Open Access Article
Open Access ArticleDevelopment of two highly sensitive and selective sensor-assisted fluorescence immunoassays for trace determination of copper residues in food samples
Ibrahim A. Darwish *a,
Zongzhi Wangb,
Ryhan J. Darlingc and
Nourah Z. Alzoman
*a,
Zongzhi Wangb,
Ryhan J. Darlingc and
Nourah Z. Alzoman a
a
aDepartment of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, P.O. Box 2457, Riyadh 11451, Saudi Arabia. E-mail: idarwish@ksu.edu.sa; Fax: +966-114676220; Tel: +966-114677348
bState Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
cDepartment of Biochemistry, Medical College of Wisconsin, Milwaukee, WI 53226, USA
First published on 2nd October 2023
Abstract
This study describes the development of two highly sensitive and selective sensor-assisted fluorescence immunoassays for the trace determination of copper ions, Cu(II) residues, in food samples. These assays were the microwell-based fluoroimmuoassay (FIA) and the kinetic exclusion assay (KinExA). FIA and KinExA were assisted by a microplate reader and a KinExA™ 3200 immunosensor, respectively. Both FIA and KinExA were developed utilizing the same antibody, capturing reagent, and fluorescence signal-generating reagent. The antibody was a mouse monoclonal antibody, designated as 8D66, that specifically recognized the Cu(II)–ethylenediaminetetraacetic acid complex (Cu(II)–EDTA) but did not recognize Cu(II)-free EDTA. The capturing reagent was Cu(II)–EDTA covalently linked to bovine serum albumin protein (Cu(II)–EDTA–BSA). The fluorescence-generating reagent was an anti-mouse IgG conjugated with fluorescein isothiocyanate (IgG–FITC). Both FIA and KinExA involved competitive binding reactions between Cu(II)–EDTA complexes, formed in the sample solution, and Cu(II)–EDTA–BSA conjugate which has been immobilized onto microwell fluorescence assay plates (in FIA) or polymethylmethacrylate beads (in KinExA) for a limited quantity of binding sites of 8D66 antibody. The conditions of both FIA and KinExA were investigated, and the optimum procedures were established. Both FIA and KinExA were validated, and all validation parameters were acceptable. Many different metal ions that are commonly encountered in food samples did not interfere with Cu(II) analysis by both FIA and KinExA. Both assays were applied to the determination of Cu(II) in food samples with satisfactory accuracy and precision. Both assays were compared favorably with inductively coupled plasma atomic emission spectroscopy. Comparative evaluation of FIA and KinExA revealed that KinExA had higher sensitivity and better precision than FIA, whereas, both assays had comparable accuracy. Both FIA and KinExA were superior to the existing atomic spectrometric methods for Cu(II). The proposed FIA and KinExA are anticipated to effectively contribute to assessing Cu(II) concentrations and controlling the exposure of humans to its potential toxicities.
Introduction
Copper is an essential trace element for the life of both mammals and plants. It plays an important role in the function of several enzymes controlling carbohydrate and lipid metabolism. Dietary copper comes from organ meats, green leafy vegetables, some fruits, and nuts. Foods, including drinking water, are considered as a potential major source of copper intake.1,2 In some countries, copper salts are added to drinking water to control the growth of algae.3 In addition, copper compounds are encountered and used on many crops that might be fed to livestock and may also exist in feed supplies and products for which no Good Agricultural Practices (GAPs) are authorized. Thus, the dietary loads highly exceed the safe trigger value for all groups of livestock. For the same reason, the residue definition for livestock products can be defined as total copper for both enforcement and risk assessment without requiring further studies. This residue definition includes copper residues arising from all forms of copper.4 Furthermore, high copper concentrations have been widely used in animal feeds as an efficient and inexpensive growth promoter.5For these reasons, human exposure to high doses of copper from foods is expected. Human exposure to copper-overload is associated with acute gastrointestinal upsets,6 severe liver necrosis, hepatic failure and carcinogenesis, culminating in Wilson's disease,7 Indian childhood cirrhosis,8,9 and idiopathic toxicosis.10 The presence of high concentrations of copper ions in vivo can change the redox system of cells causing toxicity,11 damaging DNA, proteins, and lipids.12 High concentrations of copper ions can non-specifically react with some amino acid residues leading to misfolding of proteins' structures.13 Free copper ions also compete with other substances involved in the functions of some enzymes resulting in disrupting their normal functions.14 In addition, the accumulation of copper in human body contribute to atherosclerosis and rapid cell aging posing a huge concern to public health.15–17 Furthermore, excretion of copper from livestock causes soil deterioration and water pollution which can negatively affect the growth rate of plants18–21 and pose ecological concerns.22,23
Reduction of human exposure to copper is one of the main objectives of hygienists and public authorities. Various attempts have been made to determine a safe concentration of copper in foods, and health-based safety guideline values and maximum allowable limits in human serum. These guidelines have been mandated by legislation of several countries, the World Health Organization (WHO). The Safe Drinking Water Act (section 1412) required the U.S. Environmental Protection Agency (EPA) to establish and publish maximum-contaminant-level goals and promulgate national primary food regulations for contaminants that might cause any adverse effect on human health. The Food and Nutrition Board (FNB) recommended a daily dietary copper intake for adults at 1.5–3.0 mg per day. EPA and WHO stated a maximum-residue limit (MRL) of safe copper in drinking water at 1.3 and 2 mg mL−1, respectively. This guideline was issued by the Joint Food and Agriculture Organization (FAO)/WHO Expert Committee on Food Additives.24 The European Union (EU) has established MRLs for various food products, including copper. According to Commission Regulation (EC) No. 1881/2006, the MRL for copper in most foodstuffs is set at 1 mg kg−1 (milligrams per kilogram). However, in certain specific food products like liver and other offal, the MRL can be as high as 10 mg kg−1. United States EPA sets tolerances for copper in or on food of 0.1 ppm (parts per million) for copper residues in fruits, nuts, and vegetables. The Codex Alimentarius Commission, jointly established by FAO and WHO, develops international standards for food safety. The Codex has set a general MRL of 1 mg kg−1 for copper in most food products.24
In response to these health concerns, many governmental health authorities requested researchers and administrators to conduct comprehensive studies for the effects of copper in foods on human health. The availability of appropriate sensitive and selective analytical techniques that could provide near real time data on copper levels in food samples could ultimately reduce the incidence of copper intoxication in humans. Different analytical techniques and methods are recommended for foods compliance monitoring of copper and other secondary contaminants.25–27 Of the methods, inductively coupled plasma atomic emission spectroscopy (ICP-AES) is the most commonly employed. This technique usually provides reasonable sensitivity; however, the analysis usually needs a large sample volume to ensure adequate sensitivity. In addition, the number of samples that can be analyzed is limited by the fact that samples are analyzed one at a time. Therefore, the sample turnaround time is relatively slow, and the method is not adapting to screening of large number of samples.
Immunoassays are a better alternative for elemental analysis. This methodology is attractive to local and governmental agencies because they have significant advantages over the traditional methods. They are remarkably quick, easily performed, reasonably portable to the analysis site, require minimum sample pretreatment, and have high throughput. Furthermore, studies have shown that the use of immunoassays can reduce analysis costs by 50% or even more.28 Immunoassays were successfully established in recent years for metal ions as far suitable antibodies could be generated. The generation of antibodies for metal ions involved the use of bifunctional chelating agents for metal ions and the resulting metal chelates could be covalently linked to carrier proteins. The metal chelate–protein conjugates showed distinct unique antigenic properties which could be used to generate monoclonal antibodies directed to metal ions.29,30 Microwell-based fluorescence (FIA) is one of the most dominant immunoassays. Kinetic exclusion assays (KinExA) employing KinExA™ immunosensor offered the same advantages of FIA besides the convenience of automated analysis. Fluorescence immunochromatographic assay has been described for determination of copper ions in herbal plants and soil samples;31 however, the assay has not been validated for food samples, and it suffers from the procedure convenience. KinExA has not been described yet for copper ions in food samples.
The present study describes the development of new FIA and KinExA with high sensitivity and specificity for the trace determination of Cu(II) in different food samples at concentrations as low as 0.2 and 0.05 ng mL−1 for FIA and KinExA, respectively.
Experimental
Instruments
FLx800 fluorescence microplate reader and ELx 50 automatic microplate strip washer were products of Bio-Tek Instruments Inc. (Winooski, USA). KinExA™ 3200 immunosensor was obtained from Sapidyne Instruments Inc. (Boise, ID, USA). Model MINI/18 Incubator (Genlab Ltd, Widnes, UK). Milli-Q water purification system (Labo, Millipore Ltd, Bedford, USA). UV-1601 PC double beam spectrophotometer (Shimadzu, Kyoto, Japan). Nutating mixer (Taitec, Saitama-ken, Japan).Materials
Atomic absorption spectroscopy standard metals (1000 mg L−1 in 2% HNO3) were obtained from PerkinElmer Co. (Norwalk, CT, USA). 1-(4-Isothiocyanobenzyl)-ethylenediamine-N,N,N′,N′-tetraacetic acid (ITCB-EDTA) was purchased from Dojindo Laboratories (Gaithersburg, MD, USA). Ultrapure bovine serum albumin (BSA) was purchased from Boehringer Mannheim Biochemicals (Indianapolis, IN, USA). Ethylenediaminetetraacetic acid (EDTA). 8D66 monoclonal antibody recognizing Cu(II)–EDTA was generated by fusion of spleen cells of an immunized BALB/c mouse with SP2/0-Ag14 myeloma cells. The immunogen was a protein conjugate of Cu(II)–EDTA with keyhole limpet hemocyanin protein, Cu(II)–EDTA–KLH. The IC50 values of this antibody were 0.5 and 2 ng mL−1, as determined by ELISA and KinExA, respectively. The antibody was highly specific for Cu(II)–EDTA complex among many other metal–EDTA complexes; the highest observed cross reactivity (12.9%) occurred by Zn(II)–EDTA complexes. The details for preparation and characterization of 8D66 monoclonal antibody has been submitted for publication elsewhere. Goat anti-mouse IgG conjugated with fluorescein isothiocyanate (IgG–FITC) was purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). White opaque high-binding microwell plates with flat bottom were a product of Corning/Costar, Inc. (Cambridge, MA). Polymethylmethacrylate (PMMA) beads (140–170 mesh, 98 μm) were acquired from Sapidyne Instruments Inc. (Boise, ID, USA). Metal-free disposable pipette tips were a product of Oxford Labware, Inc. (St. Louis, MO, USA). All glassware was washed with mixed HCl/HNO3 acids and liberally rinsed with purified deionized water. All plastic wares were soaked overnight in 3 M HCl and rinsed with purified deionized water before use.Preparation of Cu(II)–EDTA–BSA conjugate
Conjugate of Cu(II)–EDTA with BSA Cu(II)–EDTA–BSA was prepared by a modification of the method previously reported.32 Briefly, a solution of ITCBE (2.7 mM, in 0.1 M sodium phosphate buffer, pH 9.5). Aliquot (1 mL) of ITCBE solution was mixed with equal volume of copper nitrate solution (2.7 mM, in 50 mM phosphate buffer, pH 9.5). BSA solution (2 mL, 20 mg mL−1, in 0.1 M sodium phosphate buffer, pH 9.5) was added. The pH of the reaction mixture was maintained at pH 9.5 for 30 min by addition of KOH when necessary. The reactions were stirred overnight at 25 °C. The unreacted low molecular weight components (Cu(II) and EDTA) were removed by buffer exchange using a Centricon-30 filter that had been treated with 100 mM EDTA solution and liberally rinsed with water before use. Protein concentrations of the conjugates were determined using BCA kit,33 and the extent of substitution of free amino groups on the protein was determined by estimation of free amino groups before and after the conjugation reaction.34Analysis of Cu(II) by FIA
FIA plates were coated with Cu(II)–EDTA–BSA conjugate (2 μg mL−1, in HBS) for 2 h at 37 and blocked with BSA (2%, w/v, in HBS) for 0.5 at °C, and blocked with BSA (2%, w/v, in HBS, pH 7) for 0.5 h at 37 °C. The detailed procedures of coating and blocking were described in a previous report.37 Food samples were prepared as described in previous reports.27,35–38 and the sample solutions were conditioned by the addition of a 10% volume of HBS containing EDTA (10 mM). Aliquot of the diluted samples were mixed (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with 8D66 monoclonal antibody solution (0.25 μg mL−1 in HBS), and 50 μL of the mixture was transferred to each well of the plates. After incubation for 0.5 h at 37 °C, the plates were washed as described above. The amount of bound 8D66 antibody was quantified using adding 50 μL of IgG–FITC (1 μg mL−1), and the fluorescence signals were measured by a fluorescence microplate reader. Standard curves for Cu(II) were obtained using the same procedure on plates of the same series. Values for IC50 were calculated using the following equation:
1) with 8D66 monoclonal antibody solution (0.25 μg mL−1 in HBS), and 50 μL of the mixture was transferred to each well of the plates. After incubation for 0.5 h at 37 °C, the plates were washed as described above. The amount of bound 8D66 antibody was quantified using adding 50 μL of IgG–FITC (1 μg mL−1), and the fluorescence signals were measured by a fluorescence microplate reader. Standard curves for Cu(II) were obtained using the same procedure on plates of the same series. Values for IC50 were calculated using the following equation:| B = B0 − {(B0 − B1)[Cu(II)]/(IC50 + [Cu(II)])} |
Analysis of Cu(II) by KinExA
The features of KinExA™ 3200 immunosensor and its running have been described in a previous report.37 PMMA beads were coated with Cu(II)–EDTA–BSA (1 μg mL−1), blocked with BSA (2%, w/v, in HBS) and loaded into the flow cell of instrument by following the procedures described in a previous report.37 Food sample solutions (prepared in HBS buffer of pH 7.4) containing 10 mM EDTA were pre-equilibrated with 8D66 antibody (0.25 μg mL−1, in HBS buffer, pH 7.4). The pre-equilibrated samples (500 μL) were withdrawn by the instrument and passed through PMMA beads coated with Cu(II)–EDTA–BSA and located in the flow cell of the instrument for 120 s at a flow rate of 0.25 mL min−1. After running of the samples, any unbound reagents were washed out by passing PBS through the beads. A 500 μL of IgG–FITC (0.25 μg mL−1) was drawn and passed through the beads for 120 s at the same flow rate. Unbound IgG–FITC was subsequently removed by passing 1.5 mL of HBS through PMMA beads for 90 s at a flow rate of 1 mL min−1. The bound IgG–FITC was assessed by means of measuring the fluorescence intensity. The data were transformed from the KinExA Pro 20.0.1.26 software to a four-parameter curve fitting using Slide Write software, version 5.011 (Advanced Graphics Software, Inc., Rancho Santa Fe, CA, USA).Results and discussion
Description of FIA and KinExA
This study describes the development of FIA and KinExA for trace determination of Cu(II) in food samples. The features and steps of FIA and KinExA are illustrated in Fig. 1.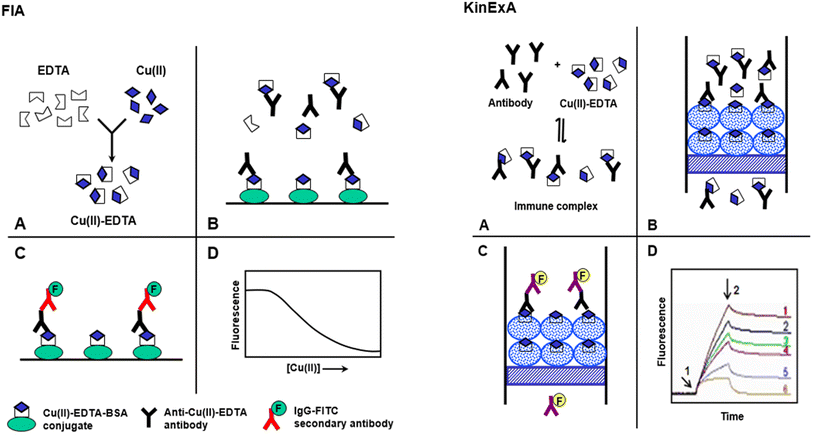 | ||
| Fig. 1 Schematic diagram for the proposed FIA and KinExA for determination of Cu(II) in food samples. | ||
FIA involved 4 main steps (A)–(D), Fig. 1. (A) Sample containing Cu(II) is diluted with buffer containing a molar excess of metal-free EDTA, to form Cu(II)–EDTA complexes. (B) The solution containing the excess EDTA and Cu(II)–EDTA complexes is mixed with antibody in microwell of FIA plates coated with Cu(II)–EDTA–BSA conjugate. The Cu(II)–EDTA complex, in solution, competes with the immobilized Cu(II)–EDTA–BSA conjugate for a limited amount of antibody binding sites. (C) After a wash step, IgG–FITC is added and allowed to bind to the anti-Cu(II)–EDTA antibody. A second wash step is applied to remove any unbound IgG–FITC. (D) Fluorescence signals are measured and plotted as the corresponding Cu(II) concentrations. The 4-parameter equation fit of the data is used for calculating the Cu(II) concentrations in the sample solutions.
KinExA is an automated flow fluorescent competitive assay; the details of KinExA™ 3200 immunosensor setup and operation have been described in a previous study.37 In KinExA, a mixture of Cu(II)–EDTA and its specific antibody is equilibrated to form immune complex (A). (B) The immune complex solution is automatically drawn by the instrument and passed rapidly through the flow cell containing the PMMA beads coated with Cu(II)–EDTA–BSA conjugate. During the flow of the mixture, the free antibody molecules bind to the immobilized Cu(II)–EDTA–BSA; however, those bound to free Cu(II)–EDTA (in solution) do not. Since the flow of the mixture is rapid, insignificant dissociation of the immune complex happens during the flow. Accordingly, the bound anti-Cu(II)–EDTA antibody molecules were kinetically excluded from the binding to the immobilized Cu(II)–EDTA–BSA. (C) The quantity of the anti-Cu(II)–EDTA antibody captured by the immobilized Cu(II)–EDTA–BSA is then quantified by IgG–FITC second antibody. The fluorescence intensity is continuously monitored by the photodiode of the KinExA™ 3200 system hardware. (D) Fluorescence-time curves (KinExAgrams) are generated. Curve 1 corresponds to zero of Cu(II) concentration; curve 6 corresponds to a saturating Cu(II) concentration. Curves 2–5 correspond to the concentrations of Cu(II) between zero and saturation. Arrow 1 indicates the introduction of IgG–FITC and arrow 2 indicates its cessation and initiation of a second buffer wash. The net fluorescence intensities measured on the PMMA beads coated with Cu(II)–EDTA–BSA are inversely correlated with Cu(II) concentrations in the original samples.
Preparation and characterization of Cu(II)–EDTA–BSA conjugate
The preparation of Cu(II)–EDTA–BSA conjugate is illustrated in Fig. 2. The bifunctional 1-(4-isothiocyanobenzyl)-ethylenediamine-N,N,N′,N′-tetraacetic acid (ITCBE) chelator was first used to form Cu(II) chelate via its tetraacetic acid groups. This chelate was coupled via reaction of the isothiocyanate group of ITCBE with the amino group residues on BSA protein. ITCBE is a modified form of EDTA that incorporates an isothiocyanate benzyl group. While both ITCBE and EDTA are chelating agents capable of forming complexes with metal ions, ITCBE offers the additional functionality and reactivity provided by the isothiocyanate benzyl group through which a protein molecule can be linked producing EDTA–protein conjugate. The use of EDTA instead will lead to chelating Cu(II) and forming Cu(II)–EDTA complex; however, the complex could not be linked to BSA protein. In order to ascertain the formation of the conjugate, colorimetric reaction involving amino groups residues of BSA was conducted on a definite concentration of the conjugate and on the same concentration of unconjugated BSA. The reaction was carried out as described in a previous report.33 The relative reduction of color signal (%) was attributed to the contribution of amino groups in the conjugation. The extent of conjugation of BSA with Cu(II)–EDTA was found to be 20.6%. It is wise to mention that this extent of conjugation was adequate for use as a capturing agent. This assumption was supported by previous study which demonstrated the successful use of protein conjugate at lower extent (15%).32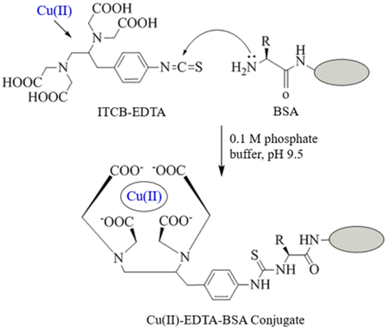 | ||
| Fig. 2 Preparation of Cu(II)–EDTA–protein conjugates. ITCB-EDTA is 1-(4-isothiocyanobenzyl)-ethylenediamine-N,N,N′,N′-tetraacetic acid. Protein was BSA and KLH. | ||
Optimization of FIA conditions
To avoid the non-specific binding of subsequently added protein reagents, the binding sites remained on the wells after coating with Cu(II)–EDTA–BSA should be blocked with an appropriate blocking reagent. Among different proteins tested, BSA was found to be the best one. Investigating the influence of BSA concentration indicated that 100 μL of BSA solution (2%, w/v) was adequate for optimum blocking of the wells, as demonstrated by the very low background signals.
| Parameter/condition | Optimum value |
|---|---|
| FIA | |
| Coating Cu(II)–EDTA–BSA conc. (μg mL−1) | 2 |
| Coating buffer | HBS |
| Coating time (h)/temperature (°C) | 2/37 |
| BSA concentration for blocking (%, w/v) | 2 |
| Blocking with BSA: time (h)/temperature (°C) | 0.5/37 |
| Concentration of EDTA (mM) | 10 |
| 8D66 antibody concentration (μg mL−1) | 0.25 |
| Binding of 8D66 antibody: time (h)/temperature (°C) | 0.5/37 |
| IgG–FITC concentration (μg mL−1) | 0.5 |
| Binding of IgG–FITC: time (h)/temperature (°C) | 1/37 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| KinExA | |
| Coated Cu(II)–EDTA–BSA (μg mL−1) | 1 |
| Volume of beads suspension (μL) | 583 |
| Flow rate of beads suspension (mL min−1) | 1 |
| Time of beads drawing (s) | 35 |
| Concentration of 8D66 antibody (μg mL−1) | 0.25 |
| Volume of Cu(II) sample (μL) | 500 |
| Time of samples drawing (s) | 120 |
| Concentration of IgG–FITC (μg mL−1) | 0.25 |
| Volume of IgG–FITC (μL) | 500 |
| Flow rate (mL min−1) | 0.25 |
Optimization of KinExA conditions and its running
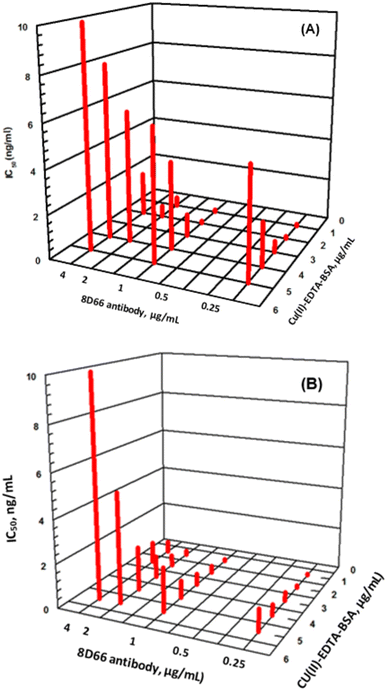
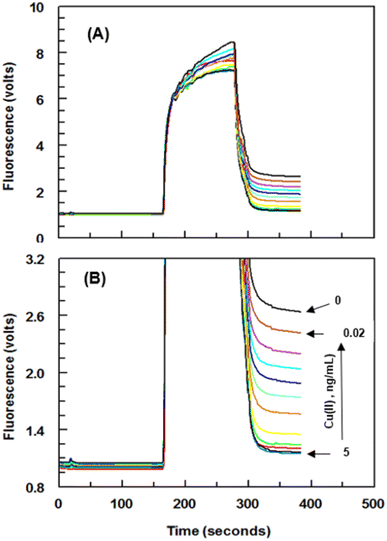
Series of experiments were carried out for establishment of the optimum concentrations of Cu(II)–EDTA–BSA conjugate required for coating onto PMMA beads and that of 8D66 antibody for effective competitive binding reactions. The optimum concentrations were those yielded the lowest IC50 value. These concentrations were 1 and 0.25 μg mL−1 for Cu(II)–EDTA–BSA and 8D66 antibody, respectively (Fig. 3B). The optimum coating of Cu(II)–EDTA–BSA on PMMA beads was achieved when the coating time was 2 h at 37 °C. The other experimental conditions were also optimized, and the optimum values are given in Table 1. Under these optimum conditions, the KinExAgram (instrument response as a function of time) for running varying concentrations of Cu(II) (0.02–5 ng mL−1) was generated (Fig. 4). The segment of the KinExAgram from 0–165 s corresponds to the background signals which generated while the flow of immune complex solution through PMMA beads located in the instrument flow cell. The segment from 166–280 s corresponds to the flow of IgG–FITC through the beads and its capturing by 8D66 antibody which has already been captured by Cu(II)–EDTA–BSA coated onto PMMA beads. This process is followed by a washing step at 281–385 s to remove any excess of IgG–FITC molecules trapped onto the beads. Accordingly, the most top curve in the KinExAgram (Fig. 4A and B) corresponds to the blank solution (0 concentration of Cu(II)), and the most bottom one corresponds to the highest Cu(II) concentration (5 ng mL−1).Validation of FIA and KinExA
Analytical recoveries studies were performed by adding various known amounts of atomic absorption grade Cu(II) to food samples. Each sample was subsequently treated and analyzed for Cu(II) content by both FIA and KinExA, as described in the Experimental section. The determined concentrations were treated according to the following equation to calculate the resulting percent recovery: percent recovery = food sample concentration/buffer sample concentration × 100. The analytical recovery was 96.4–105.2 (±1.5–5.9) and 96.7–105.1 (±1.2–5.3) for FIA and KinExA, respectively (Table 2).
| Added (ng mL−1) | Recoverya (% ± CV) | |||
|---|---|---|---|---|
| Water | Tomato paste | Bovine liver | Tea | |
| a Values are mean of 3 determinations. | ||||
| FIA | ||||
| 1.25 | 105.2 ± 5.4 | 104.3 ± 2.5 | 101.5 ± 2.7 | 104.3 ± 2.8 |
| 2.5 | 96.4 ± 4.2 | 101.5 ± 3.8 | 99.2 ± 4.5 | 102.6 ± 4.1 |
| 5 | 101.8 ± 4.1 | 99.7 ± 4.2 | 96.8 ± 5.6 | 99.8 ± 5.2 |
| 10 | 98.6 ± 5.9 | 97.8 ± 1.5 | 104.5 ± 4.8 | 97.4 ± 3.7 |
| 20 | 102.6 ± 6.2 | 101.4 ± 2.8 | 105.1 ± 3.9 | 101.6 ± 2.5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| KinExA | ||||
| 0.25 | 103.5 ± 4.2 | 102.5 ± 5.1 | 99.5 ± 3.8 | 103.5 ± 3.6 |
| 0.5 | 102.6 ± 5.3 | 101.6 ± 4.6 | 98.6 ± 2.9 | 101.9 ± 2.8 |
| 1 | 96.7 ± 4.9 | 99.8 ± 2.8 | 105.1 ± 1.7 | 100.7 ± 1.2 |
| 2 | 102.5 ± 4.8 | 97.9 ± 3.6 | 104.3 ± 4.8 | 99.2 ± 3.7 |
| 4 | 97.8 ± 4.2 | 104.1 ± 4.2 | 101.8 ± 3.4 | 102.4 ± 4.8 |
| Metal ions | FIA | KinExA | ||
|---|---|---|---|---|
| IC50a (ng mL−1) | Cross reactivityb (%) | IC50a (ng mL−1) | Cross reactivityb (%) | |
| a IC50 is the concentration of metal–EDTA complex which inhibits the color formation in the competitive immunoassay by 50%.b Calculated as IC50 [metal ion]/IC50 [Cu(II)] × 100.c Values are mean of duplicate determinations.d Other metals a were Cd(II), Pb(II), Mn(II), Mg(II), Fe(III), Al(III), Ag(I), Co(II), and Ca(II).e ND = not determined because it was more than 200 μg mL−1. | ||||
| Cu(II) | 2.4c | 100c | 0.2c | 100c |
| Zn(II) | 36.4 | 6.6 | 3.8 | 5.3 |
| Ni(II) | 192.1 | 1.3 | 18.5 | 1.08 |
| Hg(II) | 233.9 | 1.03 | 48.2 | 0.41 |
| Other metalsd | NDe | ND | NDd | ND |
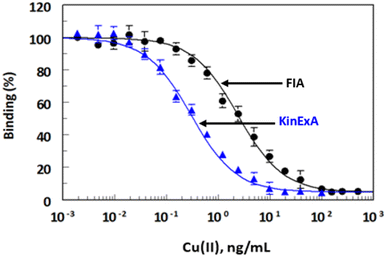
Comparison of FIA and KinExA with ICP-AES
In a series of experiments, drinking water samples were spiked with varying concentrations of Cu(II), and the samples were independently analyzed for their Cu(II) contents by FIA, KinExA, and inductively plasma coupled atomic emission spectrometry (ICP-AES). The results obtained by FIA and KinExA were plotted as a function of those measured by ICP-AES. The results were statistically compared, and it was found that concentrations measured by both FIA and KinExA correlated very well with those measured by ICP-AES. Linear regression analysis of the results yielded linear equations:| For FIA: Y = −2.32 + 0.99X (r = 0.9969) |
| For KinExA: Y = 1.85 + 1.04X (r = 0.9982) |
The high values of correlation coefficient (r), slopes (approximately 1), and low coefficient of variations (<6%) revealed the reliability (high accuracy and precision) of the results obtained by FIA and KinExA. In addition, the results confirmed the successful applicability of both FIA and KinExA for determination of Cu(II) in samples.
Comparative evaluation of FIA and KinExA
A comparative assessment of the analytical efficiency of FIA and KinExA was conducted. The results proved that KinExA had better sensitivity (lower LOD) than FIA. The better sensitivity of KinExA was due to the fact that the surface area of PMMA (∼260 mm2) is higher than that of the microwell of FIA plate (64 mm2).39 The high surface area of PMMA beads facilitated capturing of higher number of 8D66 antibody molecules and ultimately attenuated the fluorescence signals. In addition, the brief short time in which 8D66 antibody passes through the capture reagent (immobilized Cu(II)–EDTA–BSA) minimized the competitive capturing capability of Cu(II)–EDTA–BSA. Both FIA and KinExA showed comparable accuracies as recovery values were 96.4–105.2 and 96.7–103.5% for FIA and KinExA, respectively.Regarding the precision, KinExA gave better precision than FIA did, as evident from the lower variation coefficients. KinExA gave better precision because it used PMMA beads with higher surface area, and thus its precision depended on only the concentrations of 8D66 and IgG–FITC antibodies which were automatically drawn by the KinExA instrument with high precision. In FIA, the precision depended mostly on the uniformity in the quantity of Cu(II)–EDTA–BSA immobilized onto the microwells of FIA plates. These quantities are transferred manually to the wells resulting in some differences in their uniformity and accordingly the precision of the assay.
In KinExA, the reagents passed through PMMA beads with high flow rate, and accordingly the mass transport limitation, encountered in FIA, has been minimized.40,41
Advantages of FIA and KinExA
Both FIA and KinExA offered several advantages; these advantages are summarized in the following points. Both FIA and KinExA, being involved measuring fluorescent signals of strongly fluorescent labels, are highly sensitive allowing for the detection of metal ions at very low concentrations. The use of fluorescent labels in the immunoassay provides a strong signal amplification, enhancing the sensitivity of the technique. Both FIA and KinExA offered excellent selectivity, enabling the specific detection of target Cu(II) ions in presence of other metal ions. Both FIA and KinExA are high-throughput techniques that allows for simultaneous analysis of multiple samples. Both FIA and KinExA are cost-effective as both methods use small samples and cheap commercially reagents. Both FIA and KinExA, rather than other methods, did not involve extraction and/or pre-concentration procedure.Comparison of FIA and KinExA with reported methods
In literature, there several methods for detection and quantitation of Cu(II) in different food samples.38,42–48 These methods were atomic absorption spectrometry (AAS)-cloud point extraction,38 AAS-solid phase extraction,42 flame atomic spectrometry (FAS)-thin film microextraction,43 FAS-dispersive liquid–liquid microextraction,44 spectrophotometry,45,46 potentiometry,47 and ELISA.48 Most of these methods required large sample size (0.2–1 g and 8–100 mL) and involved multiple tedious steps for sample preparation such as extraction by different techniques and/or preconcentration. Comparing with these methods, the proposed FIA and KinExA, small sample size and simple sample preparation steps. In regard to the sensitivity, the proposed FIA and KinExA offered higher sensitivity than all the reported methods including ELISA.48 Since FIA involved simple batch microwell analysis with simple procedures, and KinExA involved automated flow-based analysis, they have much higher throughputs than the other reported methods which involved sequential analysis.42–48Conclusion
Two different fluorescent immunoassays were developed and validation of determination of Cu(II) in drinking water. These assays were microwell-based FIA and immunosensor-based KinExA. Validation studies of these assays confirmed their applicability for the accurate, precise, and selective determination of Cu(II) in food samples without interference from the commonly encountered other meta ions. Comparative evaluation of FIA and KinExA revealed that KinExA is superior to FIA in terms of sensitivity and precision, whereas both assays have comparable accuracy. Both assays are likely anticipated to contribute to monitoring of Cu(II) levels in foods in order significantly enhance the food's safety and decrease the risk of exposure to Cu(II)'s side effects. The use of these immunoassay systems for determination of copper in food circumvents many problems associated with the present technologies, which require sophisticated equipment in a central facility.Abbreviations
| Cu(II) | Copper ions |
| FIA | Fluoroimmunoassay |
| KinExA | Kinetic exclusion assay |
| EDTA | Ethylenediaminetetraacetic acid |
| BSA | Bovine serum albumin |
| 8D66 | Monoclonal antibody recognizing Cu(II)–EDTA complex |
| GAPs | Good agricultural practices |
| WHO | World Health Organization |
| EPA | U.S. Environmental Protection Agency |
| FNB | Food and Nutrition Board |
| FAO | Food and Agriculture Organization |
| ICP | Inductively coupled plasma |
| AES | Atomic emission spectroscopy |
| IgG–FITC | IgG conjugated with fluorescein isothiocyanate |
| PMMA | Polymethylmethacrylate beads |
| HBS | 4-(2-Hydroxyethyl)-1-piperazine ethane sulfonic acid |
| LOD | Limit of detection |
Conflicts of interest
The authors report no conflicts of interest for this work.Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSPD2023R944), King Saud University, Riyadh, Saudi Arabia. for funding this work.References
- E. J. Underwood, Trace elements in human and animal nutrition, Academic Press, New York, 4th edn, 1977, pp. 56–108 Search PubMed.
- R. A. Goyer, Toxic effects of metals, in Casarrett and Doull's toxicology: the basic science of poisons, ed. M. O. Amdur, J. Doull and C. D. Klaassen, Pergamon Press, New York, 4th edn, 1991, pp. 623–680 Search PubMed.
- G. E. Lagos, L. C. Maggi, D. Peters and F. Reveco, Model for estimation of human exposure to copper in drinking water, Sci. Total Environ., 1999, 239, 49–70 CrossRef CAS PubMed.
- Copper Development Association Inc., Uses of Copper Compounds: Agricultural Uses, available from: https://copper.org/resources/properties/compounds/agricultural.php, accessed on June 20, 2023.
- M. Sharif, M. Aziz-Ur Rahman, B. Ahmed, R. Z. Abbas and F.-U. Hassan, Copper Nanoparticles as Growth Promoter, Antioxidant and Anti-Bacterial Agents in Poultry Nutrition: Prospects and Future Implications, Biol. Trace Elem. Res., 2021, 199, 3825–3836 CrossRef CAS PubMed.
- F. Pizarro, M. Olivares, R. Uauy, P. Contreras, A. Rebelo and V. Gidi, Acute gastrointestinal effects of graded levels of copper in drinking water, Environ. Health Perspect., 1999, 107, 117–121 CrossRef CAS PubMed.
- M. L. Schilsky, Wilson disease: genetic basis of copper toxicity and natural history, Semin. Liver Dis., 1996, 16, 83–95 CrossRef CAS PubMed.
- I. H. Scheinberg and I. Steernlieb, Is non-Indian childhood cirrhosis caused by excess dietary copper?, Lancet, 1994, 344, 1002–1004 CrossRef CAS PubMed.
- A. Pandit and S. Bhave, Present interpretation of the role of copper in Indian childhood cirrhosis, Am. J. Clin. Nutr., 1996, 63, 830S–8305S CrossRef CAS PubMed.
- T. Müller, W. Müller and H. Feichtinger, Idiopathic copper toxicosis, Am. J. Clin. Nutr., 1998, 67, 1082S–1086S CrossRef PubMed.
- G. Zhong, Y. He, F. Wan, S. Wu, X. Jiang, Z. Tang and L. Hu, Effects of long-term exposure to copper on the Keap1/Nrf2 signaling pathway and Msr-related redox status in the kidneys of rats, Biol. Trace Elem. Res., 2021, 199, 4205–4217 CrossRef CAS PubMed.
- N. Husain, S. Hasan, A. A. Khan and R. Mahmood, Copper chloride inhibits brush border membrane enzymes, alters antioxidant and metabolic status and damages DNA in rat intestine: a dose-dependent study, Environ. Sci. Pollut. Res. Int., 2021, 28, 43711–43724 CrossRef CAS PubMed.
- B. Niu, B. C. Mackness, J. A. Zitzewitz, C. R. Matthews and M. L. Gross, Trifluoroethanol partially unfolds G93A SOD1 leading to protein aggregation: a study by native mass spectrometry and FPOP protein footprinting, Biochemistry, 2020, 59, 3650–3659 CrossRef CAS PubMed.
- A. Khan, J. P. Dobson and C. Exley, Redox cycling of iron by Aβ, Free Radical Biol. Med., 2006, 40, 557–569 CrossRef CAS PubMed.
- H. A. Qazmooz, H. N. Smesam, R. F. Mousa, H. K. Al-Hakeim and M. Maes, Trace element, immune and opioid biomarkers of unstable angina, increased atherogenicity and insulin resistance: results of machine learning, J. Trace Elem. Med. Biol., 2021, 64, 126703 CrossRef CAS.
- M. A. Ribeiro Sant'Ana, T. C. de Carvalho and I. F. da Silva, Concentration of heavy metals in UHT dairy milk available in the markets of Sao Luis, Brazil, and potential health risk to children, Food Chem., 2021, 346, 128961 CrossRef PubMed.
- G. Jo and T. I. Todorov, Distribution of nutrient and toxic elements in brown and polished rice, Food Chem., 2019, 289, 299–307 CrossRef CAS PubMed.
- J. Luo, T. Zhu, X. Wang, X. Cheng, Y. Yuan, M. Jin, M. B. Betancor, D. R. Tocher and Q. Zhou, Toxicological mechanism of excessive copper supplementation: effects on coloration, copper bioaccumulation and oxidation resistance in mud crab Scylla paramamosain, J. Hazard. Mater., 2020, 395, 122600 CrossRef CAS PubMed.
- O. Campana, S. L. Simpson, D. A. Spadaro and J. Blasco, Sub-lethal effects of copper to benthic invertebrates explained by sediment properties and dietary exposure, Environ. Sci. Technol., 2012, 46, 6835–6842 CrossRef CAS.
- G. A. Draghici, C. Dehelean, I. Pinzaru, D. M. Bordean, A. Borozan, A. M. Tsatsakis, L. Kovatsi and D. Nica, Soil copper uptake by land snails: a semi-field experiment with juvenile Cantareus aspersus snails, Environ. Toxicol. Pharmacol., 2019, 72, 103243 CrossRef CAS PubMed.
- P. Liao, X. Shu, M. Tang, B. Tan and Y. Yin, Effect of dietary copper source (inorganic vs. chelated) on immune response, mineral status, and fecal mineral excretion in nursery piglets, Food Agric. Immunol., 2018, 29, 548–563 CrossRef CAS.
- A. P. Perestrelo, G. Miranda, M. I. Goncalves, C. Belino and R. Ballesteros, Chronic copper sulfate poisoning, European Journal of Case Reports in Internal Medicine, 2021, 8, 002309 Search PubMed.
- K. Ghazaryan, H. Movsesyan, N. Ghazaryan and B. A. Watts, Copper phytoremediation potential of wild plant species growing in the mine polluted areas of Armenia, Environ. Pollut., 2019, 249, 491–501 CrossRef CAS.
- NIH (National Institute of Health), Office of Dietary Supplements, Copper: Fact Sheet for Health Professionals. Available from: https://ods.od.nih.gov/factsheets/Copper-HealthProfessional/, accessed on June 20, 2023.
- L. Fu and S. Shi, A novel strategy to determine the compositions of inorganic elements in fruit wines using ICP-MS/MS, Food Chem., 2019, 299, 125172 CrossRef CAS PubMed.
- E. M. Papaslioti, A. Parviainen, M. J. R. Alpiste, C. Marchesi and C. J. Garrido, Quantification of potentially toxic elements in food material by laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) via pressed pellets, Food Chem., 2019, 274, 726–732 CrossRef CAS PubMed.
- L. Hong-lin, M. Qing, Z. Xin, Y. Yu-long and T. Hua-rong, Inductively coupled plasma mass spectrometry (ICP-MS) and inductively coupled plasma optical emission spectrometer (ICP-OES)-based discrimination for the authentication of tea, Food Control, 2021, 123, 107735 CrossRef.
- F. Szurdoki, L. Jaeger, A. Harris, H. Kido, I. Wengatz, M. H. Goodrow, A. Szekacs, M. Wortberg, J. Zheng, D. W. Stoutamire, J. R. Sanborn, S. D. Gilman, A. D. Jones, S. J. Gee, P. V. Choudary and B. D. Hammock, Rapid assays for environmental and biological monitoring, J. Environ. Sci. Health, Part B, 1996, 31, 451–458 CrossRef CAS PubMed.
- D. A. Blake, R. C. Blake II, M. Khosraviani and A. R. Pavlov, Immunoassays for metal ions, Anal. Chim. Acta, 1998, 376, 13–19 CrossRef CAS.
- Y. Wang, C. Zhang and F. Liu, Antibody developments for metal ions and their applications, Food Agric. Immunol., 2020, 31, 1079–1103 CrossRef.
- H. Yang, L.-Q. Cheng, K.-R. Kang, D.-L. Hao, L.-R. Zhu, Y. Liu, J.-J. Xiang and J.-F. Huang, Rapid immunological detection of copper ions using fluorescence immunochromatographic assay, Food Agric. Immunol., 2019, 30, 60–73 CrossRef CAS.
- P. Chakrabarti, F. M. Hatcher, R. C. Blake II, P. A. Ladd and D. A. Blake, Enzyme immunoassay to determine heavy metals using antibodies to specific metal-EDTA complexes: optimization and validation of an immunoassay for soluble indium, Anal. Biochem., 1994, 217, 70–75 CrossRef CAS PubMed.
- A. F. Habeeb, Determination of free amino groups in proteins by trinitrobenzenesulfonic acid, Anal. Biochem., 1966, 14, 328–336 CrossRef CAS PubMed.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- S. Yenisoy-Karakas, Estimation of uncertainties of the method to determine the concentrations of Cd, Cu, Fe, Pb, Sn and Zn in tomato paste samples analysed by high resolution ICP-MS, Food Chem., 2012, 132, 1555–1561 CrossRef CAS.
- S. Branislav, V. Drangan, S. Zdenka, P. Nenad, J. Sasa, S. Slavoljub, B. Milica and A. Vesna, Determination of sensory properties and levels of trace elements during storage of canned meat products, J. Food Process. Preserv., 2021, 45(3), e15278 Search PubMed.
- M. M. Hamidaddin, H. AlRabiah and I. A. Darwish, Development and comparative evaluation of two immunoassay platforms for bioanalysis of crizotinib: a potent drug used for the treatment of non-small cell lung cancer, Talanta, 2019, 201, 217–225 CrossRef CAS.
- X. Guoqiang, W. Shengping, J. Xiuming, L. Xing and H. Lijun, Determination of trace copper(II) in food samples by flame atomic absorption spectrometry after cloud point extraction, Iran, J. Chem. Chem. Eng., 2021, 30, 101–107 Search PubMed.
- R. C. Blake, A. R. Pavlov and D. A. Blake, Automated kinetic exclusion assays to quantify protein binding interactions in homogeneous solution, Anal. Biochem., 1999, 272, 123–134 CrossRef CAS PubMed.
- H. Nygren and M. Stenberg, Calibration by ellipsometry of the enzyme linked immunosorbent assay, J. Immunol. Methods, 1985, 80, 15–24 CrossRef CAS PubMed.
- A. Schots, F. J. Gommers and E. Egberts, Quantitative ELISA for the detection of potato cyst nematodes in soil samples, Fundam. Appl. Nematol., 1992, 15, 55–61 Search PubMed.
- G. Xiang, Y. Zhang, X. Jiang, L. He, L. Fan and W. Zhao, Determination of trace copper in food samples by flame atomic absorption spectrometry after solid phase extraction on modified soybean hull, J. Hazard. Mater., 2010, 179, 521–525 CrossRef CAS PubMed.
- Z. Mehrani, H. Ebrahimzadeh, A. A. Asgharinezhad and E. Moradi, Determination of copper in food and water sources using poly m-phenylenediamine/CNT electrospun nanofiber, Microchem. J., 2019, 149, 103975 CrossRef CAS.
- Z. Al Othman, Y. E. Unsal, M. Habila, A. Shabaka, M. Tuzen and M. Soylak, Determination of copper in food and water by dispersive liquid-liquid microextraction and flame atomic absorption spectrometry, Anal. Lett., 2015, 48, 1738–1750 CrossRef CAS.
- S. S. Alharthi and H. M. Al-Saidi, Spectrophotometric determination of trace concentrations of copper in waters using the chromogenic reagent 4-amino-3-mercapto-6-[2-(2-thienyl)vinyl]-1,2,4-triazin-5(4H)-one: synthesis, characterization, and analytical applications, Appl. Sci., 2020, 10, 3895 CrossRef CAS.
- M. Lu, X. Fu, H. Xie, M. Liu, P. Wei, W. Zhang, Y. Xie and Y. Qi, Colorimetric determination of copper ion based on the silver-coated gold nanobipyramids, J. Food Compos. Anal., 2023, 120, 105363 CrossRef CAS.
- M. E. Mohamed, E. Y. Frag and M. E. Brawy, Rapid potentiometric sensor for determination of Cu(II) ions in food samples, Microchem. J., 2021, 164, 106065 CrossRef.
- J. Zhang, L. Xu, H. Jiang, C. Xu, W. Liu, T. Wu and H. Kuang, Development of an immunoassay for the detection of copper residues in pork tissues, Biosensors, 2021, 13, 235 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |