Aguilar, Food Technol. Biotechnol., 2017, 55, 519–530 CAS.
- D. Wu, L. Tang, R. Duan, X. Hu, F. Geng, Y. Zhang, L. Peng and H. Li, Food Chem., 2021, 347, 129052 CrossRef CAS PubMed.
- A. Sadat, M. G. Corradini and I. J. Joye, Curr. Res. Food Sci., 2022, 5, 479–490 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, Plenum Press, 1983 Search PubMed.
- D. Wu, S. Mei, R. Duan, F. Geng, W. Wu, X. Li, L. Cheng and C. Wang, Food Chem., 2020, 303, 125407 CrossRef CAS PubMed.
- H. Yang, Q. Zeng, Z. He, D. Wu and H. Li, J. Pharm. Biomed. Anal., 2020, 178, 112962 CrossRef CAS PubMed.
- D. Wu, D. Y. Liu, Y. Zhang, Z. Zhang and H. Li, Eur. J. Med. Chem., 2018, 146, 245–250 CrossRef CAS PubMed.
- S. Kitamura, K. Nakatani, T. Takaha and S. Okada, Macromol. Rapid Commun., 1999, 20, 612–615 CrossRef CAS.
- T. S. Banipal, A. Kaur, I. A. Khan and P. K. Banipal, RSC Adv., 2016, 6, 34754–34769 RSC.
- R. Goncalves, N. Mateus and V. De Freitas, J. Agric. Food Chem., 2010, 58, 11901–11906 CrossRef CAS PubMed.
- G. Rabbani, E. J. Lee, K. Ahmad, M. H. Baig and I. Choi, Mol. Pharm., 2018, 15, 1445–1456 CrossRef CAS PubMed.
- P. Chanphai, T. J. Thomas and H. A. Tajmir-Riahi, RSC Adv., 2016, 6, 53690–53697 RSC.
- H. Land and M. S. Humble, Methods Mol. Biol., 2018, 43–67 CrossRef CAS PubMed.
- J. Y. Zheng, X. Q. Zheng, L. Zhao, J. J. Yi and S. B. Cai, Curr. Res. Food Sci., 2021, 4, 543–550 CrossRef CAS PubMed.
- W. Wu, X. Hu, Z. Zeng, D. Wu, H. Li and H. Li, J. Phys. Chem. B, 2023, 127, 874–883 CrossRef CAS PubMed.
- I. Massova and P. A. Kollman, J. Am. Chem. Soc., 1999, 121, 8133–8143 CrossRef CAS.
- C. Y. Zhang, J. Zhang, H. Rao, J. Yang, X. Wang and X. Peng, Microchem. J., 2020, 159, 105529 CrossRef CAS.
- M. R. Ajmal, A. S. Abdelhameed, P. Alam and R. H. Khan, Spectrochim. Acta, Part A, 2016, 159, 199–208 CrossRef CAS PubMed.
- S. Wang, S. Dong, R. Zhang, H. Shao and Y. Liu, Process Biochem., 2014, 49, 237–243 CrossRef CAS.
- J. R. Lakowicz, Mechanisms and Dynamics of Fluorescence Quenching, Springer US, 2006, vol. 3, pp. 331–351 Search PubMed.
- G. Zhao, L. Zhu, P. Yin, J. Liu, Y. Pan, S. Wang, L. Yang, T. Ma, H. Liu and X. Liu, Food Chem., 2022, 368, 130857 CrossRef CAS PubMed.
- J. Li, R. Tian, G. Liang, R. Shi, J. Hu and Z. Jiang, Food Chem., 2021, 355, 129617 CrossRef CAS PubMed.
- X. Wu, W. He, H. Zhang, Y. Li, Z. Liu and Z. He, Food Chem., 2014, 142, 306–310 CrossRef CAS PubMed.
- C. D. Kanakis, I. Hasni, P. Bourassa, P. A. Tarantilis, M. G. Polissiou and H.-A. Tajmir-Riahi, Food Chem., 2011, 127, 1046–1055 CrossRef CAS PubMed.
- N. Tayeh, T. Rungassamy and J. R. Albani, J. Pharm. Biomed. Anal., 2009, 50, 107–116 CrossRef CAS PubMed.
- S. Li, J. Pan, X. Hu, Y. Zhang, D. Gong and G. Zhang, Process Biochem., 2020, 72 CrossRef.
- B. Liu, X. Cheng and H. Zhang, J. Pharm. Biomed. Res., 2019, 1, 53–60 Search PubMed.
- Y. Q. Li, P. Yang, F. Gao, Z. W. Zhang and B. Wu, Eur. Food Res. Technol., 2011, 233, 63–69 CrossRef CAS.
- M. Jiang, M. X. Xie, D. Zheng, Y. Liu, X. Y. Li and X. Chen, J. Mol. Struct., 2004, 692, 71–80 CrossRef.
- B. Ahmad, S. Parveen and R. H. Khan, Biomacromolecules, 2006, 7, 1350–1356 CrossRef CAS PubMed.
- Y. Wang, G. W. Zhang, J. H. Pan and D. M. Gong, J. Agric. Food Chem., 2015, 63, 526–534 CrossRef CAS PubMed.
- D. Wu, R. Duan, L. Tang, X. Hu, F. Geng, Q. Sun, Y. Zhang and H. Li, Food Chem., 2021, 359, 129960 CrossRef CAS PubMed.
- A. I. Martinez-Gonzalez, A. G. Díaz-Sánchez, R. La, C. L. Vargas-Requena, B. J. Ismael and E. Alvarez-Parrilla, Molecules, 2017, 22, 669 CrossRef PubMed.
- X. Hu, Z. Zeng, J. Zhang, D. Wu, H. Li and F. Geng, Food Chem., 2023, 405, 134824 CrossRef CAS PubMed.
- S. K. Saha, M. Murmu, N. C. Murmu, I. Obot and P. Banerjee, Surf. Interfaces, 2018, 10, 65–73 CrossRef CAS.
- B. Zhang, X. Qi, J. Mao and X. Ying, Lwt, 2020, 127 Search PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS PubMed.
- J. Angulo, P. M. Enriquez-Navas and P. M. Nieto, Chem.–Eur. J., 2010, 16, 7803–7812 CrossRef CAS PubMed.
- S. Walpole, S. Monaco, R. Nepravishta and J. Angulo, Methods Enzymol., 2019, 615, 423–451 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra06715c |
| This journal is © The Royal Society of Chemistry 2023 |