Open Access Article
Open Access ArticleUpconversion nanoparticle-based aptasensor for rapid and ultrasensitive detection of Staphylococcus aureus by low-speed centrifugation†
Na Li‡
a,
Ying Zhang‡ab,
Tiancheng Weia,
Tao Yangc,
Qing Baoc,
Qichao Chenga,
Chuanbin Mao c,
Yajun Shuai
c,
Yajun Shuai a and
Mingying Yang
a and
Mingying Yang *a
*a
aZhejiang Provincial Key Laboratory of Utilization and Innovation of Silkworm and Bee Resources, Institute of Applied Bioresource Research, College of Animal Science, Zhejiang University, Hangzhou, China. E-mail: yangm@zju.edu.cn
bSchool of Life Sciences, Westlake University, Hangzhou 310024, Zhejiang, China
cSchool of Materials Science and Engineering, Zhejiang University, Hangzhou, 310058, P. R. China
First published on 5th July 2023
Abstract
Opportunistic foodborne pathogens such as Staphylococcus aureus (S. aureus) can cause a wide variety of threats to public health. There is an urgent clinical need for a fast, simple, low-cost, and sensitive method. Here, we designed a fluorescence-based aptamer biosensor (aptasensor) for S. aureus detection using core–shell structured upconversion nanoparticles (CS-UCNPs) as a beacon. A S. aureus-specific aptamer was modified on the surface of CS-UCNPs for binding pathogens. The S. aureus bound to CS-UCNPs can then be isolated from the detection system by simple low-speed centrifugation. Thus, an aptasensor was successfully established for the detection of S. aureus. The fluorescence intensity of CS-UCNPs correlated with the concentration of S. aureus within the range of 6.36 × 102 to 6.36 × 108 CFU mL−1, resulting in the detected limit of S. aureus being 60 CFU mL−1. The aptasensor performed well in real food samples (milk) with a detection limit of 146 CFU mL−1 for S. aureus. Furthermore, we applied our aptasensor in chicken muscles for S. aureus detection, and compared it with the plate count gold standard method. There was no significant difference between our aptasensor and the plate count method within the detected limit, while the time for the aptasensor (0.58 h) was shorter than that of the plate count method (3–4 d). Therefore, we succeeded in the design of a simple, sensitive and fast CS-UCNPs aptasensor for S. aureus detection. This aptasensor system would have the potential for the detection of a wide range of bacterial species by switching the corresponding aptamer.
Introduction
Staphylococcus aureus (S. aureus), a harmful foodborne pathogen that is easily found in high-protein foods such as meat and milk, has been of wide concern in food safety.1 The infections caused by S. aureus range from mild skin and soft tissue infections (such as boils and impetigo) to more serious illnesses (such as sepsis, pneumonia, and endocarditis).2 Therefore, detecting the presence of S. aureus in food samples is the key element in reducing the risks associated with public health. Traditional S. aureus detection methods include culture-based methods3 and PCR-based methods.4 Culture-based methods remain the gold standard, but they require long incubation times, which can delay diagnosis and treatment. PCR-based methods are rapid and sensitive, but they are expensive and can produce false-positive or false-negative results. Hence, it is urgent to develop a sensitive, accurate and low-cost method for S. aureus detection.To meet these needs, several biosensors based on aptamer,5 phages,6 and antibodies7 have been designed for S. aureus detection. In particular, biosensors based on fluorescence, with their exceptional sensitivity, impressive efficiency, and swift analytic capabilities, have risen to prominence as a pervasive sensing modality in the examination and identification of low-concentration analytes. For example, Yao et al. developed a one-step fluorometric strategy in which carbon dots was used as the energy donor and gold nanoparticles as energy acceptor, the existence of S. aureus will recover the Fluorescence of the system; the LOD of the system was 10 CFU mL−1.8 Although these methods can provide accurate results, the quantum dots and fluorescence dyes used as signals have limitations in the actual sample detection, such as short fluorescence life, strong toxicity, background spontaneous fluorescence and so on.9 Lanthanide-doped upconversion nanoparticles (UCNPs), with special optical properties including large anti-Stokes shift, clear emission wavelengths and better photostability, have been considered as potential substitutes for quantum dots and organic fluorophores.10 In recent years, aptamers have been well applied in biosensors based on UCNPs due to their superior performance.11,12 Aptamers can be ssRNA or ssDNA targeting pathogenic bacteria and curl up when the 3D structure of the aptamer specifically binds to a small molecule on the surface of the pathogen.13 Furthermore, recognition elements based on ssRNA or ssDNA have advantages, such as low cost, high stability at a large range of pH and temperature, nontoxicity, etc.14
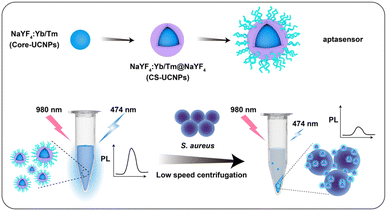
At present, most of the sensing methods of S. aureus in the detection system combined with UCNPs and aptamers are realized through Förster resonance energy transfer (FRET). The FRET process requires both energy donor and energy acceptor, and that the donor's emission spectrum and the acceptor's absorption spectrum overlap. The operation process of such an experiment is undoubtedly complicated, which is not conducive to market promotion. hence, we demonstrated a biosensor based on centrifugal separation for S. aureus detection by using core–shell UCNPs (CS-UCNPs) as signal agents and S. aureus-specific aptamer as recognition agents (Fig. 1). We first synthesized the core UCNPs. The lifetime and fluorescence signal of UCNPs are highly dependent on their upconversion efficiencies.15 To address this, we constructed UCNPs with the core–shell structure by coating core UCNPs with NaYF4 shells to have a high energy transfer efficiency.16 The functionalization of the CS-UCNPs with the S. aureus-specific aptamer resulted in an aptasensor for S. aureus detection. Compared with other FRET-based biosensors, we do not use a fluorescence quenching agent to cause fluorescence signal difference. Due to the large difference in diameter between the nanoparticles and the S. aureus (almost 30 times), it could be easy to separate them by centrifugation alone. In the absence of S. aureus, the precipitate was barely visible by low-speed centrifugation (3000 g, 5 min) and the fluorescence of the solution barely decreases. When S. aureus was present, aptasensors could bind specifically to the S. aureus. After the low-speed centrifugation (3000 g, 5 min) and separation, the precipitate (S. aureus and the binding aptasenors) cause a significant reduction in the fluorescence of the solution. Therefore, we were able to separate S. aureus by tracking the fluorescence intensity of aptasensors. Furthermore, we confirmed that the aptasensor can be used to detect S. aureus from biological samples (commercial milk).
Materials and methods
Materials and characterization
Oleic acid (OA), 1-octadecene, yttrium(III) chloride hexahydrate (YCl3·6H2O), ytterbium(III) chloride hexahydrate (YbCl3·6H2O), and thulium(III) chloride hexahydrate (TmCl3·6H2O) were bought from Sigma-Aldrich (St. Louis, USA). Methanol, sodium chloride (NaCl), and sodium hydroxide (NaOH) were purchased from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Ammonium fluoride (NH4F) was acquired from Macklin (Shanghai, China). Agar, Baird-Parker culture medium, potassium tellurite egg yolk reagent and Gram stain kit were bought from Solarbio (Beijing, China). Yeast extract and tryptone were purchased from Thermo Fisher Scientific (MA, USA). All chemicals were of analytical grade or better and used without further purification. Deionized water was used in all experiments. The nucleotide with the following sequence: 5′-SH-C6-GCAATGGTACGGTACTTCCTCGGCACGTTCTCAGTAGCGCTCGCTGGTCATCCCACAGCTACGTCAAAAGTGCACGCTACTTTGCTAA-3′ (S. aureus aptamer) and nuclease-free water were purchased from Sangon Biotechnology Inc. (Shanghai, China). Staphylococcus aureus (ATCC 29213) and Escherichia coli (ER2738) were obtained from ATCC (USA). Salmonella (isolated strain) and Enterococcus faecalis (isolated strain) were isolated and stored by our lab.Core UCNPs, core–shell UCNPs, water-soluble core–shell UCNPs, aptasensor and the specificity of aptasensor were characterized using TEM. Elemental mapping and data of core–shell UCNPs and aptasensor were obtained via Titan Chemi STEM. A full-featured fluorescence spectrometer (FLS920, Edinburgh Instruments) equipped with an external 980 nm laser at a pump power of 3 W confirmed the luminescence spectra. The ligands on the surface of core–shell UCNPs were confirmed by Fourier transform infrared spectroscopy (FTIR) according to our previous method.17 Absorption spectra were recorded on a SpectraMax M2/M2e spectrometer.
Synthesis of core–shell UCNPs
The core UCNPs were synthesized by our previously published methods with different components,18 YCl3 (0.747 M), YbCl3 (0.25 M) and TmCl3 (0.003 M). The core–shell UCNPs (CS-UCNPs) were synthesized according to a layer-by-layer method.19 Briefly, YCl3·6H2O (0.75 M), YbCl3·6H2O (0.18 M) and TmCl3·6H2O (0.003 M) in 1 mL methanol were added into the 1-octadecene (15 mL) and OA (6 mL) solvent. After stirring 750 rpm for 10 min, heated the mixture in the order of 110 °C for 30 min to remove the methanol, 150 °C for 40 min to dissolve the lanthanides, cooling down to 60 °C for 10 min, adding 2.5 mL NaOH (1 M) and 10 mL NH4F (0.4 M) slowly, vacuuming and blowing nitrogen for 3 times, heating 300 °C for 1.5 h then cooling down to room temperature. The coating shell steps are similar to steps above with mere two differences which are adding only YCl3·6H2O (0.75 M) in OA and 1-octadecene and adding prepared UCNP together with 2.5 mL NaOH (1 M) and 10 mL NH4F (0.4 M). The heating process must be done slowly. Before constructing the aptasensor, we changed the hydrophobic core–shell UCNPs into hydrophilic ones by the same acidic treatment method.20Construction of aptasensor
Core-shell UCNPs, S. aureus aptamer and HS-PEG (MW 2K) were mixed in different molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0
0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 1
0, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, 1
1000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, 1
5000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 1
000, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 and 1
0 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Here, HS-PEG binds to aptamers by forming disulfide bonds and is subsequently modified to positively charged hydrophilic UCNP to form UCNP-PEG-aptamer biosensor (aptaseneor). After stirring 1 h at 1200 rpm, the mixture was centrifuged at 8000 g for 10 min and washed with nuclease-free water. The aptasensor was collected and stored at 4 °C for further use.
000. Here, HS-PEG binds to aptamers by forming disulfide bonds and is subsequently modified to positively charged hydrophilic UCNP to form UCNP-PEG-aptamer biosensor (aptaseneor). After stirring 1 h at 1200 rpm, the mixture was centrifuged at 8000 g for 10 min and washed with nuclease-free water. The aptasensor was collected and stored at 4 °C for further use.
Detection of S. aureus by using aptasonsor
In brief, 0.1 mM aptasensor was incubated with a decreasing amount of S. aureus (start with 6.3 × 107 CFU mL−1) at 37 °C, 180 rpm, for 30 min. The mixture was centrifuged at 3000 g for 5 min. Discard the precipitate, then the upconversion fluorescence of the supernatant was measured. Different bacteria were added to the aptasensor system to replace S. aureus with the same protocols to test the specificity of the aptasensor. To detect S. aureus from real samples, purchased sterilized milk (sterilization standard followed GB25190) was polluted with different concentrations of S. aureus and then analyzed as in the previous procedures. The negative controls were sterilized milk samples. Multiple chicken samples of 25 g each were weighed and cooked at high temperatures and pressed. The sterilized milk samples and cooked chicken samples were tested according to the national standard method (GB4789.10-2016) to confirm that the cooked chicken samples did not contain S. aureus. After full homogenization in a sterile environment, 225 mL PBS buffer was added to each sample. The test was carried out using the detection method specified in the national standard method (GB4789.10-2016), and it was confirmed that the cooked chicken samples did not contain S. aureus. Artificially randomly add 1 mL of S. aureus solution of different concentrations and store at 4 °C overnight. The artificially contaminated cooked chicken samples were diluted with PBS buffer for 10-fold gradient dilution, and three consecutive groups of suitable gradient dilution samples were selected for detection by BSUNP-aptasensor and Baird-Parker plate counting method.Results and discussion
Characterization of UCNPs
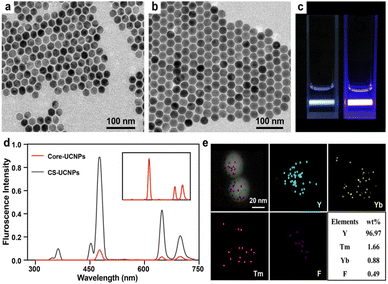
TEM images show the size and morphology of the as-prepared nanoparticles. The core UCNPs had a mean diameter of 26 nm (Fig. 2a). A thin layer of NaYF4 (3 nm) was successfully coated on the core to form a large core–shell UCNPs (CS-UCNPs), with a mean diameter of 32 nm (Fig. 2b). The CS-UCNPs emitted brighter light than core UCNPs under the same excitation by a 980 nm laser (Fig. 2c and d). The atomic compositions of CS-UCNPs were confirmed by elemental mapping (Fig. 2e), indicating that lanthanide ions (Yb3+ and Tm3+) were successfully doped in the CS-UCNPs.Construction of CS-UCNP based aptasensor
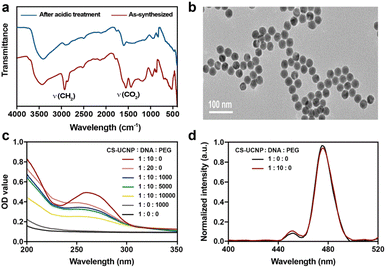
To construct an aptasensor capable of detecting S. aureus, the hydrophobic CS-UCNPs were treated with hydrochloric acid to become hydrophilic. As shown in Fig. 3a, the oleate ligands were mostly removed after the acidic treatment. However, the size and the morphology of the hydrophilic CS-UCNPs had not changed significantly compared with those before treatment (Fig. 3b). Then, we labeled the aptamer of Staphylococcus aureus on the CS-UCNP surface. The UV-vis absorbance of CS-UCNP at 260 nm confirmed the aptamer conjugation (Fig. 3c), and the best reaction molar ratio of nanoparticles and DNA was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. The upconversion luminescence of aptamer functionalized CS-UCNPs slightly decreased (Fig. 3d). Thus, these results demonstrated that the aptasensor for detecting S. aureus had been constructed successfully.
10. The upconversion luminescence of aptamer functionalized CS-UCNPs slightly decreased (Fig. 3d). Thus, these results demonstrated that the aptasensor for detecting S. aureus had been constructed successfully.
Sensitivity test of CS-UCNP based aptasensor for detecting S. aureus
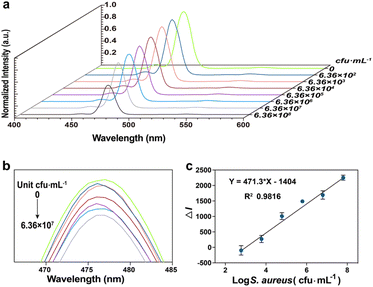
To detect S. aureus, we incubated different concentrations of S. aureus with CS-UCNP based aptasensor, Then, low-speed centrifugation was carried out to remove the S. aureus combined aptasensors. The fluorescence intensity of aptasensors decreased with the increase in the S. aureus concentration in the range from 6.36 × 102 to 6.36 × 108 CFU mL−1 (Fig. 4a, b). Importantly, the corresponding calibration plot of the decreased fluorescence intensity vs. the logarithm of S. aureus concentration presented a linear regression equation of y = 471.3x − 1404 with a square correlation coefficient of 0.98 (Fig. 4c). The limit of detection (LOD) was calculated to be 60 CFU mL−1 by the equation of LOD (KS0/S, where K is a numerical factor chosen according to the confidence level desired, S0 is the standard deviation (S. D.) of the black measurements (n = 10, K = 3)).21,22Specificity analysis of CS-UCNP-based aptasensor
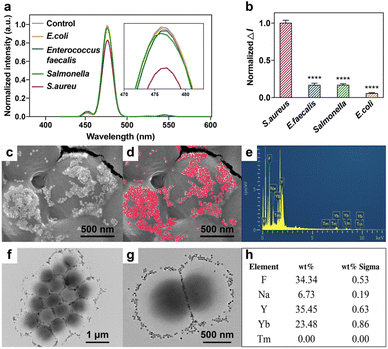
The aptamers we used have been previously reported, which had the specific property of distinguishing S. aureus from Staphylococcus epidermidis, Escherichia coli (E. coli), Streptococcus thermophilus (S. thermophilus), Enterococuus faecalis (E. faecalis), Salmonella typhimurium and Bacillus subtili.23,24 Several bacteria, including S. aureus, E. coli, E. faecalis and Salmonella were chosen to verify the selectivity of CS-UCNP based aptasensor. Fig. 5a showed the fluorescence intensity of each group. The control group means no bacteria were added to the aptasensor. As shown in Fig. 5b, other groups did not significantly decrease the fluorescence intensity except the S. aureus group. It proves that the aptamers in our research retain their original specificity. TEM and SEM images show that the CS-UCNP-based aptasensors could efficiently bind to the surface of S. aureus (Fig. 5c, d, f, and g), which is consistent with the results of fluorescence intensity. We further confirmed the binding nanoparticles were CS-UCNPs by using elemental mapping (Fig. 5e and h), suggesting that CS-UCNP can specifically bind to S. aureus.Detecting S. aureus in food samples
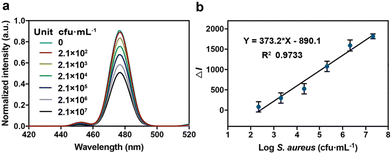
To study the performance of CS-UCNP-based aptasensors in detecting S. aureus present in foods, we chose milk as a representative sample. After the low speed of centrifugation, a gradually decreasing trend in the fluorescence intensity of the solution due to the presence of S. aureus (2.1 × 102 to 2.1 × 107 CFU mL−1) was observed (Fig. 6a). A good linearity between the decreased fluorescence intensity and the logarithm of S. aureus concentration (y = 373.2x − 890.1) was obtained (Fig. 6b). The detection limit was 146 CFU mL−1. These results indicated that the CS-UCNP-based aptasensor could work well in real samples. Furthermore, we compared the real sample (chicken muscle) detection performances of CS-UCNP-based aptasensor with the Baird-Parker plate count method, which is the golden standard detection method of S. aureus. Table 1 showed the results of the two methods. The detection results of these two methods showed a high correlation (ESI Fig. 1†), which indicated that the CS-UCNP-based aptasensor can efficiently detect S. aureus in real samples.| No. | Aptasensor (CFU g−1) | BP plate (CFU mL−1) |
|---|---|---|
| 1 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 334 334 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
| 2 | 4273 | 4270 |
| 3 | 1893 | 1870 |
| 4 | 1632 | 1620 |
| 5 | 80 | 73 |
| 6 | — | 12 |
| 7 | — | 7 |
Discussion
FRET-based biosensors are complex in design. Here we present a simplified method for experimental Staphylococcus aureus detection by centrifugation, Using UCNPs as signal agents and S. aureus-specific aptamer as recognition agents. The stability and intensity of the fluorophore are of great significance for a signal reporter, thus, we paid much attention to our core–shell structural UCNPs. By using a coating strategy, we significantly enhanced the fluorescence signal of UCNPs. Obviously, the emission waveform had been slightly changed (Fig. 2d). This phenomenon may be caused by the change of the local crystal field, since CS-UCNPs were built on the same NaYF4 host.25 To enhance the scope of our UCNP-based aptasensors, we tuned the emission of UCNPs by constructing the core–shell structure, choosing the appropriately doped ions and the concentration. Yb3+/Tm3+ has been identified as the proper coupling phonon with high upconversion efficiency.26 Meanwhile, the lattice vibrations of fluoride materials well matched the maximum phonon energy of Yb3+/Tm3+. Finally, the fluorescence intensity of the CS-UCNPs was 11.4 times higher than that of UCNPs without shell structures. We modified SH group at the 5′ of the S. aureus aptamer, so that we could easily load the aptamers onto the surface of CS-UCNPs with oleate ligands removed.27 In addition, the huge differences in size and mass between the bacteria and nanoparticles ensured the separation of them by centrifugation.Compared to other S. aureus sensors, the LOD and detection time of our aptasensor were greatly decreased (Table 2). Additionally, compared with the Baird-Parker plate count method (golden standard), the CS-UCNP-based aptasensor not only has good detection sensitivity (within the detection limit), but also costs much less time. More importantly, our aptasensor has good specificity, which may be attributed to the specific recognition between S. aureus and the aptamer. Even if the aptasensor cannot tell the difference between living and dead bacteria, it can be helpful to assess the harmfulness of S. aureus in food.
Conclusions
In summary, we successfully constructed a CS-UCNPs-based aptasensor for S. aureus detection. The core–shell structure significantly increased the fluorescence intensity, as well as the sensitivity of the aptasensor. The S. aureus bound to CS-UCNPs based aptasensor can then be isolated from the detection system by simple low-speed centrifugation. Our aptasensor was easy to be applied and showed good sensing performance in both nuclease-free water, milk, and chicken muscle samples. Furthermore, the CS-UCNP-based aptasensor can be used in a wide field due to its simplicity and low time consumption features, which provides a feasible strategy for detecting Staphylococcus aureus by the biosensor.Conflicts of interest
There are no conflicts to declare.Acknowledgements
M. Y. acknowledges the support of Zhejiang Provincial Science and Technology Plan (2021C02072-6, LY22E030004), the National Key R&D Program of China (2020YFC1806903), National Science Foundation (81871499, and 32101095), Provincial Key Laboratory Construction Plans (2020E10025), State of Sericulture Industry Technology System (CARS-18-ZJ0501), Plan of National and Zhejiang Provincial Youth Science and Technology Innovation leader ([2020]366 and 2018R52021).References
- G. Y. C. Cheung, J. S. Bae and M. Otto, Virulence, 2021, 12, 547–569 CrossRef CAS PubMed.
- N. Ahmad-Mansour, P. Loubet, C. Pouget, C. Dunyach-Remy, A. Sotto, J. P. Lavigne and V. Molle, Toxins, 2021, 13, 677 CrossRef CAS PubMed.
- X. Zhao, C.-W. Lin, J. Wang and D. H. Oh, J. Microbiol. Biotechnol., 2014, 24, 297–312 CrossRef CAS PubMed.
- A. Martinon and M. Wilkinson, J. Food Saf., 2011, 31, 297–312 CrossRef CAS.
- S. K. Samal, M. Dash, H. A. Declercq, T. Gheysens, J. Dendooven, P. Van der Voort, R. Cornelissen, P. Dubruel and D. L. Kaplan, Macromol. Biosci., 2014, 14, 991–1003 CrossRef CAS PubMed.
- X. Y. Wang, J. Y. Yang, Y. T. Wang, H. C. Zhang, M. L. Chen, T. Yang and J. H. Wang, Talanta, 2021, 221, 121668 CrossRef CAS PubMed.
- W. Gan, Z. Xu, Y. Li, W. Bi, L. Chu, Q. Qi, Y. Yang, P. Zhang, N. Gan, S. Dai and T. Xu, Biosens. Bioelectron., 2022, 199, 113860 CrossRef CAS PubMed.
- S. Yao, C. Zhao, M. Shang, J. Li and J. Wang, Food Chem. Toxicol., 2021, 150, 112071 CrossRef CAS PubMed.
- R. Renuka, N. Maroli, J. Achuth, K. Ponmalai and K. Kadirvelu, Spectrochim. Acta, Part A, 2020, 243, 118662 CrossRef PubMed.
- Q. Chen, R. Sheng, P. Wang, Q. Ouyang, A. Wang, S. Ali, M. Zareef and M. M. Hassan, Spectrochim. Acta, Part A, 2020, 241, 118654 CrossRef CAS PubMed.
- Q. Ouyang, L. Wang, W. Ahmad, Y. Yang and Q. Chen, J. Agric. Food Chem., 2021, 69, 9947–9956 CrossRef CAS PubMed.
- Y. Ma, M. Song, L. Li, X. Lao, M. C. Wong and J. Hao, Exploration, 2022, 2, 20210216 CrossRef PubMed.
- A. V. Lakhin, V. Z. Tarantul and L. V. Gening, Acta Nat., 2013, 5, 34–43 CrossRef CAS.
- M. R. Dunn, R. M. Jimenez and J. C. Chaput, Nat. Rev. Chem., 2017, 1, 1–16 CrossRef.
- X. Zhu, J. Zhang, J. Liu and Y. Zhang, Adv. Sci., 2019, 6, 1901358 CrossRef CAS PubMed.
- C. Homann, L. Krukewitt, F. Frenzel, B. Grauel, C. Wuerth, U. Resch-Genger and M. Haase, Angew. Chem., Int. Ed., 2018, 57, 8765–8769 CrossRef CAS PubMed.
- G. He, Y. Shuai, Y. Hai, T. Yang, X. Pan, Y. Liu, X. Meng, H. Yang, M. Yang and C. Mao, Mater. Today Nano, 2022, 20, 100268 CrossRef CAS.
- Z. Li, S. Lv, Y. Wang, S. Chen and Z. Liu, J. Am. Chem. Soc., 2015, 137, 3421–3427 CrossRef CAS PubMed.
- N. J. J. Johnson and F. C. J. M. van Veggel, Nano Res., 2013, 6, 547–561 CrossRef CAS.
- W. Kong, T. Sun, B. Chen, X. Chen, F. Ai, X. Zhu, M. Li, W. Zhang, G. Zhu and F. Wang, Inorg. Chem., 2017, 56, 872–877 CrossRef CAS PubMed.
- W. C. Hazeleger, W. F. Jacobs-Reitsma and H. M. W. Den Besten, Front. Microbiol., 2022, 13, 834568 CrossRef PubMed.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29(1), S49–S52 Search PubMed.
- X. Cao, S. Li, L. Chen, H. Ding, H. Xu, Y. Huang, J. Li, N. Liu, W. Cao and Y. Zhu, Nucleic Acids Res., 2009, 37, 4621–4628 CrossRef CAS PubMed.
- Q. Ouyang, Y. Yang, S. Ali, L. Wang, H. Li and Q. Chen, Spectrochim. Acta, Part A, 2021, 255, 119734 CrossRef CAS PubMed.
- S. Ye, G. Chen, W. Shao, J. Qu and P. N. Prasad, Nanoscale, 2015, 7, 3976–3984 RSC.
- G. Chen, C. Yang and P. N. Prasad, Acc. Chem. Res., 2013, 46, 1474–1486 CrossRef CAS PubMed.
- Y. Zhang, B. Duan, Q. Bao, T. Yang, T. Wei, J. Wang, C. Mao, C. Zhang and M. Yang, J. Mater. Chem. B, 2020, 8, 8607–8613 RSC.
- S.-H. Huang, Sens. Actuators, B, 2007, 127, 335–340 CrossRef CAS.
- Y. S. Lin, P. J. Tsai, M. F. Weng and Y. C. Chen, Anal. Chem., 2005, 77, 1753–1760 CrossRef CAS PubMed.
- Y. Zhang, S. Shi, J. Xing, W. Tan, C. Zhang, L. Zhang, H. Yuan, M. Zhang and J. Qiao, RSC Adv., 2019, 9, 33589–33595 RSC.
- Y.-C. Chang, C.-Y. Yang, R.-L. Sun, Y.-F. Cheng, W.-C. Kao and P.-C. Yang, Sci. Rep., 2013, 3, 1863 CrossRef PubMed.
- X. Wang, Y. Du, Y. Li, D. Li and R. Sun, J. Biomater. Sci., Polym. Ed., 2011, 22, 1881–1893 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra01555f |
| ‡ Na Li and Ying Zhang made equal contribution in this study. |
| This journal is © The Royal Society of Chemistry 2023 |