Open Access Article
Open Access ArticleSynthesis of NMN by cascade catalysis of intracellular multiple enzymes†
Wenfeng Hua,
Na Jiang,
Yifei Wu,
Cailian Zhou,
Kequan Chen and
Xin Wang
and
Xin Wang *
*
State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing 211816, Jiangsu, China. E-mail: xinwang1988@njtech.edu.cn
First published on 25th September 2023
Abstract
β-Nicotinamide mononucleotide is a biologically active nucleotide compound, and its excellent anti-aging activity is widely used in medicine and has multiple functions, making NMN have broad application prospects in the fields of nutrition, health food, and even medicine. Herein, based on the supply of the co-substrate PRPP, we designed and constructed three in vivo NMN synthesis pathways using glucose, xylose, and arabinose as raw materials and Escherichia coli as the host. The best in vivo pathway through whole-cell catalysis was identified. Then, we optimized the cell culture and catalytic conditions of the optimal path to determine the optimal conditions and ultimately obtained an NMN titer of 1.8 mM.
Introduction
β-Nicotinamide mononucleotide (NMN) is a biologically active nucleotide compound, and it is a precursor of nicotinamide adenine dinucleotide (NAD+), an atypical redox cofactor in biological metabolism.1 Studies have shown that NMN can inhibit NAD+ consuming enzymes or maintain the intracellular NAD+ level through the NAD+ recovery pathway2 and promote nerve regeneration to treat ischemic brain injury.3,4 NMN supplementation can also promote mitochondrial energy metabolism and improve cognition.5 Therefore, NMN plays an important role in brain health diseases. Simultaneously, its excellent anti-aging activity is widely used in medicine to treat age-related degenerative diseases.2,6 Because of its various functions, NMN has broad application prospects in the fields of nutrition, health food and even medicine.7In recent years, increasing attention has been paid to the synthesis methods of NMN. At present, the highest yield of NMN is obtained by applying the chemical synthesis method, such as the Ketalization reagent method.8 However, this method has problems, such as severe reaction conditions, the use of organic solvents, and poor stereoselectivity. Compared with chemical methods, the bio-based process has the advantages of high stereoselectivity, mild reaction conditions and fewer by-products. The development of biological processes for NMN synthesis is therefore highly desired.9
The biological methods for NMN production mainly include biological fermentation and enzyme catalysis. In 2020, Shoji et al. synthesized NMN by expressing heterologous NAMPT and enhancing the PRPP biosynthesis pathway.10 In the 2022 study conducted by Zhou et al., it was proposed that the use of multi-enzyme cascade catalysis to produce NMN is a very effective and convenient method.11 Using inexpensive glucose as a catalytic raw material, a large amount of the key precursor PRPP is generated through the RPPK gene to enhance the enzymatic synthesis of NMN. It is worth noting that the supply of PRPP has always been a key factor hindering the increase in NMN production.12,13 Therefore, how to accumulate a large amount of key co-substrate PRPP during the catalytic process is a problem that needs to be explored.
In this study, we designed and explored several cascaded catalytic synthesis routes to supply PRPP for NMN production. Because the co-substrate of PRPP of NMN can be synthesized through the intermediate ribulose-5-phosphate of the pentose phosphate pathway, three routes were constructed in Escherichia coli (E. coli) to supply PRPP in vivo, where glucose, xylose, and arabinose were used as raw materials. The ability to synthesize NMN through different pathways was studied, and the optimal synthesis route was determined by comparing the synthesis efficiency of NMN. Then, the cell culture and catalytic conditions were further optimized to obtain a higher level of NMN through a whole-cell bioconversion process.
Methods
Construction of plasmid
The strains and plasmids used in this chapter are listed in Table 1. E. coli BL21 (DE3) was used as the host cells. The vectors pCDFDuet and pRSFDuet were used for gene manipulation and protein expression, respectively. The details of the enzymes used in this study were extracted from the NCBI database and are summarized in Table 1.| Strains or plasmids | Description | References |
|---|---|---|
| Strains | ||
| BL21(DE3) | Used as host strain | Invitrogen |
| BL21-GP | BL21(DE3) harboring plasmid pCDFuet-zwf-pgl-gnd-rpiA/B-prs and pRSFDuet-nampt-niap-pnuc | This study |
| BL21-XP | BL21(DE3) harboring plasmid pCDFDuet- rpiA/B-prs- xylA/B-rpe and pRSFDuet-nampt-niap-pnuc | This study |
| BL21-AP | BL21(DE3) harboring plasmid pCDFDuet- rpiA/B-prs- fucI-araB and pRSFDuet-nampt-niap-pnuc | This study |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Plasmids | ||
| pCDFDuet | Expression vector, SmR, PT7, ori | This study |
| pRSFDuet | Expression vector, KmR, PT7, ori | This study |
| pCDFDuet-zwf-pgl-gnd-rpiA/B-prs | SmR, pCDFDuet harboring zwf, pgl, rpiA, rpiB and prs | This study |
| pCDFDuet-xylA/B-rpe-rpiA/B-prs | SmR, pCDFDuet harboring xylA, xylB, rpe, rpiA, rpiB and prs | This study |
| pCDFDuet-fucI-araB-rpiA/B-prs | SmR, pCDFDuet harboring fucI, araB, rpiA, rpiB and prs | This study |
| pRSFDuet-nampt-niap-pnuc | KmR, pRSFDuet harboring nampt, niap and punc | This study |
The genes used were synthesized by Nanjing Qingke Biological Company. The genes zwf, pgl, gnd, ripA, ripB and prs of E. coli MG1655 were constructed on pCDFDuet to obtain the plasmid pCDFDuet-zwf-pgl-gnd-rpiA/B-prs.
The rpiA, rpiB and prs fragments were amplified from pCDFDuet-zwf-pgl-gnd-rpiA/B-prs plasmid using ripA-F and prs-R and were constructed between the EcoRI and NcoI digestion sites of pCDFDuet-MCS-2 through homologous recombination. Xylose isomerase (xylA) and xylokinase (xylB) of E. coli MG1655 were amplified with xylA-F and xylB-R to obtain the site xylA/B fragment with NdeI and KpnI, respectively. Linearized pCDFDuet-MCS-1 and xylA/B fragments treated with NdeI and KpnI restriction endonuclease were connected with T4 ligase to obtain pCDFDuet-xylA/B. The ribose-5-phosphate isomerase (rpe) from the same source was amplified using rpe-F and rpe-R to obtain fragments with KpnI and XhoI sites, respectively. Then, pCDFDuet-xylA/B was tangent transformed using KpnI and XhoI enzymes, and the plasmid pCDFDuet-rpiA/B-prs-xylA/B-rpe was obtained by ligase.
The plasmid pCDFDuet-rpiA/B-prs was constructed in the same way as described above. AraB and D-arabinose isomerase (fucI) of E. coli MG1655 were amplified by PCR using araB-F and araB-R as well as fucI-F and fucI-R to obtain araB and fucI fragments. Then, araB-F and fucI-R were used to connect araB and fucI fragments by overlapping PCR homologous recombination and then constructed between the NdeI and BglII sites of pCDFDuet-MCS-1 through homologous recombination to obtain pCDFDuet-rpiA/B-prs-araB-fucI.
NAMPT-F and NAMPT-R were used to amplify the nicotinamide phosphoribosyltransferase gene nampt of Haemophilus ducreyi, which was constructed between the NcoI and HindIII digestion sites of pRSFDuet-MCS-1 through homologous recombination. Niap transporter from Burkholderia cepacia was amplified using NiaP-F and NiaP-R; PnuC-F and PnuC-R amplified the nicotinamide ribose transporter gene pnuc from Bacillus mycoides, constructed two fragments of niap and pnuc between the NdeI and XhoI digestion sites of pRSFDuet-MCS-2 by homologous recombination, and obtained the plasmid pRSFDuet-nampt-niap-pnuc. The specific primer sequence is shown in Table 2.
| Name | Primer sequence (5′–3′) |
|---|---|
| amn-F | AGGAGATATACCATGCCATGGATGAATAATAAGGGCT |
| amn-R | ATGCGGCCGCAAGCTCCCAAGCTTTTATCGGAACGGC |
| prs-F | AAGGAGATATACATAGGGAATTCCATATGGTGCCTGATATGA |
| prs-R | TTACCAGACTCGAGGCGGGGTACCTTAGTGTTCGAACA |
| NAMPT-F | AGGAGATATACCATGATGGACAACCTGCTGAACTACTCT |
| NAMPT-R | ATGCGGCCGCAAGCTTTACAGGGTGGTACGAGAAACCAG |
| NiaP-F | CCACCACCACCACTGATTTGTTTAACTTTAAGAAGGAGATATACCATGCCAG |
| NiaP-R | GGTTTCTTTACCAGACTCGAGCTCGAGACTTGCTTTATCAGCTGC |
| PnuC-F | ATAAGAAGGAGATATACATATGATGGTGCGTAGCCCTCTG |
| PnuC-R | CTTAAAGTTAAACAAATCAGTGGTGGTGGTGGTGGTGC |
| rpiA-F | AGGAGATATACCATGACGCAGGATGAA |
| prs-R | CGCCGAGCTCGAATTCTTAGTGTTCGAACATGGCAGAGA |
| xylA-F | GGGAATTCCATATGATGCAAGCCTATTTTGACCAGCTC |
| xylB-R | CGGGGTACCTTACGCCATTAATGGCAGAAGTTGC |
| rpe-F | CGATCGCTGACGTCGGTACCTTCTCAAGGAGAAGCGGATGAAACAGT |
| rpe-R | CTTTACCAGACTCGAGTTATTCATGACTTACCTTTGCCAGTTCACTGC |
| araB-F | AAGGAGATATACATATGATGGCGATTGCAATTGGCCT |
| araB-R | AATATGTTTTATAGAGTCGCAACGGCCTGG |
| fucI-F | TCTATAAAACATATTTTCCGAATAAAGTGAGGAATCTGTAATG |
| fucI-R | TATCCAATTGAGATCTTTAACGCTTGTACAACGGACCGTAG |
Construction of recombinant strains
The constructed plasmids pCDFDuet-zwf-pgl-gnd-rpiA/B-prs and pRSFDuet-nampt-niap-pnuc, pCDFDuet-xylA/B-rpe-rpiA/B-prs and pRSFDuet-nampt-niap-pnuc, and pCDFDuet-fucI-araB-rpiA/B-prs and pRSFDuet-nampt-niap-pnuc were co-transformed into the expression host BL21 (DE3). They were referred to as BL21-GP, BL21-XP, and BL21-AP, respectively. It was coated on LB solid medium with the final concentrations of 50 mg L−1 kanamycin (Km) and 50 mg L−1 streptomycin (Sm), and cultured at 37 °C for 10–12 hours. The expressed strain was stored in glycerol with a final concentration of 15%, and stored in an ultralow temperature refrigerator at −80 °C.Detecting the NMN production activity of the three routes
To explore the effect of the constructed three intracellular cascade catalytic synthesis NMN pathways, the co-expressed three glycerol bacteria BL21-GP, BL21-XP, and BL21-AP were activated and coated on the corresponding resistant plates to obtain single colonies. The single colonies were selected and cultured at 37 °C for 8–12 h in 5 mL of LB liquid medium with Km and Sm double antibodies. The inoculation amount was 2.5%. The bacterial solution was transferred to 100 mL of LB liquid medium with resistance for further culture. When OD600 is about 0.6∼0.8, 1‰ IPTG of inducer was added, and the bacteria were collected after induction at 18 °C for 18–20 h. Then, the cells were collected by centrifugation at 4 °C and 6000 rpm for 15 min, washed with PBS salt buffer solution, and centrifugated for later use.The detection of catalytic activity of recombinant strains was co-expressed by three pathways, and the M9 medium was used as a catalytic buffer. The specific catalytic conditions are shown in Table 3. The reaction system was maintained at 220 rpm and 30 °C for 12 h. The whole cell catalyst of the design control group was BL21 (DE3) empty bacteria.
| Metabolic pathway | Whole cell concentration (OD600) | NAM concentration (mM) | Sugar concentration (mM) |
|---|---|---|---|
| GP | 10 | 10 | Glucose 10 |
| XP | 10 | 10 | Xylose 10 |
| AP | 10 | 10 | Arabinose 10 |
| Control | 10 | 10 | Glucose 10 |
Optimization of cell culture conditions
We further optimized the expression of recombinant bacteria of the GP pathway from four aspects: induced cell concentration, IPTG addition, culture temperature and culture time. For different cell concentrations, OD600 was induced in the range of 0.3–1.2 gradient to determine the optimal concentration of induced cells. Under the optimal concentration of induced cells, the concentration of inducer IPTG (final concentration 0.25 ∼ 1.0 mM) was optimized, and the optimal addition amount of inducer IPTG was investigated. The culture temperature was optimized from 18 °C to 35 °C. Finally, the culture time was optimized to increase equally in the range of 18–24 h. One variable is maintained each time, and other conditions remain unchanged. The relative activity was determined, and the optimal expression conditions were screened out. The highest activity of intracellular recombinant bacteria under each condition was defined as 100%.Optimization of whole cell catalytic conditions
The production of NMN was optimized from four aspects: substrate concentration, catalytic reaction temperature, surfactant and whole cell catalyst concentration. First, the substrate concentration was determined. Under the premise of controlling the glucose concentration to 10 mM, the catalytic production capacity of NMN was compared with the NAM concentration of 10 mM, 25 mM, 40 mM and 55 mM. Under the condition of controlling the concentration of NAM to 10 mM, the influence of glucose was explored with different concentrations of 10 mM, 20 mM, 40 mM, and 80 mM on NMN production. Finally, the optimal substrate concentration was determined. Second, the catalytic cell concentration OD600 is maintained at 10, and 10 mM NAM and 10 mM glucose are added in the M9 medium to explore the NMN yield at 18 °C, 25 °C, 30 °C and 35 °C. Finally, the optimal catalytic temperature is confirmed. Then, under the above two optimal conditions, different surfactants, T-80, SP-80, TritonX-100 and SDS, are added to the catalytic system to make the final concentration 1% (w/v), that is, the mass concentration 1 g 100 mL−1. After confirming that the addition of T-80 affects the catalytic reaction, the optimal addition amount of surfactant is further screened. The final concentrations of T-80 added were 0.05%, 0.1%, 0.5%, 1% and 2%. After the whole cell catalytic reaction was completed, the improvement of catalytic activity compared with the original strain was determined by liquid phase detection. Finally, under the optimal reaction conditions, the NMN yield was investigated when the whole cell catalyst concentration OD600 was 10, 20, 30, 40 and 50, respectively.NMN detection method
NAM and NMN were measured using a high-performance liquid chromatography (HPLC) system (Agilent 1100 system with a UV detector) equipped with a water column Hypersil GOLD™ aQ Dim (250 mm × 4.6 mm, 5 μm, Thermo Fisher Scientific). A 0.05 mol L−1 dipotassium phosphate solution with 2% acetonitrile (v/v) was employed as the mobile phase at a flow rate of 0.6 mL min−1. The detection wavelength was 260 nm. The column was used at 30 °C. The standard samples of NMN with concentrations ranging from 0.05 mM to 0.9 mM were employed to establish the standard curve for NMN quantification in HPLC, as shown in Fig. S1A.† Similarly, standard samples of NAM with concentrations ranging from 2 mM to 10 mM were also injected into the HPLC to establish a standard curve for quantification (Fig. S1B†).Results
Design of NMN synthesis pathway catalyzed by the cascade of intracellular multiple enzymes
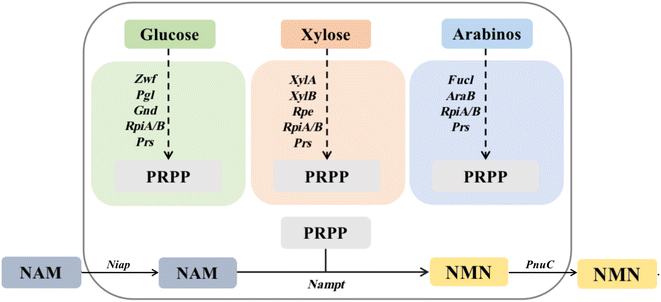
Currently, the reported routes for NMN biosynthesis mainly use NAM and PRPP as substrates to generate NMN under the catalysis of NAMPT.14 Owing to the instability of PRPP,15 it is possible to attempt to synthesize PRPP from other substrates and further generate an NMN. Therefore, based on the pentose phosphate metabolic pathway for the synthesis of PRPP, three in vivo pathways for PRPP synthesis were designed using glucose, xylose, and arabinose as raw materials. The GP pathway uses glucose as a substrate to synthesize ribose-5-phosphate using glucose-6-phosphate 1-dehydrogenase, 6-phosphate glucolactone and 6-phosphate gluconate dehydrogenase (zwf, pgl and gnd), which is then catalyzed by ribosyl-5-phosphate isomerase and ribosyl-phosphate pyrophosphate kinase (rpiA/B and prs) to synthesize PRPP. The XP pathway uses xylose as a substrate and catalyzes the synthesis of ribose-5-phosphate through xylose isomerase, xylose kinase and ribose-5-phosphate isomerase (xylA, xylB and rpe), followed by the synthesis of PRPP through rpiA/B and prs. The AP pathway uses arabinose as the substrate, catalyzes the synthesis of ribose-5-phosphate through arabinose isomerase and ribokinase (fucI and araB), and then synthesizes PRPP through rpiA/B and prs. Using PRPP as the co-substrate, NAM delivered more efficiently into cells by Niap was subsequently converted to NMN by NAMPT; then, it was transported out of cells by PnuC10 (Fig. 1).The synthesis of PRPP from different substrates is divided into three intracellular pathways, with glucose as the substrate representing the GP pathway in green: Zwf, glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49); Pgl, 6-phosphate glucosinolide enzyme (EC 3.1.1.31); Gnd, 6-phosphate gluconate dehydrogenase (EC 1.1.1.44); RpiA, ribose 5-phosphate isomerase A (EC 5.3.1.6); RpiB, ribose-5-phosphate isomerase B (EC 5.3.1.6); and Prs, ribophosphate pyrophosphate kinase (EC 2.7.6.1). Orange represents the XP pathway with xylose as the substrate: XylA, xylose isomerase (EC 5.3.1.5); XylB, xylokinase (EC 2.7.1.17); and Rpe, ribose-5-phosphate isomerase (EC 5.1.3.1). RpiA, RpiB and Prs are identical to the GP pathway. Dark blue uses arabinose as the substrate to represent the AP pathway: AraB, ribokinase (EC 2.7.1.16); and FucI, arabinose isomerase (EC 5.3.1.3). The remaining genes are the same as the GP pathway.
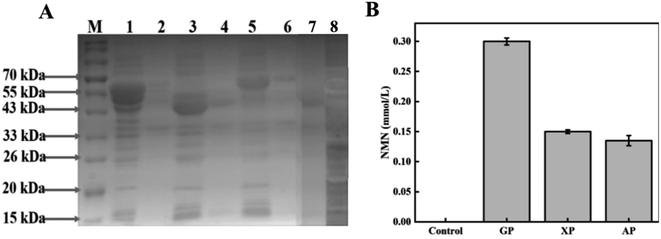
Comparison of the efficiency of the three NMN synthesis pathways
SDS-PAGE gel electrophoresis was used to verify the expression of enzymes related to the three intracellular pathways. The electrophoresis results are shown in Fig. 2A. Enzymes encoded by multiple genes in the cells can be observed to have soluble expression in their respective supernatant swimlanes. The molecular weights of zwf, pgl, and gnd in the GP pathway were 55.7, 36.3, and 51.5 kDa, respectively. The molecular weights of xylA, xylB, and rpe in the XP pathway were 49.7, 52.6, and 24.6 kDa, respectively. The molecular weights of fucI and araB in the AP pathway were 65.0, and 61.1 kDa, respectively. The molecular weights of shared rpiA, rpiB, Prs, Nampt, NiaP, and PnuC were 22.9, 16.1, 34.2, 55.6, 50.7, and 25.2 kDa, respectively.The production of NMN through the GP, XP, and AP pathways was preliminarily compared through whole-cell catalysis, and the results are shown in Fig. 2B. The whole cell catalyst of the design control group was BL21 (DE3) empty bacteria. In the control group, there was no NMN production. The production of NMN was successfully detected in all the three designed pathways, indicating the ability for NMN production using various sugars. The HPLC results of the GP pathway using glucose as the substrate are depicted in Fig. S2C,† and the final production of 0.3 mM NMN can be obtained from the standard curve illustrated in Fig. S1.† Similarly, the XP and AP pathways using xylose and arabinose as substrates produced only 0.15 mmol L−1 and 0.135 mmol L−1 of NMN, respectively. Compared with the other two pathways, NMN production based on the GP pathway was nearly twice as high, so the pathway using glucose as the substrate for NMN production was selected for subsequent optimization experiments.
Optimization of culture conditions to enhance the viability of recombinant cells via the GP pathway
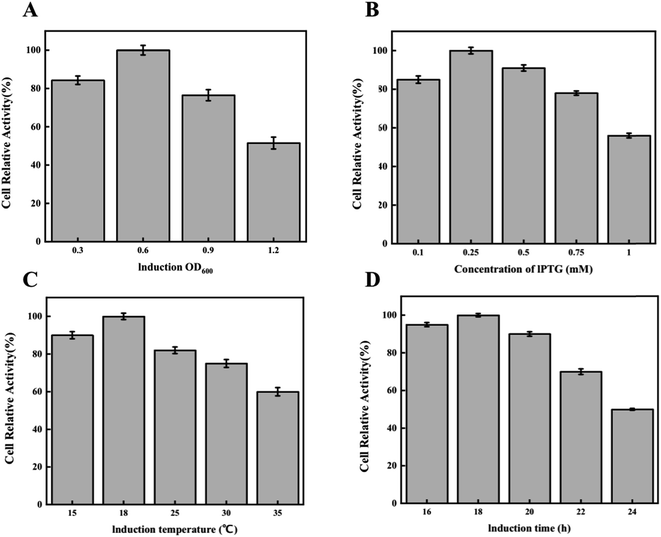
Intracellular multi-enzyme cascade catalysis is a catalytic method that utilizes the cell as the catalyst, thereby maintaining the cell in the optimal activity that is conducive to production.16,17 Therefore, the effects of cell concentration, inducer concentration, induction temperature, and culture time on recombinant cells were studied to explore the conditions for optimal cell viability.First, we investigated the effect of induction time on cell viability. The expression inducer of IPTG was added when the cells grew into an OD600 of 0.3, 0.6, 0.9, and 1.2. As shown in Fig. 3A, when IPTG was added at an OD600 of 0.6, cell viability reached its highest level. Subsequently, we compared the catalytic activity of cells when different IPTG concentrations (0.1, 0.25, 0.5, 0.75, and 1.0 mM) were used. As shown in Fig. 3B, the relative viability of the cells was highest when the inducer concentration was 0.25 mM. Next, the effects of cultivation temperature on cell viability were investigated. The results are shown in Fig. 3C. When the cultivation temperature was 18 °C, the cell viability reached the highest level. As the induction temperature increased, the catalytic activity of the cells showed a decreasing trend. Finally, we studied the effect of cultivation time on cell viability. As shown in Fig. 3D, when the cells were cultured for 18 h, cell viability reached the highest level.
Effect of catalytic conditions on NMN synthesis by the GP pathway
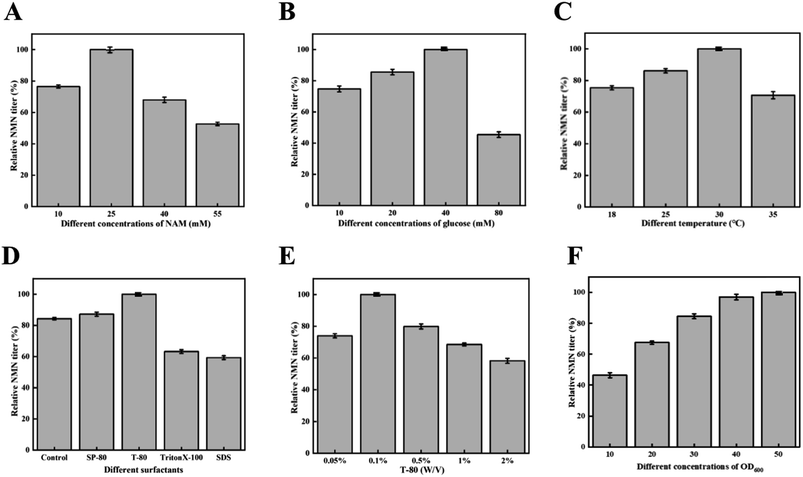
We further optimized the catalytic conditions to improve the final NMN production. First, we investigated the effects of substrate concentrations on NMN production. As shown in Fig. 4A and B, NMN production was conducted at different initial concentrations of NAM and glucose. The NMN titer reached its highest level when the concentration of NAM was 25 mM and the concentration of glucose was 40 mM. As the substrate concentration further increased, the NMN titer decreased. Based on these results, we speculated that high concentrations of NAM had an inhibitory effect on catalytic efficiency. Then, we optimized and compared the catalytic reaction temperature. As shown in Fig. 4C, when the temperature increased from 18 °C to 30 °C, the NMN titer also increased and reached a maximum level at 30 °C.In whole cell biocatalysis, in addition to external environmental factors, the barrier functions of the cell wall and membrane affect the catalytic efficiency.18 Therefore, we selected four surfactants to improve the production efficiency of NMN. As depicted in Fig. 4D, we found that the addition of surfactants had different effects on the catalytic ability of whole cells. When two surfactants, SDS and Triton X-100, were added into the whole cell system, a significant decrease in NMN production was observed. The addition of SP-80 did not show a significant effect on the whole cell catalytic system, while the addition of T-80 had a certain improvement in NMN synthesis efficiency. Therefore, T-80 was added into the reaction mixture to improve NMN production.19 Subsequently, the effect of T-80 concentrations on NMN synthesis efficiency was further studied. The results are shown in Fig. 4E, and the optimal final concentration for adding T-80 was 0.1%. The concentrations of the whole cells also have a certain impact on the final NMN production. We designed a gradient experiment to increase the concentration of catalytic cells from an OD600 of 10 to 50. The results are shown in Fig. 4F. As the concentration of catalytic cells increased, an increase in NMN production was also observed. However, when the concentration of catalytic cells increased from an OD600 of 40 to 50, the reaction reached equilibrium.
Whole cell catalytic production of NMN under optimal conditions
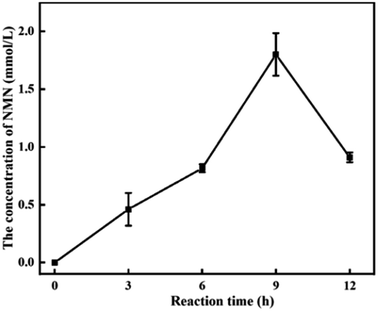
To explore NMN production under the optimal catalytic system, we conducted the experiment in a reaction mixture containing 40 of OD600, 25 mM NAM, 40 mM glucose, and 0.1% T-80. As shown in Fig. 5, after bioconversion of 9 h, NMN production reached the highest level, and 1.8 mmol L−1 (0.6 g L−1) of NMN was obtained. However, when the reaction time was extended, a decrease in NMN production was observed. We suspect that an NMN degradation pathway is present in the cells owing to the issue of production not being able to further improve. In the study conducted by Zhongshi Huang et al., it was mentioned that a portion of NMN in cells is consumed to synthesize NAD+ for bacterial growth, which is detrimental to the accumulation of NMN.14 Concurrently, they are very focused on product consumption and use gene knockout technology to knock out the Pncc of NMN to NaMN, as well as the genes NadR and UshA that convert between NMN and NR.20Discussion
NMN, as one of the key precursors of NAD+, plays a crucial role in the human body. Owing to the involvement of NAD+ in thousands of biocatalytic reactions, supplementing NMN can increase the NAD+ content of the body, thereby improving body function and delaying cell aging.21 In the study of mice conducted by Tarantini et al., it was mentioned that using NMN to treat aging mice effectively alleviates cerebral microvascular dysfunction.22 In addition, NMN can alleviate brain damage after cerebral hemorrhage (ICH). In the study by conducted Wei et al., it was found that the intake of NMN enhances the expression of protective proteins in hemoglobin, which to some extent contributes to neuroprotection during cerebral hemorrhage.23 Moreover, a large number of studies have mentioned that supplementing NMN can effectively alleviate issues related to mitochondrial energy reduction, oxidative stress, DNA damage, and inflammation.24 At present, NMN biosynthesis has attracted increasing attention.It has been mentioned in numerous studies that the accumulation of the key precursor substance PRPP in NMN synthesis is insufficient, which ultimately affects the product conversion rate.20 Currently, PRPP is mainly obtained through the process of catalyzing ATP and R-5-P to generate PRPP and by-product AMP through phosphate ribose diphosphate kinase and PRPP synthase.11 In our study, three multi-enzyme cascade catalytic pathways were constructed to explore the supply capacity of glucose, xylose, and arabinose to PRPP. The yield of NMN is not as high as glucose when xylose and arabinose are used as the only sugars; this demonstrates that PRPP can be supplied through different substrates and provides new ideas for future NMN production.
In this study, the degradation of NMN is also the reason it cannot accumulate in large quantities. In the study conducted by Huang et al., they knocked out the genes related to the conversion of nicotinamide and NMN of the host into other metabolites by applying metabolic engineering.14 The synthesis and degradation of NMN are mostly bidirectional reactions, so knocking out the synthesis and degradation genes of NR and NaMN can effectively improve the NMN titer. Simultaneously, as a key enzyme for synthesizing NMN, the activity and stability of NAMPT will also certainly affect NMN production. Therefore, its catalytic ability can be improved through the mutation modification of Nampt. In addition, the efficiency of transporting proteins is also worth paying attention to. Studies have shown that the transfer protein Niap of NAM inhibits the production of NMN. Although there is no detailed mechanism explanation, this also indicates that the selection of transfer proteins is very important. In this study, it was mentioned that the introduction of BMPnuC (from B. mycoides) increased the accumulation of NMN outside the cell. Therefore, screening for more efficient NAM importin and NMN exportin is an important factor in obtaining a higher titer NMN.
Conflicts of interest
There are no conflicts to declare.References
- Q. Shen, S. J. Zhang and Y. Z. Xue, et al., Biological synthesis of nicotinamide mononucleotide, Biotechnol. Lett., 2021, 43(12), 2199–2208 CrossRef CAS PubMed
.
- J. H. Park, A. Long and K. Owens, et al., Nicotinamide mononucleotide inhibits post-ischemic NAD+ degradation and dramatically ameliorates brain damage following global cerebral ischemia, Neurobiol. Dis., 2016, 95, 102–110 CrossRef CAS PubMed
.
- J. U. N. Yoshino and J. A. Baur, et al., NAD(+) Intermediates: the Biology and Therapeutic Potential of NMN and NR, Cell Metab., 2018, 27(3), 513–528 CrossRef CAS
.
- C. F. Lee, J. D. Chavez and L. Garcia-Menendez, et al., Normalization of NAD+ Redox Balance as a Therapy for Heart Failure, Circulation, 2016, 134(12), 883–894 CrossRef CAS
.
- X. Wang, X. Hu and Y. Yang, et al., Nicotinamide mononucleotide protects against β-amyloid oligomer-induced cognitive impairment and neuronal death, Brain Res., 2016, 1643, 1–9 CrossRef CAS
.
- Y. Takanobu, B. Jaemin and Z. Peiyong, et al., Nicotinamide Mononucleotide, an Intermediate of NAD+ Synthesis, Protects the Heart from Ischemia and Reperfusion, PLoS One, 2014, 9(6), e98972 CrossRef PubMed
.
- M. Wei, H. Sha and L. Ye, et al., Recent Advances in the Chemical Synthesis of β-Nicotinamide Mononucleotide, Curr. Org. Chem., 2022, 26, 24 Search PubMed
.
- J. E. Velasquez, P. R. Green and J. A. Wos, Method For Preparing Nicotinamide Riboside, 2017 Search PubMed.
- Q. Shen, S. J. Zhang and Y. Z. Xue, et al., Biological synthesis of nicotinamide mononucleotide, Biotechnol. Lett., 2021, 43(12), 2199–2208 CrossRef CAS
.
- S. Shoji, T. Yamaji and H. Makino, et al., Metabolic design for selective production of nicotinamide mononucleotide from glucose and nicotinamide, Metab. Eng., 2021, 65, 167–177 CrossRef CAS
.
- C. Zhou, J. Feng and J. Wang, et al., Design of an in vitro multienzyme cascade system for the biosynthesis of nicotinamide mononucleotide, Catal. Sci. Technol., 2022, 12(4), 1080–1091 RSC
.
- A. Maharjan, M. Singhvi and B. S. Kim, Biosynthesis of a Therapeutically Important Nicotinamide Mononucleotide through a Phosphoribosyl Pyrophosphate Synthetase 1 and 2 Engineered Strain of Escherichia coli, ACS Synth. Biol., 2021, 10(11), 3055–3065 CrossRef CAS PubMed
.
- W. J. Zhou, A. Tsai and D. A. Dattmore, et al., Crystal structure of E-coli PRPP synthetase, BMC Struct. Biol., 2019, 19, 7 CrossRef
.
- Z. Huang, N. Li and S. Yu, et al., Systematic Engineering of Escherichia coli for Efficient Production of Nicotinamide Mononucleotide From Nicotinamide, ACS Synth. Biol., 2022, 11(9), 2979–2988 CrossRef CAS PubMed
.
- F. Zhang, X. Sun and X. Shen, et al., Biosynthesis of allantoin in Escherichia coli via screening a highly effective urate oxidase, Biotechnol. Bioeng., 2022, 119(9), 2518–2528 CrossRef CAS PubMed
.
- K. Goldberg, K. Schroer and S. Lütz, et al., Biocatalytic ketone reduction—a powerful tool for the production of chiral alcohols—part I: processes with isolated enzymes, Appl. Microbiol. Biotechnol., 2007, 76(2), 237–248 CrossRef CAS
.
- T. Matsuda and R. Yamanaka, et al., ChemInform Abstract: Recent Progress in Biocatalysis for Asymmetric Oxidation and Reduction, ChemInform, 2009, 40(34), 513–557 CrossRef
.
- S. S. D. Silva, Surfactants, Biosurfactants, and Non-Catalytic Proteins as Key Molecules to Enhance Enzymatic Hydrolysis of Lignocellulosic Biomass, Molecules, 2022, 27(23), 8180 CrossRef
.
- J. Jannes, Effect of tween 80 on the nicotinic acid synthesis of Escherichia coli, Ann. Med. Exp. Biol. Fenn., 1956, 34(2), 206–208 CAS
.
- M. Zhou, Y. Li and H. Che, Metabolic Engineering of Bacillus licheniformis for the Bioproduction of Nicotinamide Riboside from Nicotinamide and Glucose, ACS Sustainable Chem. Eng., 2023, 11(16), 6201–6210 CrossRef CAS
.
- J. Irie and H. Itoh, Aging and homeostasis. Age-associated diseases and clinical application of NMN(Nicotinamide Mononucleotide), Clin. Calcium, 2017, 27(7), 983 CAS
.
- S. Tarantini and T. Kiss, et al., Nicotinamide mononucleotide (NMN) supplementation promotes neurovascular rejuvenation in aged mice: transcriptional footprint of SIRT1 activation, mitochondrial protection, anti-inflammatory, and anti-apoptotic effects, GeroScience, 2020, 47, 527–546 Search PubMed
.
- C. C. Wei, Y. Y. Kong and G. Q. Li, et al., Nicotinamide mononucleotide attenuates brain injury after intracerebral hemorrhage by activating Nrf2/HO-1 signaling pathway, Sci. Rep., 2017, 7(1), 717 CrossRef PubMed
.
- Y. Jiang, D. Wang and C. Zhang, et al., Nicotinamide mononucleotide restores oxidative stress-related apoptosis of oocyte exposed to benzyl butyl phthalate in mice, Cell Proliferation, 2023, 56(8), e13419 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra03857b |
| This journal is © The Royal Society of Chemistry 2023 |