Open Access Article
Open Access ArticlepH-dependent structural characteristics of lonidamine: 1H and 13C NMR study†
Pradeep Kumar Gupta† ,
Stepan Orlovskiy†,
Jeffrey Roman†,
Stephen Pickup,
David S. Nelson,
Jerry D. Glickson and
Kavindra Nath
,
Stepan Orlovskiy†,
Jeffrey Roman†,
Stephen Pickup,
David S. Nelson,
Jerry D. Glickson and
Kavindra Nath *
*
Departments of Radiology, University of Pennsylvania, Philadelphia, Pennsylvania, USA 19104. E-mail: Kavindra.Nath@pennmedicine.upenn.edu; Tel: +1-215-746-7386
First published on 3rd July 2023
Abstract
Lonidamine (LND) is an anti-cancer drug with great potential as a metabolic modulator of chemotherapy, radiotherapy, hyperthermia, and photodynamic therapy in cancer treatment. LND affects several important aspects of cancer cell metabolism: it inhibits Complex I and II of the electron transport chain (ETC) and pyruvate carriers (mitochondrial), and monocarboxylate transporters in the plasma membrane of the cell. Cancer cells are affected by changes in pH on the molecular level, and so are the drugs used to treat cancer, thus it is important to understand how pH affects their structures and LND is no exception. LND dissolves at a pH of 8.3 in tris-glycine buffer but has limited solubility at pH 7. To understand how pH affects the structure of LND, and its effect as a metabolic modulator on cancer therapy, we made up samples of LND at pH 2, pH 7, and pH 13, and analyzed these samples using 1H and 13C NMR. We looked for ionization sites to explain the behavior of LND in solution. Our results showed considerable chemical shifts between the extremes of our experimental pH range. LND was ionized at its indazole α-nitrogen, however, we did not directly observe the protonation of the carboxyl group oxygen that is expected at pH 2, which may be the result of a chemical-exchange phenomenon.
1. Introduction
Lonidamine (LND), 1-(2,4-dichlorobenzyl)-1H-indazole-3-carboxylic acid, was first tested as a potential anti-spermatogenic agent in the late 1970s.1 LND showed success in this capacity but was ultimately found to have more interesting effects in modifying aspects of cellular metabolism. LND was initially found to inhibit glycolysis through the inhibition of the glycolytic enzyme hexokinase 2, though the role of LND in this inhibition was disputed by several studies that suggested that LND did this indirectly.1–3 There was also evidence that LND inhibited the pentose phosphate pathway, as it caused a notable decrease in cellular NADPH levels that coincided with a decrease in the level of the antioxidant glutathione, which requires NADPH for proper biosynthesis.1 More importantly, LND inhibited oxidative phosphorylation, found later to occur through the inhibition of Complex I and Complex II of the electron transport chain (ETC), by inhibition of succinate–reductase activity.1,4 After the discovery of these properties, more studies were conducted to elucidate the mechanism by which LND affects its targets.Further research into LND showed that the agent inhibited the action of the mitochondrial pyruvate carrier (MPC), which transports pyruvate into the mitochondrial matrix for further oxidation into acetyl-CoA, and blocked the export of lactate out of the cell through the inhibition of monocarboxylate transporters (MCTs) in the cell's plasma membrane.4–6 LND was found to be most active on the MPC (IC50 of 2.5 μM), followed by MCTs 1, 2 and 4 (IC50 of 36–40 μM), and on Complex I and II of the ETC (IC50 of 150 μM).1 These effects downregulate the TCA cycle, cause intracellular acidification, and block the action of the ETC.1 Glycolytic enzyme and pentose phosphate pathway inhibition could be explained as consequences of the effects of LND on its three primary targets. Because cancer cells prefer the glycolytic pathway to fulfill their energetic needs, known as the Warburg effect, cancer cells will generally use more glucose and produce more lactate than normal cells. Cancer cells under LND-induced MCT inhibition will not be able to get rid of their excess lactate and will experience detrimental and selective intracellular acidification that can be combined with other therapeutic agents to enhance the treatment of malignant neoplasms.1,4,5,7
To effectively use LND in therapy it is important to understand how the agent is affected by the pH of its microenvironment. Thus, it is important to examine the chemical structure of LND and how it changes with changes in the pH. The pH-related physicochemical effects on LND were determined by means of 1H and 13C NMR spectroscopy.
2. Experimental
2.1 Materials used
Santa Cruz Biotechnology, based in Santa Cruz, California, USA, supplied our research group with LND. The LND sample was prepared by dissolving it in deuterated DMSO (dimethyl sulfoxide-d6) to a concentration of 50 mM. For the pH titrations, 1 M NaOH and 1 M HCl solutions were added, and a pH probe that was calibrated for aqueous solutions was used to measure the effective pH. A sample of 500 μL of deuterated DMSO-dissolved LND was added to ≈30 μL of aqueous solutions to titrate LND solutions. Then, a standard NMR tube with a 5 mm O.D. and 0.5 ml of this solution was used.2.2 Methods
On a vertical bore Varian spectrometer at 400 MHz and with temperature maintained at 25 °C by the temperature controller, 1H and 13C NMR experiments were carried out. The 7.11 ppm chemical shift, this corresponds with the 6-carbon proton in the dichlorobenzyl group and provides the reference point for 1H spectra. The 40.6 ppm chemical shift, which corresponds to the DMSO reference point, was used for 13C spectra for the same reason. To acquire 1H NMR spectra, the following standard measurement parameters were used: TR (repetition time) = 8.5 seconds, N (number of acquisitions) = 256, SW (spectral width) = 6400 Hz and PW (pulse width) = 9 μs (56° flip angle). To acquire 13C NMR spectra, the following measurement parameters were applied: TR = 1.5 seconds, N = 1024, SW = 24.5 kHz and PW = 6.2 μs (45° flip angle). Fourier Transform and phasing resources in NMR utility transform software (NUTS) and MestreNova were used to analyze the free induction decay (FID) files. The ChemNMR tool in ChemDraw was used to obtain the anticipated spectra.3. Results
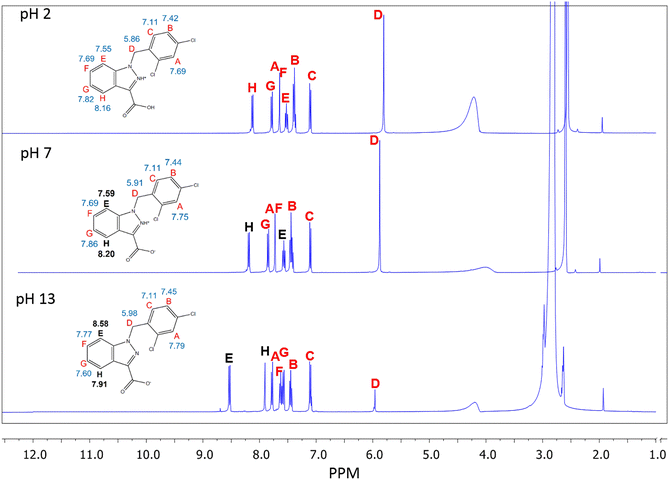
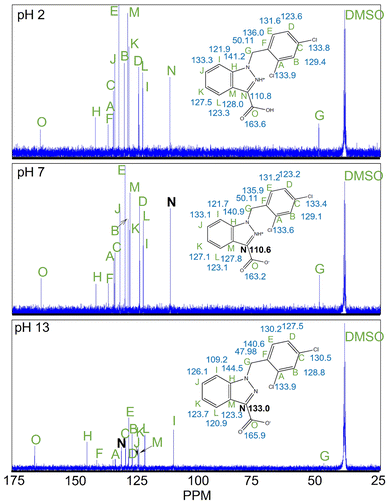
1H and 13C NMR uncovered that there were significant changes in confirmation of LND from acidic pH to basic pH. The 1H NMR shows the most noteworthy changes in shifts between pH 7 and pH 13 at position “E” and “H” (Shown Fig. 1).Additionally, the 13C NMR spectrum exhibits the greatest chemical shift change between pH 7 and pH 13 at carbon “N” (Fig. 2).
This chemical shift changed by −22.43 ppm (Table 1). In addition, 1H and 13C NMR indicate that the chemical shifts seen for LND from neutral to acidic pH change only slightly (Fig. 1 and 2). Compared to the larger changes in chemical shift that were observed between pH 2 and pH 13, all changes in chemical shift between pH 2 and pH 7 were relatively minor (Table 1).
| δ | ID | δ – δ(pH 7) | ||||
|---|---|---|---|---|---|---|
| pH 2 | pH 7 | pH 13 | pH 2 | pH 7 | pH 13 | |
| 1H | ||||||
| 5.86 | 5.91 | 5.98 | D | 0.06 | — | −0.06 |
| 7.11 | 7.11 | 7.11 | C | 0.00 | — | 0.00 |
| 7.42 | 7.44 | 7.45 | B | 0.02 | — | −0.01 |
| 7.55 | 7.59 | 8.58 | E | 0.03 | — | −0.99 |
| 7.69 | 7.75 | 7.79 | A | 0.06 | — | −0.05 |
| 7.69 | 7.75 | 7.77 | F | 0.06 | — | −0.02 |
| 7.82 | 7.86 | 7.61 | G | 0.04 | — | 0.25 |
| 8.16 | 8.20 | 7.91 | H | 0.04 | — | 0.29 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| 13C | ||||||
| 39.52 | 39.52 | 39.52 | DMSO | 0 | — | 0 |
| 50.109 | 49.834 | 47.983 | G | −0.275 | — | 1.851 |
| 110.834 | 110.603 | 133.03 | N | −0.231 | — | −22.427 |
| 121.891 | 121.675 | 109.221 | I | −0.216 | — | 12.454 |
| 123.303 | 123.095 | 120.94 | L | −0.208 | — | 2.155 |
| 123.563 | 123.206 | 127.538 | D | −0.357 | — | −4.332 |
| 127.479 | 127.13 | 123.659 | K | −0.349 | — | 3.471 |
| 128.014 | 127.791 | 123.31 | M | −0.223 | — | 4.481 |
| 129.366 | 129.114 | 128.809 | B | −0.252 | — | 0.305 |
| 131.611 | 131.187 | 130.183 | E | −0.424 | — | 1.004 |
| 133.26 | 133.141 | 126.089 | J | −0.119 | — | 7.052 |
| 133.758 | 133.401 | 130.483 | C | −0.357 | — | 2.918 |
| 133.907 | 133.55 | 133.944 | A | −0.357 | — | −0.394 |
| 136.032 | 135.868 | 140.55 | F | −0.164 | — | −4.682 |
| 141.152 | 140.936 | 144.488 | H | −0.216 | — | −3.552 |
| 163.586 | 163.281 | 165.866 | O | −0.305 | — | −2.585 |
These chemical shifts underwent changes of −0.99 ppm and 0.29 ppm, respectively (Table 1).
4. Discussion
The main focus of our laboratory and our work is to determine noninvasive biomarkers utilizing NMR imaging techniques. Our main goal in doing these pH spectroscopic studies was to determine the spectral characteristics of LND at varied pH. Although it is highly unlikely that we will be able to directly determine the changes observed in the LND spectral characteristics noninvasively (in vivo), to attempt do so, we first must determine what those characteristics are. To the best of our knowledge the structural determination of LND by NMR has never been done before.In aqueous solution, the solubility of LND dramatically decreases as the pH is lowered from 8.3 to 7.0. We hypothesize that this change in solubility results from protonation of the indazole α-nitrogen of LND to produce a zwitterion, which rapidly crystallizes. Our results support this hypothesis. In particular, the atoms which are closest to our proposed deprotonation site show the greatest change in chemical shift between pH 7 (neutral) and pH 13 (alkaline). Our 1H NMR demonstrates that the molecular environment changes most around protons “E” and “H”, while our 13C NMR demonstrates that the molecular environment changes most around carbon “N”. These two observations indicate that a substantial change must be occurring in the indazole ring of LND from neutral to alkaline pH. The proposed deprotonation of the α-nitrogen would produce changes consistent with what we observed. The second pole of the proposed zwitterion, the carboxyl group, was not directly observed. This may be because there is rapid exchange occurring with the proton in the carboxylic acid, which would make the lifetime of the fully protonated form of LND so short that NMR cannot detect it.
When addressing concerns about bioavailability of lipophilic reagents, aqueous solubility plays a major role in the pharmacodynamics of the substance regarding therapeutic effect. Sufficient concentrations of the reagent must reach the target. Details of the interactions of the compound of interest in the microenvironment related to the target become critical. Changes in pH could greatly influence the dynamic between the compound being evaluated and the site of action. It is our impression that the acute sites of action for the effects of LND have been determined (MCT; MPC, Complex I & Complex II – ubiquinone reductase).1,4–6,8,9 What has not been determined with certainty is the selectivity that has been observed for LND effects of tumor compared to normal tissue. Due to its physical properties, LND, like other lipophilic agents, is transported to its site of action by association to lipophilic sites in the blood matrix. It is interesting to speculate how pH might play a role in the differential effects seen between tumor and normal tissues. We have performed studies using 31P MRS in mouse skeletal muscle, brain, and liver demonstrating the differential effects of LND.5
Due to the Warburg effect, cancer cells exhibit increased levels of glycolysis resulting in increased production of lactic acid that leads to the corresponding acidification of the extracellular microenvironment of a tumor. This can hinder the function of immune cells, promote cancer cell migration, tissue invasion, metastasis, and upregulate cancerous stem cell phenotypes.10 Additionally, to maintain their increased intracellular energy levels, cancer cells have been shown to upregulate mechanisms that increase their intracellular pH to keep it neutral or alkaline.10–13 LND is more soluble at alkaline pH, suggesting that LND has specificity to the alkaline intracellular microenvironment of cancer cells, where it will exhibit its greatest effects in combination with other therapeutic agents as demonstrated in other studies.1,4–8 In addition, Coss, et al. have demonstrated greater levels of effect of LND in acid conditioned cells.13
Our results in this study provide context to past studies of LND and serve to elaborate on the role of pH in future studies of LND. Further speculation about the nature of the tumor acid gradient and how the confirmational pH-dependent structural changes affect LND bioavailability and LND variations in action in the acute phase of treatment seems reasonable. The mechanism for this selectivity should be further tested in both in vitro and in vivo studies using LND. Additionally, the exploration of the lack of deprotonation of the carboxylic proton at acidic pH should also be considered in the future.
5. Conclusions
Deprotonation of the LND indazole ring nitrogen occurred at an alkaline pH. This deprotonation confirms the need for careful consideration of the pH of the chemical environment when LND is used as a metabolic modulator of cancer cell therapy in future studies.Conflicts of interest
Authors have no conflict of interest.Acknowledgements
The authors would like to thank the Grant Agency of the NIH: National Institutes of Health (R01CA250102-01, R01CA228457-01A1, R01CA268601-01, R01CA172820-01A1, R01CA129544-03A1) for financial support.References
- K. Nath, L. Guo, B. Nancolas, D. S. Nelson, A. A. Shestov, S. C. Lee, J. Roman, R. Zhou, D. B. Leeper, A. P. Halestrap, I. A. Blair and J. D. Glickson, Biochim. Biophys. Acta, 2016, 1866, 151–162 CAS.
- B. A. Teicher, S. A. Holden, G. Ara and K. Menon, Int. J. Hyperther., 1995, 11, 637–645 CrossRef CAS PubMed.
- J. H. Kim, A. Alfieri, S. H. Kim, C. W. Young and B. Silvestrini, Oncology, 1984, 41(suppl. 1), 36–38 CrossRef CAS PubMed.
- K. Nath, D. S. Nelson, D. F. Heitjan, D. B. Leeper, R. Zhou and J. D. Glickson, NMR Biomed., 2015, 28, 281–290 CrossRef CAS PubMed.
- K. Nath, D. S. Nelson, A. M. Ho, S. C. Lee, M. M. Darpolor, S. Pickup, R. Zhou, D. F. Heitjan, D. B. Leeper and J. D. Glickson, NMR Biomed., 2013, 26, 98–105 CrossRef CAS PubMed.
- B. Nancolas, L. Guo, R. Zhou, K. Nath, D. S. Nelson, D. B. Leeper, I. A. Blair, J. D. Glickson and A. P. Halestrap, Biochem. J., 2016, 473, 929–936 CrossRef CAS PubMed.
- K. Nath, D. S. Nelson, M. E. Putt, D. B. Leeper, B. Garman, K. L. Nathanson and J. D. Glickson, PLoS One, 2016, 11, e0157125 CrossRef PubMed.
- L. Guo, A. A. Shestov, A. J. Worth, K. Nath, D. S. Nelson, D. B. Leeper, J. D. Glickson and I. A. Blair, J. Biol. Chem., 2016, 291, 42–57 CrossRef CAS PubMed.
- G. Cheng, Q. Zhang, J. Pan, Y. Lee, O. Ouari, M. Hardy, M. Zielonka, C. R. Myers, J. Zielonka, K. Weh, A. C. Chang, G. Chen, L. Kresty, B. Kalyanaraman and M. You, Nat. Commun., 2019, 10, 2205 CrossRef PubMed.
- C. Ward, J. Meehan, M. E. Gray, A. F. Murray, D. J. Argyle, I. H. Kunkler and S. P. Langdon, Explor Target Antitumor Ther., 2020, 1, 71–100 Search PubMed.
- P. Swietach, R. D. Vaughan-Jones, A. L. Harris and A. Hulikova, Philos. Trans. R. Soc. Lond. B Biol. Sci., 2014, 369, 20130099 CrossRef PubMed.
- R. A. Gatenby and R. J. Gillies, Nat. Rev. Cancer, 2008, 8, 56–61 CrossRef CAS PubMed.
- R. A. Coss, C. W. Storck, T. C. Wells, K. A. Kulp, M. Wahl and D. B. Leeper, Int. J. Hyperther., 2014, 30, 75–78 CrossRef CAS PubMed.
Footnote |
| † Equal first authorship |
| This journal is © The Royal Society of Chemistry 2023 |