Open Access Article
Open Access ArticleIsolation of sulfonosphingolipids from the rosette-inducing bacterium Zobellia uliginosa and evaluation of their rosette-inducing activity†
Chia-Chi Pengab,
Nils Dormanns a,
Lars Regestein
a,
Lars Regestein a and
Christine Beemelmanns
a and
Christine Beemelmanns *bc
*bc
aLeibniz Institute for Natural Product Research and Infection Biology – Hans Knöll Institute (HKI), Beutenbergstraße 11a, 07745, Jena, Germany
bAnti-infectives from Microbiota, Helmholtz-Institut für Pharmazeutische Forschung Saarland (HIPS), Campus E8.1, 66123 Saarbrücken, Germany. E-mail: christine.beemelmanns@helmholtz-hips.de
cSaarland University, 66123 Saarbrücken, Germany
First published on 14th September 2023
Abstract
The choanoflagellate Salpingoeca rosetta transitions from unicellular to multicellular forms in the presence of bacterial signaling molecules, such as sulfonosphingolipids (RIFs). We set out to characterize the abundance of RIF-like molecules within five different Bacteroidetes strains belonging to different genera. While four strains exhibited similar sulfonosphingolipid profiles with sulfobacin A as the dominant feature, the composition in Z. uliginosa differed distinctively. Targeted isolation yielded four sulfonosphingolipids, including the previously reported flavocristamide A. While none of the sulfonosphingolipids induced rosette formation, a negative impact on choanoflagellate growth and cell density was observed. In contrast, supernatant extracts of Zobellia depleted in sulfonosphingolipid-like features provoked rosette formation in S. rosetta indicating for the presence of yet another morphogenic compound class.
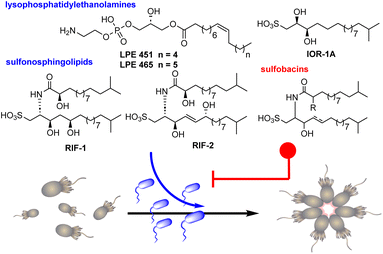
Choanoflagellates, acknowledged as the closest living relatives of animals, inhabit aquatic environments as bacterial predators and have recently emerged as a promising model system for the study of both eukaryotic development and the evolution of multicellularity.1,2 This stems from their distinct phylogenetic standing and the presence of stages resembling multicellularity in various species.3 Notably, the choanoflagellate Salpingoeca rosetta presents both unicellular and multicellular phases, including rosette formations created through successive cell divisions without the detachment of sister cells (Fig. 1).4–7 Rosettes, which develop upon detection of specific signaling molecules emitted by prey bacteria,8 have been proposed to offer a survival advantage by facilitating increased water flow, thereby boosting the rate of bacterial predation.9
The prey bacterium Algoriphagus machipongonensis significantly stimulates the predatory behavior of S. rosetta by secretion of sulfonosphingolipids (rosette-inducing factors, RIFs) RIF-1 and RIF-2, which induce the morphological transformation in a dose-dependent fashion.10–12 While each of these molecules separately triggers rosette formation in a small proportion of cells, their co-secretion fosters rosette growth in 20–30% of cells. Intriguingly, A. machipongonensis also produces two lysophosphatidylethanolamines (LPEs), which in combination with RIFs amplify rosette development in approximately 80% of all cells. Conversely, the rosette-inducing capacity is antagonized in a dose-dependent manner by the structurally related RIF-homologs such as sulfobacin D, F, and sulfonolipid IOR-1A (Fig. 1).13,14
The chemical interaction between choanoflagellates and bacteria mediated by these bacterial membrane components is representative for the inherent complex cross-kingdom communication of predator-prey relations and their chemical mediators.15,16
Sulfonosphingolipids represent an unusual class of sphingolipids featuring a sulfonic acid head group of the sphingoid base,22 and their close relationship to eukaryotic sphingolipid signaling molecules17–19 have sparked captivating questions about the prevalence and morphogenic activities of RIF-like metabolites in other prey bacteria, their role in other cross-kingdom interactions20,21 and diseases,19 as well as their biosynthetic origin.22
In this study, we pursued a high-resolution tandem mass spectrometry (HRMS2) and Global Natural Product Social Molecular Networking (GNPS)23-driven sulfonosphingolipid analyses of rosette-inducing and non-inducing strains belonging to the Bacteroidetes. Targeted isolation of sulfonosphingolipids from lipid extracts of the inducing strain Z. uliginosa led to the isolation of four Z. uliginosa specific sulfonosphingolipids, including three yet no reported derivatives. Furthermore, testing of culture extracts yielded further proof that yet another still structurally unknown small molecule class is capable of stimulating rosette formation.
Results and discussion
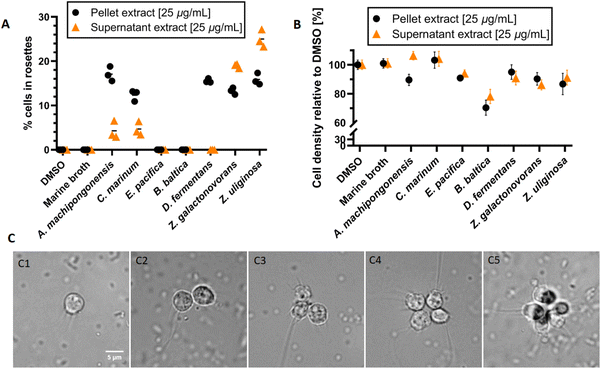
We first evaluated the effects of seven Bacteroidetes strains, including the rosette-inducing strain Algoriphagus machipongonensis PR1, Cyclobacterium marinum LMG 13164, Dyadobacter fermentans DSM 18053, Zobellia galactonovorans Dsij, Zobellia uliginosa ATCC 14397 and the non-inducing species Echinicola pacifica KMM 6172 and Belliella baltica BA134, for their capacity to induce rosette formation in a S. rosetta cell line. For this, all strains were cultivated in marine broth for three days, after which cells and supernatant were separated by centrifugation. While cell pellets were subjected to a methanolic lipid extract procedure, culture supernatants were extracted using a standardized solid-phase (C-18) extraction protocol. Obtained extracts (25 μg mL−1) were then added to a rosette-free cell culture and rosette formation monitored over a period of 48 h (Fig. 2A).Overall, cell membrane extracts from the five rosette-inducing species (A. machipongonensis, C. marinum, D. fermentans, Z. galactonovorans, Z. uliginosa) as well as enriched outer-membrane vesicles (OMVs) from A. machipongonensis (positive control) consistently triggered the formation of stable rosettes mostly without significant decrease of cell density in S. rosetta (Fig. 2B and C). In contrast, supplementation with extracts derived from either E. pacifica or B. baltica caused no observable phenotypic changes in S. rosetta.
Abundance of sulfonosphingolipids in cell membrane extracts
We then used a comparative HRMS2-based network approach to analyze the sulfonosphingolipid profile of methanolic cell membrane extracts of inducing and non-inducing Bacteroidetes species (A. machipongonensis PR1, D. fermentans DSM 18053, Z. uliginosa ATCC 14397, E. pacifica KMM 6172, B. baltica BA134, Table S1†). Although investigated strains belonged to different bacterial genera, all five strains revealed remarkably similar sulfonosphingolipid profiles (Fig. 3, S1 and S2†). Molecular ion features assigned to RIF-1 (m/z 606.442 [M–H]−) and RIF-2 (m/z 604.426 [M–H]−) were only present in very low quantities, whereas signals for the non-inducing sulfonosphingolipids such as sulfobacin D and F (m/z 590.446, and 588.430 [M–H]−) dominated the sulfonosphingolipid profiles (Fig. 3A).14 Interestingly, Z. uliginosa exhibited several unique signals with the nodes corresponding to m/z 616.462, 602.447 and 604.463 [M–H]−, sharing a sulfonosphingolipid profile nearly identical to the same genera of Z. galactonovorans (Fig. 3B), but neither showed molecular ion features assigned to RIF-1 or RIF-2.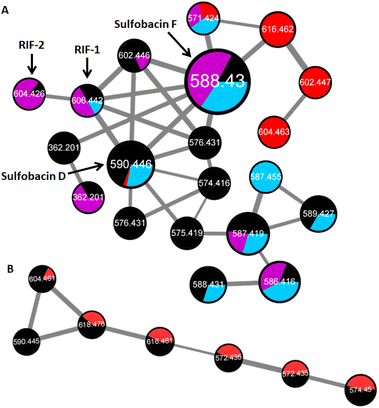 | ||
| Fig. 3 (A) HR-MS2-based GNPS cluster (negative ion mode) of cell membrane extracts with nodes for RIF-1 (m/z 606.442 [M–H]−), RIF-2 (m/z 604.426 [M–H]−), sulfobacin F (m/z 588.430 [M–H]−), sulfobacin D (m/z 590.446 [M–H]−) in A. machipongonensis PR1 (black), Z. uliginosa ATCC 14397 (red), E. pacifica KMM 6172 (purple) and B. baltica BA134 (blue). (B) HR-MS2-based GNPS cluster (negative ion mode) of cell membrane extracts in Z. uliginosa ATCC 14397 (red) and Z. galactonovorans Dsij (black). | ||
Since the cell membrane extract of Z. uliginosa demonstrated similar rosette-inducing activity as OMVs and RIF-containing extracts of A. machipongonensis, we hypothesized that RIF-like sulfonosphingolipids might trigger rosette formation. Thus, we commenced the isolation of the unique sulfonosphingolipid features (m/z 616.462, 602.447 and 604.463 [M–H]−) of Z. uliginosa using a MS-guided purification and isolation strategy.
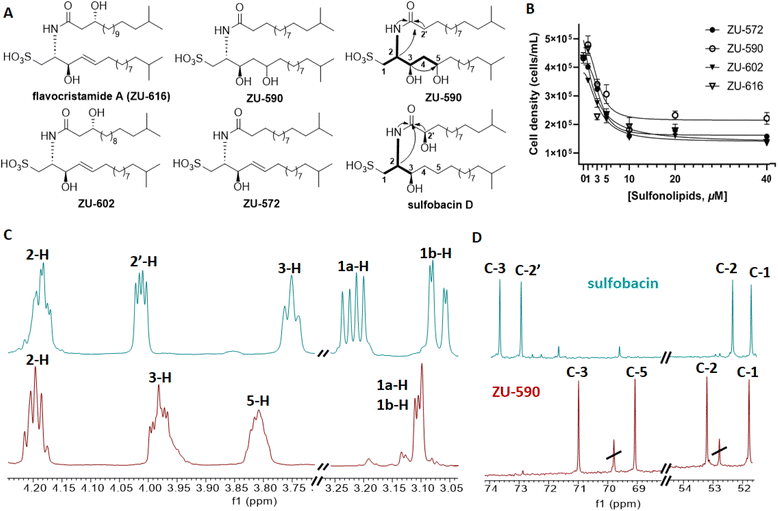
Purification of methanolic cell membrane extracts via reverse-phase chromatography yielded four sulfonosphingolipids, which were named ZU-572, ZU-590, ZU-602 and ZU-616 and showed RIF-like molecular ion features (Fig. 4A, S4–S6, Tables S2 and S3†). Comparative 1D and 2D NMR analysis revealed that compound ZU-616 exhibited the same chemical shift patterns as reported for isolated and synthetic flavocristamide A,24,25 a known DNA polymerase α inhibitor.
In contrast, isolated compounds ZU-572 and ZU-602 exhibited yet unreported molecular ion features and sulfonosphingolipid-like MS-fragmentation pattern. We then compared the 1D and 2D NMR-based chemical shifts and 2D correlations as well as HRMS/MS fragmentation patterns (major fragment of m/z 348.2214 [M–H]−) of ZU-572, ZU-602 and ZU-616, and were able to deduce that the newly isolated sulfonosphingolipids carried the same capnine base as flavocristamide A, but were acylated with (hydroxylated) iso-fatty acids of different length (Fig. 4, S5–S47†).
To verify the hypothesis, we performed methanolic acid hydrolysis of ZU-572, followed by hexane-based extraction of the methyl ester and GC-MS/MS analysis. Comparison of MS-fragmentation features with the NIST database allowed us to confidently assign the fatty acid methyl ester for ZU-572 as methyl 13-methyltetradecanoate supporting the structural hypothesis (Fig. 4).10
In contrast, ZU-590 shared the exact same mass as sulfobacin D (m/z 590.446 [M–H]−), but its 1H- and 13C-NMR chemical shifts as well as 2D NMR correlations differed from the reported sulfobacin D (Fig. 4C and D).10,14 Based on comparative 1D and 2D NMR analyses with previously synthesized and isolated sulfonosphingolipids, we were able to deduce that ZU-590 is composed of a RIF-1 like dihydroxylated capnine base, which has been acylated with an unmodified fatty acid.10
Testing for rosette-inducing activity
Subsequently, we evaluated the rosette-inducing effects of the four RIF-like sulfonosphingolipids isolated from Z. uliginosa. However, none of the isolated RIF-like sulfonosphingolipids induced rosette formation at any tested concentration. Instead, all four RIF-like sulfonosphingolipids caused a reduction in cell density (Fig. 4B) despite their structural similarity to RIF-1 and RIF-2. A similar phenomenon was observed for synthetic sulfobacin congeners and those isolated from the rosette-inducing strain C. marinum and the non-inducing species E. pacifica. Here, it can be speculated that similar to flavocristamide A, sulfobacin-like derivatives could act as DNA polymerase α inhibitors in S. rosetta; a hypothesis that warrants further investigations.14We then compared the HRMS2 profiles of both, cell membrane and supernatant extracts of Z. uliginosa to deduce potential shared molecular features (Fig. 3 and S2†). Intriguingly, sulfonosphingolipid concentrations were below detection levels in rosette-inducing supernatant extracts supporting our hypothesis, that other yet unknown bacterial-derived metabolites might be the actual morphogens. A targeted MS-analysis of enriched and semi-purified fractions also revealed that neither sphingolipids, capnine bases, siderophores, nor the algae morphogen thallusin26 were detectable within the active metabolite fractions leaving the chemical nature of the morphogen elusive.
Conclusions
Our HRMS2 based analysis of five Bacteroidetes type strains revealed highly similar sulfonosphingolipid profiles across four strains, while Z. uliginosa exhibited a similar but distinct profile with unique RIF-like sulfonosphingolipid nodes. The MS-guided isolation revealed three novel sulfonosphingolipid congeners and one known sulfonosphingolipid (flavocristamide A), which differed in the length of the fatty acid chain and the hydroxylation pattern. Evaluation of their rosette-inducing capacity uncovered that all four sulfonosphingolipids were not capable of inducing rosette formation, instead caused a reduction in cell density of S. rosetta, similar to the structurally related sulfobacins.14 The finding that cell membrane and supernatant extracts depleted of sulfonosphingolipid-like features showed still activity hints towards yet another compound class capable of inducing rosette formation. Our findings have shed more insighs will stimulate further research into diversity of bacterial signals in cross-kingdom communication, in general and predator-prey interaction of Bacteroidetes-S. rosetta in particular.Author contributions
CP contributed to conceptualization, data curation, writing – original draft, review and editing. ND contributed by formal analysis, investigation, validation and visualization as well as formal analysis. LR contributed resources, funding acquisition and investigations. CB contributed with conceptualization, formal analysis, funding acquisition, supervision, validation, visualization, writing – original draft, review and editingConflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for financial support from the German Research Foundation (DFG, BE 4799/2-1). CB greatly acknowledges funding from the European Union's Horizon 2020 research and innovation program (ERC Grant number: 802736, MORPHEUS). We would also like to thank Heike Heinecke (HKI) for recording NMR spectra.Notes and references
- T. T. Hoffmeyer and P. Burkhardt, Curr. Opin. Genet. Dev., 2016, 39, 42–47 CrossRef CAS PubMed.
- A. Woznica and N. King, Curr. Opin. Microbiol., 2018, 43, 108–116 CrossRef CAS PubMed.
- M. Carr, D. J. Richter, P. Fozouni, T. J. Smith, A. Jeuck, B. S. C. Leadbeater and F. Nitsche, Mol. Phylogenet. Evol., 2017, 107, 166–178 CrossRef PubMed.
- S. R. Fairclough, M. J. Dayel and N. King, Curr. Biol., 2010, 20, R875 CrossRef CAS PubMed.
- M. J. Dayel, R. A. Alegado, S. R. Fairclough, T. C. Levin, S. A. Nichols, K. McDonald and N. King, Dev. Biol., 2011, 357, 73 CrossRef CAS PubMed.
- S. Fairclough, Z. Chen, E. Kramer, Q. Zhen, S. Young, H. Robertson, E. Begovic, D. J. Richter, C. Russ, M. J. Westbrook, G. Manning, B. F. Lang, B. Haas, C. Nusbaum and N. King, Genome Biol., 2013, 14, R15 CrossRef PubMed.
- T. C. Levin, A. J. Greaney, L. Wetzel and N. King, eLife, 2014, 9(3), e04070 CrossRef PubMed.
- R. A. Alegado, L. W. Brown, S. Cao, R. K. Dermenjian, R. Zuzow, S. R. Fairclough, J. Clardy and N. King, eLife, 2012, 1, e00013 CrossRef PubMed.
- M. Roper, M. J. Dayel, R. E. Pepper and M. A. R. Koehl, Phys. Rev. Lett., 2013, 110, 228104 CrossRef PubMed.
- C. Beemelmanns, A. Woznica, R. A. Alegado, A. M. Cantley and N. King, J. Am. Chem. Soc., 2014, 136, 10210–10213 CrossRef CAS PubMed.
- A. Woznica, A. M. Cantley, C. Beemelmanns, E. Freinkman, J. Clardy and N. King, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 7894–7899 CrossRef CAS PubMed.
- E. V. Ireland, A. Woznica and N. King, Appl. Environ. Microbiol., 2020, 86, 029200 CrossRef PubMed.
- A. M. Cantley, A. Woznica, C. Beemelmanns, N. King and J. Clardy, J. Am. Chem. Soc., 2016, 138, 4326–4329 CrossRef CAS PubMed.
- D. Leichnitz, C.-C. Peng, L. Raguž, F. Rutaganira, T. Jautzus, L. Regestein, N. King and C. Beemelmanns, Chem.–Eur. J., 2022, 28, e202103883 CrossRef CAS PubMed.
- D. Leichnitz, L. Raguž and C. Beemelmanns, Chem. Soc. Rev., 2017, 46, 6330–6344 RSC.
- A. M. Cantley and J. Clardy, Nat. Prod. Rep., 2015, 32, 888–892 RSC.
- Y. A. Hannun and L. M. Obeid, Nat. Rev. Mol. Cell Biol., 2018, 19, 175–191 CrossRef CAS PubMed.
- J. B. Parsons and C. O. Rock, Prog. Lipid Res., 2013, 52, 249–276 CrossRef CAS PubMed.
- F. Parveen, D. Bender, S.-H. Law, V. K. Mishra, C.-C. Chen and L.-Y. Ke, Cell Sci., 2019, 8, 1573 CAS.
- P. J. Harrison, T. M. Dunnb and D. J. Campopiano, Nat. Prod. Rep., 2018, 35, 921–954 RSC.
- E. L. Johnson, S. L. Heaver, J. L. Waters, B. I. Kim, A. Bretin, A. L. Goodman, A. T. Gewirtz, T. S. Worgall and R. E. Ley, Nat. Commun., 2020, 11, 2471 CrossRef CAS PubMed.
- G. Stankeviciute, P. Tang, B. Ashley, J. D. Chamberlain, M. E. B. Hansen, A. Coleman, R. D'Emilia, L. Fu, E. C. Mohan, H. Nguyen, Z. Guan, D. J. Campopiano and E. A. Klein, Nat. Chem. Biol., 2022, 18, 305–312 CrossRef CAS PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W. T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C. C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C. C. Liaw, Y. L. Yang, H. U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. P. Boya, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P. M. Allard, P. Phapale, L. F. Nothias, T. Alexandrov, M. Litaudon, J. L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D. T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- J. Kohayashi, S. Mikami, H. Shigemori, T. Takao, Y. Shimonishi, S. Izuta and S. Yoshida, Tetrahedron, 1995, 51, 10487–10490 CrossRef.
- T. Shioiri and N. Irako, Tetrahedron, 2000, 56, 9129–9142 CrossRef CAS.
- J. F. Ulrich, M. S. Gräfe, S. Dhiman, P. Wienecke, H. D. Arndt and T. Wichard, Mar. Drugs, 2022, 20, 690 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04314b |
| This journal is © The Royal Society of Chemistry 2023 |