Open Access Article
Open Access ArticleCurcumin nanoparticles: physicochemical fabrication, characterization, antioxidant, enzyme inhibition, molecular docking and simulation studies
Qudsia Kanwal†
 *a,
Mahmood Ahmed†
*a,
Mahmood Ahmed† *b,
Muhammad Hamzaac,
Muhammad Ahmadb,
Atiq-ur-Rehmand,
Numan Yousafe,
Arshad Javaidf,
Aneela Anwar
*b,
Muhammad Hamzaac,
Muhammad Ahmadb,
Atiq-ur-Rehmand,
Numan Yousafe,
Arshad Javaidf,
Aneela Anwar g,
Iqra Haider Khanf and
Muhammad Muddassar
g,
Iqra Haider Khanf and
Muhammad Muddassar *e
*e
aDepartment of Chemistry, The University of Lahore, Lahore, Pakistan. E-mail: qudsia.kanwal@chem.uol.edu.pk
bDepartment of Chemistry, Division of Science and Technology, University of Education, College Road, Lahore, Pakistan. E-mail: mahmoodresearchscholar@gmail.com; mahmood.ahmed@ue.edu.pk
cAdditive Manufacturing Institute, Shenzhen University, China
dDepartment of Pharmacy, The University of Lahore, Lahore, Pakistan
eDepartment of Biosciences, COMSATS University Islamabad, Islamabad, Pakistan
fInstitute of Agricultural Sciences, University of the Punjab, Lahore, Pakistan
gBasic Sciences and Humanity, University of Engineering and Technology, Kala Shah Kaku Campus, Lahore, Pakistan
First published on 24th July 2023
Abstract
Curcumin is an extensively studied natural compound due to its extensive biological applications. However, there are some drawbacks linked to this compound such as poor absorption, low water-solubility, quick systemic elimination, fast metabolism, poor pharmacokinetics, low bioavailability, low penetration targeting efficacy and low stability. To overcome these drawbacks, curcumin is encapsulated in nano-carriers. In the current studies, we synthesized nanoparticles of curcumin without using nanocarriers by different methods such as nano-suspension (Cur-NSM), sonication (Cur-SM) and anti-solvent precipitation (Cur-ASP) to enhance the solubility of curcumin in water. The prepared nanoparticles were characterized by FTIR, SEM and XRD analysis. These curcumin nanoparticles were screened for their solubilities in water, DPPH scavenging, amylase, α-glucosidase and β-glucosidase enzymatic activities. The particle size of nano-curcumin was found to be in the 47.4–98.7 nm range. The reduction in particle size of curcumin dramatically increases its solubility in water to 79.2 μg mL−1, whereas the solubility of curcumin is just 0.98 μg mL−1. Cur-ASP showed the highest free radical scavenging potential (48.84 ± 0.98%) which was comparable with standard BHT (50.48 ± 1.11%) at 75.0 μg mL−1. As well, Cur-ASP showed the highest inhibition of α-amylase (68.67 ± 1.02%), α-glucosidase (58.30 ± 0.52%), and β-glucosidase (64.80 ± 0.43%) at 100 μg mL−1 which is comparable with standard drug acarbose. The greater surface area of nanoparticles exposes the various groups of curcumin for blocking the binding sites of enzymes. This strategy may be helpful in designing curcumin as a potent therapeutic agent against diabetes mellitus. Further, the molecular interactions of curcumin with α-amylase, α-glucosidase, β-glucosidase, and polyphenol oxidase were assessed by analyzing the plausible binding modes of curcumin in the binding pocket of each receptor. The best binding mode of curcumin was used to make complexes with the target proteins and their stability was confirmed by 50 ns MD simulation.
1. Introduction
Curcumin (diferuloylmethane) is a naturally occurring compound present in turmeric which demonstrates some really important medicinal properties (Curcuma longa L.). Due to its anti-inflammatory and antioxidant behavior, it has been playing a significant role in the treatment of different pathological diseases like diabetes, cancer, Alzheimer's disease, cardiovascular disease, neurological disorders, cancer, tumors and inflammatory disorders.1–6 In spite of its extraordinary pharmaceutical applications, the clinical application of curcumin is limited due to its low solubility, poor bioavailability, physico-chemical instability, poor pharmacokinetics, and fast metabolism.7–9 However, such issues can be addressed and overcome by using an effective target specific drug delivery system. To improve the pharmacokinetics and bioavailability of curcumin, nano-formulations have been reported which may be launched as a drug after effective accomplishment of pre-clinical and clinical trials.10–14Diabetes mellitus type 2 (DM) is a complex non-communicable disease linked to the dysfunction of pancreatic cells and resistance to insulin leading to postprandial hyperglycemia.15,16 Due to enhanced consumption of carbohydrate and modern lifestyle with less physical effort, this disease has become a global health challenge as well as economic burden. Due to cost effective management of DM in developing countries, the frequency may worsen, with a substantial effect on the population of developing countries. The diabetic condition, when not properly treated, develops to chronic hyperglycemia. It would increase the rate of production of non-mitochondrial and mitochondrial reactive oxygen species (ROS). This phenomenon helps to accelerate the activation of protein kinase C (PKC) isoforms, polyol pathway flux, hexosamine pathway flux and advanced glycation end products involved in the hyper glycaemia-induced oxidative damage followed by, enhanced ROS production. This enhanced ROS has negative impact on insulin regulation signaling resulting in insulin resistance, impaired glucose tolerance, mitochondrial dysfunction and cell dysfunction.17–19 Reduction of insulin gene expression takes place due to the interaction of ROS with pancreatic cells and due to low level of antioxidant enzymes, hence results in reduction in insulin secretion.20 One of the major therapeutic targets recently reported for type 2 DM management is to control the glucose reabsorption in intestine. For this purpose, inhibition of glucose hydrolyzing enzymes like amylase and glucosidase is necessary.21–23 The alpha-amylase (-1,4-glucan-4-glucanohydrolases) is the most prominent enzyme as secretory product of salivary gland and that of pancreas. It is responsible for the conversion of mixture of complex carbohydrates to oligosaccharides and disaccharides by hydrolysis. The oligosaccharides produced were further hydrolyzed by glucosidase to terminal glucose monosaccharide units. The existing inhibitors of alpha-amylase and glucosidase enzymes used for the clinical treatment of DM have wide range of side effects such as diarrhea hypoglycemia, bowel bloating and flatulence which limits their use.24 There is, therefore, an urgent need to look for less toxic alternatives for the management of DM. Dietary intake from herbal source with high antioxidants potentials has exhibited a wonderful beneficial impact on pancreatic cells in diabetic condition by deferring or stopping beta cells dysfunction against glucose toxicity.25,26
Within the context of above study, a comparative mechanistic approach should be applied to know that how the curcumin nano-formulations enhance the therapeutic activity as compared to native curcumin. How the nano-curcumin may control the progression, recurrence, metastases, and disorder of diseases. The nano-curcumin may provide greater surface area to expose its OH groups on the surface of molecule. Thus, individual molecule becomes significantly solubilized by solvation after developing hydrogen bonding with water molecules to enhance the solubility and hence bioavailability of these compounds. Hence, a lower amount of dose will be required for target specific curcumin nanoparticles delivery to reduce the side effects of a specific dose and thus to restrict the binding sides of enzymes to reduce the glucose toxicity. Additionally, current developments have shown that a nano-formulation of drug controls the micro-environment and provides a better therapeutic index. Hence, by developing nano-curcumin would solve many diseases related problems and being the part of our diet may help to facilitate patient by reducing cost, and side effects, and by increasing potential benefits. The overall aim of this study is the fabrication of curcumin nanoparticles by different methods to enhance the solubility of curcumin in water and to make it bioavailable for human body. Furthermore, the present study investigates the antioxidant and antidiabetic activities, and in silico studies were performed to check the role of curcumin as ligands against the α-amylase, α-glucosidase, β-glucosidase, and polyphenol oxidase enzymes by molecular docking studies. The stability of curcumin with the said enzymes was also confirmed by analyzing the MD trajectories generated by 50 ns simulation.
2. Experimental
2.1. Chemicals
HPLC grade ethanol and n-hexane (Panreac, Barcelona, Spain) were acquired from Hajvery Scientific Store, Lahore-Pakistan. The GenPure (Thermo Scientific, USA) water system was used to prepare ultrapure water (18 MΩ cm resistivity).2.2. Fabrication of curcumin nanoparticles
Curcumin nanoparticles were prepared using three methods for its enhanced solubility such as evaporation through nano-suspension method (NSM), sonication method (SM) and anti-solvent precipitation method (ASP). The particles obtained from these methods were named as Cur-NSM, Cur-SM and Cur-ASP respectively.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) was used. This suspension mixture was subjected to a rapid evaporation under vacuum using rotary evaporator. The obtained suspension was freeze-dried under vacuum.
15) was used. This suspension mixture was subjected to a rapid evaporation under vacuum using rotary evaporator. The obtained suspension was freeze-dried under vacuum.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 v/v ratio of ethanol
10 v/v ratio of ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water. Nano suspension of curcumin was obtained, which was subjected to centrifugation at the rate of 20
water. Nano suspension of curcumin was obtained, which was subjected to centrifugation at the rate of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The curcumin nanoparticles were vacuum dried after filtration.
000 rpm for 15 min. The curcumin nanoparticles were vacuum dried after filtration.2.3. Characterization of nano-curcumin
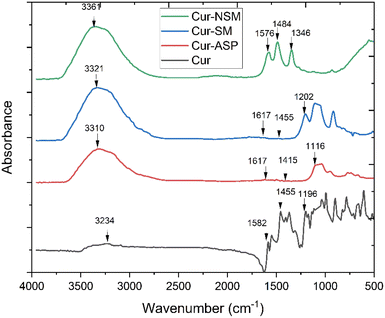
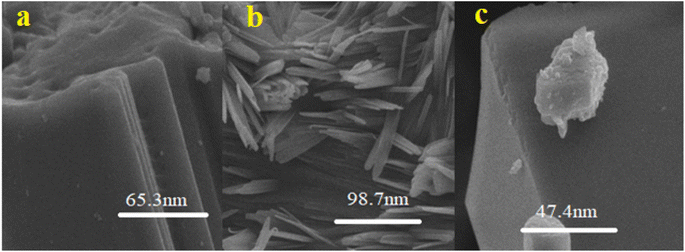
Nano-curcumin were characterized for their particles size and surface morphology using SEM, XRD, FTIR and UV/Visible spectrophotometry. The absorbance spectra were recorded using UV-visible spectrophotometry (U-2910, Hitachi, Japan) ranging from of 200 to 800 nm wavelengths. X-ray powder diffractometer (Bruker D8 ADVANCE Eco) was used to record diffraction pattern with Kα (Cu) radiation (λ = 0.154 nm). These radiations were incident at 1° angle with interval of 2θ, FTIR (Bruker alpha-II) spectrophotometer with smart diamond crystal ATR was used to scan samples of nano-curcumin at wavenumber ranging from 4000 to 500 cm−1 to identify the chemical composition and bonding present. SEM machine (HITACHI S-3400N) was employed for surface morphological investigation of newly synthesized nanoparticles. The SEM instrument was operated at emission current of 60 μA and acceleration voltage was kept as 10 kV.2.4. Solubility studies of nano-curcumin in water
Saturated solution of each nano-curcumin (Cur-NSM, Cur-SM and Cur-ASP) as well of pure curcumin (Cur) was prepared separately in deionized water (30 mL) under constant stirring at room temperature for 24 h. After 24 h, the mixture was centrifuged for the removal of undissolved particles followed by filtration using PTFE membrane (Merck, Germany). Each filtrate was diluted with equal amount of deionized water. The amount of dissolved curcumin was determined using UV/Visible spectrophotometer (U-2910, Hitachi, Japan) at 422 nm.2.5. 1,1-Diphenyl-2-picrylhydrazyl (DPPH)-2,2-diphenyl-1-picrylhydrazyl free radical scavenging assay
Free radical scavenging activity (FRSA) was calculated using methanol solution of DPPH.29–31 15–75 μm mL−1 of each nano-curcumin sample was mixed with 3.0 mL of methanolic solution of DPPH (0.1 mM). After a vigorous shaking, the solution was allowed to stand for 30 min at room temperature in the dark. The absorbance was measured at 515 nm and scavenging activity was calculated using the following equation.Butylated hydroxytoluene (BHT) was used as positive control, whereas methanol was used as negative control. BHT as well as nano-curcumin having FRSA ≥50% were examined in concentration range of 15.0, 30.0, 45.0, 60.0, and 75.0 μg mL−1 in DMSO to evaluate their 50% inhibitory concentration (IC50) in μg mL−1. The assay was performed in triplicate and IC50 was calculated by using GraphPad Prism 7.0 Software (Graph Pad Software Inc.).
2.6. α-Amylase inhibitory assay
Each nano-curcumin sample in a concentration ranged 20–100 μg mL−1 was prepared in DMSO and phosphate buffer (0.02 M, pH 6.9, 500 μL) containing of α-amylase (0.5 mg mL−1) were incubated for 10 min at 25 °C whereas acarbose was used as standard drug in same manner. The blank has same volume of phosphate buffer (0.02 M) instead of nano-curcumin. After incubation substrate solution (starch 1%, 500 μL) in phosphate buffer (0.02 M) was added to reaction mixture and incubated again for 10 min at 25 °C and the reaction was stopped by addition of reagent 3,5-dinitrosalicylic acid (1 mL). Afterwards the reaction mixture was placed in water bath for 5 min in water bath at 100 °C and after cooling to room temperature the mixture was diluted with 10 mL of water. Absorbance was measured at λmax 540 nm using UV/Vis spectrophotometer. Alpha-amylase inhibitory activity of different samples and the standard α-amylase inhibitor (acarbose) were calculated as followsThe assay was performed in triplicate and IC50 was calculated by using GraphPad Prism 7.0 Software (Graph Pad Software Inc.).
2.7. α-Glucosidase inhibition assay
This method was adopted with slight modification in reported method.32 Rat intestinal acetone powder (1 g) was added to phosphate buffer (100 mL, pH 7.0, 0.1 M). The sonication of reaction mixture was carried out for 30 min, which was followed by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C for 10 min.). The obtained supernatant was named as maltase enzyme/rat intestinal α-glucosidase. Each nano-curcumin sample in a concentration ranged from 20 to 100 μg mL−1 and solutions were made in DMSO. The phosphate buffers (0.1 M, pH 7.0, 100 μL) containing α-glucosidase (20 μL) and maltase solution (100 μL, 37 mM) were incubated for 30 min at 37 °C, whereas acarbose was used as standard drug in same manner. The blank has same volume of phosphate buffer (0.1 M) instead of nano-curcumin. After incubation, substrate solution (p-nitrophenyl glucopyranoside, 25 μL) was added to reaction mixture and incubated again for 5 min at 37 °C. Absorbance was measured at λmax of 400 nm using UV/Vis spectrophotometer. α-Glucosidase inhibitory activity of different samples and the standard α-glucosidase inhibitor (acarbose) were calculated as follows
000 rpm, 4 °C for 10 min.). The obtained supernatant was named as maltase enzyme/rat intestinal α-glucosidase. Each nano-curcumin sample in a concentration ranged from 20 to 100 μg mL−1 and solutions were made in DMSO. The phosphate buffers (0.1 M, pH 7.0, 100 μL) containing α-glucosidase (20 μL) and maltase solution (100 μL, 37 mM) were incubated for 30 min at 37 °C, whereas acarbose was used as standard drug in same manner. The blank has same volume of phosphate buffer (0.1 M) instead of nano-curcumin. After incubation, substrate solution (p-nitrophenyl glucopyranoside, 25 μL) was added to reaction mixture and incubated again for 5 min at 37 °C. Absorbance was measured at λmax of 400 nm using UV/Vis spectrophotometer. α-Glucosidase inhibitory activity of different samples and the standard α-glucosidase inhibitor (acarbose) were calculated as followsThe assay was performed in triplicate and IC50 was calculated by using GraphPad Prism 7.0 Software (Graph Pad Software Inc.).
2.8. β-Glucosidase inhibition assay
Each nano-curcumin sample in a concentration range of 20–100 μg mL−1 was prepared in DMSO and phosphate buffer (pH 5, 400 μL) containing β-glucosidase (200 μL, 20 mg mL−1) were incubated for 10 min at 37 °C, whereas acarbose was used as standard drug in the same manner. The blank has same volume of phosphate buffer (0.02 M) instead of nano-curcumin. After incubation, substrate solution (p-nitrophenyl-β-D-glucopyranoside 200 μL, 0.2 mg mL−1) in phosphate buffer was added to reaction mixture and incubated again for 30 min at 37 °C. Absorbance was measured at λmax of 410 nm using UV/Vis spectrophotometer. β-Glucosidase inhibitory activity of different samples and the standard β-glucosidase inhibitor (acarbose) were calculated as followsThe assay was performed in triplicate and IC50 was calculated by using GraphPad Prism 7.0 Software (Graph Pad Software Inc.).
2.9. Molecular docking and simulation studies
The molecular interactions of curcumin with α-amylase (PDB ID: 7TAA), α-glucosidase (PDB ID: 2QMJ), β-glucosidase (PDB ID: 2XWE), and polyphenol oxidase (PDB ID: 2Y9X) were analyzed by molecular docking studies. The co-crystal structures of target proteins were preprocessed in Schrodinger Maestro33 using Protein Preparation Wizard. The structures were refined by adding hydrogens, creating disulfide, assigning bond orders, and by removing water molecules. Then PROPKA34 was used to optimize hydrogen bonds at pH 7.0. The structures were then minimized by applying OPLS_2005 forcefield. To conduct site specific docking, a 3D grid at specific coordinates i.e., (7TAA: X = 38.85, Y = 39.69, Z = 30.46), (2QMJ: X = −20.82, Y = −6.64, Z = −5.24), (2XWE: X = −33.21, Y = −13.53, Z = 19.53), and (2Y9X: X = −10.02, Y = −28.02, Z = −43.6) was generated. The curcumin was prepared by using LigPrep tool by applying OPLS_2005 forcefield. Finally, the standard precision mode of glid was used to dock the ligand with target proteins.The binding poses of curcumin with each target were analyzed and best mode was selected for protein–ligand stability analysis at 50 ns MD simulation using VMD35 and NAMD36 tools. The input topology and parameter files were prepared by Ambertools 21,37 while the ligand topology files were generated by Antechamber. The missing hydrogens in protein structures were added by using the LeaP program of ambertools. Each solvation of each complex was done in a box of 10 Å containing TIP3P water model.38 The systems were further neutralized by adding counter Na+ and Cl− ions. The energy clashes of the complexes were removed by minimizing them using ff14SB and GAFF force fields for protein and ligand, respectively.39 Finally, the solvation systems were subjected to three additional equilibrations at 200, 250, and 300 K. The equilibrated systems were then subjected to production run for 50 ns simulation and the MD trajectories were saved at every 2 ps interval. The trajectories were analyzed by BIO3D package40 of R.
3. Results and discussion
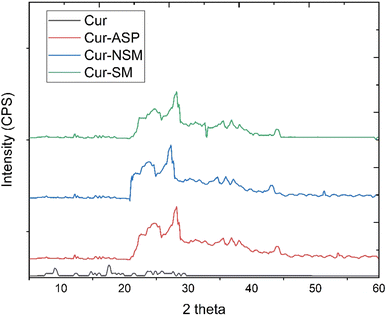
3.1. Characterization of nano-curcumin
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group conjugated with double bonded ethylene carbon vibrations. The peaks appeared at 1346 cm−1 and at 1116 cm−1 originate from the vibrational frequencies (stretching) of Csp2–O and Csp3–O bonds, respectively.
O group conjugated with double bonded ethylene carbon vibrations. The peaks appeared at 1346 cm−1 and at 1116 cm−1 originate from the vibrational frequencies (stretching) of Csp2–O and Csp3–O bonds, respectively.
3.2. Solubility studies of nano-curcumin in water
The solubility studies of nano-curcumin were compared with pure curcumin, molar absorptivity was used to determine the solubility of nano-curcumin in water by using Lambert–Beer law (A = εcl). The results revealed that the Cur-ASP has maximum molar absorptivity which indicated its higher solubility than the Cur-NS and Cur-SM (Table 1). The reduction in particle size of curcumin increases its solubility drastically and the results of our study are inconsistent with the reported ones.41,42| Sample | Molar absorptivity (mol−1 cm−1) | Absorbance | Concentration (mol L−1) | Solubility (μg mL−1) |
|---|---|---|---|---|
| Cur-NSM | 6.18 × 103 | 0.837 | 1.21 × 10−4 | 74.0 |
| Cur-SM | 6.58 × 103 | 1.246 | 7.15 × 10−5 | 46.7 |
| Cur-ASP | 6.66 × 103 | 1.432 | 2.14 × 10−4 | 79.2 |
| Curcumin | 4.900 × 104 | 0.132 | 2.68 × 10−6 | 0.98 |
The nano-sized curcumin particles promote dissolution due to their larger surface area and hence to increase their aqueous solubility. Some previously reported studies show similar trends too where the efficacy and solubility of nanoparticles was enhanced by reducing the particle size of the active ingredients to nanoparticles size. The reduced particles size gives rise to a better wettability because of an increased specific area and thus solubility and dissolution rate of curcumin nanoparticles increases dramatically.10,28
3.3. 1,1-Diphenyl-2-picrylhydrazyl (DPPH)-2,2-diphenyl-1-picrylhydrazyl free radical scavenging assay
DPPH free radical scavenging assay was employed to determine the antioxidant potential of nano-curcumin, curcumin, and standard (butylated hydroxytoluene, BHT). Free radical scavenging efficacies (Fig. 4a) of all the nano-curcumin showed a dose-dependent gradual increase at 15.0–75.0 μg mL−1. The IC50 of each of nano-curcumin, curcumin, and standard were also determined (Fig. 4b). The results revealed that all the nano-curcumin and curcumin tested had a highest DPPH free radical scavenging activity at the maximum concentration (75.0 μg mL−1). Cur-ASP showed the highest free radical scavenging potential (48.84 ± 0.98%) as compared to Cur-NSM (48.28 ± 1.45%), Cur-SM (47.33 ± 1.38%), and curcumin (45.63 ± 0.78%) at the concentration of 75 μg mL−1 whereas BHT showed the DPPH free radical scavenging activity of 50.48 ± 1.11%.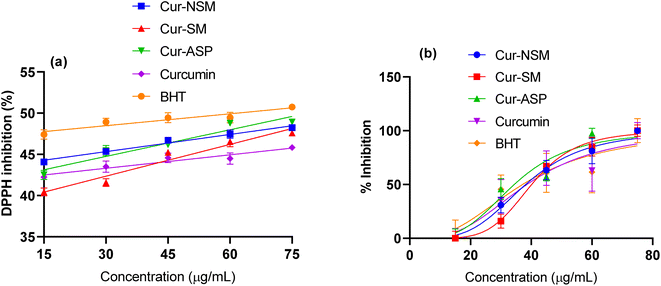 | ||
| Fig. 4 1,1-Diphenyl-2-picrylhydrazyl (DPPH)-2,2-diphenyl-1-picrylhydrazyl radical scavenging activities of (a) Cur-NSM, Cur-SM, Cur-ASP, curcumin, BHT and (b) IC50. | ||
Moreover, Cur-ASP was found to be most active nano-curcumin with an IC50 of 37.05 μg mL−1 followed by Cur-SM (IC50 = 39.57 μg mL−1) and Cur-NSM gave the lowest anti-free radical activity with an IC50 of 40.59 μg mL−1 (Fig. 4b). For comparative purposes, BHT was used as antioxidant standards and showed an interesting anti-free radical activity with an IC50 of 36.15 μg mL−1. In our study, curcumin nanoparticles of different sizes provide increased surface area to expose the functional phenolic groups more effectively, which makes them able to effectively display their free radical scavenging ability.
Curcumin exhibits potential biological activities to fight against chronic diseases and can act on several molecular pathways. However, the antioxidant feature of curcumin is considered as the most important one. Oxidative stress is a result of an imbalance between the elimination and production of ROS and can be related to many chronic diseases and aging process.43 The properties of curcumin responsible for the removal of reactive nitrogen and oxygen, regulation of different enzymes and metal chelation are because of the action of curcumin on markers of oxidative stress. This proves that curcumin acts as a potential antioxidant.44 Various functional groups are present in curcumin. The phenolic functional groups trap electron to deter the production of H2O2 and scavenge the superoxide radicals, the β-diketo group that produces metal–ligand complexation, and carbon to carbon double bonds. Thus, the molecule shows some unique antioxidant characteristics.45 Some researchers reported antioxidant activities of curcumin versus oxidative stress caused by diabetes mellitus. The study was performed on cochlear fibroblasts in rat models of diabetes mellitus and the authors concluded that an increased expression of superoxide dismutase causes curcumin to confer antioxidant protection.46 The antiradical power of an antioxidant can be determined with the DPPH˙ method by measuring a decrease in absorbance of DPPH˙ at 515 nm. An antioxidant scavenges the DPPH˙ by donating a hydrogen to form a stable DPPH˙ molecule and the absorbance decreases as a result. The molecules, in the radical form, give an absorbance at 515 nm which disappears after acceptance of hydrogen or an electron from an antioxidant compound to become a diamagnetic stable molecule.47
As Fig. 5 shows, the heptadienone links between the two methoxyphenol rings contain a highly activated C-atom in the keto form of curcumin. An H-atom can easily be abstracted by curcumin from this C-atom. However, the abstraction of H-atom from the phenolic ring is much difficult because of an intramolecular H-bonding of phenolic hydrogen atoms to the adjacent methoxy groups. Theoretical calculations show that b is the most stable one among the intermediates (a–c) (Fig. 5). While the value of calculated formation energy (ΔH) for b is −42.05 kcal mol−1, this value was found as 39.45 kcal mol−1 for “a” and 54.70 kcal mol−1 for “c”. An H-atom is easily abstracted by DPPH radicals from the free hydroxyl group that is responsible for the exceptional antioxidant properties of curcumin. As a result, the reaction of DPPH radicals in alcoholic medium disappears by curcumin, while the phenolic portion of curcumin appears as an electron donor reaction site. The ability of curcumin to donate an electron is analyzed from the measurement of one-electron-transfer to DPPH radicals. The tert-butoxyl [(CH3)3CO˙] radicals were also used to investigate the H-atom transfer reactions of curcumin and same results were obtained with the same mechanism observation of these radicals scavenging. In addition, the donation of hydrogen atom from β-diketone moiety to a lipid peroxyl radical or a lipid alkyl was marked as a potential antioxidant action of curcumin.47,48
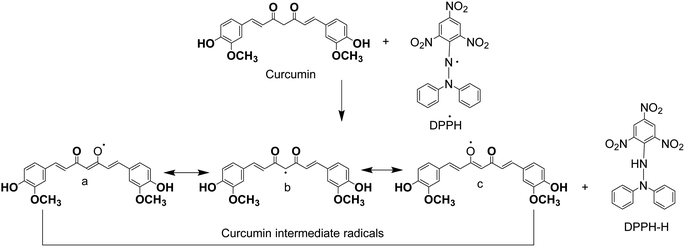 | ||
| Fig. 5 Proposed reaction of curcumin with 1-diphenyl-2-picrylhydrazyl (DPPH)-2,2-diphenyl-1-picrylhydrazyl. | ||
3.4. α-Amylase inhibitory assay
Nano-curcumin, curcumin, and standard (acarbose) were subjected to α-amylase inhibitory assay. The α-amylase inhibitory values (Fig. 6a) of all the nano-curcumin showed a dose-dependent gradual increase in the concentration range of 20.0–100.0 μg mL−1. The IC50 of each of nano-curcumin, curcumin, and standard were also determined (Table 2). The results revealed that all the nano-curcumin and curcumin tested had a highest α-amylase inhibition activity at the maximum concentration (100.0 μg mL−1). Cur-ASP showed the highest α-amylase inhibition potential (68.67 ± 1.02%) as compared to Cur-NS (67.85 ± 0.45%) and Cur-SM (55.67 ± 1.11%) at the concentration of 100 μg mL−1. Curcumin and acarbose showed the α-amylase inhibition of 52.84 ± 0.41% and 68.74 ± 0.28% respectively.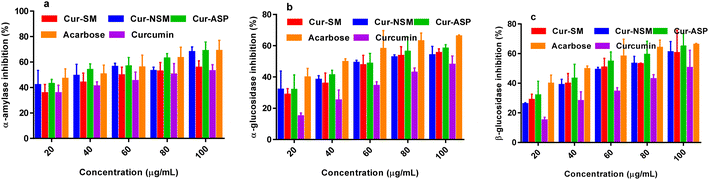 | ||
| Fig. 6 α-Amylase inhibition (a) α-glucosidase inhibition (b) β-glucosidase inhibition (c) of Cur-NSM, Cur-SM, Cur-ASP, curcumin, and acarbose. | ||
| Sample | IC50 (μg mL−1), mean ± SD | ||
|---|---|---|---|
| α-Amylase | α-Glucosidase | β-Glucosidase | |
| Cur-NSM | 51.54 ± 0.54 | 49.10 ± 0.22 | 47.69 ± 0.34 |
| Cur-SM | 50.29 ± 0.78 | 48.51 ± 0.29 | 49.05 ± 0.19 |
| Cur-ASP | 49.61 ± 0.61 | 47.62 ± 0.48 | 47.13 ± 0.21 |
| Curcumin | 52.79 ± 1.02 | 52.61 ± 0.92 | 51.92 ± 0.14 |
| Acarbose | 48.57 ± 0.23 | 45.76 ± 0.13 | 45.55 ± 0.21 |
Cur-ASP was found to be most active nano-curcumin with an IC50 of 49.61 ± 0.61 μg mL−1 followed by Cur-SM (IC50 = 50.29 ± 0.78 μg mL−1) and Cur-NSM gave the inhibitory activity with an IC50 of 51.54 ± 0.54 μg mL−1 (Table 2). For comparative purposes, acarbose was used as reference standards, which showed α-amylase inhibition activity with an IC50 of 48.57 ± 0.23 μg mL−1.
The main cause of most of the DM complications in patients is hyperglycemia. Inhibition of complex polysaccharides hydrolysis by pancreatic α-amylase is one of the most effective strategies to manage DM. Nano-curcumin is a potential agent to manage the DM because it shows excellent inhibition effect against α-amylase.49
3.5. α-Glucosidase inhibition assay
Nano-curcumin, curcumin, and standard (acarbose) were subjected to α-glucosidase inhibitory assay. The α-glucosidase inhibitory values (Fig. 6b) of all the nano-curcumin showed a dose-dependent gradual increase at 20.0–100.0 μg mL−1. The results revealed that all the nano-curcumin and curcumin tested had a highest α-glucosidase inhibition activity at the maximum concentration (100.0 μg mL−1). Cur-ASP showed the highest α-glucosidase inhibition potential (58.30 ± 0.52%) as compared to Cur-NSM (53.98 ± 1.12%) and Cur-SM (55.44 ± 1.11%) at the concentration of 100 μg mL−1. Curcumin and acarbose showed α-glucosidase inhibition of 47.87 ± 0.31% and 66.04 ± 0.18% respectively. The IC50 of each of nano-curcumin, curcumin, and standard were also determined (Table 2). Cur-ASP was found to be most active nano-curcumin with an IC50 of 47.62 ± 0.48 μg mL−1 followed by Cur-SM (IC50 = 48.51 ± 0.29 μg mL−1) and Cur-NSM gave the inhibitory activity with an IC50 of 49.10 ± 0.22 μg mL−1 (Table 2). For comparative purposes, acarbose was used as reference standards and showed α-glucosidase inhibition activity with an IC50 of 45.76 ± 0.13 μg mL−1.In glucose metabolism, α-glucosidase is highly important as it breaks α-glucopyranoside bond as well as breaks oligosaccharides and disaccharides into absorbable monosaccharide units that ultimately maintain the after meal availability of glucose and degree of postprandial hyperglycemia. Due to their site (intestine) directed action, α-glucosidase inhibitors are charming targets to develop nontoxic, safer, and potent antidiabetic agents.50 The nano-curcumin demonstrated promising inhibition results versus α-glucosidase which indicates that curcumin is a potential candidate for the treatment of DM.
3.6. β-Glucosidase inhibition assay
β-Glucosidase inhibition assay was also performed for all the nano-curcumin, curcumin, and standard (acarbose). The β-glucosidase inhibitory values (Fig. 6c) of all the nano-curcumin also showed a dose-dependent gradual increase at 20.0–100.0 μg mL−1. The results showed that all the nano-curcumin and curcumin tested had a highest β-glucosidase inhibition activity at the maximum concentration (100.0 μg mL−1). Cur-ASP showed the highest β-glucosidase inhibition potential (64.80 ± 0.43%) as compared to Cur-NSM (60.98 ± 0.92%) and Cur-SM (60.44 ± 0.81%) at the concentration of 100 μg mL−1. Curcumin and acarbose showed β-glucosidase inhibition of 50.38 ± 0.11% and 66.05 ± 0.47% respectively. The IC50 of each of nano-curcumin, curcumin, and standard were also determined (Table 2). Cur-ASP was found to be most active nano-curcumin with an IC50 of 47.13 ± 0.21 μg mL−1 followed by Cur-SM (IC50 = 49.05 ± 0.19 μg mL−1) and Cur-NSM gave the inhibitory activity with an IC50 of 47.69 ± 0.34 μg mL−1 (Table 2). For comparative purposes, acarbose was used as reference standards and showed α-glucosidase inhibition activity with an IC50 of 45.55 ± 0.21 μg mL−1.In the human body, acidic β-glucosidase can act as a catalyst in the hydrolysis of glucosylceramide as a lysosomal enzyme. Lysosomal storage disorders can occur due to accumulation of glucosylceramide as a result of dysfunction and misfolding of the acidic β-glucosidase activity. In various cancers such as hepatocellular and breast cancers, some enhanced levels of enzyme were observed especially during advanced stages of above diseases and thus resistance of tumors increases against chemotherapy or drugs. Curcumin is a famous and powerful anticancer agent and nano-curcumin showed exceptional results to inhibit β-glucosidase. Nano-curcumin could give anticancer effects via targeting the β-glucosidase in the treatment of cancer.51
The inhibition activity is closely related to the particle size. Greater the particle size, lower will be the activity. If the particle size is smaller, the surface area increases which makes the functional groups available for H-bonding become more significant while bulging on the surface and there is more opportunity for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, OCH3, OH groups present in the structure of curcumin to create more H-bonding and some other groups having hydrophobic linkage may become important to generate overall bonding in such a way it may change the symmetry of enzyme thus making it unavailable for it substrate to bind and hydrolyze to release glucose to blood. In this way it is an indirect strategy to control glucose level in blood by controlling α-amylase and α-glucosidase enzyme. As well to use it as anticancer agent to control the β-glucosidase.
O, OCH3, OH groups present in the structure of curcumin to create more H-bonding and some other groups having hydrophobic linkage may become important to generate overall bonding in such a way it may change the symmetry of enzyme thus making it unavailable for it substrate to bind and hydrolyze to release glucose to blood. In this way it is an indirect strategy to control glucose level in blood by controlling α-amylase and α-glucosidase enzyme. As well to use it as anticancer agent to control the β-glucosidase.
3.7. Molecular docking and simulation studies
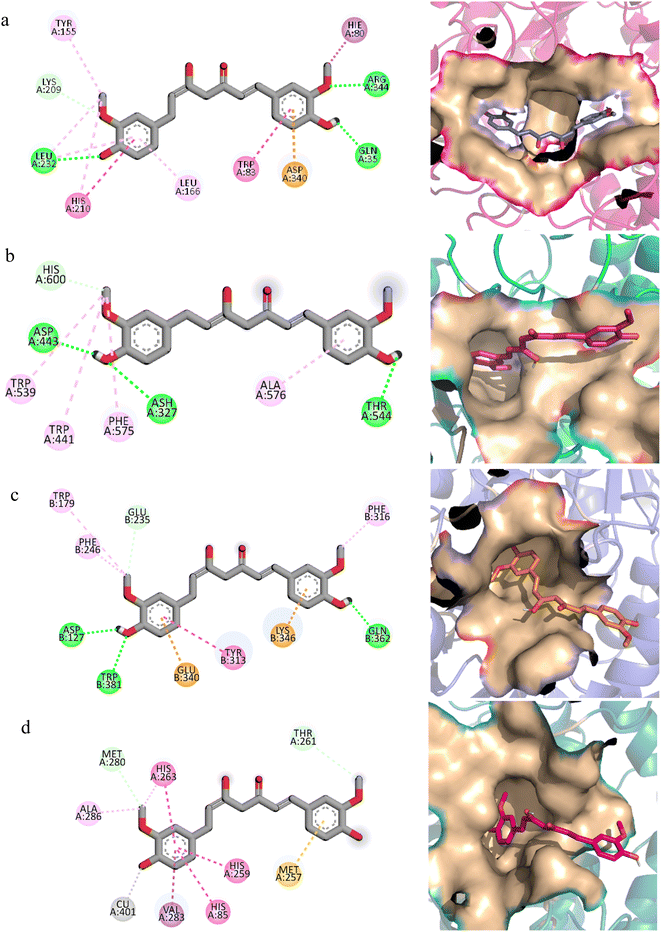
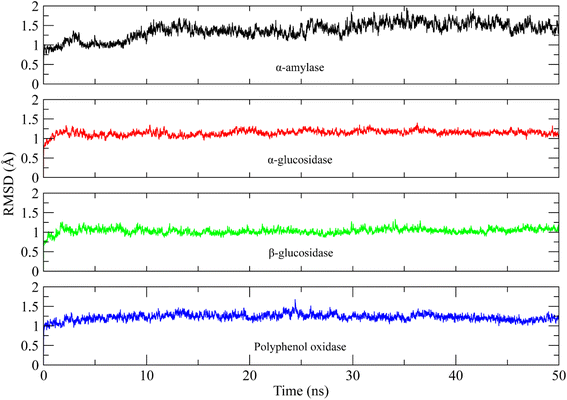
The molecular interactions of curcumin with the target proteins were analyzed by docking studies. Several docked poses of curcumin were generated into the binding sites and among them, one pose was selected based on the interactions and plausible binding mode. Curcumin made 3 hydrogen bonds with Gln35, Leu232, Arg344, one Pi–sigma bond with His80, one Pi–anion bond with Asp340, and was involved in hydrophobic interactions with Trp83, Tyr155, Leu166, and His210 residues of α-amylase Fig. 7a. Similarly, curcumin made three hydrogen bonds with Ash327, Asp443, and Thr544 of α-glucosidase while made four hydrophobic interactions with Trp441, Trp539, Phe575, and Ala576 Fig. 7b. In case of β-glucosidase, curcumin made 3 hydrogen bonds with Asp127, Gln362, Trp381, one Pi–Pi stacking with Tyr313, two Pi-cation and anion bonds with Glu340 and Lys346, and three hydrophobic interactions with Trp179, Phe246, and Phe310 Fig. 7c. Lastly, it one Pi–sigma bond with Val283, one Pi–sulfur bond with Met257, one hydrophobic interaction with Ala286, and three Pi–Pi stackings with His85, His259, and His 263 of polyphenol oxidase Fig. 7d.In order to find the stability of protein–ligand complex, the RMSD of protein backbone atoms complexed with curcumin was calculated over the simulation time (Fig. 8).52 It can be observed that three complexes (of α-glucosidase, β-glucosidase, polyphenol oxidase) equilibrated at 2 ns and then attained stability in RMSD values in the range of 1–1.25 Å throughout the simulation with minor deviations in polyphenol oxidase (green) during 20 to 25 ns where it increased to ∼1.5 Å but attained previous confirmation at 30 ns. The α-amylase complex (black) equilibrated at 5 ns and attained a value of 1 Å till ∼8 ns. The RMSD value then gradually increased to ∼1.5 Å at 10 ns and remained in the range of 1.25–1.5 Å till 30 ns. The RMSD increased to ∼2 Å after 30 ns and remained in this range till 40 ns and then attained the previous value of ∼1.25 Å during 40 to 50 ns period. The small deviations in RMSD values indicated that the complexes were stable during simulation.
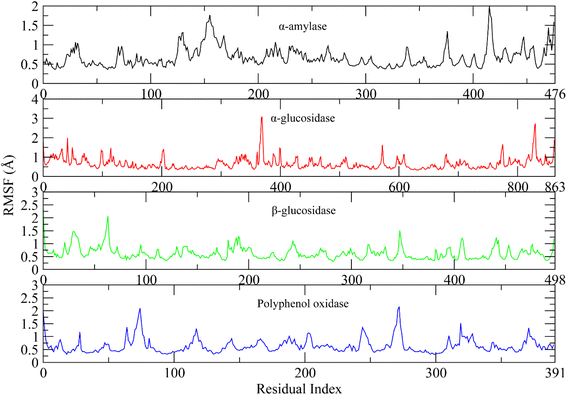
The root mean square fluctuations describe the flexible residues of the protein.53 The higher RMSF value shows the flexible regions such as loops while lower value indicates the rigid residues such as helices and sheets. The RMSF plots of studied systems are shown in Fig. 9. The RMSF plots of all complexes showed that the protein residues did not show major fluctuations. Only loop regions showed minor fluctuations. The residues from 120 to 160 and 410 to 420 showed minor fluctuations in α-amylase complex. The residues from 370 to 380 and 820 to 830 in α-glucosidase, 60 to 70 in β-glucosidase, and 70 to 80, 260 to 280 in polyphenol oxidase showed minor variations. The other residues did not show such fluctuations indicating the rigid portion of the proteins.
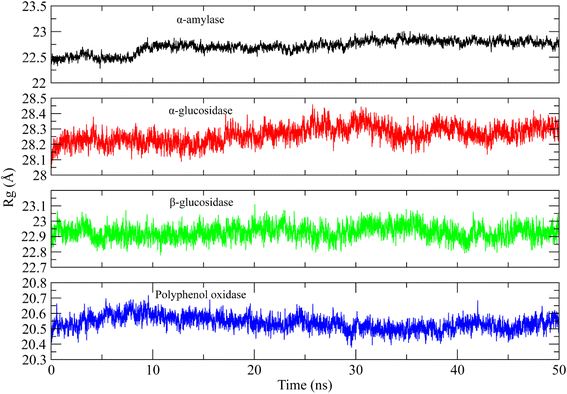
Similarly, the radius of gyration (Rg) was calculated to check the compactness of the protein when the ligands were bound to it.54 The higher Rg values indicate the unfolding events in protein structure during simulation. The Rg plots of the systems are shown in Fig. 10. It can be observed that the Rg value of α-amylase complex was in range of 22.5 Å till 8 ns and then gradually increased to ∼22.75 Å at 10 ns and then remained in the same range till the end of simulation. The Rg value of α-glucosidase complex was in the range of 28.2 to 28.3 Å till 20 ns and then slightly increased to 28.4 Å during 20 to 30 ns interval and then attained the previous values during 35 to 50 ns. The Rg value of β-glucosidase was remained in the range of 22.9 to 23 Å throughout the simulation time. Lastly, the Rg value of polyphenol oxidase complex was high in first 20 ns as compared to last 30 ns and attained a value in range of 20.5 Å towards the end of simulation. The stable Rg values showed that the proteins did not undergo any unfolding event during simulation when bound to the curcumin.
4. Conclusion
Curcumin is nontoxic and very cost-effective polyphenol with wide range of medicinal properties. It is extracted from curcuma longa. Several in vitro, in vivo, and clinical trial investigations reported in literature have provided evidence for the bioactive role of curcumin in the prevention and treatment of various human diseases. Curcumin nano-formulations have markedly exhibited their potential in inhibition of the enzymes responsible for diabetic mellitus. A structure activity relationship has been established using molecular docking studies for the relation of alpha amylase and alpha glucosidase enzymes inhibition with that of curcumin. Cur-ASP has smaller size (47.4 nm) than that of Cur-MS (98.7 nm) and Cur-NSM (65 nm). It has inhibited the DPPH free radical, and all three enzymes responsible for diabetes. It means that smaller the size, more surface is exposed with functional groups would be provided which has more opportunity to bind and inhibit the binding sites of enzymes more easily as compared to the native curcumin. It has been confirmed that curcumin nano-formulations improve curcumin bioavailability by enhancing its water solubility and are systemically safe. It would be more available to biological system when more soluble in water. However, testing of these formulations as therapeutic modalities is highly desirable and is crucial for future clinical trials and for human use. In addition, implementation of curcumin nano-formulations as enzyme inhibitors exhibited even more than parallel inhibition results to that of the standard drug used. In present study, we have shown the alpha-amylase inhibition (AAI) as well as α glucosidase inhibition and antioxidant activity of pure and modified curcumin for the treatment of DM. Curcumin being a magical molecule exhibited a promising antioxidant as well as antidiabetic potential that can be explored in the development of novel drugs for oxidative stress-related diabetes management. A weak AAI suggests other possible therapeutic targets in the management of DM. Our studies partially confirmed the traditional use of curcumin modified to different nano-sizes using different fabrication techniques to enhance its water solubility and thus bioavailability for diabetes management in the region. Since the oxidative stress has a key role in the development of DM. Increased oxidative species production and reduced antioxidant capacity have been repeatedly shown in subjects with DM. Owing to detrimental effects of oxidative stress on the development of DM and the progression of its vascular complications, antioxidant therapy has been considered a potentially effective approach to treat the DM. Further, the molecular docking studies revealed that the plausible binding modes of curcumin with α-amylase, α-glucosidase, β-glucosidase, and polyphenol oxidase showed better molecular interactions and make stable complexes with the studied enzymes when explored by 50 ns long MD simulation.Data availability
All data generated or analysed during this study are included in this published article.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors received no specific funding for this work.References
- M. Ahmed, A. Khaleeq, R. Huma and S. Munir, Spectrosc. Lett., 2017, 50, 432–439 CrossRef CAS
.
- M. Ahmed, M. A. Qadir, M. I. Shafiq, M. Muddassar, Z. Q. Samra and A. Hameed, Arabian J. Chem., 2019, 12, 41–53 CrossRef CAS
.
- M. Ahmed, M. A. Qadir, M. I. Shafiq, M. Muddassar, A. Hameed, M. N. Arshad and A. M. Asiri, Acta Pharm., 2017, 67, 385–395 CrossRef PubMed
.
- M. Ahmed, M. A. Qadir, A. Hameed, M. Imran and M. Muddassar, Chem. Biol. Drug Des., 2018, 91, 338–343 CrossRef CAS PubMed
.
- M. Ahmed, M. A. Qadir, A. Hameed, M. N. Arshad, A. M. Asiri and M. Muddassar, Bioorg. Chem., 2018, 76, 218–227 CrossRef CAS PubMed
.
- G. H. Shin, J. Li, J. H. Cho, J. T. Kim and H. J. Park, J. Food Sci., 2016, 81, N494–N501 CrossRef CAS PubMed
.
- M. Ahmed, M. A. Qadir, A. Hameed, M. N. Arshad, A. M. Asiri and M. Muddassar, Biochem. Biophys. Res. Commun., 2017, 490, 434–440 CrossRef CAS PubMed
.
- M. Ahmed, M. A. Qadir and M. I. Shafiq, Lat. Am. J. Pharm., 2017, 36, 1789–1795 CAS
.
- M. Ahmed, S. Ahmad and M. Irfan, Sep. Sci. Plus, 2021, 4, 118–127 CrossRef CAS
.
- R. K. Basniwal, H. S. Buttar, V. Jain and N. Jain, J. Agric. Food Chem., 2011, 59, 2056–2061 CrossRef PubMed
.
- T. Gupta, J. Singh, S. Kaur, S. Sandhu, G. Singh and I. P. Kaur, Front. bioeng. biotechnol., 2020, 8, 879 CrossRef PubMed
.
- H. Yavarpour-Bali, M. Ghasemi-Kasman and M. Pirzadeh, Int. J. Nanomed., 2019, 14, 4449 CrossRef CAS PubMed
.
- A. Ubeyitogullari and O. N. Ciftci, Sci. Rep., 2019, 9, 1–11 CrossRef PubMed
.
- G. M. Abu-Taweel, M. F. Attia, J. Hussein, E. M. Mekawi, H. M. Galal, E. I. Ahmed, A. A. Allam and M. E. El-Naggar, Biomed. Pharmacother., 2020, 131, 110688 CrossRef CAS PubMed
.
- L.-J. Yan, J. Diabetes Res., 2014, 2014, 137919 Search PubMed
.
- F. Zaccardi, D. R. Webb, T. Yates and M. J. Davies, Postgrad. Med. J., 2016, 92, 63–69 CrossRef CAS PubMed
.
- D. E. Ejike, M. A. Adam, O. S. Sheu, P. Nganda, E. Iliya, D. A. Moses and O. O. Alfred, J. Diabetes Endocrinol., 2018, 9, 11–19 CrossRef
.
- A. A. Oroojan, N. Chenani and M. An’aam, Int. J. Hepatol., 2020, 2020, 5890378 Search PubMed
.
- V. Solmaz, H. Köse Özlece, H. A. Eroglu, H. Aktuğ, O. Erbaş and D. Taşkıran, Neurol. Res. Int., 2017, 2017, 5952149 Search PubMed
.
- R. P. Robertson and J. S. Harmon, FEBS Lett., 2007, 581, 3743–3748 CrossRef CAS PubMed
.
- P. Dandekar, S. Ramkumar and A. RaviKumar, Stud. Nat. Prod. Chem., 2021, 70, 381–410 CAS
.
- L. Sim, K. Jayakanthan, S. Mohan, R. Nasi, B. D. Johnston, B. M. Pinto and D. R. Rose, Biochemistry, 2010, 49, 443–451 CrossRef CAS PubMed
.
- J. Wu, B. Hu, S. Lu, R. Duan, H. Deng, L. Li, L. He, Y. Zhao, J. Wang and Z. Yu, Carbohydr. Res., 2022, 511, 108478 CrossRef CAS PubMed
.
- B. Salehi, A. Ata, N. V. Anil Kumar, F. Sharopov, K. Ramirez-Alarcon, A. Ruiz-Ortega, S. Abdulmajid Ayatollahi, P. Valere Tsouh Fokou, F. Kobarfard and Z. Amiruddin Zakaria, Biomolecules, 2019, 9, 551 CrossRef PubMed
.
- Y. Naveen, A. Urooj and K. Byrappa, J. Herb. Med., 2021, 28, 100436 CrossRef
.
- M. M. Yallapu, P. K. B. Nagesh, M. Jaggi and S. C. Chauhan, AAPS J., 2015, 17, 1341–1356 CrossRef CAS PubMed
.
- M. Kakran, N. G. Sahoo, I.-L. Tan and L. Li, J. Nanopart. Res., 2012, 14, 1–11 CrossRef
.
- S. S. Hettiarachchi, S. P. Dunuweera, A. N. Dunuweera and R. G. Rajapakse, ACS Omega, 2021, 6, 8246–8252 CrossRef CAS PubMed
.
- M. Ahmed, M. Imtiaz Shafiq, A. Khaleeq, R. Huma, M. Abdul Qadir, A. Khalid, A. Ali and A. Samad, J. Chem., 2016, 2016, 8072305–8072310 Search PubMed
.
- M. Ahmed, A. Khaleeq and S. Ahmad, World Appl. Sci. J., 2014, 30, 1664–1667 CAS
.
- M. Ahmed, S. K. Shahzadi, M. A. Qadir, A. I. Durrani, S. Ahmad, I. KHAN, A. Munir and A. Khaleeq, Lat. Am. J. Pharm., 2020, 39, 372–375 CAS
.
- M. Ezati, F. Ghavamipour, H. Adibi, K. Pouraghajan, S. S. Arab, R. H. Sajedi and R. Khodarahmi, Spectrochim. Acta, Part A, 2022, 121806 Search PubMed
.
- L. J. S. S. Schrödinger, Schrödinger, LLC, New York, NY, 2017, 2017(2), pp. 2017–2011 Search PubMed
.
- M. O. Kim, S. E. Nichols, Y. Wang and J. A. J. McCammon, J. Comput. Aided Mol. Des., 2013, 27, 235–246 CrossRef CAS PubMed
.
- W. Humphrey, A. Dalke and K. J. J. Schulten, J. Mol. Graph., 1996, 14, 33–38 CrossRef CAS PubMed
.
- J. C. Phillips, D. J. Hardy, J. D. Maia, J. E. Stone, J. V. Ribeiro, R. C. Bernardi, R. Buch, G. Fiorin, J. Hénin and W. J. T. Jiang, J. Chem. Phys., 2020, 153, 044130 CrossRef CAS PubMed
.
- D. A. Case, H. M. Aktulga, K. Belfon, I. Ben-Shalom, S. R. Brozell, D. Cerutti, T. Cheatham, V. W. D. Cruzeiro, T. Darden and R. E. Duke, Amber 2021: Reference Manual, 2021 Search PubMed
.
- W. Jorgensen and J. J. J. C. P. Chandrasekhar, J. Chem. Phys., 1983, 79, 926 CrossRef CAS
.
- Y. Duan, C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo and T. J. Lee, J. Comput. Chem., 2003, 24, 1999–2012 CrossRef CAS PubMed
.
- B. J. Grant, L. Skjærven and X. Q. J. P. S. Yao, Protein Sci., 2021, 30, 20–30 CrossRef CAS PubMed
.
- S. Alam, J. J. Panda and V. S. Chauhan, Int. J. Nanomedicine, 2012, 7, 4207 CAS
.
- M. Ghosh, A. T. Singh, W. Xu, T. Sulchek, L. I. Gordon and R. O. Ryan, Nanomedicine, 2011, 7, 162–167 CrossRef CAS PubMed
.
- M. Pulido-Moran, J. Moreno-Fernandez, C. Ramirez-Tortosa and M. Ramirez-Tortosa, Molecules, 2016, 21, 264 CrossRef PubMed
.
- K. Jakubczyk, A. Drużga, J. Katarzyna and K. Skonieczna-Żydecka, Antioxidants, 2020, 9, 1092 CrossRef CAS PubMed
.
- G. C. Jagetia and G. K. Rajanikant, Antioxidants, 2015, 4, 25–41 CrossRef CAS PubMed
.
- M. L. Del Prado-Audelo, I. H. Caballero-Florán, J. A. Meza-Toledo, N. Mendoza-Muñoz, M. González-Torres, B. Florán, H. Cortés and G. Leyva-Gómez, Biomolecules, 2019, 9, 56 CrossRef PubMed
.
- T. Ak and İ. Gülçin, Chem.-Biol. Interact., 2008, 174, 27–37 CrossRef CAS PubMed
.
- S. M. Abbas, N. Ahmad, U. A. Rana, S. U.-D. Khan, S. Hussain and K.-W. Nam, Electrochim. Acta, 2016, 212, 260–269 CrossRef CAS
.
- D. Stojkovic, M. Smiljkovic, A. Ciric, J. Glamoclija, L. Van Griensven, I. C. Ferreira and M. Sokovic, S. Afr. J. Bot., 2019, 120, 100–103 CrossRef CAS
.
- A. Mushtaq, U. Azam, S. Mehreen and M. M. Naseer, Eur. J. Med. Chem., 2023, 249, 115119 CrossRef CAS PubMed
.
- X. Liu, F. Li, L. Su, M. Wang, T. Jia, X. Xu, X. Li, C. Wei, C. Luo and S. Chen, Bioorg. Chem., 2022, 127, 106016 CrossRef CAS PubMed
.
- K. Sargsyan, C. Grauffel and C. Lim, J. Chem. Theory Comput., 2017, 13, 1518–1524 CrossRef CAS PubMed
.
- R. Martinez, A. Blasina, J. F. Hallin, W. Hu, I. Rymer, J. Fan, R. L. Hoffman, S. Murphy, M. Marx and G. Yanochko, PLoS One, 2015, 10, e0138616 CrossRef PubMed
.
- M. Y. Lobanov, N. Bogatyreva and O. J. M. B. Galzitskaya, Mol. Biol., 2008, 42, 623–628 CrossRef CAS
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |