Open Access Article
Open Access ArticleHydrocarbon degradation strategy and pyoverdine production using the salt tolerant Antarctic bacterium Marinomonas sp. ef1.†
Marco Zannotti *ad,
Kesava Priyan Ramasamyb,
Valentina Loggia,
Alberto Vassallo
*ad,
Kesava Priyan Ramasamyb,
Valentina Loggia,
Alberto Vassallo c,
Sandra Pucciarellicd and
Rita Giovannetti
c,
Sandra Pucciarellicd and
Rita Giovannetti *ad
*ad
aChemistry Interdisciplinary Project, School of Science and Technology, Chemistry Division, University of Camerino, 62032 Camerino, Italy. E-mail: marco.zannotti@unicam.it; rita.giovannetti@unicam.it
bDepartment of Ecology and Environmental Science, Umeå University, Umeå, 901 87, Sweden
cSchool of Biosciences and Veterinary Medicine, Biosciences and Biotechnology Division, University of Camerino, 62032 Camerino, Italy
dIridES s.r.l., Via Via Gentile III da Varano n° 1, 62032, Camerino, Italy
First published on 26th June 2023
Abstract
One of the most concerning environmental problems is represented by petroleum and its derivatives causing contamination of aquatic and underground environments. In this work, the degradation treatment of diesel using Antarctic bacteria is proposed. Marinomonas sp. ef1 is a bacterial strain isolated from a consortium associated with the Antarctic marine ciliate Euplotes focardii. Its potential in the degradation of hydrocarbons commonly present in diesel oil were studied. The bacterial growth was evaluated in culturing conditions that resembled the marine environment with 1% (v/v) of either diesel or biodiesel added; in both cases, Marinomonas sp. ef1 was able to grow. The chemical oxygen demand measured after the incubation of bacteria with diesel decreased, demonstrating the ability of bacteria to use diesel hydrocarbons as a carbon source and degrade them. The metabolic potential of Marinomonas to degrade aromatic compounds was supported by the identification in the genome of sequences encoding various enzymes involved in benzene and naphthalene degradation. Moreover, in the presence of biodiesel, a fluorescent yellow pigment was produced; this was isolated, purified and characterized by UV-vis and fluorescence spectroscopy, leading to its identification as a pyoverdine. These results suggest that Marinomonas sp. ef1 can be used in hydrocarbon bioremediation and in the transformation of these pollutants in molecules of interest.
1. Introduction
In the 21st century, the industrial development with the need to develop/improve new goods to achieve the best quality of life, inevitably, led to important changes in the natural environment due to contamination problems. Among these, one of the most widespread is caused by petroleum and its derivates which can eventually end up in the aquatic and underground environment, causing the contamination of aquifers and sources. Nowadays, the necessity of petroleum and fuels is still high, even though humankind is trying to move towards the increased use of renewable energy. Transport of petroleum hydrocarbons may result in accidental spills causing drastic damage to the environment, especially marine ecosystems. After accidental spills, hydrocarbons can evaporate, migrate, or be adsorbed by soils and organisms.1Fuels, whose production represents most of the crude oil processed, are the petroleum products most commonly released into the environment and the largest quantity comes from those that are used to power vehicles or that are transported as goods. Among these, diesel is particularly difficult to handle, as it is composed of many compounds of different chemical structure and biodegradability;2 moreover, it comes in liquid form, exhibiting much slower degradation due to its low evaporation rate compared with other fuel petroleum derivatives.
Diesel fuel is composed mainly by saturated, unsaturated, and aromatic hydrocarbons. The molecular composition of diesel consists in aliphatic (74%) and aromatic hydrocarbons (24%) – including polycyclic aromatic hydrocarbons (PAHs) such as naphthalene – and other compounds (2%).1,3 The number of the carbon chains in diesel is between 11 and 25. Linear alkanes are less toxic than branched alkanes, that in turn are less toxic than cyclic and aromatic hydrocarbons. Aromatic hydrocarbons are identified as carcinogenic, toxic, and mutagenic for living organisms, with a dangerous effect on the nervous system and excretory system.4,5 Diesel in water ecosystems reduces light penetration and oxygen solubilization, it is bioaccumulated in the fat tissue of the living organisms, it decreases the fertility, and it inhibits plant growth when present at high concentration. Biodiesel refers to a vegetable oil- or animal fat-based diesel fuel consisting of long-chain alkyl (methyl, ethyl, or propyl) esters. Biodiesel is composed by fatty acid esters typically made by a reaction between lipids (e.g., vegetable oil, soybean oil) and an alcohol. Biodiesel can be used alone or blended with petrol diesel in any proportions.6
Fuels and petroleum derivates spills must be removed from the environment, since their toxicity and harmful impact on living organism; the remediation technologies used to restore the environment after hydrocarbons pollution in water include physical, chemical, and biological treatments. Among physical methods, barriers are commonly used to isolate the pollutant compounds in water.1 Chemical treatments involve absorption materials like activated carbons that can immobilize and remove hydrocarbons, and emulsifiers can also be applied due to their ability to form oil–water emulsion, reducing the interfacial area.1 Biological treatments, or bioremediation, involve microorganisms able to use hydrocarbons as carbon and energy source,7 transforming them into less toxic and non-harmful compounds. In this process, bacteria use the hydrocarbons as carbon and energy source.1,7 Several examples of bacteria are reported for their ability to degrade diesel,8 such as Rhodococcus sp.,9 Pseudomonas sp.,9 Acinetobacter sp.,10 Bacillus and Alcanivorax;11 in the latter case, bacteria produce biosurfactants that increase the solubility of diesel in water facilitating the bioremediation process.11
Usually, biodegradation rate decreases at lower temperatures; for this reason, bioremediation technology has moved towards the use of Antarctic bacteria for their ability to grow and survive at low temperatures.12 The biodegradation is probably the most environmentally friendly treatment since it is simple to apply, inexpensive, as the major costs are related to the bacterial culturing processes.13
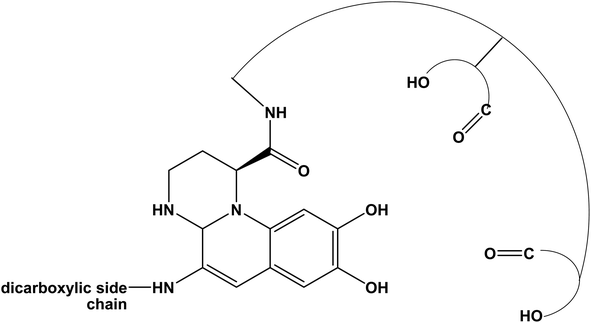
It is interesting also to note that, when these bacteria face stress conditions or essential element deficiency, they can produce fluorescent pigments. In fact, in nature, bacteria and fungi present the ability to produce a class of natural molecules called siderophores, that show high affinity for iron and metals.14,15 Different classes of siderophores can be produced by bacteria, such as carboxylates, catecholates, hydroxamates, phenolates, and pyoverdines;16 the last ones, in response of iron limitation, are commonly synthesized by Pseudomonas.17–22 A typical pyoverdine molecule consists in three distinct structures: (i) the fluorescent chromophore related to the di-hydroxyquinoline structure, (ii) a peptide chain consisting in 6 to 12 amino acids connected to the chromophore, and (iii) a dicarboxylic acid, that usually is succinic or malic acid, or their corresponding monoamide analog (i.e., glutamic and α-ketoglutaric acid, respectively), bound to the amino group present in the chromophore.20,23,24 Several pyoverdines differ only for the acid moiety bound to the amino group.25–28 The ability of chelation of the pyoverdine can be used for several medical applications, as example in diagnostic for the detection of infection;29 the siderophore, also, demonstrated to be a promising target for the detection of pathogens.30 In addition, its features make pyoverdine suitable for agriculture purposes, as example to provide plants nutrients by triggering the mobilization of specific metals ions;31 moreover, the ability to bind metals can be used for bioremediation and sensor application32,33 and its fluorescence behavior can be also exploited for the detection of organic molecules.34
In this paper, it is reported the use of the Antarctic bacteria Marinomonas sp.35 for the diesel and biodiesel degradation. This bacterial strain, able to live in extreme temperature conditions, was isolated from a consortium associated to the Antarctic psychrophilic ciliate Euplotes focardii.36,37 The COD analysis confirmed that Marinomonas sp. ef1 can degrade diesel fuel at both 4 and 22 °C. Furthermore, this strain can grow, also, in presence of biodiesel and, at the same time, can produce a fluorescent pigment that was characterized by UV-vis and fluorescence spectroscopy showing the identified pigment, which has typical features of pyoverdine molecule.
2. Experimental section
2.1 Strain culturing conditions and pigment extraction
The bacterial strain used in this study was isolated from the Antarctic psychrophilic ciliate Euplotes focardii and named Marinomonas sp. ef1, as reported in the previous work.38 It was grown in LB medium (5 g per L NaCl, 5 g per L yeast extract, 10 g per L tryptone) or on LB Agar plates (the recipe is as that of LB with the addition of 15 g per L bacto agar).To monitor the growth in presence of diesel and biodiesel, 500 μL of Marinomonas culture at stationary phase were washed with a saline medium (SM) composed of 35 mM Na2HPO4, 20 mM KH2PO4, 400 mM NaCl, 10 mM NH4NO3, 1 mM MgSO4, and 8 mM KCl, and then inoculated into 50 mL of the same SM, as artificial seawater, supplemented with 1% (v/v) commercial diesel oil or with 1% (v/v) biodiesel oil. Marinomonas sp. ef1 growing in SM without diesel was used as negative control. The growth was monitored under shaking conditions at 200 rpm in a water bath set at 22 °C and 4 °C, by measuring the optical density at 600 nm (OD600) using a NanoDrop (Thermo Scientific™) spectrophotometer. Three replicates for each test were measured.
Marinomonas sp. ef1 cells were also incubated with 1% (v/v) of biodiesel in saline medium at 22 °C for a time period of 6 days; the cultures were centrifuged for 10 min at 3000 rpm (Beckmann J2-21) and filtrated by PTFE sterile ReliaPrep Syringe™ filters (0.22 μm, Ahlstrom) to completely remove bacterial cells from samples by pelleting in order to obtain a water solution containing a yellow pigment.
All the reagents were purchased from Sigma-Aldrich and used without further purification.
2.2 Chemical oxygen demand (COD) determination
COD represent a proper parameter for the evaluation of the amount of diesel organic content before and after the bacterial degradation process. COD evaluation of Marinomonas sp. ef1 cultures growing in SM supplemented with 1% (v/v) of diesel was performed every 24 hours for six days; in parallel, the optical density at 600 nm of each sample was analyzed. The cultures were centrifuged for 10 min at 3000 rpm (Beckmann J2-21) and filtrated by PTFE sterile ReliaPrep Syringe™ filters (0.22 μm, Ahlstrom) to completely remove bacterial cells from samples. 500 μL of the samples were diluted with Milli-Q water to a final volume of 25 mL and COD was determined by adding 0.5 g of HgSO4, to remove Cl− interference, and 2.5 mL of 96% H2SO4. Then, 0.5 g of Ag2SO4, as catalyst, and 12.5 mL of 0.25 N K2Cr2O7 were added. After adding 25 mL of H2SO4 (96%) the samples were boiled for 3 hours. When the samples were cooled, 2–3 drops of ferroin like indicator were added and COD evaluated by titration with a solution of 0.25 N FeSO4(NH4)2SO4·6H2O till the change from blue/green to red. The same procedure was repeated for only Milli-Q water as blank. The COD was evaluated and reported in mg O2 per L using the following equation:| COD (mg O2 per L) = [(m1 − m2)N × 8 × 1000]/V |
All reagents were purchased from Carlo Erba reagents and used without further purification.
2.3 Purification and characterization of fluorescence pigment
The 1% (v/v) of biodiesel solution containing the yellow pigment formed after incubation with Marinomonas sp. ef1 cells was centrifuged for 10 min at 3000 rpm (Beckmann J2-21) to remove bacterial cells. The supernatant was collected and filtrated by using a PTFE sterile ReliaPrep Syringe™ filters (0.22 μm, Ahlstrom) for further investigations.The pH values of yellow pigment samples were adjusted by the addition of 1 M NaOH and 1 M HCl (Carlo Erba) to the solution and measured by using a pH-meter electrode CDC401 (Hach).
At each pH value, the UV-vis and fluorescence spectra of the yellow pigment were recorded by using Agilent Cary 8454 UV-Vis Diode Array spectrophotometer and an Ocean HDX fluorimeter (Ocean Insight), equipped with a monochromatic laser at 365 nm. The Fe(III) complex of the yellow pigment was spectrophotometrically monitored in presence of 6 μM FeCl3 (Sigma Aldrich).
2.4 Marinomonas sp. ef1 genome analysis
Prediction of the gene clusters encoding for secondary metabolites in the genome of Marinomonas sp. ef1 (available under the BioProject accession number PRJNA388049) was performed using the online software antiSMASH (v 6.1.1),39 selecting “strict” as detection strictness.3. Results and discussions
3.1 Marinomonas sp. ef1 genome analysis
John et al. (2020) analyzed the genome of Marinomonas sp. ef1 (available under the BioProject accession number PRJNA388049) and they reported the presence of genes encoding 2-keto-4-pentenoate hydratase (mhpD) and benzene 1,2-dioxygenase subunits alpha (bedC1) and beta (bnzB). We further identified additional genomic sequences encoding enzymes such as naphthalene and salicylate hydroxylases (nagG and nagH), predicted to be involved in the degradation of aromatic compounds.40Salicylate is an important intermediate in the bacterial degradation of polycyclic aromatic hydrocarbons and salicylate hydroxylases play essential roles in linking the peripheral and ring-cleavage catabolic pathways.41 The genes encoding enzymes involved in degradation of benzene were also investigated. In detail, genes encoding for catechol 1,2-dioxygenase (catA), muconate cycloisomerase 1 (catB), muconolactone D-isomerase (catC), 3-oxoadipate enol-lactonase (catD), 3-oxoadipate CoA-transferase subunits alpha (catI) and beta (catJ), two beta-ketoadipyl-CoA thiolases (catF or pcaF), and 3-ketoacyl-CoA thiolase (fadA) were found. According to KEGG pathway,42 this set of genes might link the degradation of benzene to the TCA cycle, through the final formation of acetyl-CoA and succinyl-CoA. Spatial proximity in the genome sequence and/or partial overlapping of the coding sequences suggest that these genes might be organized in two operons (i.e., catBCA and catIJFD).
Finally, we found in the Marinomonas sp. ef1 genome the catechol 1,2-dioxygenase, homogentisate 1,2-dioxygenase, and protocatechuate 3,4-dioxygenase coding genes which may be involved in the metabolism of diesel degradation.43 The ability of this organism to breakdown aromatic compounds ensures the uptake of carbon from alternative sources, a characteristic that might have helped this bacterium to survive in the high selective Antarctic habitat.
3.2 Diesel degradation ability by Marinomonas sp. ef1
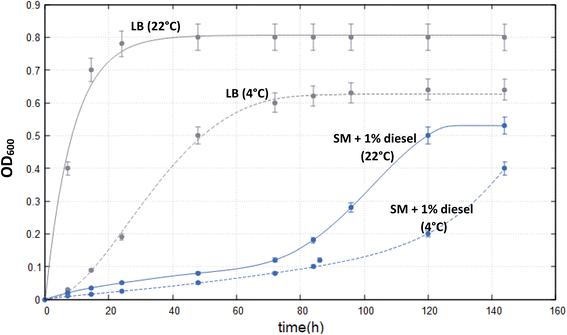
The ability of Marinomonas sp. ef1 to grow in presence of hydrocarbons was preliminarily checked at 4 °C, in saline medium (SM), in presence of 1% (v/v) diesel oil as a sole carbon source, during a period of 5 days. As it is shown in Fig. S1,† presence of diesel oil sustained the bacterial growth; indeed, the sample containing diesel showed an increased turbidity, due to cellular growth, with respect to the negative control lacking diesel. Therefore, the growth of Marinomonas sp. ef1 was monitored by measuring the OD at 600 nm at two incubation temperature (i.e., 4 °C and 22 °C) under shacking conditions in LB medium (used as control) and SM supplemented with 1% (v/v) of commercial diesel. These two temperatures were chosen because 22 °C is the optimal growth temperature of Marinomonas sp. ef1 in LB medium (John et al., 2020), while 4 °C mimics the temperature of its natural habitat (i.e., Antarctic Sea water).Growth curves (Fig. 1) showed that the two cultures of Marinomonas sp. ef1 in SM supplemented with 1% of diesel reached the plateau after 5 days at 22 °C and 6 days at 4 °C. As expected, when growing in LB medium, the plateau was reached earlier, just after 24 h in the case of 22 °C and after 3 days at 4 °C. These results clearly demonstrated that Marinomonas sp. ef1 could use diesel as sole carbon source to growth at both 22 °C and 4 °C, although the growth rate was slower in comparison to the control cultures.
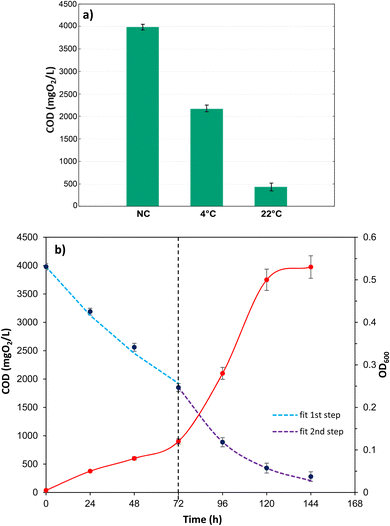
To determine the biodegradation capacity of Marinomonas sp. ef1, the COD values were measured in cultures grown in presence of diesel at 22 °C and 4 °C, after 6 days of incubations. As it is possible to observe in the Fig. 2a, COD values agreed with the growth performances observed previously (Fig. 1); indeed, cultures of Marinomonas sp. ef1 growing at 22 °C had a lower COD in comparison to the culture at 4 °C. In detail, at 22 °C the COD value was reduced by 87% when compared to the negative control, whereas at 4 °C the COD reduction was 54%, confirming that Marinomonas sp. ef1 metabolized the hydrocarbons present in diesel more efficiently at 22 °C.
Thus, COD and OD600 values were measured daily in cultures incubated at 22 °C in presence of diesel. As reported in Fig. 2b, the COD values decreased constantly during the experiment, whereas the OD600 increased, demonstrating the ability of Marinomonas sp. ef1 to use hydrocarbons of diesel as substrates.
The OD intensity is proportional to the cell density as reported in the eqn (1):
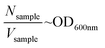 | (1) |
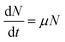 | (2) |
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N − ln N − ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N0 = μt N0 = μt
| (3) |
According to these equations, the eqn (3) can be expressed as:
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OD − ln OD − ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OD0 = μt OD0 = μt
| (4) |
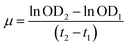 | (5) |
Specifically, during the lag phase the specific growth rate for Marinomonas sp. ef1 cultures at 22 °C in presence of 1% (v/v) of diesel is equal to 0.0182 h−1, whereas for the exponential phase the specific growth rate is 0.0297 h−1 (Table 1).
| Specific growth rate (lag phase) | 0.0182 h−1 |
| Specific growth rate (exp phase) | 0.0297 h−1 |
| k1 (step 1), R2 | 0.0101 h−1, 0.995 |
| k2 (step 2), R2 | 0.0304 h−1, 0.999 |
Taking into considerations the consumption of the substrate, represented by the hydrocarbons present in the diesel and measured as COD values, in these conditions the diesel degradation follows two steps (Fig. 2b); the slowest step is between t0 and 3 days (72 hours), while the second faster step is between 3 and 5 day (120 hours), that corresponds to the exponential growth of the bacteria. Both the degradation steps follow a first order kinetics44 that can be described as follow:
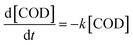 | (6) |
[COD] = [COD]0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e(−kt) e(−kt)
| (7) |
In Fig. S2,† the plot of ln[COD]/[COD]0 versus time (h), describes the two different steps during the diesel degradation, corresponding to two different degradation kinetic constants as reported in Table 1. In Fig. 2b the dash lines represent the experimental fitting by using the calculated kinetic constants showing a good estimation of the process. In Table 1 the specific growth rate and the kinetic results are resumed.
3.3 Biodiesel degradation ability by Marinomonas sp. ef1
The growth of Marinomonas sp. ef1 was also estimated at 22 °C in SM in presence of 1% (v/v) biodiesel as a sole carbon source by measuring the OD600; the growth curve reached the plateau after 4 days of incubation, as reported in Fig. 3.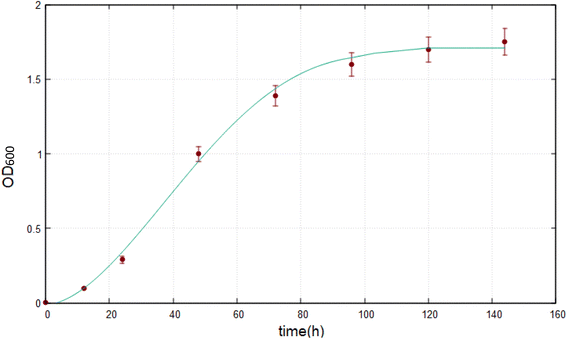 | ||
| Fig. 3 Growth curve of Marinomonas sp. ef1 in SM supplemented with 1% (v/v) of biodiesel at 22 °C. The experimental data represents a mean ± standard deviation of three replicates. | ||
In presence of 1% (v/v) of biodiesel at 22 °C, the lag phase is very short and the exponential growth is faster as demonstrated by the calculated specific growth (eqn (5)) that, for the exponential phase, is 0.0639 h−1; this value is much higher than Marinomonas sp. ef1 cultivated in the same condition with commercial diesel.
In this case, the evaluation of the COD parameter, to confirm the biodiesel degradation by Marinomonas sp. ef1 was not possible, because of the formation of a yellow pigment in the cultures that impaired COD measurements. Indeed, in presence of biodiesel, Marinomonas sp. ef1 cultures turned to yellow and a fluorescence was observed when they were exposed to UV rays; these features were probably due to the production of a secondary metabolite and the amount of this yellow compound was also proportional to the bacterial growth and to the percentage of biodiesel added to the sample, as shown in Fig. S3.†
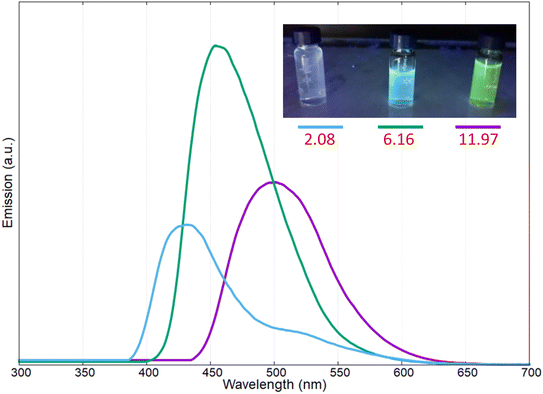
The yellow pigment was isolated as reported in the experimental section and characterized by UV-vis and fluorescence spectroscopy at different pH values. In Fig. 4 the UV-vis spectra of the pigments at three different pH values are reported (i.e., pH: 2.08, 6.16, and 11.97); in this case the intermediate pH is that of the extracted pigments from the bacterial culture.
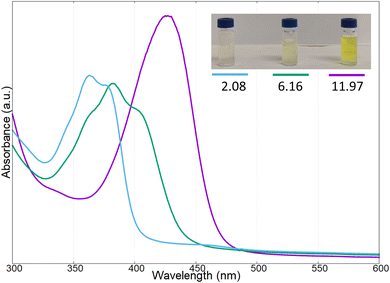 | ||
| Fig. 4 UV-vis spectra of the pigment produced by Marinomonas sp. ef1, at different pH values (2.08, 6.16, 11.97). | ||
As reported in Fig. 4, at acidic pH, the solution containing the pigment showed a very light-yellow colour, almost transparent; the related UV-vis spectrum presented two adsorption bands at 366 and 380 nm. Increasing the pH to 6.16, the band at 366 nm decreased in intensity, and a new absorption band was present at 410 nm; at this pH value, the colour of the solution was light-yellow. Reaching a strong basic pH (11.97), the shoulder at 366 nm and the band at around 380 nm disappeared completely, while the band at 410 nm increased and red-shifted to obtain a broadened absorption band at 430 nm; in these conditions the colour of the solution became bright yellow.
From these results, and in agreement with literature, it is plausible to propose that this secondary compound produced by the Marinomonas sp. ef1 is a pyoverdine molecule, which is a fluorescent molecule due to the presence of the di-hydroxyquinoline structure (Fig. 5).19,45
The spectral changes of the pyoverdine can be attributed to the protonation and deprotonation of –OH groups of the dicarboxylic acid, and of the catecholate group present in the di-hydroxyquinoline structure.18,19,46
As reported in the Fig. 6, also the fluorescence emission of the pigment changed as function of the pH; specifically, at acid pH, the fluorescence emission was centered at 431 nm (violet emission), and red shifted at 458 nm at intermediate pH (blue light emission) and at around 500 nm at basic pH (yellow emission).47–49
To further confirm that the pigment produced was the siderophore pyoverdine, the ability to strongly bind Fe(III), even at very low concentration, was evaluated, as reported in the case of other pyoverdine molecules. The hydroxyl groups of the dihydroxyquinoline, carboxyl acid and amino acids groups on the peptide chain of the pyoverdine allow this molecule to capture very strongly and rapidly Fe(III).18,50
In this context, the pyoverdine at pH 6.16, in presence of 6 μM of FeCl3, showed a colour change from pale-yellow to orange after only 60 seconds, due to the formation of stable complex with Fe(III). The UV-vis spectral variations reported in Fig. 7 showed a red shift of the main bands with an increasing of absorbance at around 400 nm; at this pH, a broad ligand-to-metal charge transfer band at higher wavelength (470 nm) and a small band at 550 nm were observed.18,21,45
At genomic level, the analysis with the online software antiSMASH (v. 6.1.1)39 allowed the identification of four genome regions containing secondary metabolite biosynthetic gene clusters (Table 2). Noteworthy, one of the predicted secondary metabolites biosynthetic regions (1.1 in Table 2) was classified as a nonribosomal peptide synthetase (NRPS) cluster, sharing a 38% similarity with that of the siderophore turnerbactin from the bacterium Teredinibacter turnerae T7901. This region includes two genes predicted to encode NRPS (indicated as core biosynthetic genes, in red, in Fig. 8), reported to be responsible for pyoverdine synthesis in Pseudomonas.22 This cluster may represent a good candidate for pyoverdine biosynthesis in Marinomonas sp. ef1.
| Contig | Region | Type | Position | Most similar known cluster | |||
|---|---|---|---|---|---|---|---|
| From | To | Name | Type | Similarity | |||
| 1 | Region 1.1 | NRPS | 292.233 | 359.796 | Turnerbactin | NRP | 38% |
| 6 | Region 6.1 | Betalactone | 240.314 | 264.144 | |||
| 8 | Region 8.1 | Acyl-amino-acids | 53.961 | 115.232 | |||
| 9 | Region 9.1 | Ectoine | 91.642 | 102.031 | Ectoine | Other | 83% |
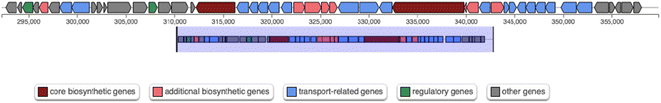 | ||
| Fig. 8 Scheme of region 1.1 predicted by antiSMASH and containing a putative NRPS gene cluster (Table 1). | ||
The biosynthesis of pyoverdine by Marinomonas sp. ef1 in presence of biodiesel may be a response to iron deficiency in the environment;17 in contrast, this production was not observed in the case of cultures containing diesel.
4. Conclusions
In this study, the Antarctic bacterium Marinomonas sp. ef1 was used successfully for diesel and biodiesel degradation. Marinomonas sp. ef1 showed a different behavior when the culture grew with 1% (v/v) of diesel or biodiesel. The hydrocarbons present in the composition of diesel can be used by the bacteria as carbon source permitting them to reproduce and growing in this extreme condition. The bacterial growth monitored by the optical density at 600 nm (OD600) demonstrate the ability of the Antarctic bacterium to grow better and more rapidly at 22 °C instead of 4 °C, even if at this temperature, the bacteria, in presence of diesel, were able to grow. The degradation of diesel was monitored by the evaluation of COD parameter, showing a reduction about 87% of COD at 22 °C, whereas the COD reduction at 4 °C was 54%. In addition, a kinetic study demonstrated that the diesel degradation at 22 °C follows two different steps, the first slower and the second faster, both following a first order kinetics with different degradation rate.When the Marinomonas sp. ef1 was cultivated with 1% (v/v) of biodiesel, the bacterium always showed the capability to reproduce itself, and in this case, more rapidly with respect to the same condition with commercial diesel. In addition, in presence of biodiesel the bacterium was stimulated to synthesize a yellow secondary metabolite, that showed the typical absorption and fluorescence profile of the pyoverdine siderophore. The capability of Marinomonas sp. ef1 to exploit alternative carbon sources probably contributes to its survival and that one of its host E. focardii under the nutrient limitations in the Antarctic environment; in fact the pyoverdine biosynthesis may be a strategy to scavenge Fe(III) ions that are essential for basic metabolism and to face iron deficiency in the Antarctic environment. In this case the presence or the absence of pyoverdine represent an indication of the presence of biodiesel or diesel, respectively.
Conflicts of interest
There are no conflicts to declare.References
- J. E. Vidonish, K. Zygourakis, C. A. Masiello, G. Sabadell and P. J. J. Alvarez, Thermal Treatment of Hydrocarbon-Impacted Soils: A Review of Technology Innovation for Sustainable Remediation, Engineering-Prc, 2016, 2, 426–437 CAS.
- D. K. Chaudhary, R. Bajagain, S.-W. Jeong and J. Kim, Insights into the biodegradation of diesel oil and changes in bacterial communities in diesel-contaminated soil as a consequence of various soil amendments, Chemosphere, 2021, 285, 131416 CrossRef CAS.
- F. Y. Liang, M. M. Lu, T. C. Keener, Z. F. Liu and S. J. Khang, The organic composition of diesel particulate matter, diesel fuel and engine oil of a non-road diesel generator, J. Environ. Monit., 2005, 7, 983–988 RSC.
- D. N. Das and N. Ravi, Influences of polycyclic aromatic hydrocarbon on the epigenome toxicity and its applicability in human health risk assessment, Environ. Res., 2022, 213 CrossRef CAS PubMed.
- F. Yang, Q. Q. Zhang, H. R. Guo and S. C. Zhang, Evaluation of cytotoxicity, genotoxicity and teratogenicity of marine sediments from Qingdao coastal areas using in vitro fish cell assay, comet assay and zebrafish embryo test, Toxicol. in Vitro, 2010, 24, 2003–2011 CrossRef CAS.
- J. L. Zhu, Z. Wang and R. N. Li, Experimental study on particle microstructure of diesel/biodiesel blend fuels measured by SAXS and HRTEM techniques, Process Saf. Environ. Prot., 2022, 165, 680–693 CrossRef CAS.
- M. Salari, V. Rahmanian, S. A. Hashemi, W. H. Chiang, C. W. Lai, S. M. Mousavi and A. Gholami, Bioremediation Treatment of Polyaromatic Hydrocarbons for Environmental Sustainability, Water-Sui, 2022, p. 14 Search PubMed.
- L. C. Goveas, S. Nayak and R. Selvaraj, Concise review on bacterial degradation of petroleum hydrocarbons: Emphasis on Indian marine environment, Bioresour. Technol. Rep., 2022, 19, 101136 CrossRef CAS.
- K. Rehman, A. Imran, I. Amin and M. Afzal, Inoculation with bacteria in floating treatment wetlands positively modulates the phytoremediation of oil field wastewater, J. Hazard. Mater., 2018, 349, 242–251 CrossRef CAS PubMed.
- Y. Y. Cai, R. K. Wang, P. H. Rao, B. C. Wu, L. L. Yan, L. J. Hu, S. Park, M. Ryu and X. Y. Zhou, Bioremediation of Petroleum Hydrocarbons Using Acinetobacter sp. SCYY-5 Isolated from Contaminated Oil Sludge: Strategy and Effectiveness Study, Int. J. Environ. Res. Public Health, 2021, 18, 819 CrossRef CAS PubMed.
- F. E. Khalid, Z. S. Lim, S. Sabri, C. Gomez-Fuentes, A. Zulkharnain and S. A. Ahmad, Bioremediation of Diesel Contaminated Marine Water by Bacteria: A Review and Bibliometric Analysis, J. Mar. Sci. Eng., 2021, 9, 155 CrossRef.
- H. S. Yap, N. N. Zakaria, A. Zulkharnain, S. Sabri, C. Gomez-Fuentes and S. A. Ahmad, Bibliometric Analysis of Hydrocarbon Bioremediation in Cold Regions and a Review on Enhanced Soil Bioremediation, Biology, 2021, 10, 354 CrossRef CAS PubMed.
- S. Bala, D. Garg, B. V. Thirumalesh, M. Sharma, K. Sridhar, B. S. Inbaraj and M. Tripathi, Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment, Toxics, 2022, 10, 484 CrossRef CAS PubMed.
- M. Saha, S. Sarkar, B. Sarkar, B. K. Sharma, S. Bhattacharjee and P. Tribedi, Microbial siderophores and their potential applications: a review, Environ. Sci. Pollut. Res., 2016, 23, 3984–3999 CrossRef CAS PubMed.
- C. E. Lankford and B. R. Byers, Bacterial Assimilation of iron, CRC Crit. Rev. Microbiol., 1973, 2, 273–331 CrossRef CAS.
- Z. Roskova, R. Skarohlid and L. McGachy, Siderophores: an alternative bioremediation strategy?, Sci. Total Environ., 2022, 819, 153144 CrossRef CAS PubMed.
- R. C. Hider and X. L. Kong, Chemistry and biology of siderophores, Nat. Prod. Rep., 2010, 27, 637–657 RSC.
- A. M. Albrechtgary, S. Blanc, N. Rochel, A. Z. Ocaktan and M. A. Abdallah, Bacterial Iron Transport - Coordination Properties of Pyoverdin Paa, a Peptidic Siderophore of Pseudomonas-Aeruginosa, Inorg. Chem., 1994, 33, 6391–6402 CrossRef CAS.
- H. Boukhalfa, S. D. Reilly, R. Michalczyk, S. Iyer and M. P. Neu, Iron(III) coordination properties of a pyoverdin siderophore produced by Pseudomonas putida ATCC 33015, Inorg. Chem., 2006, 45, 5607–5616 CrossRef CAS PubMed.
- H. Budzikiewicz, M. Schafer, D. U. Fernandez and J. M. Meyer, Structure proposal for a new pyoverdin from Pseudomonas sp PS 6.10, Z. Naturforsch., C: J. Biosci., 2006, 61, 815–820 CrossRef CAS PubMed.
- A. Bultreys, I. Gheysen, B. Wathelet, H. Maraite and E. de Hoffmann, High-performance liquid chromatography analyses of pyoverdin siderophores differentiate among phytopathogenic fluorescent Pseudomonas species, Appl. Environ. Microbiol., 2003, 69, 1143–1153 CrossRef CAS PubMed.
- M. J. Calcott, J. G. Owen, I. L. Lamont and D. F. Ackerley, Biosynthesis of Novel Pyoverdines by Domain Substitution in a Nonribosomal Peptide Synthetase of Pseudomonas aeruginosa, Appl. Environ. Microbiol., 2014, 80, 5723–5731 CrossRef PubMed.
- P. Visca, F. Imperi and I. L. Lamont, Pyoverdine siderophores: from biogenesis to biosignificance, Trends Microbiol., 2007, 15, 22–30 CrossRef CAS PubMed.
- H. Wei and L. Aristilde, Structural characterization of multiple pyoverdines secreted by two Pseudomonas strains using liquid chromatography-high resolution tandem mass spectrometry with varying dissociation energies, Anal. Bioanal. Chem., 2015, 407, 4629–4638 CrossRef CAS PubMed.
- J. M. Meyer, C. Gruffaz, V. Raharinosy, I. Bezverbnaya, M. Schafer and H. Budzikiewicz, Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method, BioMetals, 2008, 21, 259–271 CrossRef CAS PubMed.
- M. Hannauer, M. Schafer, F. Hoegy, P. Gizzi, P. Wehrung, G. L. A. Mislin, H. Budzikiewicz and I. J. Schalk, Biosynthesis of the pyoverdine siderophore of Pseudomonas aeruginosa involves precursors with a myristic or a myristoleic acid chain, FEBS Lett., 2012, 586, 96–101 CrossRef CAS PubMed.
- G. Ghssein and Z. Ezzeddine, A Review of Pseudomonas aeruginosa Metallophores: Pyoverdine, Pyochelin and Pseudopaline, Biology, 2022, 11, 1711 CrossRef CAS PubMed.
- J. G. Owen and D. F. Ackerley, Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a, BMC Microbiol., 2011, 11, 218 CrossRef CAS PubMed.
- M. Petrik, E. Umlaufova, V. Raclavsky, A. Palyzova, V. Havlicek, H. Haas, Z. Novy, D. Dolezal, M. Hajduch and C. Decristoforo, Imaging of Pseudomonas aeruginosa infection with Ga-68 labelled pyoverdine for positron emission tomography, Sci. Rep., 2018, 8, 15698 CrossRef PubMed.
- B. Ciui, M. Tertiş, A. Cernat, R. Săndulescu, J. Wang and C. Cristea, Finger-Based Printed Sensors Integrated on a Glove for On-Site Screening Of Pseudomonas aeruginosa Virulence Factors, Anal. Chem., 2018, 90, 7761–7768 CrossRef CAS PubMed.
- C. Ferret, J. Y. Cornu, M. Elhabiri, T. Sterckeman, A. Braud, K. Jezequel, M. Lollier, T. Lebeau, I. J. Schalk and V. A. Geoffroy, Effect of pyoverdine supply on cadmium and nickel complexation and phytoavailability in hydroponics, Environ. Sci. Pollut. Res., 2015, 22, 2106–2116, DOI:10.1007/s11356-014-3487-2.
- K. Yin, Y. Wu, S. Wang and L. Chen, A sensitive fluorescent biosensor for the detection of copper ion inspired by biological recognition element pyoverdine, Sens. Actuators, B, 2016, 232, 257–263 CrossRef CAS.
- R. Nosrati, S. Dehghani, B. Karimi, M. Yousefi, S. M. Taghdisi, K. Abnous, M. Alibolandi and M. Ramezani, Siderophore-based biosensors and nanosensors; new approach on the development of diagnostic systems, Biosens. Bioelectron., 2018, 117, 1–14 CrossRef CAS PubMed.
- K. Yin, W. Zhang and L. Chen, Pyoverdine secreted by Pseudomonas aeruginosa as a biological recognition element for the fluorescent detection of furazolidone, Biosens. Bioelectron., 2014, 51, 90–96 CrossRef CAS PubMed.
- M. S. John, J. A. Nagoth, M. Zannotti, R. Giovannetti, A. Mancini, K. P. Ramasamy, C. Miceli and S. Pucciarelli, Biogenic Synthesis of Copper Nanoparticles Using Bacterial Strains Isolated from an Antarctic Consortium Associated to a Psychrophilic Marine Ciliate: Characterization and Potential Application as Antimicrobial Agents, Mar. Drugs, 2021, 19, 263 CrossRef CAS PubMed.
- S. Pucciarelli, R. R. Devaraj, A. Mancini, P. Ballarini, M. Castelli, M. Schrallhammer, G. Petroni and C. Miceli, Microbial Consortium Associated with the Antarctic Marine Ciliate Euplotes focardii: An Investigation from Genomic Sequences, Microb. Ecol., 2015, 70, 484–497 CrossRef CAS PubMed.
- M. Mozzicafreddo, S. Pucciarelli, E. C. Swart, A. Piersanti, C. Emmerich, G. Migliorelli, P. Ballarini and C. Miceli, The macronuclear genome of the Antarctic psychrophilic marine ciliate Euplotes focardii reveals new insights on molecular cold adaptation, Sci. Rep., 2021, 11, 18782 CrossRef CAS PubMed.
- M. S. John, J. A. Nagoth, K. P. Ramasamy, P. Ballarini, M. Mozzicafreddo, A. Mancini, A. Telatin, P. Lio, G. Giuli, A. Natalello, C. Miceli and S. Pucciarelli, Horizontal gene transfer and silver nanoparticles production in a new Marinomonas strain isolated from the Antarctic psychrophilic ciliate Euplotes focardii, Sci. Rep., 2020, 10, 10218 CrossRef CAS PubMed.
- K. Blin, S. Shaw, A. M. Kloosterman, Z. Charlop-Powers, G. P. van Wezel, M. H. Medema and T. Weber, antiSMASH 6.0: improving cluster detection and comparison capabilities, Nucleic Acids Res., 2021, 49, W29–W35 CrossRef CAS PubMed.
- J. S. Seo, Y. S. Keum and Q. X. Li, Bacterial Degradation of Aromatic Compounds, Int. J. Environ. Res. Public Health, 2009, 6, 278–309 CrossRef CAS PubMed.
- T. Fang and N. Y. Zhou, Purification and characterization of salicylate 5-hydroxylase, a three-component monooxygenase from Ralstonia sp strain U2, Appl. Microbiol. Biotechnol., 2014, 98, 671–679 CrossRef CAS PubMed.
- KEGG PATHWAY Database, https://www.genome.jp/entry/ko00362, accessed March 2023 Search PubMed.
- KEGG PATHWAY Database, https://www.genome.jp/dbget-bin/www_bget?ko00623, accessed March 2023 Search PubMed.
- R. Giovannetti, E. Rommozzi, C. A. D'Amato and M. Zannotti, Kinetic Model for Simultaneous Adsorption/Photodegradation Process of Alizarin Red S in Water Solution by Nano-TiO2 under Visible Light, Catalysts, 2016, 6, 84 CrossRef.
- R. Xiao and W. S. Kisaalita, Purification of Pyoverdines of Pseudomonas-Fluorescens-2-79 by Copper-Chelate Chromatography, Appl. Environ. Microbiol., 1995, 61, 3769–3774 CrossRef CAS PubMed.
- T. Palanche, S. Blanc, C. Hennard, M. A. Abdallah and A. M. Albrecht-Gary, Bacterial iron transport: Coordination properties of azotobactin, the highly fluorescent siderophore of Azotobacter vinelandii, Inorg. Chem., 2004, 43, 1137–1152 CrossRef CAS PubMed.
- J. Greenwald, F. Hoegy, M. Nader, L. Journet, G. L. A. Mislin, P. L. Graumann and I. J. Schalk, Real time fluorescent resonance energy transfer visualization of ferric pyoverdine uptake in pseudomonas aeruginosa – a role for ferrous iron, J. Biol. Chem., 2007, 282, 2987–2995 CrossRef CAS PubMed.
- M. M. Naik and S. K. Dubey, Lead-Enhanced Siderophore Production and Alteration in Cell Morphology in a Pb-Resistant Pseudomonas aeruginosa Strain 4EA, Curr. Microbiol., 2011, 62, 409–414 CrossRef CAS PubMed.
- A. Bashir, T. Tian, X. Yu, C. Meng, M. Ali and L. Li, Pyoverdine-Mediated Killing of Caenorhabditis elegans by Pseudomonas syringae MB03 and the Role of Iron in Its Pathogenicity, Int. J. Mol. Sci., 2020, 21, 2198 CrossRef CAS PubMed.
- T. C. Johnstone and E. M. Nolan, Beyond iron: non-classical biological functions of bacterial siderophores, Dalton Trans., 2015, 44, 6320–6339 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra02536e |
| This journal is © The Royal Society of Chemistry 2023 |