Open Access Article
Open Access ArticleIdentification of peptide epitopes of the gp120 protein of HIV-1 capable of inducing cellular and humoral immunity†
Jazmín García-Machorroa,
Mara Gutiérrez-Sánchez b,
Diego Alexander Rojas-Ortegab,
Martiniano Belloc,
Sergio Andrade-Ochoade,
Sebastián Díaz-Hernándezc,
José Correa-Basurto
b,
Diego Alexander Rojas-Ortegab,
Martiniano Belloc,
Sergio Andrade-Ochoade,
Sebastián Díaz-Hernándezc,
José Correa-Basurto *c and
Saúl Rojas-Hernández
*c and
Saúl Rojas-Hernández *b
*b
aLaboratorio de Medicina de Conservación, Escuela Superior de Medicina, Instituto Politécnico. Plan de San Luis y Díaz Mirón s/n, Col. Casco de Santo Tomas, Delegación Miguel Hidalgo, C.P. 11340, Ciudad de México, Mexico
bLaboratorio de Inmunobiología Molecular y Celular, Sección de Estudios de Posgrado e Investigación, Escuela Superior de Medicina, Instituto Politécnico Nacional, México City, Mexico. E-mail: saulrohe@yahoo.com.mx
cLaboratorio de Diseño y Desarrollo de Nuevos Fármacos e Innovación Biotécnológica (Laboratory for the Design and Development of New Drugs and Biotechnological Innovation), Escuela Superior de Medicina, Instituto Politécnico Nacional. Plan de San Luis y Díaz Mirón s/n, Col. Casco de Santo Tomas, Delegación Miguel Hidalgo, C.P. 11340, Ciudad de México, Mexico. E-mail: corrjose@gmail.com
dFacultad de Ciencias Químicas, Universidad Autónoma de Chihuahua, Circuito Universitario S/N, 31125 Chihuahua, México
eEscuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Prolongación de Carpio y Plan de Ayala S/N, Colonia Santo Tomas, 11340 Ciudad de México, Mexico
First published on 20th March 2023
Abstract
The Human Immunodeficiency Virus (HIV-1) causes Acquired Immunodeficiency Syndrome (AIDS) and a high percentage of deaths. Therefore, it is necessary to design vaccines against HIV-1 for the prevention of AIDS. Bioinformatic tools and theoretical algorisms allow us to understand the structural proteins of viruses to develop vaccines based on immunogenic peptides (epitopes). In this work, we identified the epitopes: P1, P2, P10, P27 and P30 from the gp120 protein of HIV-1. These peptides were administered intranasally alone or with cholera toxin (CT) to BALB/c mice. The population of CD4+, CD8+ T lymphocytes and B cells (CD19/CD138+, IgA+ and IgG+) from nasal-associated lymphoid tissue, nasal passages, cervical and inguinal nodes was determined by flow cytometry. In addition, anti-peptides IgG and IgA from serum, nasal and vaginal washings were measured by ELISA. The results show that peptides administered by i.n. can modulate the immune response of T and B lymphocyte populations, as well as IgA and IgG antibodies secretion in the different sites analyzed. In conclusion, bioinformatics tools help us to select peptides with physicochemical properties that allow the induction of the humoral and cellular responses that depend on the peptide sequence.
Introduction
Acquired immunodeficiency syndrome (AIDS) in humans is caused by two retroviruses, the human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2).1 They share a large number of similarities, including replication, transmission and clinical symptoms.2 However, HIV-1 is responsible for most of the HIV infections worldwide causing 680![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2020 and 37.7 million people living with HIV worldwide.3 It is important to mention that more than 80% of people with HIV-1 were infected by mucosal exposure.4 Due to the advance of the pandemic, the development of a vaccine against the virus has become a priority for scientists. However, the HIV genetic variability yields multiple genetic subtypes that make vaccine development difficult.5 The most genetic variability occurs in the surface gp120 and gp41 envelope (Env) glycoproteins that mediate virus entry into the T lymphocytes of host cells.6,7 Gp120 is very important for virus infection due to its initial recognition of some receptors such as DC-SIGN, heparan-sulfate proteoglycans and CD4 from T lymphocytes of the host.8 One of the proposed mechanisms to prevent viral entry into host CD4+ cells and consequently avoid the infection, is through the blockage of gp120-CD4 complex formation.9 In addition, the generation of neutralizing antibodies has attracted considerable attention, since they can compete with CD4 receptors, by joining the interface of both external and internal domains of the gp120 CD4 binding site (CD4bs).10 Several studies have demonstrated that neutralizing antibodies are able to block the gp120 making this glycoprotein interesting for developing safe and effective vaccines for HIV.4,11 However, to date, efforts to design and develop of HIV vaccines under traditional methods have not been successful.12 At present, there are in silico strategies which allow the identification of possible epitopes (immunogenic peptides) from the linear sequence of proteins.13 In addition, docking and molecular dynamics (MD) simulations are capable to discriminate among several epitopes based on differences in affinity and peptide–protein complex stability on the major histocompatibility complex (MHC) both MHC-I and MHC-II.14,15 In addition, the Quantitative Structure Activity-Relationship (QSAR) analysis allows obtaining the peptide properties that could aid to predict potential affinity between peptides on the MHC-I groove.16 Rodríguez-Fonseca and collaborators evaluated in silico dendrimer-G4-PAMAM-peptide complexes using three-dimensional (3D) models of the gp120 from HIV-1 that were intranasally administered, either peptides alone or complexes to female BALB/c mice. They determined that the peptides were immunogenic at systemic and mucosal levels (nasal and vaginal), and G4-PAMAM dendrimer–peptide complexes had better IgG and IgA response in serum and nasal washes.17
000 deaths in 2020 and 37.7 million people living with HIV worldwide.3 It is important to mention that more than 80% of people with HIV-1 were infected by mucosal exposure.4 Due to the advance of the pandemic, the development of a vaccine against the virus has become a priority for scientists. However, the HIV genetic variability yields multiple genetic subtypes that make vaccine development difficult.5 The most genetic variability occurs in the surface gp120 and gp41 envelope (Env) glycoproteins that mediate virus entry into the T lymphocytes of host cells.6,7 Gp120 is very important for virus infection due to its initial recognition of some receptors such as DC-SIGN, heparan-sulfate proteoglycans and CD4 from T lymphocytes of the host.8 One of the proposed mechanisms to prevent viral entry into host CD4+ cells and consequently avoid the infection, is through the blockage of gp120-CD4 complex formation.9 In addition, the generation of neutralizing antibodies has attracted considerable attention, since they can compete with CD4 receptors, by joining the interface of both external and internal domains of the gp120 CD4 binding site (CD4bs).10 Several studies have demonstrated that neutralizing antibodies are able to block the gp120 making this glycoprotein interesting for developing safe and effective vaccines for HIV.4,11 However, to date, efforts to design and develop of HIV vaccines under traditional methods have not been successful.12 At present, there are in silico strategies which allow the identification of possible epitopes (immunogenic peptides) from the linear sequence of proteins.13 In addition, docking and molecular dynamics (MD) simulations are capable to discriminate among several epitopes based on differences in affinity and peptide–protein complex stability on the major histocompatibility complex (MHC) both MHC-I and MHC-II.14,15 In addition, the Quantitative Structure Activity-Relationship (QSAR) analysis allows obtaining the peptide properties that could aid to predict potential affinity between peptides on the MHC-I groove.16 Rodríguez-Fonseca and collaborators evaluated in silico dendrimer-G4-PAMAM-peptide complexes using three-dimensional (3D) models of the gp120 from HIV-1 that were intranasally administered, either peptides alone or complexes to female BALB/c mice. They determined that the peptides were immunogenic at systemic and mucosal levels (nasal and vaginal), and G4-PAMAM dendrimer–peptide complexes had better IgG and IgA response in serum and nasal washes.17
Therefore, in order to have potential candidates for HIV-1 vaccines, in this work we focused to identify gp120 epitopes by combining bioinformatic epitope predictors, molecular docking, molecular dynamics (MD) simulations and QSAR studies. Once finished the in silico analyses, the peptides were synthesized and administered intranasally alone or with cholera toxin to BALB/c mice to assess their ability to induce CD4+ or CD8+ positive cells, plasma cells, IgG and IgA-specific antibodies.
Materials and methods
Theoretical procedure
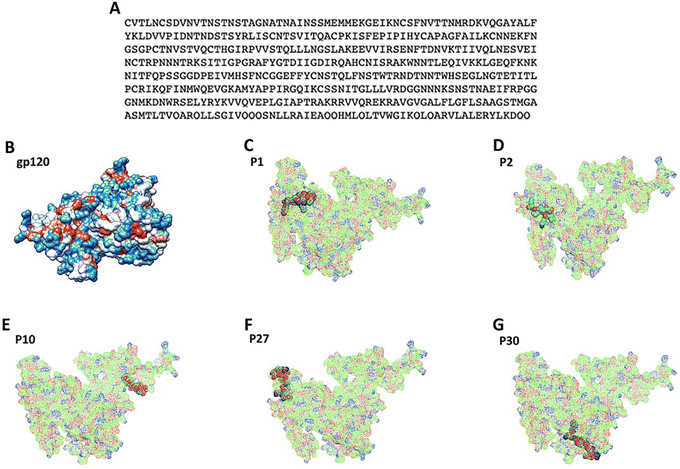
Homology modeling. The Swiss-Model (http://swissmodel.expasy.org/) server was used for the automatic homology modeling19–21 of the consensus sequence (Fig. 1A). The built 3D structures obtained were completed using the Modeller program (http://www.salilab.org/modeller/).22 The 3D structure was subjected to analysis by the RAMPAGE Ramachandran (http://eds.bmc.uu.se/ramachan.html) server in order to validate the structure quality.23
| QSAR models | |
|---|---|
a EHOMO = energy of the HOMO orbital; Qtot = total absolute charge; Alog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = Ghose–Crippen octanol–water partition coefficient; AMR = molar refractivity; ICR = radial centric information index; Hy = hydrophilic factor; I = ionization potential. P = Ghose–Crippen octanol–water partition coefficient; AMR = molar refractivity; ICR = radial centric information index; Hy = hydrophilic factor; I = ionization potential. |
|
| Model 1 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 = −0.32477(EHOMO) + 0.31845(Qtot) − 0.023(AMR) − 0.17143(Alog IC50 = −0.32477(EHOMO) + 0.31845(Qtot) − 0.023(AMR) − 0.17143(Alog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) − 2.31748 P) − 2.31748 |
| Model 2 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 = 0.34514(EHOMO) − 5.08622(ICR) + 0.61229(Qtot) − 0.23163(Hy) − 7.56378 IC50 = 0.34514(EHOMO) − 5.08622(ICR) + 0.61229(Qtot) − 0.23163(Hy) − 7.56378 |
| Model 3 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50 = −0.41182(I) + 4.77837(ICR) − 0.31823(Qtot) − 0.13378(Alog IC50 = −0.41182(I) + 4.77837(ICR) − 0.31823(Qtot) − 0.13378(Alog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) − 11.18058 P) − 11.18058 |
The search space included β-folded chains and α-helix chains of MHC following the previously published procedure.24 A box 70 × 100 × 90 Å with a grid spacing 0.375 Å3 was generated. Docking parameters used were 100 runs, with 100 million energy evaluations for each test, and a population size of 100 individuals.26 Peptide was treated as flexible. Results were analyzed using Autodock Tools software version 1.5.0 (https://autodocksuite.scripps.edu/adt/, February 2023) and figures were prepared with the Chimera software.27
Experimental procedures
The immunization schedule was carried out as was described by Rodríguez-Fonseca et al., 2019,17 briefly groups of 6 mice were immunized intranasal (i.n.), briefly mice were lightly anesthetized with ethyl ether, and subsequently applied doses with 30 μg of the peptide alone (P1, P2, P10, P27 and P30, respectively) or co-administered with CT, in total there were 3 immunizations on days 1, 7 and 14. The control mice received 30 μL of PBS.
For the analysis of the percentage of B cells and IgG and IgA plasma cells, the antibodies (BD biosciences) were used: CD19 PE, CD138 APC and IgA FITC and IgG FITC. The cells were stained according to the BD Bioscience protocol for the detection of intracellular staining. The signal intensities were measured and analyzed by means of a FACSAria flow cytometer (Becton Dickinson) for the performance of relative fluorescence. 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were collected in the FSC/SSC point diagram.
000 events were collected in the FSC/SSC point diagram.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution and nasal and vaginal washings 1
100 dilution and nasal and vaginal washings 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 dilution (100 μL) for all groups. Subsequently, the plates were washed with PBS-T and incubated with 100 μL goat anti-mouse IgG (Thermo scientific), anti-mouse IgA (Zymed Laboratories, San Francisco, CA) and anti-mouse IgM (Pierce, Rockford, IL) were added at a 1
2 dilution (100 μL) for all groups. Subsequently, the plates were washed with PBS-T and incubated with 100 μL goat anti-mouse IgG (Thermo scientific), anti-mouse IgA (Zymed Laboratories, San Francisco, CA) and anti-mouse IgM (Pierce, Rockford, IL) were added at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6000, 1
6000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 and 1
500 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 dilution respectively, the antibodies were incubated for 2 h at 37 °C. The plates were washed with PBS-T and the enzymatic reactions were started adding 100 μL o-phenylenediamine 0.5 mg mL−1 in phosphate-citrate buffer 50 mM, pH = 5.2 in the presence of H2O2. After 15 min, the reactions were stopped with 50 μL of 2.5 M H2SO4 and the absorbance at 492 nm (A492) was measured in a Multiscan Ascent (Thermo Labsystems) microplate reader.
3000 dilution respectively, the antibodies were incubated for 2 h at 37 °C. The plates were washed with PBS-T and the enzymatic reactions were started adding 100 μL o-phenylenediamine 0.5 mg mL−1 in phosphate-citrate buffer 50 mM, pH = 5.2 in the presence of H2O2. After 15 min, the reactions were stopped with 50 μL of 2.5 M H2SO4 and the absorbance at 492 nm (A492) was measured in a Multiscan Ascent (Thermo Labsystems) microplate reader.Results
Bioinformatic results
126 full sequences for gp120 protein of HIV-1 (from group M subtype B) were found at NCBI (ESI:† Fig. S1), being this subtype B the most predominant in the developed countries of the world, such as the United States and European countries,30 and also, the most disseminated variant.31 These protein sequences were submitted to multiple alignment sequence analysis obtaining a consensus sequence (Fig. 1A). This consensus sequence was used to build the 3D model (Fig. 1B and S2A†) which corresponds to report by Liu et al.32 The Ramachandran analysis showed that 98% of the amino acids are in favorable regions (ESI:† Fig. S2B). Then, the consensus sequence was submitted to MHC-I epitope predictors (ProPred, MCHPred and IEDB) obtaining 30 peptides (Table 2).| Peptides | MCHPred | Predep | IEDB | |||
|---|---|---|---|---|---|---|
| No. | Sequence | Position | IC50 | Confidence | Energy | Percentile |
| P1 | YRLISCNTS | 76 | 23.77 | 0.78 | −0.64 | 57.5 |
| P2 | FYKLDVVPI | 59 | 0.55 | 1 | −4.38 | 88 |
| P3 | VQLNESVEI | 171 | 405.51 | 0.44 | −2.63 | 77 |
| P4 | VVQVEPLGI | 375 | 182.81 | 0.89 | −4.09 | 15.5 |
| P5 | IKQLQARVL | 460 | 17.5 | 1 | −1.15 | 70 |
| P6 | IRPVVSTQL | 139 | 70.47 | 1 | −1.27 | 74.5 |
| P7 | IVQQQSNLL | 435 | 7.33 | 1 | −3.33 | 17.15 |
| P8 | IRQAHCNIS | 209 | 20.99 | 0.89 | 1.16 | 71.5 |
| P9 | VITQACPKI | 45 | 74.82 | 0.89 | −4.05 | 72.5 |
| P10 | LGFLSAAGS | 407 | 4.97 | 1 | −3.98 | 86 |
| P11 | FFYCNSTQL | 264 | 32.36 | 1 | −4.02 | 29.5 |
| P12 | LRAIEAQQH | 443 | 181.55 | 0.89 | 0.49 | 62.5 |
| P13 | VPIDNTNDS | 65 | 241.55 | 0.89 | −0.67 | 48 |
| P14 | IRGQIKCSS | 322 | 72.61 | 0.89 | −0.33 | 82 |
| P15 | MLQLTVWGI | 452 | 17.99 | 0.89 | −4.96 | 16 |
| P16 | YKLDVVPID | 60 | 231.21 | 0.89 | −2.04 | 79 |
| P17 | LNGTETITL | 291 | 671.43 | 0.89 | −1.69 | 43.5 |
| P18 | YKVVQVEPL | 373 | 34.91 | 1 | −1.56 | 46.5 |
| P19 | FAILKCNNE | 108 | 1.61 | 0.89 | −0.79 | 58.5 |
| P20 | VVQREKRAV | 392 | 192.31 | 0.78 | −2.12 | 50.5 |
| P21 | LISCNTSV | 78 | 17.46 | 1 | −3.48 | 28.5 |
| P22 | LERYLKDQQ | 470 | 79.98 | 0.67 | 0.63 | 90.5 |
| P23 | LLSGIVQQQ | 431 | 51.4 | 1 | −1.73 | 59.5 |
| P24 | LSGIVQQQS | 432 | 318.42 | 1 | 1.44 | 28.5 |
| P25 | LTVWGIKQL | 445 | 127.94 | 0.89 | −2.22 | 46.5 |
| P26 | VWGIKQLQA | 457 | 260.62 | 1 | −4.15 | 36.5 |
| P27 | FNVTTNMRD | 42 | 6.53 | 1 | −2.19 | 68.5 |
| P28 | MTLTVQARQ | 422 | 37.5 | 0.89 | −1.72 | 30 |
| P29 | YAPPIRGQI | 318 | 6.32 | 1 | −2.5 | 72.5 |
| P30 | FNSTWTRND | 273 | 3.73 | 1 | 1.16 | 81 |
Also, the consensus sequence submitted to MHC-II epitope predictors identified some epitopes predicted for MHC-I, ProPred: P1, P2 and P10 (ESI:† Fig. S3), MCH2Pred: P10 and IEDB: P2, P3, P13, P14, P29 (Table 3). In addition, ElliPRO identified some B-cell epitopes (Table 4) from the 3D structure.13
| Peptides | ProPred | MHC2Pred | IEDB | |||
|---|---|---|---|---|---|---|
| No. | Sequence | Position | Score | IC50 | Confidence | Percentile |
| a WA = without activity. | ||||||
| P1 | YRLISCNTS | 76 | 58.3 | 903.65 | 0.89 | 31.12 |
| P2 | FYKLDVVPI | 59 | 57.2 | WA | WA | 89.62 |
| P3 | VQLNESVEI | 171 | 34 | 3243.4 | 0.89 | 88.01 |
| P4 | VVQVEPLGI | 375 | 30.83 | 1364.58 | 0.78 | 18.25 |
| P5 | IKQLQARVL | 460 | 26.5 | 3715.35 | 0.89 | 4.16 |
| P6 | IRPVVSTQL | 139 | 24 | 1174.9 | 1 | 28.42 |
| P7 | IVQQQSNLL | 435 | 36.67 | 4742.42 | 0.89 | 9.45 |
| P8 | IRQAHCNIS | 209 | 39.77 | 1534.62 | 1 | 76.18 |
| P9 | VITQACPKI | 85 | 32.83 | 2747.89 | 1 | 62.18 |
| P10 | LGFLSAAGS | 407 | 54.09 | 9.68 | 1 | 2.91 |
| P11 | FFYCNSTQL | 264 | 31.09 | 2249.05 | 0.89 | 15.4 |
| P12 | LRAIEAQQH | 443 | 36.04 | 2971.67 | 0.89 | 32.89 |
| P13 | VPIDNTNDS | 65 | 36.36 | 933.25 | 0.78 | 81.48 |
| P14 | IRGQIKCSS | 322 | 33.71 | 2415.46 | 0.89 | 86.37 |
| P15 | MLQLTVWGI | 452 | 31.33 | 4965.92 | 1 | 20.96 |
| P16 | YKLDVVPID | 60 | 28.65 | 1224.62 | 1 | 76.36 |
| P17 | LNGTETITL | 291 | 2.64 | 1279.38 | 0.89 | 72.08 |
| P18 | YKVVQVEPL | 373 | 59.48 | 1364.58 | 0.78 | 13.37 |
| P19 | FAILKCNNE | 108 | 32.58 | WA | WA | 54.81 |
| P20 | VVQREKRAV | 392 | 19.28 | WA | WA | 65.58 |
| P21 | LISCNTSV | 78 | 3.37 | 446.68 | 1 | 45.17 |
| P22 | LERYLKDQQ | 470 | 1.63 | 2857.59 | 1 | 43.79 |
| P23 | LLSGIVQQQ | 431 | 1.76 | 2259.44 | 1 | 28.79 |
| P24 | LSGIVQQQS | 432 | 0.8 | 843.33 | 0.89 | 28.79 |
| P25 | LTVWGIKQL | 445 | 1.86 | 847.23 | 0.89 | 32.72 |
| P26 | VWGIKQLQA | 457 | 2.26 | 3715.35 | 0.89 | 32.72 |
| P27 | FNVTTNMRD | 42 | 38.09 | 3126.08 | 0.89 | 64.64 |
| P28 | MTLTVQARQ | 422 | 11.6 | 3435.58 | 1 | 35.38 |
| P29 | YAPPIRGQI | 318 | 16.59 | 2857.59 | 0.89 | 86.37 |
| P30 | FNSTWTRND | 273 | 22.34 | WA | WA | 76.14 |
| No. | Peptides | Position | Score |
|---|---|---|---|
| P31 | RQLLSGIVQ(P24)QQSNLLRAIEAQQHMLQLTVWGI(P15)KQL(P25)QARVLALERYLKDQQ(P22) | 423 | 0.923 |
| P32 | FNVTTNMRD(P27)KVQGAYALFYKLDVVPI(P2) | 43 | 0.866 |
| P33 | GAYALFYKLDVVPI(P2)DNT | 55 | 0.833 |
| P34 | LISCNTSV(P21) | 79 | 0.825 |
| P35 | YLKDQQ(P22) | 474 | 0.989 |
| P36 | RVLALER | 467 | 0.981 |
| P37 | LTVWGIKQL(P25)QA | 456 | 0.96 |
| P38 | QQSNLL | 439 | 0.911 |
| P39 | LSGIVQ(P24) | 433 | 0.878 |
| P40 | MTLTVQARQ(P28)L | 423 | 0.821 |
Finally, the PaPROC server allow us to discard some peptides that suffer degradation (ESI:† Table S1). As it is known, antibodies must be able to reach the peptides on the 3D structure of gp120 selecting those peptides exposed on protein surface (Fig. 1C–G).
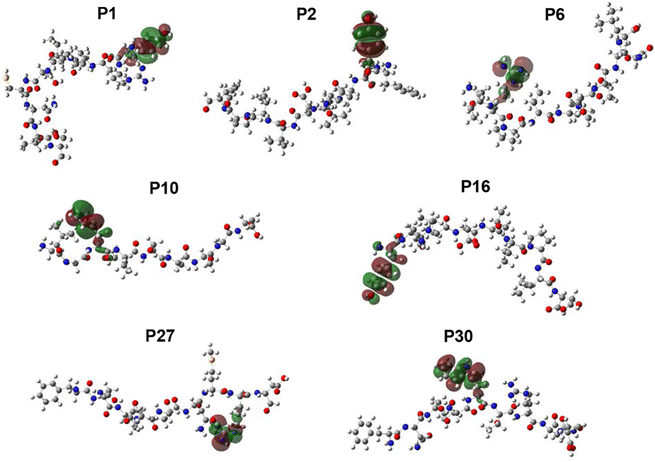
After all bioinformatic analyses described, five peptides were identified as promissory: P2 (FYKLDVVPI), P6 (IRPVVSTQL), P10 (LGFLSAAGS), P16 (YKLDVVPID) and P30 (FNSTWTRND). These peptides were submitted to QSAR using a mathematical prediction model.33 Values of descriptors calculated for the five peptides exposed in the gp120 protein can be observed in ESI† Table S2, while in Table 5 the prediction of IC50 is found and the HOMO–LUMO densities (Fig. 2). The QSAR studies showed P2 as the most promissory according to the IC50 values more favored according to model 1 and 3 (Table 5).
| Peptides | Model 1 | Model 2 | Model 3 | ||||
|---|---|---|---|---|---|---|---|
| No. | Sequence | log | IC50 | log | IC50 | log | IC50 |
| P2 | FYKLDVVPI | −1.06 | 0.09 | 0.60 | 4.03 | −0.90 | 0.13 |
| P6 | IRPVVSTQL | 0.52 | 3.34 | 2.39 | 243.27 | 1.57 | 36.79 |
| P10 | LGFLSAAGS | 0.41 | 2.55 | 1.63 | 42.29 | 1.43 | 27.06 |
| P16 | YKLDVVPID | 1.01 | 10.26 | 1.86 | 72.46 | 1.89 | 78.25 |
| P30 | FNSTWTRND | 0.25 | 1.77 | 0.52 | 3.32 | 1.80 | 62.51 |
 | ||
| Fig. 2 Map of Highest Occupied Molecular Orbital (HOMO) of the peptides exposed in the three-dimensional structures of the HIV-1 gp120 protein. | ||
Experimental results
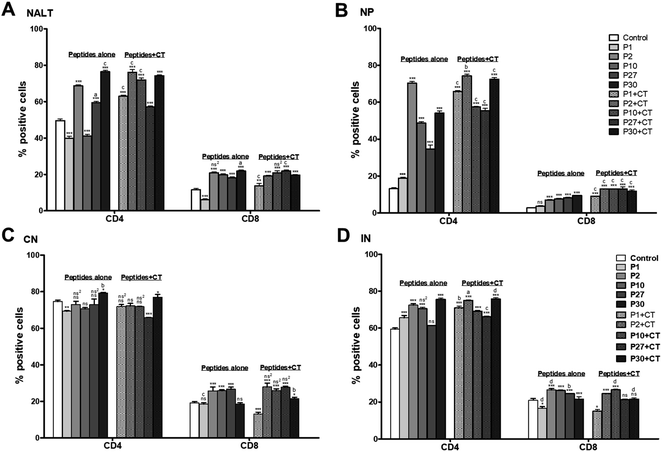
Regarding CD8+ T lymphocytes, there was a significant increase (P < 0.001) for all the peptides, except P1, which was significantly lower than the control group (P < 0.001). When the peptides were administered with CT, there was a significant increment in all the cases compared to the control group (P < 0.001), being P27 which had the highest CD8+ T cell levels compared to the other peptides (P < 0.001) (Fig. 3, Panel A).
Regarding to the lymphocyte phenotype in NP, the percentage of CD4+ lymphocytes was significantly higher with the peptides alone and peptides co-administered with CT compared to the control group (P < 0.001). P2 showed a significantly higher increment compared to the rest of the peptides, either given alone or with CT (P < 0.001). However, it was significantly higher co-administered with CT than alone (P < 0.01). Like CD4+ cells, CD8+ T lymphocytes population showed a significant increment with peptides administered alone and peptides co-administered with CT compared to the control group (P < 0.001), except for P1 treatment, which did not show a significant difference with the control group. Particularly, P2, P10, and P27 co-administrated with CT had a significantly higher increment compared with the other peptides (P < 0.001) (Fig. 3, Panel B).
The percentage of CN lymphocytes show that CD4+ T lymphocytes decreased with the peptides alone or even with CT. Except, P30 alone or with CT which significantly increased the percentage of CD4+ compared to the control group (P < 0.05). P30 alone was significantly higher than P30 co-administered with CT (P < 0.05). Concerning CD8+ T lymphocytes, when P2, P10, and P27 were administered alone or with CT, they showed a significant increment in the cells percentage (P < 0.001) compared to the control group (Fig. 3, Panel C).
The percentage for CD4+ T lymphocytes with P1, P2, P10, and P30 administered alone increased compared to the control group (P < 0.001). P27 did not show a significant difference respect to the control. Instead, all the peptides co-administered with CT increased significantly compared to the control group (P < 0.001). Emphasizing P30 alone or with CT which showed a significantly higher increment than other peptides. For CD8+ T lymphocytes, most of the peptides either alone or co-administered with CT promoted a significant increment compared to the control group (P < 0.001), except for P27 with CT and P30 with CT which did not present a significant difference. While P2 alone or co-administrated with CT decreased compared to the control group (P < 0.05) (Fig. 3, Panel D).
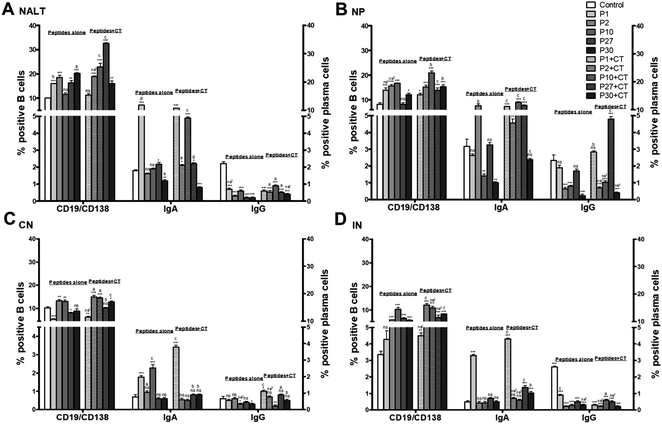
The results obtained for NALT show a significant increase (P < 0.001) for CD19/CD138 positive cells after immunization with the peptides alone (except P10) compared to the control group. When the peptides were co-administered with CT, most of the peptides showed a significant increment compared to the control group (P < 0.001), only P1 with CT did not have a significant difference compared to the control group. However, P27 with CT was significantly higher than the other peptides (P < 0.001). Concerning IgA positive plasma cells, P1 alone significantly increased the percentage of cells compared to the control (P < 0.001). Finding that P1 was significantly higher than the rest of the peptides (P < 0.001). On the other hand, the administration of the peptides either alone or with CT significantly decreases the percentage of IgG positive plasma cells compared to the control group (P < 0.001) (Fig. 4, Panel A).
Regarding to the peptide immunization in the NP, P1, P2, and P10 had a significant increment in the percentage of cells positive for CD19/CD138 (P < 0.001) compared to the control group. P10 with CT was significantly higher than the other peptides (P < 0.001). The analyses of IgA positive plasma cells showed that only P2 significantly increased the percentage of cells (P < 0.001) compared to the control group. While peptides with CT in most cases increased the percentage of IgA compared to the control group (P < 0.001), except for P30 with CT where no significant difference was found. P10 with CT was significantly higher than all the peptides (P < 0.001). The immunization effect in the positive IgG cell percentage was completely different than it was for IgA cells where the intranasal administration with the peptides alone decreased the percentage of IgG cells in most cases (P < 0.001). Only P27 showed a significant increment of IgG positive cells compared to the control and the other peptides (P < 0.001) (Fig. 4, Panel B).
The percentage of B lymphocytes in CN show that P2 and P10 significantly increased the cells (P < 0.01). P30 did not show a significant difference, P1 (P < 0.001), and P27 (P < 0.05) showed a significant decrease compared with the control group. Interestingly, the administration of P2 and P10 with CT also significantly increased the percentage of CD19/CD138 positive cells compared to the control group (P < 0.001) and compared to the other peptides (P < 0.05). In the case of IgA-positive cells, the immunization with P1 or P10 significantly increased the percentage (P < 0.001); while P2, P27, and P30 did not show a significant difference to the control group. When the peptides were co-administered with CT, only P1 showed a significant increase compared to the control group, and it was significantly higher than the other peptides (P < 0.001). Regarding the percentage of plasma cells positive for IgG, the immunization with the peptides alone or with CT did not modify the percentage of cells, except for P1 co-administered with CT, which was the treatment that significantly increased the IgG-positive cells compared to the control and the other peptides (P < 0.05) (Fig. 4, Panel C).
In addition, the results for the B lymphocytes in IN show that P1 did not have a significant difference to the control group, most of the peptides either alone or with CT were significantly higher than the control group (P < 0.001), however, P1 with CT did not have a significant difference to the control group. P2 with CT was significantly higher than all the other peptides (P < 0.001). Regarding the populations of plasma cells positive for IgA, only P1 induced a significant increase in the percentage of cells (P < 0.001), while the other peptides did not induce significant differences. P1, P27, and P30 with CT significantly increased the percentage of cells compared with the control group. P1 with CT was significantly higher than all the other peptides either alone or with CT (P < 0.001). The IgG-positive plasma cells populations significantly decreased with the administration of peptides alone or peptides with CT compared to the control group (P < 0.001) (Fig. 4, Panel D).
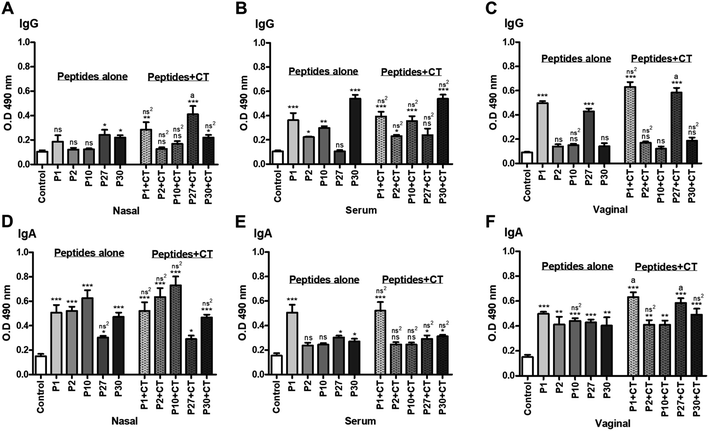
The specific IgG in the serum shown that most of the peptides administered alone were capable to increment this antibody compared to the control where P1 and P30 had a difference of P < 0.001; while P2 with P < 0.05 and P10 with P < 0.01. In contrast, P27 did not present a significant difference to the control group. The administration of peptides with CT showed the same pattern as those administrated alone, finding that there was not a significant difference compared to the treatment with peptides alone (Fig. 5, Panel B).
Regarding to the specific IgG in vaginal washes, only P1 and P27 alone or co-administered with CT had a significant increase in anti-P1 and anti-P27 IgG levels compared to the control group (P < 0.001), and such levels were significantly higher than the other peptides (P < 0.05) (Fig. 5, Panel C).
The presence of specific IgA in nasal washes shown that peptides alone increase the specific IgA anti-peptides compared to the control group (P < 0.001). Although immunization with P27 had a slight increase, this was still significant (P < 0.05). IgA levels from immunized mice with peptides plus CT showed a similar pattern where a significant difference was found compared to the control mice. It is important to mention that in both cases, immunization with peptides alone as well as with CT had a similar IgA response, however, there are no significant differences between them (Fig. 5, Panel D).
About the specific IgA anti-peptides in serum, the P1 increased anti-P1 IgA levels compared with the control group (P < 0.001). P27 and P30 immunization slightly increased IgA anti-P27 and anti-P30 compared to the control group (P < 0.05). The administration of P1 plus CT also showed that anti-P1 IgA levels was significantly increased compared to the control group (P < 0.001). Although P27 + CT and P30 + CT slightly increase anti-P27, and anti-P30 IgA levels compared to the control group (P < 0.05). Again, we can observe that no significant differences were found in the IgA response between the groups of mice immunized with the peptides alone or those co-administered with CT (Fig. 5, Panel E).
Finally, the specific IgA from vaginal washes show an increment in those mice immunized with P1, P10, P27 (P < 0.001); P2 and P30 (P < 0.01) compared to the control group. When P1, P27 and P30 (P < 0.001), P2 and P10 (P < 0.01) peptides were co-administered with CT, there was also a significant increment of IgA levels compared to the control group. Particularly, P1 with CT and P27 with CT were significantly higher than all the other peptides (P < 0.05) (Fig. 5, Panel F).
Discussion
The bioinformatics studies have become very important for the development of vaccines because the prediction of immunogenic peptides facilitates the design of friendly vaccines as has been performed for tuberculosis,34 Taenia solium,35 and even for cancer.36 The peptide predictors reduce development costs and prevent unwanted responses produced with vaccines derived from attenuated pathogens, either alive or dead.37 In this study there was used the consensus sequence (Fig. 1A) of full gp120 protein obtained after multiple alignment analyses. The consensus sequence was used for building the 3D structure and predicting epitopes to identify antigens (Ag) capable to induce humoral and cellular responses for vaccination purposes. The peptide prediction for MHC-I and MHC-II binding regions yielded several lineal epitopes, and the non-lineal prediction was achieved by ElliProt identifying P2 into peptides P32 and P33 (Table 2). Moreover, the 3D structure visualization of gp120 showed five exposed predicted epitopes which can be recognized by antibodies (Fig. 1C–G).13The QSAR model, which is based on predicting IC50 values, allows us to classify peptides that show affinity or not for the major histocompatibility complex (MHC), taking into account that lower IC50 has higher affinity.38 We previously reported results for calculating the IC50 of HIV-1 immunogenic peptides.24 In this work we use this model to calculate the IC50 of the peptides P2, P10, P27 and P30, obtaining outstanding IC50 values, however, the P2 was the better with a predicted IC50 of 0.09 mM and 0.13 mM for model 1 and model 3, respectively (Table 5), which could be explained as a consequence of its richness in electrons (Fig. 2), which favors its affinity to couple to MHC-I.24,38 In addition, P2 and P10 have Tyr and Phe residues (Fig. 2) that could play an important role in the stabilization of macromolecular complexes due to their aromatic properties.39 In addition, their spatial conformation of these peptides could suggest better recognition by antibodies generated through immunization.40
Peptides predicted and analyzed in silico (3D protein exposed, IC50 by QSAR) allow us to select the most promissory peptides (P1, P2, P10, P27 and P30) to be chemically synthesized and tested under experimental procedures. The target peptides were administered by intranasal immunization to female BALB/c mice, alone or with CT as adjuvant. Several studies have shown that intranasal immunization is effective in bringing together a local and systemic response. It is due to lymphocytes located at intranasal tissue can migrate by lymphatic system to other mucosal compartments reaching the respiratory tract, the gastric and genital tract,41 where lymphocytes perform effector functions such as cytokine release and antibody secretion.42–44 In addition, CT immunization as an adjuvant has been shown to modulate humoral responses, as well as modify populations of B and T lymphocytes, macrophages, and dendritic cells from various sites such as NALT, CN, NP, and spleen.29,45,46
These studies reinforce the results that we found since after intranasal immunization with the peptides (P1, P2, P10, P27, and P30) co-administered with CT increased the phenotype of CD4+ and CD8+ T lymphocytes in NALT, CN and IN compared to the control and immunized groups (Fig. 3).
It is important to note the presence of CD4+ or CD8+ T cells on these mentioned sites, may suggest that antigen-presenting cells travel to the distal mucosal compartments to generate a reservoir of cells from memory that can act when infection occurs as previously reported,47 and furthermore, according to Wu et al., 1997,48 the nasal mucous membranes drain better in the cervical ganglia thanks to the permeability achieved with the use of CT as an adjuvant.
Once dendritic cells (DCs) captured the antigen from lumen antigen or lamina proper of nasal, they travel to the nearest lymphoid nodule including the NALT and cervical ganglia, where they would be presenting the antigen to the T lymphocytes, thus initiating the inducing response.48 In addition, the effector cells both T and B lymphocytes which express integrins (α4β1, α4β7, etc.) as well as integrin receptors (CCR9, CCR10, etc.) on their surface that allow them to migrate to more distant nodules as the inguinal ganglia.49
The increase in the percentages of CD4+ and CD8+ T cells (particularly with P30 and P2 treatments) in NALT, CN and IN compared to the control group, suggests that the peptides are being captured by APC and processed for subsequent presentation to T lymphocytes. Then, the cell proliferation is carried out clonal by T cells, to have a favorable impact on the immune response, this could possibly have a positive effect on the challenge against virus infection, but more studies are needed to prove.
On the other hand, it is worth mentioning that the increase in B lymphocytes in the mice groups immunized with P2 and P10 in CN, as well as the increment in these B cells in all the mice groups immunized with each of the peptides (P1, P2, P10, P27 and P30) in IN, compared to the control group (Fig. 4 Sections C and D). These results suggests that Naive B lymphocytes were activated by the presence of the antigen and migrated to secondary lymphoid tissues. Activation may be mediated by dependent T antigens, a co-response with CD4 T lymphocytes, where in the germinal centers become plasma cells (PC) or memory cells, and there will also be a change immunoglobulin class.50 PCs are terminally differentiated in plasmatic cells that provide protective immunity by producing antigen-specific antibodies.51
Therefore, the percentage of PC was analyzed, either positive for IgA or IgG. Populations of IgA positive cells show different behaviors according to the analyzed tissues (NALT, NP, CN and IN) and are the responses are depending of the groups immunized with the peptides alone or co-administered with CT (Fig. 4, items A–D). P1 + CT increases the percentage of positive cells in all sites analyzed, except in the nostrils compared to control. In the case of IgG positive PC, a decrease in the cellular percentage of all groups of immunized mice was obtained in most of the analyzed sites regarding control; except in the nostrils, where P27 + CT increased the percentage compared to control (Fig. 4, items A–D). These results allowed us to question whether IgA and IgG were soluble in the serum of immunized mice. Furthermore, several reports suggest that immunization of soluble proteins with CT stimulates Th1/Th2 responses.52 Therefore, the presence of specific IgA and IgG antibodies against the designed peptides of HIV-1 gp120 in serum, nasal and vaginal washes was measured (Fig. 5, items A–F). In the case of IgG, there was observed that the administered peptides (P1, P2, P10, P27 and P30, respectively) did not show a significant difference with the control group in most cases (Fig. 5, items A–C); but when they were co-administered with CT, P1, and P27 result in an increased production of specific antibodies in nasal and vaginal lavages regarding control. While in serum, there was a significant increase in antibodies against the peptides being P30 with the best response either administered either alone or P30 + CT (Fig. 5B). Antibodies may participate in the activation of cellular effector functions of the innate response, such as antibody-mediated cytotoxicity or phagocytosis; it is mediated by the recognition of antibodies bound to the antigen by Fcγ receptors present in immune cells such as macrophages, B cells, PC and neutrophils.53 In the immune analysis of the RV144 HIV-1 vaccine efficacy trial conducted by Haynes et al., 2012,54 the estimated efficacy was significantly correlated with the binding of IgG antibodies to variable regions 1 and 2 (V1V2) of HIV-1 envelope proteins (Env) and the binding of IgA antibodies in plasma to Env, both mediated by the binding of antibodies against Fc receptors. This supports the idea that the FcR-binding Ig-HIV immunocomplex plays a critical role in protecting against HIV infection.
The tissues of the mucosa, such as the nasal or vaginal mucosa, contain IgA, although they can also contain IgG antibodies. The presence of IgG in the mucous membranes could be explained by the presence of the FcRn receptor, which mediates the transepithelial transport of IgG in various tissues, one of them the nasal mucosa.55 The PCs in the lamina propria of the mucosa are mainly responsible to produce local IgA. Its epithelial transport is attributed to the presence of pIgR in the epithelial cells of the mucosa, which carries IgA from the basolateral region of the cells to the apical region.56 The IgA and IgG titers in various tissues indicate that when the peptides were immunized alone or with the adjuvant, there was an elevation in the levels of specific antibodies (Fig. 5, items A–F). Antibodies present in mucous membranes and serum could have a neutralizing role for the HIV-1 virus and thus prevent infection. However, there are no long-term antibody protection findings in immunized patients.57 Studies are needed to verify the protection that peptide-specific antibodies can provide against HIV infection. Furthermore, finding an increment of IgG in groups of immunized mice in serum suggests that a systemic response against peptides is being achieved, this is also important since these antibodies could act against the spread of the virus to the lymph nodes.
As previously was mentioned, HIV infection has been shown to cause dysregulation of the B lymphocyte population, significant memory cell loss, and therefore also decreased antibody production;58,59 Therefore, it is a great result that the intranasal immunization with peptides from HIV-1 gp120 increase in IgA (in nasal and vaginal lavages) and IgG (in serum) (Fig. 5, items A–F). This suggests that the peptides can mount a local and systemic response when they are immunized intranasally and adjuvants with CT.
It is worth mentioning that the immune response induced by P2 does correspond with the results of the QSAR studies, which is a promising peptide to be a vaccine candidate (Table 5), however, we can say based on the experimental results that we found that all the peptides evaluated (P1, P2, P10, P27 and P30) are immunogenic, but this depended on the cell populations found of T and B lymphocytes, as well as IgA and IgG positive cells in the analyzed site, whether NALT, NP, CN and IN, as well as the levels of IgA and IgG antibodies in sera and nasal washes (Fig. 3–5). Therefore, the search and design of HIV-1 gp120 peptides using bioinformatics tools such as epitope prediction, docking and molecular dynamics simulations, as well as 3D structure analysis docking, binding site prediction and simulation are tools of great importance in the design of promising vaccines under a guide of strategies that could elicit a protective immune response against HIV.
Conclusions
Bioinformatics is useful in the prediction of epitopes with immunogenic potential; however, it is necessary to test in a biological system the induction of immune response (humoral and cellular response) since bioinformatic models are a mathematical approximation of some variables found in the organism, but do not consider all of them. In this work the information coincides for the peptides (P1, P2, P10, P27 and P30) studied that modify the response of both CD4 and CD8 lymphocytes as well as the response of IgA and IgG antibodies and this response was greater when the peptides were co-administered with CT. Another important aspect that is worth highlighting is that intranasal immunization is capable of inducing responses of both IgA and IgG antibodies, effector CD4 and CD8 T lymphocytes as well as cells positive for IgA and IgG locally (NALT and lamina propria) as well as in other mucosal compartments such as CN and IN. These strategies for the design of vaccines must be able not only to induce immunogenic responses but also to be immunoprotective; therefore, the use of bioinformatics tools is convenient for the choice of immunogenic epitopes, the immunization route, and the use of adjuvants.Abbreviations
| AIDS: | Acquired immunodeficiency syndrome |
| APC: | Allophycocyanin |
| AuNPs: | Gold nanoparticles |
| CN: | Cervical nodes |
| CT: | Cholera toxin |
| FITC: | Fluorescein isoTioCyanat |
| HIV: | Human immunodeficiency virus |
| IN: | Inguinal nodes |
| mAbs: | Monoclonal antibodies |
| NALT: | Lymphoid tissue associated with nose |
| NP: | Nasal passages |
| PBS: | Phosphate-buffered saline |
| P1: | Peptide 1 |
| P2: | Peptide 2 |
| P10: | Peptide 10 |
| P27: | Peptide 27 |
| P30: | Peptide 30 |
| PE: | Phycoerythrin |
| PerCP: | Peridinin-chlorophyll proteins |
Ethical approval
This study was conducted under licence according with the Mexican federal regulations for animal experimentation and care (ESM, ESM.CICUAL-07/23-06-2017, NOM-062-ZOO-1999, Ministry of Agriculture, Mexico City, Mexico).Author contributions
Authors Saúl Rojas-Hernández, Jazmín García-Machorro and José Correa-Basurto conceived and planned the research. Author Mara Gutiérrez-Sánchez and Diego Alexander Rojas-Ortega carried out the experiments. Authors Saúl Rojas-Hernández, Jazmín García-Machorro, José Correa-Basurto and Mara Gutiérrez-Sánchez, steered the experiments. Authors Martiniano Bello, Sergio Andrade-Ochoa, Sebastián Díaz-Hernández carried out the bioinformatic studies. Authors Saúl Rojas-Hernández, Jazmín García-Machorro, José Correa-Basurto and Mara Gutiérrez-Sánchez analyzed data and conducted statistical analyses. Author Saúl Rojas-Hernández, Jazmín García-Machorro, José Correa-Basurto, Mara Gutiérrez-Sánchez and Diego Alexander Rojas-Ortega wrote the manuscript. All authors read and approved the manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The study was funded by the following grants: CIENCIA DE FRONTERA CONACYT 265230, PAPIIT-UNAM IN224519, IPN-SIP 20221248, CONACYT-CB-254600, CB-A1-S-21278, APN-782 and COFAA-SIP/IPN.References
- P. M. Sharp and B. H. Hahn, Cold Spring Harbor Perspect. Med., 2011, 1, a006841 CrossRef PubMed.
- S. Nyamweya, A. Hegedus, A. Jaye, S. Rowland-Jones, K. L. Flanagan and D. C. Macallan, Rev. Med. Virol., 2013, 23, 221–240 CrossRef CAS PubMed.
- T. Getaneh, A. Negesse, G. Dessie and M. Desta, J. Clin. Tuberc. Other Mycobact. Dis., 2022, 27, 100310 CrossRef CAS PubMed.
- M. S. Cohen, Y. Q. Chen, M. McCauley, T. Gamble, M. C. Hosseinipour, N. Kumarasamy, J. G. Hakim, J. Kumwenda, B. Grinsztejn, J. H. Pilotto, S. V. Godbole, S. Mehendale, S. Chariyalertsak, B. R. Santos, K. H. Mayer, I. F. Hoffman, S. H. Eshleman, E. Piwowar-Manning, L. Wang, J. Makhema, L. A. Mills, G. de Bruyn, I. Sanne, J. Eron, J. Gallant, D. Havlir, S. Swindells, H. Ribaudo, V. Elharrar, D. Burns, T. E. Taha, K. Nielsen-Saines, D. Celentano, M. Essex and T. R. Fleming, N. Engl. J. Med., 2011, 365, 493–505 CrossRef CAS PubMed.
- J. R. Mascola and D. C. Montefiori, Annu. Rev. Immunol., 2010, 28, 413–444 CrossRef CAS PubMed.
- L. A. Araújo and S. E. Almeida, Viruses, 2013, 5, 595–604 CrossRef PubMed.
- V. Yoon, M. Fridkis-Hareli, S. Munisamy, J. Lee, D. Anastasiades and L. Stevceva, Curr. Med. Chem., 2010, 17, 741–749 CrossRef CAS PubMed.
- J. S. McLellan, M. Pancera, C. Carrico, J. Gorman, J.-P. Julien, R. Khayat, R. Louder, R. Pejchal, M. Sastry, K. Dai, S. O'Dell, N. Patel, S. Shahzad-ul-Hussan, Y. Yang, B. Zhang, T. Zhou, J. Zhu, J. C. Boyington, G.-Y. Chuang, D. Diwanji, I. Georgiev, Y. Do Kwon, D. Lee, M. K. Louder, S. Moquin, S. D. Schmidt, Z.-Y. Yang, M. Bonsignori, J. A. Crump, S. H. Kapiga, N. E. Sam, B. F. Haynes, D. R. Burton, W. C. Koff, L. M. Walker, S. Phogat, R. Wyatt, J. Orwenyo, L.-X. Wang, J. Arthos, C. A. Bewley, J. R. Mascola, G. J. Nabel, W. R. Schief, A. B. Ward, I. A. Wilson and P. D. Kwong, Nature, 2011, 480, 336–343 CrossRef CAS PubMed.
- C. B. Wilen, J. C. Tilton and R. W. Doms, Cold Spring Harbor Perspect. Med., 2012, 2, 1–13 Search PubMed.
- S. Zolla-Pazner, Nat. Rev. Immunol., 2004, 4, 199–210 CrossRef CAS PubMed.
- J. P. Moore, P. W. Parren and D. R. Burton, J. Virol., 2001, 75, 5721–5729 CrossRef CAS PubMed.
- B. F. Haynes, Curr. Opin. Immunol., 2015, 35, 39–47 CrossRef CAS PubMed.
- J. Ponomarenko, H. H. Bui, W. Li, N. Fusseder, P. E. Bourne, A. Sette and B. Peters, BMC Bioinf., 2008, 9, 514 CrossRef PubMed.
- C. Cárdenas, A. Bidon-Chanal, P. Conejeros, G. Arenas, S. Marshall and F. J. Luque, J. Comput.-Aided Mol. Des., 2010, 24, 1035–1051 CrossRef PubMed.
- J. M. Khan and S. Ranganathan, Immunome Res., 2010, 6(1), S2 CrossRef PubMed.
- D. Flower, H. McSparron, M. Blythe, C. Zygouri, D. Taylor, P. Guan, S. Wan, P. Coveney, V. Walshe, P. Borrow and I. Doytchinova, Novartis Found. Symp., 2003, 254, 102–120 CAS.
- R. A. Rodríguez-Fonseca, M. Bello, M. de Los Muñoz-Fernández, J. Luis Jiménez, S. Rojas-Hernández, M. J. Fragoso-Vázquez, M. Gutiérrez-Sánchez, J. Rodrigues, N. Cayetano-Castro, R. Borja-Urby, O. Rodríguez-Cortés, J. García-Machorro and J. Correa-Basurto, Colloids Surf., B, 2019, 177, 77–93 CrossRef PubMed.
- C. Gille and C. Frommel, Bioinformatics, 2001, 17, 377–378 CrossRef CAS PubMed.
- K. Arnold, L. Bordoli, J. Kopp and T. Schwede, Bioinformatics, 2006, 22, 195–201 CrossRef CAS PubMed.
- F. Kiefer, K. Arnold, M. Kunzli, L. Bordoli and T. Schwede, Nucleic Acids Res., 2009, 37, D387–D392 CrossRef CAS PubMed.
- M. C. Peitsch, Bio/Technology, 1995, 13, 658–660 CAS.
- A. Sali and T. L. Blundell, J. Mol. Biol., 1993, 234, 779–815 CrossRef CAS PubMed.
- S. C. Lovell, I. W. Davis, W. B. Arendall 3, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson and D. C. Richardson, Proteins, 2003, 50, 437–450 CrossRef CAS PubMed.
- S. Andrade-Ochoa, J. García-Machorro, M. Bello, L. M. Rodríguez-Valdez, C. A. Flores-Sandoval and J. Correa-Basurto, J. Biomol. Struct. Dyn., 2018, 36, 2312–2330 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- P. K. Loyola, R. Campos-Rodriguez, M. Bello, S. Rojas-Hernandez, M. Zimic, M. Quiliano, V. Briz, M. A. Munoz-Fernandez, L. Tolentino-Lopez and J. Correa-Basurto, Immunol. Res., 2013, 56, 44–60 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- R. G. Bellini, A. P. Guimaraes, M. A. Pacheco, D. M. Dias, V. R. Furtado, R. B. de Alencastro and B. A. Horta, J. Mol. Graphics Modell., 2015, 60, 34–42 CrossRef CAS PubMed.
- M. M. Carrasco-Yepez, R. Campos-Rodríguez, A. A. Reséndiz-Albor, C. Peña-Juárez, A. Contis-Montes de Oca, I. M. Arciniega-Martínez, P. Bonilla-Lemus and S. Rojas-Hernandez, Parasite Immunol., 2018, 40, 1–10 CrossRef PubMed.
- M. Santerre, Y. Wang, S. Arjona, C. Allen and B. E. Sawaya, AIDS Rev., 2019, 21, 76–83 Search PubMed.
- D. M. Junqueira and S. E. Almeida, Virology, 2016, 495, 173–184 CrossRef CAS PubMed.
- S. Q. Liu, S. X. Liu and Y. X. Fu, J. Mol. Model., 2008, 14, 857–870 CrossRef CAS PubMed.
- M. A. Toropova, A. M. Veselinović, J. B. Veselinović, D. B. Stojanović and A. A. Toropov, Comput. Biol. Chem., 2015, 59, 126–130 CrossRef CAS PubMed.
- D. Ortega-Tirado, E. I. Niño-Padilla, A. A. Arvizu-Flores, C. Velazquez, C. Espitia, C. J. Serrano, J. A. Enciso-Moreno, A. Sumoza-Toledo and A. Garibay-Escobar, Mol. Immunol., 2020, 125, 123–130 CrossRef CAS PubMed.
- M. Zimic, A. H. Gutiérrez, R. H. Gilman, C. López, M. Quiliano, W. Evangelista, A. Gonzales, H. H. García and P. Sheen, Bioinformation, 2011, 6, 271–274 CrossRef PubMed.
- J. Liu, M. Fu, M. Wang, D. Wan, Y. Wei and X. Wei, J. Hematol. Oncol., 2022, 15, 28 CrossRef PubMed.
- R. J. Malonis, J. R. Lai and O. Vergnolle, Chem. Rev., 2020, 120, 3210–3229 CrossRef CAS PubMed.
- S. Corbet, H. V. Nielsen, L. Vinner, S. Lauemoller, D. Therrien, S. Tang, G. Kronborg, L. Mathiesen, P. Chaplin, S. Brunak, S. Buus and A. Fomsgaard, J. Gen. Virol., 2003, 84, 2409–2421 CrossRef CAS PubMed.
- M. Jirásek, M. Rickhaus, L. Tejerina and H. L. Anderson, J. Am. Chem. Soc., 2021, 143, 2403–2412 CrossRef PubMed.
- G. L. Ramírez-Salinas, J. García-Machorro, S. Rojas-Hernández, R. Campos-Rodríguez, A. C. de Oca, M. M. Gomez, R. Luciano, M. Zimic and J. Correa-Basurto, Arch. Virol., 2020, 165, 891–911 CrossRef PubMed.
- S. Rojas-Hernández, M. A. Rodríguez-Monroy, R. López-Revilla, A. A. Reséndiz-Albor and L. Moreno-Fierros, Infect. Immun., 2004, 72, 4368–4375 CrossRef PubMed.
- M. Carrasco-Yepez, R. Campos-Rodriguez, I. Lopez-Reyes, P. Bonilla-Lemus, A. Y. Rodriguez-Cortes, A. Contis-Montes de Oca, A. Jarillo-Luna, A. Miliar-Garcia and S. Rojas-Hernandez, Exp. Parasitol., 2014, 145, S84–S92 CrossRef CAS PubMed.
- A. Jarillo-Luna, L. Moreno-Fierros, R. Campos-Rodríguez, M. A. Rodríguez-Monroy, E. Lara-Padilla and S. Rojas-Hernández, Parasite Immunol., 2008, 30, 31–38 CrossRef CAS PubMed.
- N. Lycke, Nat. Rev. Immunol., 2012, 12, 592–605 CrossRef CAS PubMed.
- R. F. Rolando Alberto, B. Martiniano, R. H. Saúl, G. M. Jazmín, G. S. Mara, E. P. Alan Rubén, F. V. Manuel Jonathan, M. M. Juan Vicente and C. B. José, RSC Adv., 2020, 10, 20414–20426 RSC.
- I. Jabbal-Gill, J. Drug Targeting, 2010, 18, 771–786 CrossRef CAS PubMed.
- H. Y. Wu, E. B. Nikolova, K. W. Beagley and M. W. Russell, Immunology, 1996, 88, 493–500 CrossRef CAS PubMed.
- H. Y. Wu, E. B. Nikolova, K. W. Beagley, J. H. Eldridge and M. W. Russell, Infect. Immun., 1997, 65, 227–235 CrossRef CAS PubMed.
- E. J. Kunkel and E. C. Butcher, Immunity, 2002, 16, 1–4 CrossRef CAS PubMed.
- J. R. Mora and U. H. von Andrian, Mucosal Immunol., 2008, 1, 96–109 CrossRef CAS PubMed.
- X. Wang, G. L. Hao, B. Y. Wang, C. C. Gao, Y. X. Wang, L. S. Li and J. D. Xu, Cell Biosci., 2019, 9, 26 CrossRef PubMed.
- H. P. Jones, L. M. Hodge, K. Fujihashi, H. Kiyono, J. R. McGhee and J. W. Simecka, J. Immunol., 2001, 167, 4518–4526 CrossRef CAS PubMed.
- F. Nimmerjahn and J. V. Ravetch, Nat. Rev. Immunol., 2008, 8, 34–47 CrossRef CAS PubMed.
- B. F. Haynes, P. B. Gilbert, M. J. McElrath, S. Zolla-Pazner, G. D. Tomaras, S. M. Alam, D. T. Evans, D. C. Montefiori, C. Karnasuta, R. Sutthent, H. X. Liao, A. L. DeVico, G. K. Lewis, C. Williams, A. Pinter, Y. Fong, H. Janes, A. DeCamp, Y. Huang, M. Rao, E. Billings, N. Karasavvas, M. L. Robb, V. Ngauy, M. S. de Souza, R. Paris, G. Ferrari, R. T. Bailer, K. A. Soderberg, C. Andrews, P. W. Berman, N. Frahm, S. C. De Rosa, M. D. Alpert, N. L. Yates, X. Shen, R. A. Koup, P. Pitisuttithum, J. Kaewkungwal, S. Nitayaphan, S. Rerks-Ngarm, N. L. Michael and J. H. Kim, N. Engl. J. Med., 2012, 366, 1275–1286 CrossRef CAS PubMed.
- S. Heidl, I. Ellinger, V. Niederberger, E. E. Waltl and R. Fuchs, Protoplasma, 2016, 253, 1557–1564 CrossRef CAS PubMed.
- R. E. Horton and G. Vidarsson, Front. Immunol., 2013, 4, 200 Search PubMed.
- H. Billings, B. D. Wines, W. B. Dyer, R. J. Center, H. M. Trist, S. J. Kent and P. M. Hogarth, AIDS Res. Hum. Retroviruses, 2019, 35, 842–852 CrossRef CAS PubMed.
- A. De Milito, C. Mörch, A. Sönnerborg and F. Chiodi, AIDS, 2001, 15, 957–964 CrossRef CAS PubMed.
- H. Longwe, S. Gordon, R. Malamba and N. French, BMC Infect. Dis., 2010, 10, 280 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra08160a |
| This journal is © The Royal Society of Chemistry 2023 |