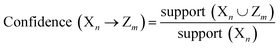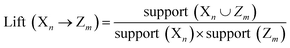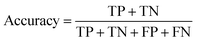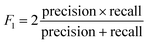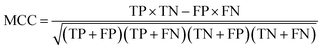Open Access Article
Open Access ArticleSequence rules for gold-binding peptides
Jose Isagani B. Janairo *
*
Department of Biology, De La Salle University, 2401 Taft Avenue, Manila, 0922, Philippines. E-mail: jose.isagani.janairo@dlsu.edu.ph
First published on 12th July 2023
Abstract
Metal-binding peptides play a central role in bionanotechnology, wherein they are responsible for directing growth and influencing the resulting properties of inorganic nanomaterials. One of the key advantages of using peptides to create nanomaterials is their versatility, wherein subtle changes in the sequence can have a dramatic effect on the structure and properties of the nanomaterial. However, precisely knowing which position and which amino acid should be modified within a given sequence to enhance a specific property can be a daunting challenge owing to combinatorial complexity. In this study, classification based on association rules was performed using 860 gold-binding peptides. Using a minimum support threshold of 0.035 and confidence of 0.9, 30 rules with confidence and lift values greater than 0.9 and 1, respectively, were extracted that can differentiate high-binding from low-binding peptides. The test performance of these rules for categorizing the peptides was found to be satisfactory, as characterized by accuracy = 0.942, F1 = 0.941, MCC = 0.884. What stands out from the extracted rules are the importance of tryptophan and arginine residues in differentiating peptides with high binding affinity from those with low affinity. In addition, the association rules revealed that positions 2 and 4 within a decapeptide are frequently involved in the rules, thus suggesting their importance in influencing peptide binding affinity to AuNPs. Collectively, this study identified sequence rules that may be used to design peptides with high binding affinity.
Introduction
The use of peptides as capping agents is an effective technique for producing size-controlled and functional nanomaterials. This method is deeply rooted in natural systems, where proteins are mainly responsible for the formation of highly ordered mineralized structures with remarkable properties.1 In this biomimetic technique, the peptide binds to the surface of the growing nanoparticle and therefore influences its growth2 as well as the resulting properties. Considering that peptides play a crucial role in the outcome of nanomaterial production, extensive research has been devoted to understanding how peptide properties relate to nanomaterial synthesis. Previous studies have shown that both intrinsic and extrinsic factors govern peptide-mediated synthesis of inorganic nanomaterials. Intrinsic factors are related to properties inherent to the peptide, such as sequence,3 topology,4 assembly,5,6 and conformation7 of the peptide. On the other hand, extrinsic factors are related to the nanomaterial synthesis reaction conditions, such as the type of buffer8 used and pH.9Among these, the amino acid composition of the peptide was one of the earliest factors to be studied.10,11 These studies revealed that subtle changes in the primary structure can have drastic effects on the size, shape, and properties of the material. Building on these results, it was later determined that the position at which the residue substitutions were made had a large impact on the nanomaterials. For instance, it was demonstrated that substituting residues 6 and 11 for the Pd4 palladium-binding peptide substantially altered the catalytic activity of the produced Pd nanoparticles.12 For the AG3 gold-binding peptide, changing the amino acid based on the fourth13 and ninth14 positions can have a huge impact on the structure and arrangement of the produced AuNPs. These studies underscore the importance of both amino acid modifications and the correct position at which these modifications are carried out to tune the properties of the peptide and the created materials. However, determining these factors can be extremely challenging because of the numerous possible combinations that need to be explored. Despite this daunting challenge, understanding how specific sequence variations can impact the nanomaterial synthesis process may lead to the discovery of sequence patterns that can be used for the rational design of metal-binding peptides possessing desired properties, such as strong binding affinity. This will also facilitate a deeper and more precise understanding on the level of the amino acid on interactions that exist between biomolecules and inorganics. Machine learning (ML) is a promising approach to address the complexity of the problem, wherein ML can uncover and understand these sequence patterns from a given dataset. In particular, data mining can be used to establish associations between the primary structure and property of the peptide.
In this study, classification based on association rules (CBA) was implemented on a dataset composed of decapeptides and their experimentally determined binding affinities for gold nanoparticles (AuNPs). The analysis presents a series of sequence rules that can be used to categorize peptides based on whether they strongly or poorly bind to AuNPs. Based on these rules, specific amino acids and positions within the decapeptide were identified that exhibited strong associations with binding affinity.
Methodology
The decapeptides and their experimentally determined binding affinities for AuNPs were obtained from the data of Tanaka et al.15 The dataset featured 1720 decapeptides which were then divided into quartiles based on their binding affinity. The training and test datasets (n = 860) used in this study included decapeptides belonging to the first quartile to represent high-binding peptides (Class A) and those that belong to the fourth quartile to represent low-binding peptides (Class B). The dataset was divided into 70/30 proportions for the training and test sets. Each position in the decapeptide (P1–P10) was treated as a variable, and the amino acid in each position was considered as an item. Association rules were extracted from the training set using the a priori algorithm in the ArulesCBA package.16 The minimum confidence threshold for the rules was set at 0.9, whereas the minimum support was varied from 0.01 to 0.05. The validity of the rules was evaluated using lift. These metrics were calculated using the following formula:where support (Xn∪Zm) represents the frequency of occurrence of both amino acids X at position n and Z at position m together.
The rules were pruned using the M2 method. The rules are then used for classification; for example, when categorizing a case that has not been encountered before, the initial rule that matches the case determines its classification. If there are no rules that apply to the case, it is assigned to the default class, which is Class B. Rule confidence is the primary criterion in determining rule priority followed by support. The performance of the extracted rules in classifying the decapeptides in the test set was assessed using accuracy, F1, and the Matthews Correlation Coefficient (MCC) as the main metrics for evaluating the performance of the rules. These performance metrics were calculated using the following formula:
where TP = true positive, TN = true negative, FP = false positive, FN = false negative. All R packages and their dependents used in the study were executed in R version 4.1.0 (ref. 17) running in a MacOS environment. The dataset and R script used in the study are freely available at https://github.com/jijanairo/sequencerules.
Results and discussion
The data used in this study were obtained from an array-based colorimetric assay, which is related to the binding affinity of decapeptides to AuNP.15 The study involved individually incubating 1720 decapeptides with AuNPs, and the change in color was used as a measure of the peptide binding strength for gold. In this study, only the first and fourth quartiles were used because using the entire dataset did not yield any rules that could distinguish high-binding affinity peptides from low-binding affinity peptides. This suggests that a high degree of heterogeneity exists, necessitating a more focused and narrower search strategy. As anticipated, association rules were extracted from the decapeptides in the first and fourth quartiles. Thus, even if the final dataset used was relatively small (n = 860) compared to the original dataset, meaningful insights were still obtained and can still provide value as long as the limitations of the model are recognized and taken into consideration. In addition, a dataset composed of 860 peptides is still a substantial population within the context of metal-binding peptides, as data availability is typically scarce owing to constraints in the acquisition of data. Thus, past studies that created ML models for metal-binding peptides used datasets composed of less than 100 peptides.18–20The goal of CBA is to identify a rule in the form of an association that is valuable, providing insights about the dataset that were previously unknown and likely difficult to explicitly express. Contextualizing this goal into the present research translates into searching the dataset of decapeptides to find patterns in the amino acid composition and the peptide position that are associated with strong binding affinity. Association rule mining is a machine learning data mining technique that extracts rules from a given set of data, which can then be used for classification.16 Association rules have been previously used to study protein composition, such as in the identification of hydrophobic sequence motifs associated with a particular secondary structure.21 In another example, association rule mining was applied to protein sequences to uncover global associations in the amino acid composition of known proteins.22 Thus, the goal of association rule mining and CBA, which are data mining techniques, is to uncover valuable patterns within the data. This study is the first instance in which CBA will be used to study metal-binding peptides and aims to determine sequence motifs associated with strong binding affinity for AuNPs. This is an important step towards rational peptide design, which remains a challenge for bionanotechnology.23
The support or frequency of occurrence of the rules is an important factor to be considered when association rules within a given set of transactions are extracted.24 Support quantifies how frequently an itemset appears in the dataset; in this case, how often a specific amino acid appears in a particular position in the dataset. On the other hand, confidence quantifies the reliability of predicting a specific decapeptide into either class when a particular amino acid in a given position is observed in the input data. A higher confidence value indicates stronger association between an amino acid in a given position and the peptide category (Class A or B). Therefore, a high confidence value suggests that the presence of a particular amino acid at a given position increases the likelihood of the corresponding peptide category. The minimum support was first tuned to identify the most suitable threshold that balances classification performance and the number of rules generated.
As shown in Table 1, a low support cutoff generates many rules, leading to overfitting. However, a relatively high support limits the number of rules generated, leading to poor classification performance. A minimum support threshold of 0.035 was selected because it appears to be the best compromise between rule complexity as measured by the number of extracted rules, and classification performance. Nineteen rules associated with high-binding affinity peptides were identified, whereas 11 for low-binding affinity peptides (Table 2). All the identified rules are considered important because the lift values are greater than 1 and the confidence is greater than 0.9. Lift is a measure of rule strength because it indicates the extent to which the right-hand side (RHS) of the rule occurs when the left-hand side (LHS) is present. Thus, a lift greater than one indicates a high degree of association, which is not attributed to chance between the LHS of the rule and the RHS. Conversely, a lift value of less than 1 suggests a negative association, whereas lift = 1 indicates no association or independence between the LHS and the RHS.
| Minimum support | ||||||
| 0.01 | 0.02 | 0.03 | 0.035 | 0.04 | 0.05 | |
| Training set (n = 602) | ||||||
| Accuracy | 1 | 1 | 0.982 | 0.968 | 0.89 | 0.836 |
| F1 | 1 | 1 | 0.982 | 0.968 | 0.88 | 0.818 |
| MCC | 1 | 1 | 0.963 | 0.937 | 0.794 | 0.684 |
| Test set (n = 258) | ||||||
| Accuracy | 0.981 | 0.981 | 0.954 | 0.942 | 0.814 | 0.783 |
| F1 | 0.981 | 0.981 | 0.954 | 0.941 | 0.7838 | 0.748 |
| MCC | 0.962 | 0.962 | 0.907 | 0.884 | 0.654 | 0.589 |
| Number of association rules | 59 | 45 | 35 | 30 | 22 | 10 |
| Association rule | Support | Confidence | Lift | |
|---|---|---|---|---|
| 1 | {P1 = W, P10 = W} → {Class = A} | 0.0764 | 1 | 2 |
| 2 | {P2 = W, P3 = K} →{Class = A} | 0.0598 | 1 | 2 |
| 3 | {P4 = W, P8 = G} →{Class = A} | 0.0465 | 1 | 2 |
| 4 | {P2 = W, P7 = H} → {Class = A} | 0.0449 | 1 | 2 |
| 5 | {P1 = H, P2 = W} → {Class = A} | 0.0415 | 1 | 2 |
| 6 | {P2 = W, P4 = W} → {Class = A} | 0.0415 | 1 | 2 |
| 7 | {P2 = W, P6 = M} → {Class = A} | 0.0382 | 1 | 2 |
| 8 | {P2 = T, P10 = W} → {Class = A} | 0.0365 | 1 | 2 |
| 9 | {P4 = Q, P9 = W} → {Class = A} | 0.0365 | 1 | 2 |
| 10 | {P5 = W, P10 = W} → {Class = A} | 0.0465 | 0.966 | 1.931 |
| 11 | {P5 = E, P7 = H} → {Class = A} | 0.0449 | 0.964 | 1.929 |
| 12 | {P2 = W, P9 = A} → {Class = A} | 0.0432 | 0.963 | 1.926 |
| 13 | {P5 = W, P6 = E} → {Class = A} | 0.0365 | 0.957 | 1.913 |
| 14 | {P4 = W, P7 = Q} → {Class = A} | 0.0349 | 0.955 | 1.909 |
| 15 | {P8 = W} → {Class = A} | 0.0914 | 0.948 | 1.897 |
| 16 | {P1 = W} → {Class = A} | 0.148 | 0.947 | 1.894 |
| 17 | {P5 = K, P6 = W} → {Class = A} | 0.0415 | 0.926 | 1.852 |
| 18 | {P6 = M, P8 = G} → {Class = A} | 0.0398 | 0.923 | 1.846 |
| 19 | {P3 = Q, P10 = Q} → {Class = A} | 0.0332 | 0.909 | 1.818 |
| 20 | {P5 = H} → {Class = B} | 0.0631 | 1 | 2 |
| 21 | {P10 = R} → {Class = B} | 0.0565 | 1 | 2 |
| 22 | {P9 = K} → {Class = B} | 0.0498 | 1 | 2 |
| 23 | {P5 = T} → {Class = B} | 0.0432 | 1 | 2 |
| 24 | {P8 = R} → {Class = B} | 0.0431 | 1 | 2 |
| 25 | {P1 = S} → {Class = B} | 0.0432 | 1 | 2 |
| 26 | {P7 = R} → {Class = B} | 0.0432 | 1 | 2 |
| 27 | {P10 = L} → {Class = B} | 0.0399 | 1 | 2 |
| 28 | {P6 = R} → {Class = B} | 0.0382 | 1 | 2 |
| 29 | {P3 = R} → {Class = B} | 0.0365 | 1 | 2 |
| 30 | {} → {Class = B} | 0.5 | 0.5 | 1 |
The extracted rules highlight the importance of tryptophan for a high-binding affinity peptide. This was evidenced by the prevalence of tryptophan in the extracted rules at multiple positions, as well as its combination with other amino acids. Eighty-four percent of the rules for categorizing peptides into Class A involve tryptophan (Fig. 1). The importance of this residue in AuNP synthesis is known for its ability to reduce Au3+ ions,25,26 wherein increasing the number of tryptophan residues in a gold biomineralization peptide leads to an increase in reducing efficiency.27 However, the binding affinity data for the decapeptides used in this study were obtained using already formed AuNPs. This suggests that the importance of tryptophan in gold-binding peptides is due to its contribution to binding to the AuNP surface and not for its ability to reduce Au ions and influence nucleation. This is consistent with the knowledge that tryptophan can bind to AuNPs through its indole, carboxyl, and amino group.28,29 Moreover, when the tryptophan residue of the gold-binding peptide AuBP1 (WAGAKRLVLRRE) was changed to alanine, the binding affinity of the peptide for gold, as determined through QCM measurements substantially decreased.30 These studies confirm that tryptophan is directly involved in anchoring the biomolecule onto the metal surface.
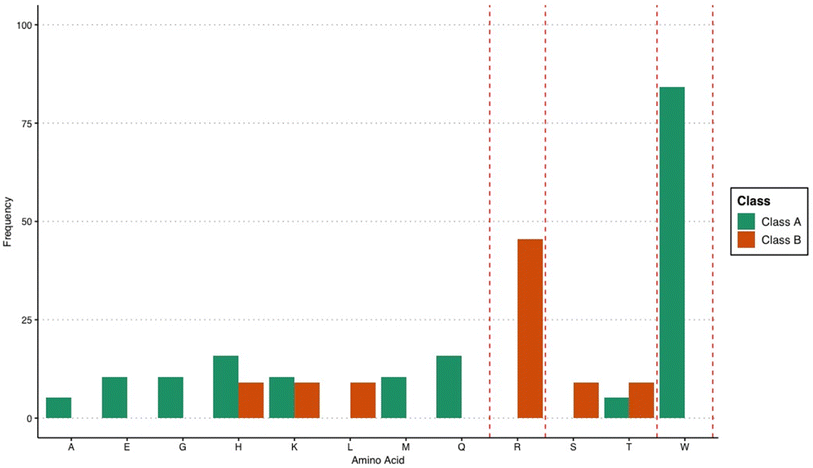 | ||
| Fig. 1 Frequency of amino acids in the association rules that classify peptides into either Class A or B. | ||
Among the rules that classify peptides into Class B, 46% involve arginine. Notably, the rules that classify peptides into Class A did not involve arginine (Fig. 1). The strong association of arginine with low-binding affinity peptides was unexpected, considering that this amino acid is known to bind strongly with gold surfaces.31,32 However, the exact location of arginine within the decapeptide may have a stronger contribution, leading to an association with weaker binding affinity for AuNP. While amino acid composition is an important factor in determining the ability of peptides to bind to inorganic surfaces, the orientation of the peptide during binding is equally important. Peptides have very different structures in the unbound state compared to the bound state33 which demonstrates the importance of peptide structure and orientation in relation to binding. In addition, the manner in which the peptide is bound to the surface of the nanoparticle also affects the resulting properties of the nanomaterial.3 Therefore, the rules for low-binding peptides may suggest that the placement of arginine at these specific locations can influence peptide structure, leading to decreased affinity for gold. This is highly plausible considering that it was observed before that the precise placement of arginine within a given sequence can have long-range effects on the peptide structure, especially on helical motifs.34 Related to this observation, this study identified positions within the decapeptide sequence that appeared to be critical in influencing the binding behavior of the peptide. Positions 2 and 4 were frequently present in the mined association rules for differentiating Class A peptides from B (Fig. 2). The exact reason why these identified positions influence the classification of the peptides based on binding affinity requires further examination and should be explored in future studies. However, it can be postulated that these positions are important for anchoring the peptide onto the gold surface. The structure and orientation of bound metal-binding peptides are dictated by the anchoring points, wherein the contributing residues are always in direct contact with the surface.35 As has been shown in previous studies, specific positions within a sequence are important for the binding affinity of the biomolecule. For the AYSSGAPPMPPF gold-binding peptide for instance, it was discovered that the second, ninth, and twelfth residues are anchoring points onto the gold surface,36 and changing the amino acid composition at any of these locations can be used to modulate the binding affinity. For the AuBP1 peptide, the C- and N-termini appear to play an integral role in the peptide-binding process because point mutations at these locations severely incapacitated the peptide to bind to the gold surface.30 Apart from influencing the anchoring behavior of peptides, specific mutations within the sequence can also alter gold-binding behavior by varying the interaction of the peptide with the solvent.37 Considering the importance of the location and correct placement of amino acids within a sequence, the present study identified precise locations within a decapeptide that can be further explored to evaluate how they can be used to optimize peptide properties.
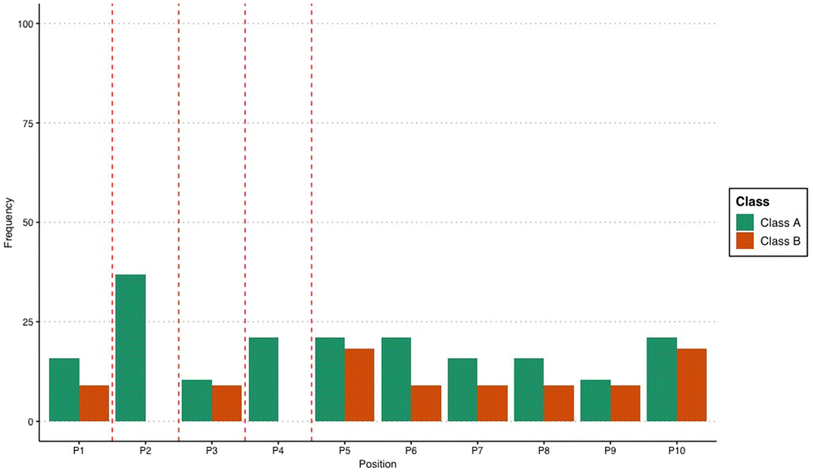 | ||
| Fig. 2 Frequency of the positions within the decapeptide in the association rules that classify peptides into either Class A or B. | ||
Identifying “hotspots” in a peptide sequence can be immensely difficult owing to combinatorial complexity. Using machine learning in the form of association rule mining, an alternative method for the discovery of important positions and amino acids that are associated with binding affinity is presented. Although ML models have been created in the past to study metal-binding peptides,18–20,38,39 this is the first attempt to use CBA to study their sequence patterns. Previous ML applications have focused on identifying the desired physicochemical properties of peptides associated with high binding affinity. While the results of these analyses have shed light on the general features of metal-binding peptides, the sequence rules presented in this study offer insights that are more specific and precise regarding the amino acid composition and location. The results of this study offer a starting point for the design and optimization of gold-binding peptides.
As with any model, the sequence rules discovered through data mining have inherent limitations. The first limitation is the applicability of the rules to decapeptides. Since the rules are based on amino acid composition and position, they are only relevant to decapeptides. This limitation is due to the scarcity of available data in this domain, which is why external validation was not conducted. Best effort was exerted to find reported decapetides with high and low binding affinities. Unfortunately, the search did not yield suitable data for external validation. Be that as it may, the findings presented still offer novel insights on how the peptide sequence can influence binding behavior to gold surfaces.
Conclusion
This work presents association rules based on experimental results, which can aid in the rational design of peptides with a strong affinity for gold surfaces. The results identified key positions and important residues among the decapeptides that bind strongly to gold. Specifically, the rules discovered sequence patterns that suggest tryptophan and arginine are important in differentiating high-binding from low-binding affinity peptides. In addition, the rules identified positions 2 and 4 within a decapeptide as hotspots or are frequently associated with the extracted rules. Collectively, these rules can satisfactorily discriminate between high-binding and low-binding decapeptides, as characterized by various performance metrics, which include accuracy, F1, and MCC. Accuracy refers to proportion of correctly classified peptides out of the total number peptides. F1 gives a glimpse of the overall effectiveness of the classifier since it combines precision and recall in a single metric. MCC is a holistic measures of classification performance since it relies on all quadrants of the confusion matrix. Overall, the sequence rules suggest that the presence of amino acids known to bind strongly to AuNP is not sufficient to ensure that the peptide will also exhibit strong interactions with AuNPs. It is also important to consider the precise placement and order of these residues within the metal-binding peptide sequence.These results provide the groundwork for further exploration to fully elucidate why these positions are frequently involved in the rules, and to pinpoint the role of tryptophan and arginine residues in AuNP synthesis. For example, the peptide with the highest binding affinity in the dataset could be used as a model to analyze the effects of systematically changing the amino acid at positions 2 and 4. In addition, molecular dynamics simulations can be carried out to derive insights on the impact of tryptophan and arginine residues within the decapeptide on its conformation. Similarly, DFT calculations can also be employed to study the rules from a reactivity perspective. These are exciting points of inquiry which can shed light on these research questions and may lead us closer to the rational design of metal-binding peptides.
Conflicts of interest
There are no conflicts of interest to declare.References
- J. I. B. Janairo, Peptide-Mediated Biomineralization, Springer, Singapore, 2016, DOI:10.1007/978-981-10-0858-0.
- R. Coppage, J. M. Slocik, B. D. Briggs, A. I. Frenkel, H. Heinz, R. R. Naik and M. R. Knecht, Crystallographic Recognition Controls Peptide Binding for Bio-Based Nanomaterials, J. Am. Chem. Soc., 2011, 133(32), 12346–12349, DOI:10.1021/ja203726n.
- Y. Li, Z. Tang, P. N. Prasad, M. R. Knecht and M. T. Swihart, Peptide-Mediated Synthesis of Gold Nanoparticles: Effects of Peptide Sequence and Nature of Binding on Physicochemical Properties, Nanoscale, 2014, 6(6), 3165–3172, 10.1039/c3nr06201e.
- J. I. B. Janairo, T. Sakaguchi, K. Hara, A. Fukuoka and K. Sakaguchi, Effects of Biomineralization Peptide Topology on the Structure and Catalytic Activity of Pd Nanomaterials, Chem. Commun., 2014, 50, 9259–9262, 10.1039/c4cc04350b.
- A. D. Merg, J. Slocik, M. G. Blaber, G. C. Schatz, R. Naik and N. L. Rosi, Adjusting the Metrics of 1-D Helical Gold Nanoparticle Superstructures Using Multivalent Peptide Conjugates, Langmuir, 2015, 31(34), 9492–9501, DOI:10.1021/acs.langmuir.5b02208.
- K. Tomizaki, K. Kishioka, H. Kobayashi, A. Kobayashi, N. Yamada, S. Kataoka, T. Imai and M. Kasuno, Roles of Aromatic Side Chains and Template Effects of the Hydrophobic Cavity of a Self-Assembled Peptide Nanoarchitecture for Anisotropic Growth of Gold Nanocrystals, Bioorg. Med. Chem., 2015, 23(22), 7282–7291, DOI:10.1016/j.bmc.2015.10.027.
- N. Choi, L. Tan, J. Jang, Y. M. Um, P. J. Yoo and W.-S. Choe, The Interplay of Peptide Sequence and Local Structure in TiO2 Biomineralization, J. Inorg. Biochem., 2012, 115, 20–27, DOI:10.1016/j.jinorgbio.2012.05.011.
- J. I. B. Janairo and K. Sakaguchi, Effects of Buffer on the Structure and Catalytic Activity of Palladium Nanomaterials Formed by Biomineralization, Chem. Lett., 2014, 43(8), 1315–1317, DOI:10.1246/cl.140405.
- A. Tofanello, É. G. A. Miranda, I. W. R. Dias, A. J. C. Lanfredi, J. T. Arantes, M. A. Juliano and I. L. Nantes, PH-Dependent Synthesis of Anisotropic Gold Nanostructures by Bioinspired Cysteine-Containing Peptides, ACS Omega, 2016, 1(3), 424–434, DOI:10.1021/acsomega.6b00140.
- R. Coppage, J. M. Slocik, M. Sethi, D. B. Pacardo, R. R. Naik and M. R. Knecht, Elucidation of Peptide Effects That Control the Activity of Nanoparticles, Angew. Chem., Int. Ed., 2010, 49(22), 3767–3770, DOI:10.1002/anie.200906949.
- R. Coppage, J. M. Slocik, B. D. Briggs, A. I. Frenkel, R. R. Naik and M. R. Knecht, Determining Peptide Sequence Effects That Control the Size, Structure, and Function of Nanoparticles, ACS Nano, 2012, 6(2), 1625–1636, DOI:10.1021/nn204600d.
- R. Coppage, J. M. Slocik, H. Ramezani-Dakhel, N. M. Bedford, H. Heinz, R. R. Naik and M. R. Knecht, Exploiting Localized Surface Binding Effects to Enhance the Catalytic Reactivity of Peptide-Capped Nanoparticles, J. Am. Chem. Soc., 2013, 135(30), 11048–11054, DOI:10.1021/ja402215t.
- S. Mokashi-Punekar, S. C. Brooks, C. D. Hogan and N. L. Rosi, Leveraging Peptide Sequence Modification to Promote Assembly of Chiral Helical Gold Nanoparticle Superstructures, Biochemistry, 2021, 60(13), 1044–1049, DOI:10.1021/acs.biochem.0c00361.
- S. C. Brooks, R. Jin, V. C. Zerbach, Y. Zhang, T. R. Walsh and N. L. Rosi, Single Amino Acid Modifications for Controlling the Helicity of Peptide-Based Chiral Gold Nanoparticle Superstructures, J. Am. Chem. Soc., 2023, 145(11), 6546–6553, DOI:10.1021/jacs.3c00827.
- M. Tanaka, S. Hikiba, K. Yamashita, M. Muto and M. Okochi, Array-Based Functional Peptide Screening and Characterization of Gold Nanoparticle Synthesis, Acta Biomater., 2017, 49, 495–506, DOI:10.1016/j.actbio.2016.11.037.
- M. Hahsler, I. Johnson, T. Kliegr and J. Kuchař, Associative Classification in R: Arc, ArulesCBA, and RCBA, R J., 2019, 11(2), 254–267 CrossRef.
- R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2021 Search PubMed.
- N. Du, M. R. Knecht, M. T. Swihart, Z. Tang, T. R. Walsh and A. Zhang, Identifying Affinity Classes of Inorganic Materials Binding Sequences via a Graph-Based Model, IEEE/ACM Trans. Comput. Biol. Bioinf., 2015, 12(1), 193–204, DOI:10.1109/TCBB.2014.2321158.
- J. I. B. Janairo, Predictive Analytics for Biomineralization Peptide Binding Affinity, Bionanoscience, 2019, 9(1), 74–78, DOI:10.1007/s12668-018-0578-4.
- J. I. B. Janairo, K. B. Aviso, M. A. B. Promentilla and R. R. Tan, Enhanced Hyperbox Classifier Model for Nanomaterial Discovery, AI, 2020, 1(2), 299–311, DOI:10.3390/ai1020020.
- D. Stelle, M. C. Barioni and L. P. Scott, Using Data Mining to Identify Structural Rules in Proteins, Appl. Math. Comput., 2011, 218(5), 1997–2004, DOI:10.1016/j.amc.2011.07.011.
- N. Gupta, N. Mangal, K. Tiwari and P. Mitra, Mining Quantitative Association Rules in Protein Sequences, Data Mining: Theory, Methodology, Techniques, and Applications, ed. G. J. Williams and S. J. Simoff, Springer Berlin Heidelberg, Berlin, Heidelberg, 2006, pp. 273–281, DOI:10.1007/11677437_21.
- J. I. B. Janairo, T. Sakaguchi, K. Mine, R. Kamada and K. Sakaguchi, Synergic Strategies for the Enhanced Self-Assembly of Biomineralization Peptides for the Synthesis of Functional Nanomaterials, Protein Pept. Lett., 2018, 25, 4–14, DOI:10.2174/0929866525666171214110206.
- B. Liu, Y. Ma and C.-K. Wong, Classification Using Association Rules: Weaknesses and Enhancements, in Data Mining for Scientific and Engineering Applications, ed. R. L. Grossman, C. Kamath, P. Kegelmeyer, V. Kumar and R. R. Namburu, Springer US, Boston, MA, 2001, pp. 591–605, DOI:10.1007/978-1-4615-1733-7_30.
- C. J. Munro, Z. E. Hughes, T. R. Walsh and M. R. Knecht, Peptide Sequence Effects Control the Single Pot Reduction, Nucleation, and Growth of Au Nanoparticles, J. Phys. Chem. C, 2016, 120(33), 18917–18924, DOI:10.1021/acs.jpcc.6b06046.
- P. R. Selvakannan, S. Mandal, S. Phadtare, A. Gole, R. Pasricha, S. D. Adyanthaya and M. Sastry, Water-Dispersible Tryptophan-Protected Gold Nanoparticles Prepared by the Spontaneous Reduction of Aqueous Chloroaurate Ions by the Amino Acid, J. Colloid Interface Sci., 2004, 269(1), 97–102, DOI:10.1016/S0021-9797(03)00616-7.
- M. Ozaki, S. Yoshida, M. Oura, T. Tsuruoka and K. Usui, Effect of Tryptophan Residues on Gold Mineralization by a Gold Reducing Peptide, RSC Adv., 2020, 10(66), 40461–40466, 10.1039/d0ra07098j.
- P. Joshi, V. Shewale, R. Pandey, V. Shanker, S. Hussain and S. P. Karna, Tryptophan–Gold Nanoparticle Interaction: A First-Principles Quantum Mechanical Study, J. Phys. Chem. C, 2011, 115(46), 22818–22826, DOI:10.1021/jp2070437.
- S. Hussain and Y. Pang, Surface Geometry of Tryptophan Adsorbed on Gold Colloidal Nanoparticles, J. Mol. Struct., 2015, 1096, 121–128, DOI:10.1016/j.molstruc.2015.05.001.
- M. A. Nguyen, Z. E. Hughes, Y. Liu, Y. Li, M. T. Swihart, M. R. Knecht and T. R. Walsh, Peptide-Mediated Growth and Dispersion of Au Nanoparticles in Water via Sequence Engineering, J. Phys. Chem. C, 2018, 122(21), 11532–11542, DOI:10.1021/acs.jpcc.8b02392.
- A. V. Verde, J. M. Acres and J. K. Maranas, Investigating the Specificity of Peptide Adsorption on Gold Using Molecular Dynamics Simulations, Biomacromolecules, 2009, 10(8), 2118–2128, DOI:10.1021/bm9002464.
- M. Sethi and M. R. Knecht, Understanding the Mechanism of Amino Acid-Based Au Nanoparticle Chain Formation, Langmuir, 2010, 26(12), 9860–9874, DOI:10.1021/la100216w.
- P. A. Mirau, R. R. Naik and P. Gehring, Structure of Peptides on Metal Oxide Surfaces Probed by NMR, J. Am. Chem. Soc., 2011, 133(45), 18243–18248, DOI:10.1021/ja205454t.
- W. R. Fiori, K. M. Lundberg and G. L. Millhauser, A Single Carboxy-Terminal Arginine Determines the Amino-Terminal Helix Conformation of an Alanine-Based Peptide, Nat. Struct. Biol., 1994, 1(6), 374–377, DOI:10.1038/nsb0694-374.
- H. Heinz, B. L. Farmer, R. B. Pandey, J. M. Slocik, S. S. Patnaik, R. Pachter and R. R. Naik, Nature of Molecular Interactions of Peptides with Gold, Palladium, and Pd - Au Bimetal Surfaces in Aqueous Solution, J. Am. Chem. Soc., 2009, 131(16), 9704–9714 CrossRef CAS PubMed.
- Z. Tang, J. P. Palafox-Hernandez, W.-C. Law, Z. E. Hughes, M. T. Swihart, P. N. Prasad, M. R. Knecht and T. R. Walsh, Biomolecular Recognition Principles for Bionanocombinatorics: An Integrated Approach To Elucidate Enthalpic and Entropic Factors, ACS Nano, 2013, 7(11), 9632–9646, DOI:10.1021/nn404427y.
- D. A. Cannon, N. Ashkenasy and T. Tuttle, Influence of Solvent in Controlling Peptide-Surface Interactions, J. Phys. Chem. Lett., 2015, 6(19), 3944–3949, DOI:10.1021/acs.jpclett.5b01733.
- J. I. B. Janairo, A Machine Learning Classification Model for Gold-Binding Peptides, ACS Omega, 2022, 7(16), 14069–14073, DOI:10.1021/acsomega.2c00640.
- Z. E. Hughes, M. A. Nguyen, J. Wang, Y. Liu, M. T. Swihart, M. Poloczek, P. I. Frazier, M. R. Knecht and T. R. Walsh, Tuning Materials-Binding Peptide Sequences toward Gold- and Silver-Binding Selectivity with Bayesian Optimization, ACS Nano, 2021, 15(11), 18260–18269, DOI:10.1021/acsnano.1c07298.
| This journal is © The Royal Society of Chemistry 2023 |