Open Access Article
Open Access ArticleCovalent functionalization of graphene sheets for plasmid DNA delivery: experimental and theoretical study
Mohyeddin Assali *a,
Naim Kittana
*a,
Naim Kittana *b,
Ismail Badran
*b,
Ismail Badran c and
Safa Omaria
c and
Safa Omaria
aDepartment of Pharmacy, Faculty of Medicine and Health Sciences, An-Najah National University, Nablus, Palestine. E-mail: m.d.assali@najah.edu
bDepartment of Biomedical Sciences, Faculty of Medicine and Health Sciences, An-Najah National University, Nablus, Palestine. E-mail: naim.kittana@najah.edu
cDepartment of Chemistry, Faculty of Sciences, An-Najah National University, Nablus, Palestine
First published on 2nd March 2023
Abstract
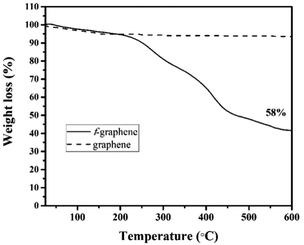
Several approaches, including plasmid transfection and viral vectors, were used to deliver genes into cells for therapeutic and experimental purposes. However, due to the limited efficacy and questionable safety issues, researchers are looking for better new approaches. Over the past decade, graphene has attracted tremendous attention in versatile medical applications, including gene delivery, which could be safer than the traditional viral vectors. This work aims to covalently functionalize pristine graphene sheets with a polyamine to allow the loading of plasmid DNA (pDNA) and enhance its delivery into cells. Graphene sheets were successfully covalently functionalized with a derivative of tetraethylene glycol connected to polyamine groups to improve their water dispersibility and capacity to interact with the pDNA. The improved dispersibility of the graphene sheets was demonstrated visually and by transmission electron microscopy. Also, it was shown by thermogravimetric analysis that the degree of functionalization was about 58%. Moreover, the surface charge of the functionalized graphene was +29 mV as confirmed by zeta potential analysis. The complexion of f-graphene with pDNA was achieved at a relatively low mass ratio (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The incubation of HeLa cells with f-graphene loaded with pDNA that encodes enhanced green fluorescence protein (eGFP) resulted in the detection of fluorescence signal in the cells within one hour. f-Graphene showed no toxic effect in vitro. Density functional theory (DFT) and quantum theory of atoms in molecules (QTAIM) calculations revealed strong binding with ΔH298 = 74.9 kJ mol−1. QTAIM between the f-graphene and a simplified model of pDNA. Taken together, the developed functionalized graphene could be used for the development of a new non-viral gene delivery system.
1). The incubation of HeLa cells with f-graphene loaded with pDNA that encodes enhanced green fluorescence protein (eGFP) resulted in the detection of fluorescence signal in the cells within one hour. f-Graphene showed no toxic effect in vitro. Density functional theory (DFT) and quantum theory of atoms in molecules (QTAIM) calculations revealed strong binding with ΔH298 = 74.9 kJ mol−1. QTAIM between the f-graphene and a simplified model of pDNA. Taken together, the developed functionalized graphene could be used for the development of a new non-viral gene delivery system.
1. Introduction
Gene therapy is emerging as a new class of therapeutics for the treatment of many inherited and acquired incurable diseases. It is a rapidly growing and promising field of medical research that involves the delivery of genes that encode proteins aimed to correct specific deficits within diseased cells.1 Viral vectors have been used successfully over the past few decades to deliver genes into cells for research purposes. Although a few viral vector-based drugs recently received approval from the regulatory authorities to treat certain types of cancers and monogenic diseases, there are still real concerns regarding their safety due to potential in vivo mutagenicity, the probability to induce severe immune reactions, and several other health hazards. Therefore, there is a demand for developing safer alternatives.2–4Carbon-based nanomaterials possess interesting physical, mechanical, and chemical properties that enable their versatile use in various biomedical fields.5–7 Graphene sheets constitute a class of carbon allotropes with a two-dimensional (sheet-like) structure with a large surface area that is proposed to load a large number of therapeutic molecules, rendering it as an attractive candidate for drug and gene delivery.8 Graphene and its derivatives have been used for biomedical purposes like drug delivery, gene delivery, biosensing, bioimaging, tissue engineering, and cancer therapy.9 However, graphene is hydrophobic in nature and tends to form aggregates in most solvents.10 Therefore, the potential application of graphene in the biological field implies the necessity to improve its dispersibility, especially in water, to be compatible with biological environments. An important approach to improve the dispersibility of graphene in water is by covalent or non-covalent functionalization with polar functional groups, likewise, the loading of drugs and nucleic acids.11–13 Non-covalent functionalization could be achieved by polymer wrapping and adsorption of surfactants or small aromatic molecules. It usually involves decorating functional species onto graphene sheets via π–π stacking, hydrophobic attraction, hydrogen bonding and/or electrostatic interactions.14,15 However, the noncovalent approach suffers from low stability. On the other hand, covalent functionalization provides higher water dispersibility and stability which involves the formation of covalent adducts with the sp2 carbon structures in graphene.16,17 One of the successful and effectively developed functionalization approaches is the electrophilic substitution of aryl diazonium salt on the surface of the graphene sheet which provides tremendous applications in various fields.18–21 Herein, we used this method to covalently functionalize graphene sheets with adequate molecules of a hydrophilic linker (derived from tetraethylene glycol) to improve the water dispersibility of the graphene. In addition, to load plasmid DNA (pDNA), it is necessary to introduce tetramine spermine, which is a cationic linker on the surface of the graphene sheets. Tetramine spermine is a synthetic polyamine that showed efficient and safe gene delivery to the cells without the incidence of the immune response.22–25 The resulting functionalized graphene (Scheme 1) was loaded with pDNA that encodes enhanced green fluorescence protein (eGFP) as a model to test its capacity to deliver genetic materials into cells. In this work, we aim to covalently functionalize the graphene sheets with the appropriate linkers to load the plasmid DNA and deliver the resulting complex to the cancer cells to achieve appropriate gene transfection. Moreover, the complexation of the functionalized graphene with the pDNA has been studied by quantum theoretical calculations.
2. Materials and methods
2.1. Reagents and instrumentation
All reagents were used as received without further purification. Propiolic acid, L-ascorbic acid sodium salt, anhydrous copper sulphate (CuSO4), trifluoroacetic acid (TFA), 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium tetrafluoroborate (TBTU), and tetraethylene glycol were purchased from Alfa Aesar Company, England. Graphene nano-powder was purchased from Nanostructured & Amorphous Materials, Inc. (NanoAmor), USA. Sodium azide, 4-nitrobenzylamine hydrochloride, N,N-diisopropylethylamine (DIPEA), di-t-butyl dicarbonate (BOC2O), isoamyl nitrite were purchased from Sigma-Aldrich Company, Germany. Column chromatography was used for purification steps. Thin layer chromatography was used to observe the reaction and was purchased from DC-Fertigfolien ALUGERAM®SIL G/UV254, MACHEREY NAGEL Company, Germany. A centrifuge (Zentrifugen, Germany) and a sonicator (Digital Ultrasonic Cleaner) were used to dissolve and disperse the functionalized graphene. For the gel electrophoresis test, agarose powder, ethylenediaminetetraacetic acid (EDTA), trizma powder, and sodium dodecyl sulfate (SDS) were purchased from Sigma-Aldrich Company, Germany. Plasmid eGFP was gifted by Professor Nabieh Ayob, Haifa. For the biological test, phosphate-buffered saline, fetal bovine serum 10% medium FBS, trypsin–EDTA solution 1X, and RPMI medium were purchased from Sigma-Aldrich Company, Germany. A transmission electron microscope (TEM) (FEI Company, Netherlands) was used to analyse the size and morphology of the graphene sheets. Ultraviolet-visible data was recorded on a 7315 Spectrophotometer, Jenway, UK. Nuclear Magnetic Resonance spectra were obtained using Bruker Avance 500 spectrometer, (Switzerland). Thermogravimetric analysis (TGA) spectra were recorded by (STA 409 PC Luxx®, NETZSCH) under nitrogen (100 cc min−1) with a heating rate of 20 °C min−1 in a range of 25–600 °C. Zeta potential was measured by NanoBrook Omni, Brookhaven Instruments Corporation, USA. A fluorescence microscope (Olympus) was used to detect the GFP protein.2.2. Synthesis and characterization
All the synthetic procedures and gene experiments were prepared at An-Najah University laboratories. NMR, TEM, and TGA measurements were conducted at the University of Jordan.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH
MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NH4OH 10
NH4OH 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1) to provide triethylenetetramine-Boc compound (1) (yield: 73%, 533.8 mg). Rf: 0.48 (DCM
0.1) to provide triethylenetetramine-Boc compound (1) (yield: 73%, 533.8 mg). Rf: 0.48 (DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH
MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NH4OH) (10
NH4OH) (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1). 1H NMR (500 MHz, CDCl3): δ 3.35–3.20 (bm, 10H, 4CH2N & CH2NH), 2.68 (bt, 2H, CH2NH2), 1.41 (s, 27H, C(CH3)3), 1.35 (s, 2H, NH2).
0.1). 1H NMR (500 MHz, CDCl3): δ 3.35–3.20 (bm, 10H, 4CH2N & CH2NH), 2.68 (bt, 2H, CH2NH2), 1.41 (s, 27H, C(CH3)3), 1.35 (s, 2H, NH2).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH) (15
MeOH) (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain compound (5) as an oily product (yield 75.2%, 261.4 mg). FTIR: 3345 (NH), 2928 (CH2), 2105 (N3), 1688 (CO) cm−1. 1H NMR (500 MHz, CDCl3): δ 3.96 (s, 2H, C
1) to obtain compound (5) as an oily product (yield 75.2%, 261.4 mg). FTIR: 3345 (NH), 2928 (CH2), 2105 (N3), 1688 (CO) cm−1. 1H NMR (500 MHz, CDCl3): δ 3.96 (s, 2H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CONH), 3.68–3.64 (m, 8H, 4CH2O), 3.39–3.24 (m, 16H, 4CH2N, 2CH2NH, COC
2CONH), 3.68–3.64 (m, 8H, 4CH2O), 3.39–3.24 (m, 16H, 4CH2N, 2CH2NH, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH2N3 & COCH2C
2CH2N3 & COCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2O), 2.2 (t, 2H, J = 4.4 Hz, CH2N3), 1.44 (s, 27H, C(CH3)3).
2O), 2.2 (t, 2H, J = 4.4 Hz, CH2N3), 1.44 (s, 27H, C(CH3)3).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (1
EtOAc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to obtain the product as a yellowish powder. The yield was (339 mg, 67%). 1H NMR (500 MHz, CDCl3): δ 8.20 (d, 2H, J = 4.9 Hz, Ph), 7.44 (d, 2H, J = 4.8 Hz, Ph), 6.42 (bs, 1H, NH), 4.57 (d, 2H, J = 5.9 Hz, C
2) to obtain the product as a yellowish powder. The yield was (339 mg, 67%). 1H NMR (500 MHz, CDCl3): δ 8.20 (d, 2H, J = 4.9 Hz, Ph), 7.44 (d, 2H, J = 4.8 Hz, Ph), 6.42 (bs, 1H, NH), 4.57 (d, 2H, J = 5.9 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2NH), 2.84 (s, 1H, C
2NH), 2.84 (s, 1H, C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CH). 13C NMR (125.7 MHz, CDCl3): δ 159.0, 152.3, 147.6, 128.4, 124.1, 75.4, 74.4, 43.1.
CH). 13C NMR (125.7 MHz, CDCl3): δ 159.0, 152.3, 147.6, 128.4, 124.1, 75.4, 74.4, 43.1.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (2
H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), then zinc powder (977 mg, 14.9 mmol) and ammonium chloride (194.2 mg, 3.63 mmol) were added to the reaction and stirred under reflux for 4 h. The reaction was filtrated then washed with ethyl acetate then evaporated to yield a yellowish product with a yield of 300 mg, 88.5%. Rf: 0.70 (Hex/EtOAc 1
1), then zinc powder (977 mg, 14.9 mmol) and ammonium chloride (194.2 mg, 3.63 mmol) were added to the reaction and stirred under reflux for 4 h. The reaction was filtrated then washed with ethyl acetate then evaporated to yield a yellowish product with a yield of 300 mg, 88.5%. Rf: 0.70 (Hex/EtOAc 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). 1H NMR (500 MHz, CDCl3): δ 7.28 (t, 1H, J = 7.8 Hz, NHCO), 7.08 (d, 2H, J = 8.8 Hz, Ph), 6.64 (d, 2H, J = 8.5 Hz, Ph), 5.63 (s, 2H, NH2), 4.38 (d, 2H, J = 5.6 Hz, C
2). 1H NMR (500 MHz, CDCl3): δ 7.28 (t, 1H, J = 7.8 Hz, NHCO), 7.08 (d, 2H, J = 8.8 Hz, Ph), 6.64 (d, 2H, J = 8.5 Hz, Ph), 5.63 (s, 2H, NH2), 4.38 (d, 2H, J = 5.6 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2NH), 3.72 (s, 1H, C
2NH), 3.72 (s, 1H, C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CH). 13C NMR (125.7 MHz, CDCl3): δ 148.4, 145.2, 130.9, 129.3, 129.0, 115.3, 73.2, 72.2, 43.4.
CH). 13C NMR (125.7 MHz, CDCl3): δ 148.4, 145.2, 130.9, 129.3, 129.0, 115.3, 73.2, 72.2, 43.4.2.3. Complexion of f-graphene (10) with pEGFP-c1
f-Graphene was mixed with the plasmid DNA (pDNA) pEGFP-C1 at different N/P ratios; 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 7.5
1, 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 10
1, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (N/P ratio defined as the molar ratio of nitrogen in f-graphene to phosphate group in the DNA (N/P ratio)). To guarantee the consistency of the signal intensity on the gel, the amount of the pDNA was fixed while the amount of f-graphene was increased according to the intended N/P ratio. The mixtures of f-graphene and pDNA were mixed vigorously and then were then incubation for 45 min at 37 °C. Equal volumes of each sample were loaded per well of 0.8% agarose gel submerged in TAE buffer. The electrophoresis was run at a voltage of 110 volts for 30 min. The gel was visualized by a UV PhotoDoc-It imaging system.
1 (N/P ratio defined as the molar ratio of nitrogen in f-graphene to phosphate group in the DNA (N/P ratio)). To guarantee the consistency of the signal intensity on the gel, the amount of the pDNA was fixed while the amount of f-graphene was increased according to the intended N/P ratio. The mixtures of f-graphene and pDNA were mixed vigorously and then were then incubation for 45 min at 37 °C. Equal volumes of each sample were loaded per well of 0.8% agarose gel submerged in TAE buffer. The electrophoresis was run at a voltage of 110 volts for 30 min. The gel was visualized by a UV PhotoDoc-It imaging system.
2.4. Plasmid DNA release assay
An efficient release of pDNA from the pDNA–f-graphene is essential for the successful expression of the transfected genes.29 Therefore as a model, this possibility was investigated by using the nonionic surfactant sodium dodecyl sulfate (SDS) aqueous solution at a concentration that was added to the pDNA–graphene complex.30 Afterward, the samples were subjected to agarose gel electrophoresis.2.5. In vitro plasmid pEGFP-C1 transfection experiment
For in vitro transfection experiments, HeLa cells were cultured in RPMI medium at standard laboratory conditions at a density of 6 × 104 cells per well of 48-well plates. The complex pDNA–f-graphene, which was prepared as a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (containing a total of 3 μg pDNA), was added per well, and the culture plates were incubated for 24 h, then 250 μl of fresh RPMI containing 6.2 μl of 0.01% Tween was added. In parallel, a control experiment was done in wells without cells to rule out the autofluorescence of the test compound. After that, the HeLa cells were examined by fluorescence microscopy to check for the expression of the enhanced green fluorescent protein (eGFP).
1 ratio (containing a total of 3 μg pDNA), was added per well, and the culture plates were incubated for 24 h, then 250 μl of fresh RPMI containing 6.2 μl of 0.01% Tween was added. In parallel, a control experiment was done in wells without cells to rule out the autofluorescence of the test compound. After that, the HeLa cells were examined by fluorescence microscopy to check for the expression of the enhanced green fluorescent protein (eGFP).
2.6. Cell viability test
The cells were seeded in a plastic plate (96 well) in a density of 5 × 103 per well overnight then the cells were incubated for 3 h with the f-graphene at a concentration equivalent to that used in the transfection experiment (4.32 mg ml−1). After that, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) solution was added to each well and the plate was incubated for 4 h in the cell culture incubator and the absorbance was measured at a wavelength of 490 nm.2.7. Theoretical calculations
To investigate the bonding interactions involved in this work, the graphene sheets were first optimized using the BAND engine in Amsterdam Modelling Suite (AMS) 2022.31,32 Starting from the reported graphene's structure, a single sheet of 11 × 11 rings was constructed, and optimized, allowing the optimization along the XY-plane under periodic boundary conditions. The optimization was done using the Perdew–Burke–Ernzerhof (PBE) exchange-correlation functional with Grimme's dispersion (PBE-D),33,34 and the double-ζ basis set (DZP). This level of theory has demonstrated good performance in similar previous studies.35–38 Scalar relativistic effects were treated with the ZORA approach,39–41 and a large frozen core. All geometry optimizations were completed with an energy cut-off of 0.0001 Ha, and a gradient convergence of 0.005 Ha Å−1. In the next step, the linker functional group (cf. Scheme 1) was attached to the graphene surface, and its interaction with a simplified DNA sequence was studied under the same level of theory. During this geometry optimization, the coordinates of the graphene surface atoms were constrained while all other atoms were relaxed.The binding energy between the f-graphene/linker and the DNA functional groups was calculated by:
| ΔH298 (reaction) = ΔH298 (products) + ΔH298 (reactants) | (1) |
| H298 = SPE + ZPE + Hcorr. | (2) |
Finally, the quantum theory of atoms in molecules (QTAIM) calculations were done on the optimized system from the previous step, using the ADF engine in AMS.42,43 The criteria for determining the type of electrostatic interactions from the QTAIM calculations were based on previous publications.44 Further details on the calculations can be found in previous studies.37,45,46
3. Results and discussion
3.1. Synthesis and functionalization of graphene sheets
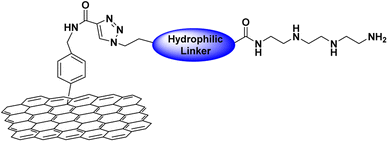
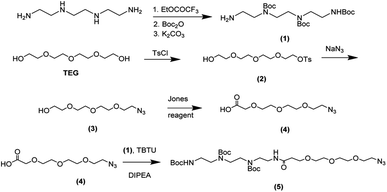
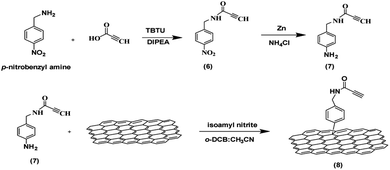
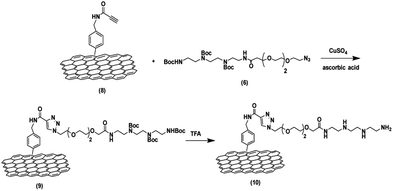
The synthesis of the functionalized graphene was achieved in multi-steps. Beginning with the synthesis of the tetramine-Boc linker (1) that was connected to another linker (4) derived from tetraethylene glycol to improve the water dispersibility of the graphene sheets and to form a complex with the plasmid DNA through electrostatic interaction. So, the synthesis of tetramine-Boc (1) was achieved by selective protection of one of the primary amine groups of the triethylenetetramine by ethyl trifluoroacetate at −78 °C, followed by the protection of the other amine groups by the addition of di tert-butyl dicarbonate (Boc2O) to obtain the fully protected amine groups. After that, the trifluoroacetyl group was cleaved at basic pH using potassium carbonate then recrystallization of the product to obtain the pure triethylenetetramine-Boc (1). The second linker (4) was synthesized starting from tetraethylene glycol (TEG) by converting one of the hydroxyl groups to tosyl to obtain the OH-TEG-OTs (2). Then, the tosyl group was reacted with sodium azide in ethanol to obtain the OH-TEG-N3 (3). Finally, compound (3) was oxidized by Jone's reagent to obtain the required linker (4). After that, the tetramine-Boc (1) was connected with linker (4) through an amidation reaction using TBTU as a coupling agent and DIPEA as a base to obtain compound (5), as shown in Scheme 2.The bonding between the synthesized N3-TEG-tetramine-Boc (5) and the graphene sheets was achieved through a click reaction, which is a copper-catalyzed azide–alkyne cycloaddition reaction to form a triazole ring. This reaction is facile and rapid with a high yield that was discovered by Fokin, Sharpless, and Meldal.47–49 As the azide linker was successfully synthesized, the alkyne group needs to be attached to the graphene sheet. N-(4-Nitrobenzyl)propiolamide (6) was prepared by the formation of an amide linkage between p-nitrobenzyl amine and propiolic acid using TBTU and DIPEA to get compound (6) followed by the reduction of the nitro group to amine group using zinc and ammonium chloride in presence of 10% MeOH to produce compound (7). The covalent functionalization of graphene sheets was carried out through electrophilic aromatic substitution using diazonium salt.50 Therefore, the amine group of compound (7) was reacted with isoamyl nitrite to form the diazonium salt in the presence of o-DCB, and acetonitrile as solvents. Followed by the addition of graphene sheets to get the functionalized graphene (8) terminated with the alkyne group as shown in Scheme 3.
After the synthesis of graphene–alkyne (8). It was effectively bonded with compound (5) through a click reaction. In this experiment, a click reaction was employed by using anhydrous CuSO4 and ascorbic acid as catalysts, which dissolved in distilled H2O and DCM to obtain f-graphene (9). In a final step, a deprotection reaction was conducted to obtain free amine groups in the presence of trifluoroacetic acid (TFA) in DCM to obtain the required graphene-TEG-tetramine (10) as shown in Scheme 4. To determine the amount of free amine loaded on the surface of graphene sheets, the Kaiser test was conducted to obtain a total amine-loaded amount of 0.135 mmol per gram of graphene which confirms the successful functionalization of the graphene sheets with the tetraamine linker.
3.2. Characterization of functionalized graphene
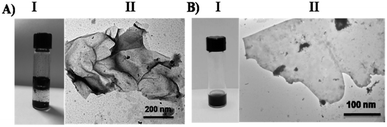 | ||
| Fig. 1 (A) (I) A vial of pristine graphene mixed with water; (II) TEM image of pristine graphene. (B) (I) A vial of f-graphene (10); (II) TEM image of f-graphene (10). | ||
Moreover, zeta potential measurement was used to determine the net charge on the surface of the nanomaterials and provide additional information on the physical stability of the formed suspension.51,52 Therefore, the zeta potential obtained for the functionalized graphene was +29 mV, which indicates a positive charge surrounding the surface of the graphene sheet due to the tetramine functionalization. This positive charge will increase the stability due to the repulsion effect and prevent the aggregation and precipitation behavior of the pristine graphene.
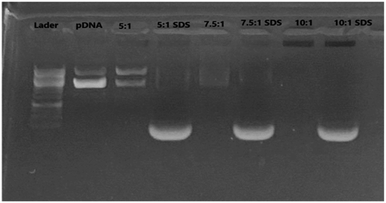
3.3. Evaluation of graphene–pDNA complex formation and dissociation
Agarose gel electrophoresis was employed to investigate the formation of pDNA–f-graphene complex at various N/P ratios (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 7.5
1, 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 10
1, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). This would be indicated by a retard of pDNA migration through an agarose gel. At the same time, pDNA–f-graphene complex formation should be reversible as the pDNA should be released inside the cells to be available for the expression of the molecular machinery of the cells. To investigate this capability, the pDNA–f-graphene complex was incubated with SDS anionic surfactant that is expected to dissociate pDNA from graphene. As shown in Fig. 3, compared to the others, the N/P ratio of 10
1). This would be indicated by a retard of pDNA migration through an agarose gel. At the same time, pDNA–f-graphene complex formation should be reversible as the pDNA should be released inside the cells to be available for the expression of the molecular machinery of the cells. To investigate this capability, the pDNA–f-graphene complex was incubated with SDS anionic surfactant that is expected to dissociate pDNA from graphene. As shown in Fig. 3, compared to the others, the N/P ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 completely inhibited pDNA migration. At the same time whenever pDNA–f-graphene complex was pre-incubated with SDS, a pDNA band was observed on the agarose gel. Taken together, the use of the 10
1 completely inhibited pDNA migration. At the same time whenever pDNA–f-graphene complex was pre-incubated with SDS, a pDNA band was observed on the agarose gel. Taken together, the use of the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio could result in a successful formation of a reversible complex. The formation of this complex is supported by the findings of the theoretical calculations, as discussed later in section 3.6.
1 N/P ratio could result in a successful formation of a reversible complex. The formation of this complex is supported by the findings of the theoretical calculations, as discussed later in section 3.6.
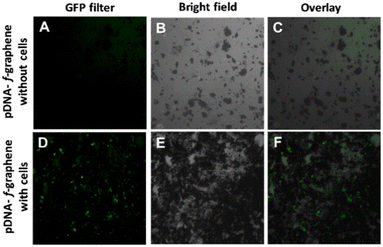
3.4. Transfection of HeLa cells with pDNA–f-graphene complex
To evaluate the potential for the expression of genes carried by pDNA in vitro, HeLa cells were incubated with the pDNA–f-graphene complex prepared at the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio. The used pDNA contains a reporter gene encoding for enhanced green fluorescent protein (EGFP), which is traditionally used to evaluate the efficiency of transfection.56 The transfection was evaluated by fluorescence microscopy after incubating the cells with the test compounds for 1 h. Cell culture wells containing pDNA–f-graphene without cells were also imaged to rule out autofluorescence (negative control). As shown in the lower panel of Fig. 4, a green signal was observed in many of the cells incubated with pDNA–f-graphene complex, something that was not observed in the absence of cells (upper panel), which indicates a successful transfection of the pDNA in the HeLa cells, and a successful expression of the carried-on genes. More details on the exact interaction between the pDNA and the graphene complex are to be discussed in section 3.6.
1 N/P ratio. The used pDNA contains a reporter gene encoding for enhanced green fluorescent protein (EGFP), which is traditionally used to evaluate the efficiency of transfection.56 The transfection was evaluated by fluorescence microscopy after incubating the cells with the test compounds for 1 h. Cell culture wells containing pDNA–f-graphene without cells were also imaged to rule out autofluorescence (negative control). As shown in the lower panel of Fig. 4, a green signal was observed in many of the cells incubated with pDNA–f-graphene complex, something that was not observed in the absence of cells (upper panel), which indicates a successful transfection of the pDNA in the HeLa cells, and a successful expression of the carried-on genes. More details on the exact interaction between the pDNA and the graphene complex are to be discussed in section 3.6.
3.5. Cell viability assay
Pristine carbon nanoparticles were reported in the literature to be cytotoxic for different types of cells.57 Therefore, it was necessary to test whether this risk could be reduced by the implemented functionalization and the respective increase in hydrophilicity. The toxicity of f-graphene at an N/P ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was investigated by using the MTS test on HeLa cells, as shown in Fig. 5. Using similar transfection conditions as in the experiments described above, there was no change in the percentage of cell viability between the control cells and those incubated with f-graphene, which indicates no cytotoxicity for f-graphene under these conditions.
1 was investigated by using the MTS test on HeLa cells, as shown in Fig. 5. Using similar transfection conditions as in the experiments described above, there was no change in the percentage of cell viability between the control cells and those incubated with f-graphene, which indicates no cytotoxicity for f-graphene under these conditions.
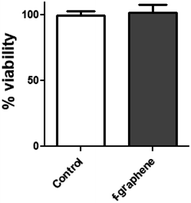 | ||
| Fig. 5 Investigation of the effect of f-graphene on the viability of HeLa cells by using the MTS test, Student's t-test was used to compare the means, α = 0.05, n = 5. | ||
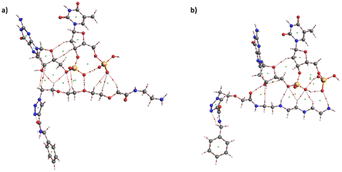
3.6. Bonding interactions based on theoretical calculations
In the previous sections, we showed that the graphene/tetramine linker was bound strongly with pDNA through electrostatic interaction. To better understand this interaction, we studied a molecular system made of the tetramine linker bonded to a graphene sheet, and two nucleobases (guanine–thymine) that resemble the pDNA. The theoretical study was done as described in the experimental section (2.7).First, the tetramine linker/graphene structure was optimized under periodic boundary conditions, as shown in Fig. 6. This serves as a benchmark before adding the unit resembling the pDNA. Next, the optimization was repeated by placing the G–T base pair next to the linker. The optimization was done in two stages, with the G–T pair placed on opposite sides of the linker; the ether (a) and amine (b) parts, as shown in Fig. 7. The geometry of the entire system reflects that the G–T pair is well aligned along either side of the tetramine linker, as originally hypothesized in this work. Furthermore, it appears that strong interaction exists between the G–T base and the linker, due to the close bond distances between the two. For instance, the OH⋯O bond distance between the linker and the base is 1.32 Å in Fig. 7a, and 1.57 Å in Fig. 7b. This strongly suggests the formation of hydrogen bonding between the two moieties. The binding energy between the linker and the G–T base pair was calculated using eqn (1) in the theoretical section in terms of standard enthalpy change at 298 K (ΔH298). The value was discovered to be 74.9 kJ mol−1, which occurs within the scale of intermolecular attractions.
 | ||
| Fig. 6 Two different views for the optimized structure of graphene/tetramine linker as obtained at PBE-D/DZP level of theory. | ||
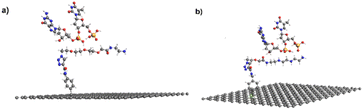 | ||
| Fig. 7 Two optimized structures of the graphene/tetramine linker with the guanine-thymine nucleobases. (a) From the ether side, and (b) from the amine side. Obtained at the PBE-D/DZP level of theory. | ||
To further confirm this type of interaction, we performed QTAIM calculations on the optimized structure as previously described in the theoretical section (2.7). The QTAIM theory, also known as Bader's theory, locates critical points (CP) in molecular structures by using the topology of the electron density (ρ) and its Laplacian (∇2ρ) structures.44,58 For each CP, the calculations produce other parameters, such as the local potential energy density (V(b)), local gradient kinetic energy density (G(b)), and total energy density (H(b)), that can be used to reveal the nature of bonding between two atoms. The strength of QTAIM lies in its description of the CP's located between molecules–known as bond critical points (BCP). The latter can be classified as either a shared interaction (e.g., covalent and polar) or a closed-shell interaction (e.g., hydrogen bond, ionic, van der Waals).
Fig. 8 depicts the QTAIM topology of the tetramine linker/G-T base at the PBE-D/DZP level of theory. The linker is aligned along the ether side (Fig. 8a) and the amine side (Fig. 8b). The QTAIM parameters for the two geometers, a and b, are also listed in Table 1. All the BCP in the table are characterized with a rank and signature of (3, −1), indicating an intermolecular interaction. The electron densities are on the scale of 0.01 to 0.1 Hartree, which is a typical range for intermolecular bonding (covalent bonds have higher values). All Laplacian values in Table 1 are positive and in the range of 10−2 Harreee, indicating van der Waals interaction.44,58
| CP # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Geometry a | ||||||||||
| Rho | 0.018 | 0.008 | 0.005 | 0.120 | 0.008 | 0.010 | 0.011 | 0.008 | 0.006 | 0.008 |
| Laplacian | 0.070 | 0.028 | 0.020 | 0.083 | 0.028 | 0.039 | 0.044 | 0.032 | 0.025 | 0.031 |
| Gb | 0.015 | 0.006 | 0.004 | 0.097 | 0.006 | 0.008 | 0.009 | 0.006 | 0.005 | 0.006 |
| Vb | −0.013 | −0.004 | −0.003 | −0.174 | −0.004 | −0.006 | −0.007 | −0.005 | −0.003 | −0.004 |
| Hb | 0.002 | 0.001 | 0.001 | −0.077 | 0.001 | 0.002 | 0.002 | 0.002 | 0.001 | 0.002 |
| ½V(b) (kJ mol−1) | 17.4 | 5.4 | 3.4 | 228.4 | 5.4 | 7.7 | 9.2 | 6.0 | 4.4 | 5.8 |
| V/G | 0.9 | 0.7 | 0.7 | 1.8 | 0.7 | 0.8 | 0.8 | 0.7 | 0.7 | 0.7 |
| CP # | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Geometry b | |||||||
| Rho | 0.011 | 0.009 | 0.012 | 0.006 | 0.061 | 0.013 | 0.010 |
| Laplacian | 0.044 | 0.038 | 0.047 | 0.024 | 0.181 | 0.048 | 0.038 |
| Gb | 0.009 | 0.008 | 0.010 | 0.005 | 0.058 | 0.010 | 0.008 |
| Vb | −0.007 | −0.006 | −0.008 | −0.003 | −0.070 | −0.008 | −0.006 |
| Hb | 0.002 | 0.002 | 0.002 | 0.001 | −0.012 | 0.002 | 0.002 |
| ½ V(b) (kJ mol−1) | 8.6 | 7.3 | 10.1 | 4.2 | 92.1 | 10.7 | 7.4 |
| V/G | 0.8 | 0.7 | 0.8 | 0.7 | 1.2 | 0.8 | 0.7 |
One particular BCP, #4, has a higher Laplacian value of 0.083 Harreee. This BCP corresponds to the OH⋯H bond between the linker and the G–T base pair as shown in Fig. 8a. A useful criterion for identifying the nature of a BCP is the ratio of potential (V) and kinetic energy (G) densities. If the ratio is less than one, the bond is considered a pure closed shell (i.e., ionic, H-bonding, VDV). Otherwise, the bond is considered a regularly closed shell (i.e. covalent). In the case of BCP #4, the V/G ratio is 1.8, and the bond energy, on the other hand, can be considered as half the value of Vb.59 Table 1 shows a value of 228.4. The V/G ratio and relatively high bond energy value of BCP #4 highly suggest that it is a strong VDV bond.
A similar observation can be observed from Table 1 for BCP #5. The Laplacian, bond energy, and V/G ratio for this point are 0.181 H, 92.1 kJ mol−1, and 1.2, respectively. These parameters suggest it is a strong H-bond, based on the above criteria.
Thus, there are 17 BCPs between the tetramine linker and the G–T base, with two of them referring to strong bonding. This finding is in concert with the experimental results described in this work.
4. Conclusions
Graphene sheets were covalently functionalized with the appropriate linkers to improve their water dispersibility and to provide a positive charge on their surface. The TEM image of the f-graphene (10) confirms the functionalization and the separation of the graphene sheets with a degree of functionalization of 58% as obtained by TGA. Moreover, the zeta potential showed a value of +29 mV which indicates good water dispersibility with the decoration of the positive charge on the surface of the graphene sheet. Agarose gel electrophoresis with and without SDS demonstrated a successful formation of a reversible complex of pDNA with f-graphene, with an optimum N/P ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In vitro studies demonstrated the capacity of this system to deliver DNA materials into cells, as illustrated by the detection of EGFP signal in HeLa cells treated with pDNA–graphene complex comparing with control. This was achieved without affecting the viability of the cells. In addition, quantum theoretical calculations suggested a strong binding between the pDNA, represented by a G-T base pair, and the functionalized graphene. The binding energy, in terms of enthalpy change (ΔH298), was determined to be 74.9 kJ mol−1. QTAIM calculations were used to further investigate the interactions, which revealed strong VDV and H-bond interactions. The development of f-graphene for gene delivery systems has great potential to establish a new family of drugs that might improve the treatment of many difficult diseases such as cancer and different types of genetic disorders.
1. In vitro studies demonstrated the capacity of this system to deliver DNA materials into cells, as illustrated by the detection of EGFP signal in HeLa cells treated with pDNA–graphene complex comparing with control. This was achieved without affecting the viability of the cells. In addition, quantum theoretical calculations suggested a strong binding between the pDNA, represented by a G-T base pair, and the functionalized graphene. The binding energy, in terms of enthalpy change (ΔH298), was determined to be 74.9 kJ mol−1. QTAIM calculations were used to further investigate the interactions, which revealed strong VDV and H-bond interactions. The development of f-graphene for gene delivery systems has great potential to establish a new family of drugs that might improve the treatment of many difficult diseases such as cancer and different types of genetic disorders.
Author contributions
The authors confirm their contribution to the paper as follows: study conception and design: M. Assali, N. Kittana. Theoretical calculations and modelling: I. Badran; perform the experiments: S. Omari; data analysis and validation, M. Assali, N. Kittana and I. Badran. Draft manuscript preparation: M. Assali, N. Kittana, and I. Badran. All authors reviewed the results and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the Association of Arab Universities for the fund. S. Omari thanks the Faculty of Graduate Studies at An-Najah National University to facilitate the achievement of this work.60 The authors acknowledge Mrs. Duaa Qattan for TEM analysis and Hamdi Mango center for TGA analysis.Notes and references
- J. Reiser, X.-Y. Zhang, C. S. Hemenway, D. Mondal, L. Pradhan and V. F. La Russa, Expert Opin. Biol. Ther., 2005, 5, 1571–1584 CrossRef CAS PubMed.
- D. K. Chellappan, N. S. Sivam, K. X. Teoh, W. P. Leong, T. Z. Fui, K. Chooi, N. Khoo, F. J. Yi, J. Chellian and L. L. Cheng, Biomed. Pharmacother., 2018, 108, 1188–1200 CrossRef CAS PubMed.
- M. Vincent, I. de Lázaro and K. Kostarelos, Gene Ther., 2017, 24, 123 CrossRef CAS PubMed.
- J. T. Bulcha, Y. Wang, H. Ma, P. W. L. Tai and G. Gao, Signal Transduction Targeted Ther., 2021, 6, 53 CrossRef CAS PubMed.
- P. R. Riley and R. J. Narayan, Curr. Opin. Biomed. Eng., 2021, 17, 100262 CrossRef CAS PubMed.
- I. Badran and M. O. Al-Ejli, ChemistrySelect, 2022, 7, e202202976 CrossRef CAS.
- I. W. Almanassra, A. D. Manasrah, U. A. Al-Mubaiyedh, T. Al-Ansari, Z. O. Malaibari and M. A. Atieh, J. Mol. Liq., 2020, 304, 111025 CrossRef CAS.
- D. G. Papageorgiou, I. A. Kinloch and R. J. Young, Prog. Mater. Sci., 2017, 90, 75–127 CrossRef CAS.
- J. Shi and Y. Fang, in Graphene, Elsevier, 2018, pp. 215–232 Search PubMed.
- R. K. Layek and A. K. Nandi, Polymer, 2013, 54, 5087–5103 CrossRef CAS.
- A. Lopez and J. Liu, Advanced Intelligent Systems, 2020, 2, 2000123 CrossRef.
- W. Yu, L. Sisi, Y. Haiyan and L. Jie, RSC Adv., 2020, 10, 15328–15345 RSC.
- G.-h. Yang, D.-d. Bao, H. Liu, D.-q. Zhang, N. Wang and H.-t. Li, J. Inorg. Organomet. Polym. Mater., 2017, 27, 1129–1141 CrossRef CAS.
- V. Georgakilas, J. N. Tiwari, K. C. Kemp, J. A. Perman, A. B. Bourlinos, K. S. Kim and R. Zboril, Chem. Rev., 2016, 116, 5464–5519 CrossRef CAS PubMed.
- J. A. Mann and W. R. Dichtel, J. Phys. Chem. Lett., 2013, 4, 2649–2657 CrossRef CAS.
- C. Wetzl, A. Silvestri, M. Garrido, H. L. Hou, A. Criado and M. Prato, Angew. Chem., Int. Ed., 2022, e202212857 Search PubMed.
- J. Sturala, J. Luxa, M. Pumera and Z. Sofer, Chem.–Eur. J., 2018, 24, 5992–6006 CrossRef CAS PubMed.
- J. R. Lomeda, C. D. Doyle, D. V. Kosynkin, W.-F. Hwang and J. M. Tour, J. Am. Chem. Soc., 2008, 130, 16201–16206 CrossRef CAS PubMed.
- A. Sinitskii, A. Dimiev, D. A. Corley, A. A. Fursina, D. V. Kosynkin and J. M. Tour, ACS Nano, 2010, 4, 1949–1954 CrossRef CAS PubMed.
- H. Zhang, E. Bekyarova, J.-W. Huang, Z. Zhao, W. Bao, F. Wang, R. C. Haddon and C. N. Lau, Nano Lett., 2011, 11, 4047–4051 CrossRef CAS PubMed.
- M. Assali, M. Almasri, N. Kittana and D. Alsouqi, ACS Biomater. Sci. Eng., 2019, 6, 112–121 CrossRef PubMed.
- T. Dewa, T. Asai, Y. Tsunoda, K. Kato, D. Baba, M. Uchida, A. Sumino, K. Niwata, T. Umemoto, K. Iida, N. Oku and M. Nango, Bioconjugate Chem., 2010, 21, 844–852 CrossRef CAS PubMed.
- P. A. Puchkov and M. A. Maslov, Pharmaceutics, 2021, 13, 920 CrossRef CAS PubMed.
- P. A. Puchkov, K. A. Perevoshchikova, I. A. Kartashova, A. S. Luneva, T. O. Kabilova, N. G. Morozova, M. A. Zenkova and M. A. Maslov, Russ. J. Bioorg. Chem., 2017, 43, 561–569 CrossRef CAS.
- H. M. Ghonaim, S. Li and I. S. Blagbrough, Pharm. Res., 2009, 27, 17–29 CrossRef PubMed.
- M. Assali, A. N. Zaid, N. Kittana, D. Hamad and J. Amer, Nanotechnology, 2018, 29, 245101 CrossRef PubMed.
- M. Assali, N. Kittana, S. Alhaj-Qasem, M. Hajjyahya, H. Abu-Rass, W. Alshaer and R. Al-Buqain, Sci. Rep., 2022, 12, 12062 CrossRef CAS PubMed.
- M. Assali, N. Kittana, S. Dayyeh and N. Khiar, Nanotechnology, 2021, 32, 205101 CrossRef CAS PubMed.
- S. R. Dave and X. Gao, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2009, 1, 583–609 CAS.
- J. M. Benns, J.-S. Choi, R. I. Mahato, J.-S. Park and S. W. Kim, Bioconjugate Chem., 2000, 11, 637–645 CrossRef CAS PubMed.
- G. te Velde, F. M. Bickelhaupt, E. J. Baerends, C. Fonseca Guerra, S. J. A. van Gisbergen, J. G. Snijders and T. Ziegler, J. Comput. Chem., 2001, 22, 931–967 CrossRef CAS.
- P. H. T. Philipsen, G. te Velde, E. J. Baerends, J. A. Berger, P. L. de Boeij, M. Franchini, J. A. Groeneveld, E. S. Kadantsev, R. Klooster, F. Kootstra, M. C. W. M. Pols, P. Romaniello, M. Raupach, D. G. Skachkov, J. G. Snijders, C. J. O. Verzijl, J. A. Celis Gil, J. M. Thijssen, G. Wiesenekker, C. A. Peeples, G. Schreckenbach and T. Ziegler, BAND 2022.1, SCM, Theoretical Chemistry, Vrije Universiteit, Amsterdam, The Netherlands, http://www.scm.com Search PubMed.
- S. Grimme, S. Ehrlich and L. Goerigk, J. Comput. Chem., 2011, 32, 1456–1465 CrossRef CAS PubMed.
- J. P. Perdew and Y. Wang, Phys. Rev. B, 2018, 98, 079904 CrossRef.
- J. Sun, A. Ruzsinszky and J. P. Perdew, Phys. Rev. Lett., 2015, 115, 036402 CrossRef PubMed.
- I. V. Lebedeva, A. V. Lebedev, A. M. Popov and A. A. Knizhnik, Comput. Mater. Sci., 2017, 128, 45–58 CrossRef CAS.
- I. Badran, N. S. Riyaz, A. M. Shraim and N. N. Nassar, Comput. Theor. Chem., 2022, 1211, 113689 CrossRef CAS.
- A. Förster and L. Visscher, J. Chem. Theory Comput., 2021, 17, 5080–5097 CrossRef PubMed.
- E. v. Lenthe, E. J. Baerends and J. G. Snijders, J. Chem. Phys., 1993, 99, 4597–4610 CrossRef.
- E. v. Lenthe, E. J. Baerends and J. G. Snijders, J. Chem. Phys., 1994, 101, 9783–9792 CrossRef.
- E. v. Lenthe, A. Ehlers and E.-J. Baerends, J. Chem. Phys., 1999, 110, 8943–8953 CrossRef.
- J. I. Rodríguez, R. F. W. Bader, P. W. Ayers, C. Michel, A. W. Götz and C. Bo, Chem. Phys. Lett., 2009, 472, 149–152 CrossRef.
- J. I. Rodríguez, J. Comput. Chem., 2013, 34, 681–686 CrossRef PubMed.
- P. L. A. Popelier and P. Popelier, Atoms in Molecules: An Introduction, Prentice Hall, 2000 Search PubMed.
- I. Badran, A. Rauk and Y. Shi, J. Phys. Chem. A, 2019, 123, 1749–1757 CrossRef CAS PubMed.
- I. Badran and Y. J. Shi, J. Phys. Chem. A, 2015, 119, 590–600 CrossRef CAS PubMed.
- H. C. Kolb and K. B. Sharpless, Drug Discovery Today, 2003, 8, 1128–1137 CrossRef CAS PubMed.
- C. W. Tornøe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- C. D. Doyle and J. M. Tour, Carbon, 2009, 47, 3215–3218 CrossRef CAS.
- S. Bhattacharjee, J. Controlled Release, 2016, 235, 337–351 CrossRef CAS PubMed.
- M. Assali and N. Zohud, Drug Dev. Res., 2020, 82, 448–457 CrossRef PubMed.
- F. Liu, M. Wang, Y. Chen and J. Gao, J. Solid State Chem., 2019, 276, 100–103 CrossRef CAS.
- H. Y. Nan, Z. H. Ni, J. Wang, Z. Zafar, Z. X. Shi and Y. Y. Wang, J. Raman Spectrosc., 2013, 44, 1018–1021 CrossRef CAS.
- T. Ozawa, Bull. Chem. Soc. Jpn., 1965, 38, 1881–1886 CrossRef CAS.
- L. Peng, W. Xiong, Y. Cai, Y. Chen, Y. He, J. Yang, J. Jin and H. Li, Bioengineered, 2017, 8, 225–231 CrossRef CAS PubMed.
- X. Guo and N. Mei, J. Food Drug Anal., 2014, 22, 105–115 CrossRef CAS PubMed.
- R. F. Bader, Acc. Chem. Res., 1985, 18, 9–15 CrossRef CAS.
- M. J. Javan, Comput. Theor. Chem., 2021, 1205, 113440 CrossRef.
- O. Safa, Msc., An-Najah National University, 2018.
| This journal is © The Royal Society of Chemistry 2023 |