Open Access Article
Open Access ArticleSynthesis, characterization, molecular docking, and antimicrobial activities of dinuclear nickel(II), palladium(II), and platinum(IV) complexes†
Reem M. A. Ebrahim *ad,
Abubakar Abdelbagib,
Yousif Sulfab*c,
Omer Abdalla Ahmed Hamdic,
Samah A. Shokrib and
Elmugdad A. Alid
*ad,
Abubakar Abdelbagib,
Yousif Sulfab*c,
Omer Abdalla Ahmed Hamdic,
Samah A. Shokrib and
Elmugdad A. Alid
aBiotechnology Department, Africa City of Technology, Khartoum, Sudan. E-mail: reemaaboalsoud@gmail.com
bPharmaceutical Microbiology Department, Faculty of Pharmacy, Al-Neelain University, Khartoum, Sudan
cChemistry Department, Faculty of Science and Technology, Al-Neelain University, Khartoum, Sudan. E-mail: yousifsulfab@yahoo.com
dChemistry Department, Faculty of Science, Sudan University of Science and Technology, Khartoum, Sudan
First published on 14th September 2023
Abstract
New nickel(II), palladium(II), and platinum(IV) complexes were synthesized by reacting the metal ions with benzidinedioxime in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole ratio. The CHN elemental analysis, spectroscopic analyses, and powder X-ray diffraction (PXRD) results showed that two Ni(II) and two Pd(II) ions coordinated to two benzidinedioxime ligands via the nitrogen atoms of both oxime groups and the two azomethine nitrogen atoms. In the case of the dinuclear platinum(IV) complex, however, each Pt(IV) is coordinated with the two oxygen atoms of the oxime group and the two azomethine nitrogen atoms of the ligand. Both elemental analyses and PXRD indicated that the complex ions of Ni(II) and Pt(IV) have distorted octahedral geometry, whereas Pd(II) has a square planar geometry. Molecular docking studies showed that the nickel(II) complex is the most potent dual DHPS/DHFR bacterial inhibitor. The receptor of the DHPS enzyme (3ZTE) showed the best interaction with the nickel(II) complex when compared to a receptor of the DHFR enzyme (3FRB). All the synthesized complexes and ligand exhibited significant results against PS. Aeruginous than their corresponding SMX–TMP drug. Among the three synthesized complexes, the nickel(II) complex possessed the highest antimicrobial activities against tested microorganisms.
1 mole ratio. The CHN elemental analysis, spectroscopic analyses, and powder X-ray diffraction (PXRD) results showed that two Ni(II) and two Pd(II) ions coordinated to two benzidinedioxime ligands via the nitrogen atoms of both oxime groups and the two azomethine nitrogen atoms. In the case of the dinuclear platinum(IV) complex, however, each Pt(IV) is coordinated with the two oxygen atoms of the oxime group and the two azomethine nitrogen atoms of the ligand. Both elemental analyses and PXRD indicated that the complex ions of Ni(II) and Pt(IV) have distorted octahedral geometry, whereas Pd(II) has a square planar geometry. Molecular docking studies showed that the nickel(II) complex is the most potent dual DHPS/DHFR bacterial inhibitor. The receptor of the DHPS enzyme (3ZTE) showed the best interaction with the nickel(II) complex when compared to a receptor of the DHFR enzyme (3FRB). All the synthesized complexes and ligand exhibited significant results against PS. Aeruginous than their corresponding SMX–TMP drug. Among the three synthesized complexes, the nickel(II) complex possessed the highest antimicrobial activities against tested microorganisms.
Introduction
Schiff base ligands have played essential roles as excellent chelating agents due to a lone pair of electrons in an sp2 hybridized orbital of the nitrogen atom in the azomethine group.1–3 Schiff base complexes have different physical and chemical properties and pharmacological activities than their corresponding ligands. The variation of properties of their complexes depends on the type of metal used.4–7In recent years, Schiff base transition metal complexes played essential roles in different fields such as agriculture,8–10 pharmaceutics,11–13 and industry.14,15 They have biological properties, such as antibacterial, antifungal, anticancer, and herbicidal applications.16–20 The chemical structural and functional groups of Schiff base ligands play an essential role in the mechanism of several enzymes. For example, aminotransferase enzymes use pyridoxal phosphate as a cofactor.21
Dinuclear Schiff base transition metal complexes comprise two transition metal ions in the same molecular entity.22–24 The magnetic properties of dinuclear complexes can differ significantly from those of mononuclear complexes. These new properties depend on the nature and magnetic interaction between the metal ions through the bridging ligands.22–24 Several dinuclear complexes having N4O4 Schiff-base ligands have been reported, and the antibacterial activity of these complexes screened against Gram-positive and Gram-negative strains.25–27 Compared to standard drugs, the complexes most potent activity was observed against all tested strains.27–30
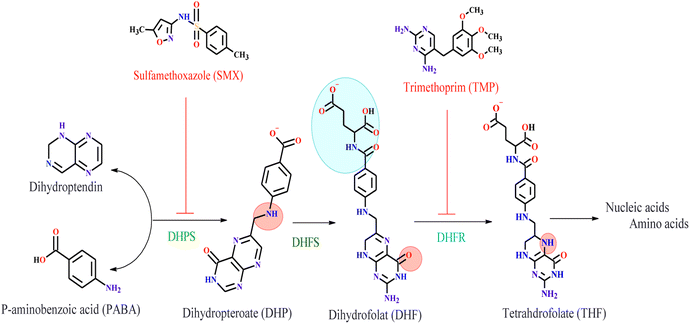
The Sulfamethoxazole–Trimethoprim (SMX–TMP) antibiotic drug is an inhibitor of two enzyme families involved in synthesizing tetrahydrofolate (THF). These are dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR). Sulfamethoxazole (SMX) binds to bacterial dihydropteroate synthase (DHPS) to the conversion of para-aminobenzoic acid (PABA) to dihydropteroate (DHP) in the process of THF formation. DHPS inhibition leads to defective thymidine biosynthesis and slows or blocks folic acid biosynthesis.31,32 The second enzyme is Dihydrofolate Reductase (DHFR) is a vital antibiotic target in folic acid biosynthesis.33 It catalyzes the production of nicotinamide adenine dinucleotide phosphate (NADPH) by the reduction of dihydrofolate (DHF) to tetrahydrofolate (THF).32,34,35 Trimethoprim (TMP) is a drug used to treat urinary tract infections (UTIs). It inhibits bacterial DHFR.32,36 TMP is highly effective in S. aureus DHFR (SaDHFR) than in human DHFR, which leads to preferential inhibition of bacterial folic acid synthesis.31,32 The key enzymes involved in bacterial folic acid biosynthesis and the antibiotics that inhibit them are shown in Fig. 1. The dual targeting of two enzymes made SMX–TMP an excellent choice for treating bacterial infections. However, this has resulted in the development of antimicrobial resistance,32 encouraging us to synthesize new compounds containing dinuclear metal ions and test them as bacterial inhibitors for DHPS and DHFR enzymes.
Materials and methods
Materials
Benzidine (99%, Riedel-de-Haen AG, Germany), diacetylmonoxime (99%, Trust Chemical Laboratories, India). Platinum(IV) chloride (assay = 99.99%, Onyxment, Poland). Nickel(II) chloride hexahydrate (assay = 98.0%, Loba Chemie, Spain). Palladium(II) acetate (assay = 98%, Sigma-Aldrich, Germany). Ethanol (99.9%, Duksan, Germany). Methanol (assay = 99.8%, Sigma-Aldrich, Germany). Diethyl ether (97.5%, Alpha Chemika, India). Dimethyl sulphoxide (99.9, Alpha Chemika, India). Glacial acetic acid (85%, Alpha Chemika, India) were used as received.Synthesis of the benzidinedioxime ligand (N2,N2′-biylbis(N3-hydroxybutane-2,3-diimine))
Benzidine (2.00 g, 10 mmol) was dissolved in 50 mL of absolute hot ethanol, and 2,4 butene-dione monoxime (2.22 g, 20 mmol) was dissolved in 20 mL of absolute ethanol. A few drops of glacial acetic acid were added to the mixture. The mixture was refluxed by stirring for 8 hours. The precipitate was filtrated, washed with cold ethanol and ether, and left to dry in the air.Synthesis of the metal complexes
The metal complexes were prepared by adding a hot solution of the appropriate metal salt (2.5 mmol) in ethanol to a hot benzidinedioxime ligand (2.5 mmol) solution. The resulting mixture was stirred under reflux for 20 hours, and the complexes precipitated. The precipitate was filtered off and washed with cold ethanol and diethyl ether several times.Elemental analysis measurement
The elemental analyses of benzidinedioxime and its complexes were determined using Flash EA 1112 CHN.1H-Nuclear magnetic resonance (1HNMR)
The 1H NMR spectrum was obtained for the dioxime benzidine and complexes solution in DMSO D6 (0.05 mM) using a Bruker DPX 250 and 300 MHz spectrometer. Standard pulse sequences were used for the 1H one-bond and long-range HMBC spectra.Electronic properties
The electronic spectra were obtained using a Shimadzu 160 UV-visible spectrophotometer in the range (of 200–1000 nm) using a quartz cell of 1.0 cm length with a concentration of 1.43 mmol L−1 of samples in dimethyl sulfoxide at 25 °C.Powder X-ray diffraction (PXRD)
The powders' crystalline phases and unit cell parameters were determined by PXRD using a Shimadzu 7000 X-ray diffractometer. At room temperature, data were collected for a 2θ range of 20–80° at a step size of 0.02 and a 5 seconds count time.Molecular docking
The molecular docking studies were carried out using Molecular Operating Environment (MOE 2014). All the minimizations were performed with MOE until an RMSD gradient of 0.01 kcal mol−1 Å−1 with an MMFF94X force field was achieved, and the partial charges were automatically calculated. Docking simulations were performed using the crystal structure of DHPS enzyme (PDB ID![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3TYE) in complex to XTZ and the crystal structure of DHFR enzyme (PDB ID
3TYE) in complex to XTZ and the crystal structure of DHFR enzyme (PDB ID![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3FRB) in complex to TOP, which was obtained from Protein Data Bank. Finally, the optimized poses were ranked using the London DG free-energy estimates. Docking poses were visually inspected, and interactions with binding pocket residues were analyzed.
3FRB) in complex to TOP, which was obtained from Protein Data Bank. Finally, the optimized poses were ranked using the London DG free-energy estimates. Docking poses were visually inspected, and interactions with binding pocket residues were analyzed.
Antimicrobial activity
The antimicrobial potency of 10 mg mL−1 of synthesized compounds, trimethoprim sulfumethaxol, and Gentamycin was evaluated by cup plate method according to Seeley et al. 1975.37 Using many standards and isolated microorganisms, two Gram-positive bacteria: Staphylococcus aureus, ATCC25923, and Bacillus subtilis MCTC8236. Four Gram negative bacteria, Pseudomonas aeruginosa ATCC9017, Proteus Vulgaris ATCC6380, Salmonella typhimurium (isolate), and Escherichia coli ATCC25922 and fungi as candid albicans ATCC4430373. All strains were provided by the National Research Centre (NRC); in Khartoum, Sudan.Results and discussion
Synthesis
The yield of benzidinedioxime (3Z,2′E,3′Z)-N2,N2′-biylbis (N3-hydroxybutane-2,3-diimine) is found to be 73% and has a melting point of 242 °C. Benzidinedioxime is soluble in ethanol and DMSO.The nickel(II), palladium(II), and platinum(IV) complexes are intensely colored and stable in the solid form. Their m.p. is >360 °C. The yield of the complex of nickel(II), palladium(II), and platinum(IV) were found to be 58.0%, 83.64%, and 85.15%, respectively. All complexes are partially soluble in ethanol and are soluble in DMSO.
Fourier transform-infrared (FT-IR) analyses of ligand and complexes
FT-IR spectroscopy measurements were performed to examine the functional groups present in the ligand in the range of 4000 cm−1 to 400 cm−1.The FT-IR spectra of the benzidinedioxime are shown in Table 1. This spectrum confirms the proposed structure. After the condensation reaction of benzidine, the new peak at 1616 cm−1 indicating imine CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond appears; these assignments are in good agreement with theoretical and literature data.38–41
N bond appears; these assignments are in good agreement with theoretical and literature data.38–41
| Benzidinedioxime | Complex of Ni(II) | Complex of Pd(II) | Complex of Pt(IV) | Functional groups |
|---|---|---|---|---|
| Wave number (cm−1) | Wave number (cm−1) | Wave number (cm−1) | Wave number (cm−1) | |
| 3365, 3296 | 3433, 3336 | 3329, 3199 | — | O–H stretching vibrations of the oxime group |
| 2775 | 3032 | 3043 | 3198 | Stretching vibrations of saturated C–H |
| 1617 | 1612 | 1619 | 1607 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibrations of imine N stretching vibrations of imine |
| 1463 | 1489 | 1567 | 1496 | C–H bending vibration |
| 1362, 1258, 1121 | 1378, 1288, 1233 | 1499, 1381, 1278 | 1404, 1404, 1187 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations of the aromatic ring C stretching vibrations of the aromatic ring |
| 1017 | 1173 | 1204 | 1013 | C–C stretching vibration |
| 970 | 824 | 820 | 821 | C–N stretching vibration |
Elemental analysis measurement
The H, C, and N percentages in the ligand and complexes have been determined by CHN elemental analyzer. The results are within the accepted ±5.0% error margin. The complexes have 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 metal to ligand stoichiometry. The proposed formulae and elemental analysis data of the ligand and complexes are shown in Table 2.
1 metal to ligand stoichiometry. The proposed formulae and elemental analysis data of the ligand and complexes are shown in Table 2.
| Compounds | Experimental percentage (%) | Calculation percentage (%) | ||||
|---|---|---|---|---|---|---|
| C | H | N | C | H | N | |
| C20H22N4O2 | 70.77 | 6.43 | 15.14 | 68.5 | 6.29 | 16.00 |
| [Ni2(C20H22N4O2)2](Cl)4 (H2O)6 | 46.85 | 5.47 | 10.97 | 48.29 | 5.63 | 11.27 |
| Pd2(C20H22N4O2)2(CH3CO2)4 | 53.92 | 5.32 | 10.12 | 53.49 | 5.20 | 10.4 |
| Pt2(C20H20N4O2)2(Cl4)(H2O)4 | 40.08 | 4.12 | 8.90 | 40.10 | 4.01 | 9.36 |
Nuclear magnetic resonance
The palladium(II) complex appears to have a high-intensity peak for protons of phenyl groups as compared to the complexes of nickel(II) and platinum(IV) that due to the different shapes of the crystal, hydrogen bonds, and symmetric and asymmetric structure of the complexes.48 The change of chemical shift value and intensity of protons of aromatic complexes aromatic rings indicate the formation of coordinate bonds between para-imine groups with metals. The disappearance of protons of oxime groups indicates the coordination of deportation of the oxime groups by metal ions or the formation of hydrogen bonds.
Powder X-ray diffraction
The crystal structure of complexes bis-benzidinedioxime was determined by powder X-ray diffraction. Pattern peaks of bis-benzidinedioxime complexes display that almost all the crystals were grown in the same phase. The crystals obtained from complexes are shown to be weak diffracting and fail to provide high-resolution reflection data. As a consequence of Fig. 5ESI,† only partial structure determination was possible. The bond length and angles' accuracy has determined the crystal structure's quality. The brood peak indicated a small crystallization size in the nano-crystalline scale range.The crystal properties of benzidinedioxime and dinuclear complexes were reported in Table 3. The metal center of palladium (53) has little distortion of square planar geometry, while palladium (54) has a high distortion of square planar geometry.
| Properties | Benzidinedioxime | Bis-benzidinedioxime nickel(II) | Bis-bezidinedioxime palladium(II) | Bis-benzidinedioxime platinum(IV) |
|---|---|---|---|---|
| Space group | P1 (1) | P 21 3 | P -1 | P 41 3 2 |
| Symmetry operations | Centrosymmetric | Non-centrosymmetric | Centrosymmetric | Non-centrosymmetric |
| Lattice type | Triclinic | Cubic | Triclinic | Cubic |
| Cell volume (°A3) | 983.920 | 1022.021 | 1942.981 | 995.875 |
| Alpha | 98.853 | 90.000 | 90.705 | 90.000 |
| Beta | 91.032 | 90.000 | 93.634 | 90.000 |
| Gamma | 88.809 | 90.000 | 101.998 | 90.000 |
| A/°A | 9.96474 | 10.073 | 8.175 | 9.986 |
| B/°A | 9.84346 | 10.073 | 14.844 | 9.986 |
| C/°A | 992![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847 847 |
10.073 | 16.408 | 9.986 |
The atoms central of nickel(II) and platinum(IV) have slight distortion octahedral coordination geometry defined by two chelating atoms (benzidinedioxime ligand) and two monodentate ligands (di aqua ligands) at a complex of nickel(II) and dichloro ligands at a complex of platinum shown in Fig. 2. The selected bonds and angles of the complex of Ni(II), Pd(II), and Pt(IV) are represented in Tables 4 and 5, respectively.
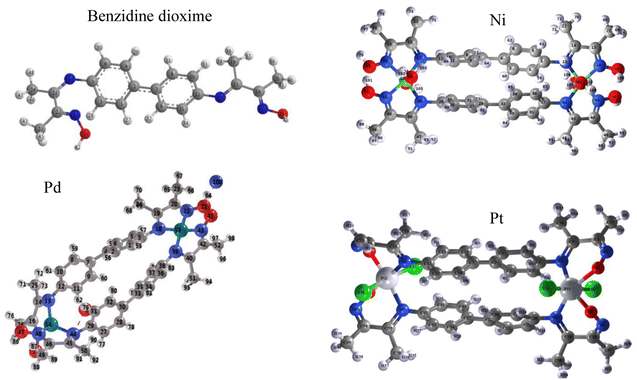 | ||
| Fig. 2 PXRD crystal structure of benzidinedioxime, bis-benzidinedioxime nickel(II), bis-benzidinedioxime palladium(II), and bis-benzidinedioxime platinum(IV). | ||
| Atom | Bond distance (°A) | Atom | Bond distance (°A) | Atom | Bond distance (°A) |
|---|---|---|---|---|---|
| Ni52-O57 | 1.86 | Pd53-N22 | 1.76 | N17-Pt54 | 2.03 |
| Ni52-O58 | 1.86 | Pd53-N39 | 1.81 | N44-Pt54 | 2.03 |
| Ni53-O54 | 1.86 | Pd53-N43 | 1.77 | N7-Pt27 | 1.93 |
| Ni53-O55 | 1.86 | Pd53-N18 | 1.81 | N34-Pt27 | 2.15 |
| N13-Ni52 | 1.83 | Pd54-N13 | 2.026 | O21-Pt27 | 1.93 |
| N16-Ni52 | 1.82 | Pd54-N17 | 2.008 | O48-Pt27 | 1.86 |
| N42-Ni52 | 1.83 | Pd54-N44 | 2.032 | O26-Pt54 | 1.87 |
| N45-Ni52 | 1.82 | Pd54-N48 | 2.015 | O53-Pt54 | 1.87 |
| N41-Ni53 | 1.82 | Pt54-Cl55 | 2.42 | ||
| N17-Ni53 | 1.91 | Pt54-Cl56 | 2.42 | ||
| N20-Ni53 | 1.89 | Pt27-Cl57 | 2.34 | ||
| N38-Ni53 | 1.91 | Pt27-Cl58 | 2.39 |
| Atom | Bond angles (°A3) | Atom | Bond angles (°A3) | Atom | Bond angles (°A3) |
|---|---|---|---|---|---|
| Ni52-N16-O56 | 125.01 | N13-Pd54-N44 | 125.74 | N7-Pt27-O21 | 86.48 |
| N16-Ni52-N42 | 160.31 | N13-Pd54-N48 | 127.76 | N7-Pt27-N34 | 103.93 |
| N16-Ni52-N5 | 88.99 | N17-Pd54-N44 | 126.59 | N7-Pt27-O48 | 171.57 |
| N16-Ni52-O57 | 85.83 | N17-Pd54-N48 | 109.67 | N7-Pt27-Cl57 | 81.12 |
| N16-Ni52-O58 | 101.76 | N18-Pd53-N39 | 91.64 | N7-Pt27-Cl58 | 92.14 |
| N17-Ni53-N20 | 82.8 | N18-Pd53-N43 | 174.05 | N17-Pt54-O26 | 88.39 |
| N17-Ni53-N38 | 111.98 | N22-Pd53-N39 | 174.15 | N17-Pt54-Cl55 | 85.86 |
| N17-Ni53-N41 | 162.69 | N22-Pd53-N43 | 90.69 | N17-Pt54-Cl56 | 89.54 |
| N17-Ni53-O54 | 92.76 | N13-Pd54-N17 | 85.96 | O21-Pt27-N34 | 167.65 |
| N17-Ni53-O55 | 79.98 | N18-Pd53-N22 | 89.45 | O21-Pt27-O48 | 85.73 |
| N41-Ni53-O54 | 99.12 | N39-Pd53-N43 | 88.82 | O21-Pt27-Cl57 | 91.95 |
| N41-Ni53-O55 | 90.41 | N44-Pd54-N48 | 84.98 | O21-Pt27-Cl58 | 96.16 |
| N42-Ni52-N45 | 83.8 | N17-Pt54-N44 | 97.01 | ||
| N42-Ni52-O57 | 77.69 | N17-Pt54-O53 | 174.61 | ||
| N52-Ni52-O58 | 96.0 | O26-Pt54-N44 | 172.24 | ||
| N20-Ni53-N38 | 162.24 | O26-Pt54-O53 | 87.5 | ||
| N20-Ni53-N41 | 84.57 | O26-Pt54-Cl55 | 92.58 | ||
| N20-Ni53-O54 | 90.28 | O26-Pt54-Cl56 | 91.75 | ||
| N20-Ni53-O55 | 99.15 | N34-Pt27-O48 | 83.45 | ||
| N38-Ni53-N41 | 82.72 | N34-Pt27-Cl57 | 83.27 | ||
| N38-Ni53-O54 | 79.57 | N34-Pt27-Cl58 | 90.15 | ||
| N38-Ni53-O55 | 93.32 | O48-Pt27-Cl57 | 95.91 | ||
| N45-Ni52-O57 | 101.27 | O48-Pt27-Cl58 | 91.91 | ||
| N45-Ni52-O58 | 85.96 | N44-Pt54-O53 | 86.75 | ||
| O57-Ni52-O58 | 169.69 | N44-Pt54-Cl55 | 82.3 | ||
| N13-Ni52-N16 | 84.25 | N44-Pt54-Cl56 | 93.86 | ||
| N13-Ni52-N2 | 108.05 | Cl57-Pt27-Cl58 | 169.13 | ||
| N13-Ni52-N45 | 160.5 | O53-Pt54-Cl55 | 90.85 | ||
| N13-Ni52-O57 | 96.48 | O53-Pt54-Cl56 | 94.07 | ||
| N13-Ni52-O58 | 77.58 | Cl55-Pt54-Cl56 | 173.59 |
Electronic properties of ligand and complexes
Ligand and complexes scanned in the range 200–1000 nm.The spectrum shows many peaks due to electronic, vibration, and rotation transitions. All compounds have a strong peak at 300 nm indicating high electronics transition in the UV region Fig. 3. The spectra showed two peaks at around 300–350 nm due to n → σ* and n → π* electronic transitions. The absorption in the 400 nm is due to π → π* electronic transition of the unsaturated conjugated compound Fig. 3.
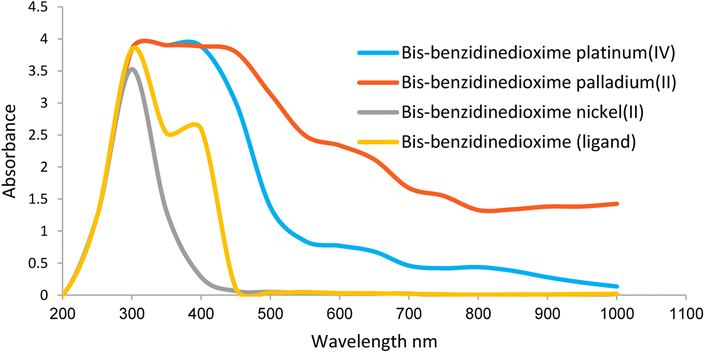 | ||
| Fig. 3 Electronic properties of benzidinedioxime, bis-benzidinedioxime nickel(II), bis benzidinedioxime palladium(II) and bis-benzidinedioxime platinum(IV). | ||
Molecular docking
Molecular docking studies using a crystal structure of DHPS (PDB ID![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3TYE) and DHFR (PDB ID
3TYE) and DHFR (PDB ID![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3FRB) obtained from the Protein Data Bank server. The result demonstrated that the benzidinedioxime, bis-benzidinedioxime nickel(II), bis-benzidinedioxime palladium(II), and bis-benzidinedioxime platinum(IV) have higher activity and lower root mean square deviation (RMSD) value against DHFR than DHPS, as shown in Table 6. Furthermore, the bis-benzidinedioxime nickel(II) complex showed the best potent inhibitor against both enzymes.
3FRB) obtained from the Protein Data Bank server. The result demonstrated that the benzidinedioxime, bis-benzidinedioxime nickel(II), bis-benzidinedioxime palladium(II), and bis-benzidinedioxime platinum(IV) have higher activity and lower root mean square deviation (RMSD) value against DHFR than DHPS, as shown in Table 6. Furthermore, the bis-benzidinedioxime nickel(II) complex showed the best potent inhibitor against both enzymes.
| Compounds | Binding energy (S) and RMSD- refine of dock benzidinedioxime and complexes with 3FRB (DHFR enzyme) | Binding energy (S) and RMSD- refine of dock benzidinedioxime and complexes with 3TYE (DHBS enzyme) | ||
|---|---|---|---|---|
| S | RMSD-refine | S | RMSD-refine | |
| Bis-benzidinedioxime nickel(II) | −21.23 | 1.874 | −22.00 | 1.779 |
| Bis-benzidinedioxime palladium(II) | −18.80 | 1.694 | −19.16 | 2.952 |
| Bis-benzidinedioxime platinum(IV) | −16.13 | 1.286 | −14.49 | 1.559 |
| Benzidinedioxime | −10.47 | 1.340 | −9.386 | 1.346 |
| XTZ | — | — | −14.054 | 1.608 |
| TMP | −11.27 | 1.217 | — | — |
| Gentamycin | −17.63 | 1.894 | −15.91 | 1.749 |
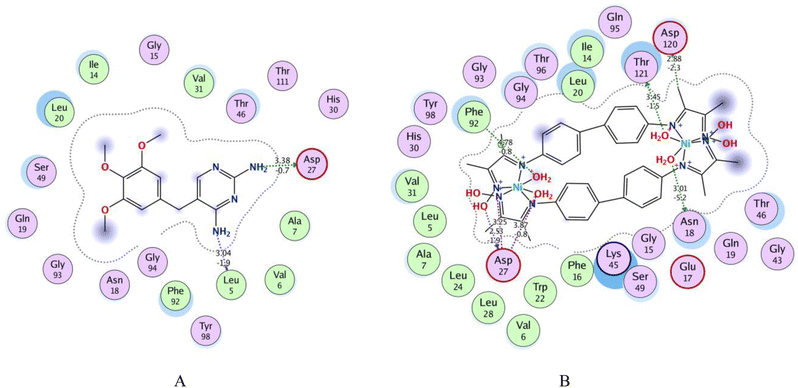
Fig. 4 shows 2D interactions of the co-crystallized Trimethoprim (TMP) ligand and complexes compound within the active site of DHFR. 2D interaction of the TMP ligand displays that the two amino groups at the pyrimidine create hydrogen bond donors with Asp 27 and Leu 5. Thr 46, Val 31, and Ser 49 are attached to the TMP ligand by Vander Waal interaction. The closed contour indicated the lower solvent exposure to terminal atoms Fig. 4(A).
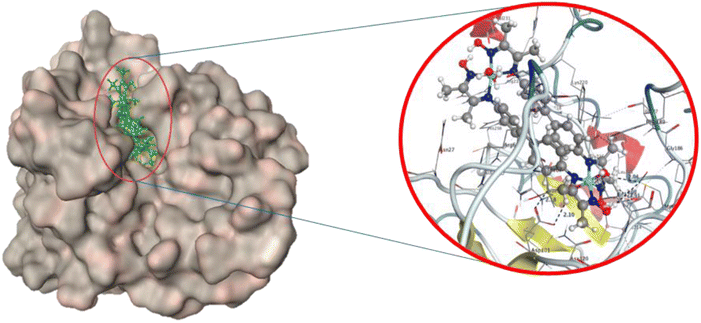
Fig. 4B and 5 show that bis-benzidinedioxime nickel(II) can fit into a DHFR pocket with four hydrogen bond donors. One of the donor hydrogen bonds was created between the methyl group and Asp 120. In contrast, three hydrogen bond donors formed between Thr121, Asn 18, and Asp 27 with the water molecular, which bonded by a coordinated bond with the transition metal (nickel(II)). The acceptor hydrogen bond is produced amongst the oxygen atom at the aqua ligand with Asp 120. The imine group bonded by an ionic bond with Asp 27. Arene–H connected with bis-benzidinedioxime nickel(II) with Phe 92. Additionally, Vander Waal interactions formed between complex and hydrophobic residues. Losing the contour around a compound indicated that the intermediate solvent interacted with terminal atoms. The docking revealed that the bis-benzidinediooxime of palladium(II) interacts with Asn 18, and Asp 27 amino acid residues of DHFR enzyme via donor hydrogen bonds, which formed between the phenyl ring and hydroxyl group respectively. Arene–H linked with phenyl ring and Thr 46 as well as many of Vander Waal interactions formed between Asn 18, Glu 17, Thr 46, Lys 45, Gly 94, Leu 20, Ser 49. Phe 16, Ile 50, Leu 28, Phe 92, Val 31, Val 6, Ala 7, Ile 14, Thr 121 and Asp 120 Fig. 6ESI(D).† Missing the contour around a compound indicated that the intermediate solvent was exposed to terminal atoms. The bis-benzidinedioxime platinum(IV) interacts with the active site via an ionic bond that is formed between the imine group and Asp 120 and a donor hydrogen bond between the benzene ring and chloride ion with Asp 120 and Ile 14 respectively. Arene–H is linked between the benzene ring and Asn 18, and another arene–H is coupled between the alkyl group and Phe 92. The platinum(IV) also provides a molecular bulk necessary for Vander Waal interactions with Lys 45, Asp 120, Thr 121, Asn 18, Ser 49, Ile 14, Leu 20, Ile 50, Leu 28, Tyr 98, Phe 92, Val 31, Ala 7, Gly 94, Thr 96, Gln 95 and Thr 46 Fig. 6ESI(E).† Losing the contour around a compound indicated that the intermediate solvent interacted with terminal atoms. The oxime group of benzidinedioxime has formed a donor hydrogen bond with Asp 120 furthermore, Vander Waal interactions formed between benzidinedioxime and Thr 121, Ile 14, Thr 46, Ser 49, Ile 50, Leu 28, Val 31, Leu 54, Phe 92, Leu 20, and Asn 18 Fig. 6ESI(F).† Losing the contour around a compound indicated that the intermediate solvent interacted with terminal atoms.
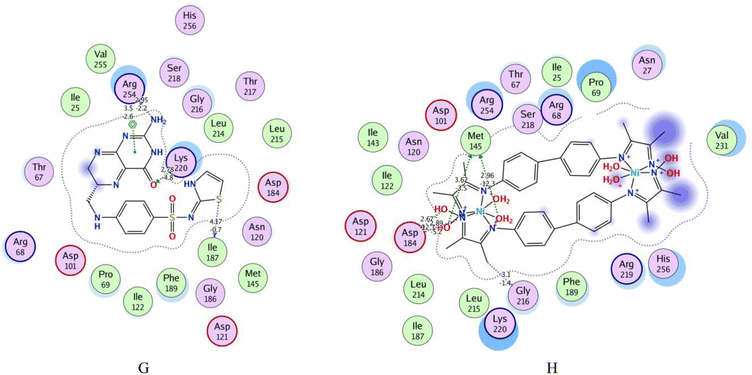
4-[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl) methyl] amino-N-(1,3-thiazol-2-yl) benzenesulfonamide (XTZ) ligand has strong interactions with the dummy atoms of the protein as shown in Fig. 6(G). The 2D interaction of the XTZ ligand displays that the crystalline ligand XTZ formed a donor hydrogen bond by the Arg 254 with an amino group and Ile 187 with a sulfur atom at the thiazole ring; furthermore, the acceptor hydrogen bond formed between the XTZ ligand with the active site. The carbonyl group at the benzene ring has a moderately strong acceptor hydrogen bond to Lys 220. Arene–H interaction formed with the benzene ring and Arg 254. Lys 220, Gly 216, Phe 189, and Arg 68 residues are interfaced with the side chains of the XTZ ligand by Vander Waal forces.
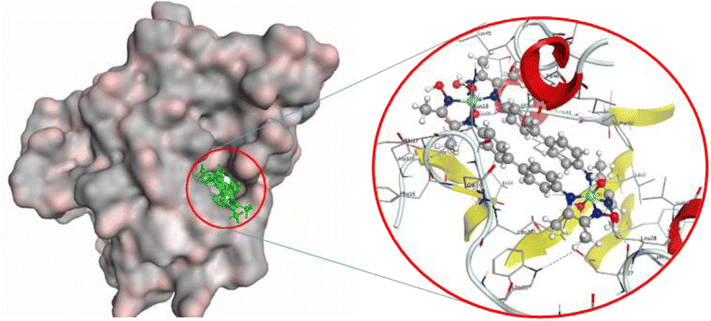
The 2D and 3D interactions of bis-benzidinedioxime nickel(II) within the active site of DHPS are shown in Fig. 6(H) and 7. The co-crystallized bis-benzidinedioxime nickel II with 3TYE has formed four hydrogen bonds between Asp 218, Gly 216, and Met 145. An ionic bond is generated between the oxime group's oxygen atoms with Asp 18. The region around the alkyl chains in the terminal of the molecule is a hydrophobic surface in the proximity of “greasy” residues, which are appeared in light green. The polar surface of bis-benzidinedioxime nickel(II) has more closely affiliated with polar or ionic residues. The loosening of contour and the high solvent exposure of the terminal atoms indicate that the oxime and alkyl groups at the end of the ligand (right) are far out of the active site. The bis-benzidinedioxime palladium(II) showed a strong binding pattern with the DHPS enzyme. The 2D interaction palladium(II) complex with an active site exhibited the ionic bond formed between the hydroxyl group and Asp 184. Both alkyl and oxime groups of complex interacted with the Asp 101, Met 145, and Gly 216 via donor hydrogen bonds. The rest of the amino acid residues Lys 220, Ser 221, Arg 68, Pro 69, Val 231, Arg 219, His 256, Ser 218, Arg 254, Gly 216, Asp 184, Asn 120, Ile 122, Thr 67 and Phe 189 formed Vander Waal interactions with palladium(II) complex Fig. 7ESI(I).† Losing the contour around a compound indicated that the intermediate solvent interacted with terminal atoms. The phenyl ring in bis-benzidinedioxime platinum(IV) engaged with Ser 221 by arene–H interaction, and also the platinum(IV) complex formed Vander Waal interactions with Ser 218, Arg 219, Lys 220, His 256, Ser 221, Gly 188, Pro 69, Phe 71, Phe 189, Arg 68, Arg 254 and Thr 67 Fig. 7ESI(J).† Losing the contour around a compound indicated that the intermediate solvent interacted with terminal atoms. The benzidinedioxime interacts with DHPS via connected arene H interaction between the phenyl group and lys 220. The amino acid residues Thr 67, Phe189, Arg 254, Ile 122, Asp 184, Asn 120, Pro 69, Lys 220, and Ser 221 interacted Vander Waal interactions with benzidinedioxime Fig. 7ESI(K).†
2D interaction of palladium(II) complex, platinum(IV), and benzidinedioxime with DHFR pocket and DHPS Pocket are available in Fig. 6ESI and 7,† respectively.
Biological activity
The synthesized ligand and complexes screened for antimicrobial activities against different bacterial species and fungi. All complexes have higher biological activities against most of the tested microorganisms than the free ligand, confirming that the transition metals increase the activity of the compounds against them.26,49–51 The high antimicrobial activity of the complexes as compared to the benzidinedioxime might be due to their highest lipophilicity. This causes an increase in their penetration ability through the lipid membrane, which could block or inhibit the growth of the microorganism.52–54 The nickel(II) complex showed the highest antibacterial activity against microorganisms of all the tested complexes. On the other hand, the platinum(IV) complex showed the lowest activity against microorganisms, as summarized in Table 7.| Bacterial and fungi | Diameter of the inhibition zone (mm) | |||||
|---|---|---|---|---|---|---|
| Ni(II) | Pd(II) | Pt(IV) | L | Drugs | ||
| Gentamycin | SMX–TMP | |||||
| E. coli | 17.3 ± 1.8 | 22.3 ± 2.2 | 0 | 15.0 ± 0.00 | 27 | 32 |
| P. vulgaris | 15.7 ± 0.89 | 10.7 ± 2.2 | 0 | 0 | 25 | 35 |
| S. Typhimurium | 16.3 ± 1.1 | 18.3 ± 1.1 | 14.0 ± 1.0 | 12.7 ± 1.8 | 26 | 35 |
| PS. areuginosa | 17.3 ± 11 | 14.0 ± 0.0 | 0 | 14.7 ± 0.4 | 27 | 0 |
| S. aruse | 17.7 ± 0.4 | 0 | 0 | 14.3 ± 1.6 | 25 | 33 |
| B. Subtilis | 19.0 ± 0.7 | 16.7 ± 2.2 | 19.3 ± 0.9 | 16.7 ± 2.2 | 26 | 21 |
| Candida | 16.3 ± 1.1 | 15.0 ± 0.7 | 13.3 ± 0.44 | 14.3 ± 1.1 | 25 | 33 |
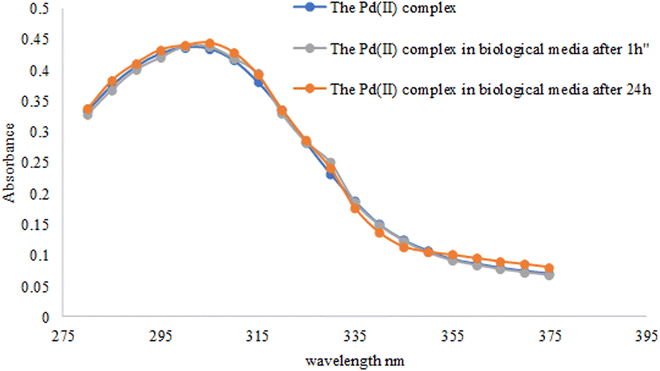
All synthesized complexes and a ligand exhibit a significant result against PS. Aeruginous than their corresponding SMX–TMP drugs54 Table 7. The study of palladium(II) complex stability in biological media showed no difference between the three scans, the complex alone and the complex in LB media after 1 h and 24 h. These results indicated the high stability of palladium(II) in biological media Fig. 8.
Conclusion
The complex compounds were prepared and characterized using elemental analysis, 1HNMR, UV-VIS spectrophotometer, scanning electron microscopy, and powder X-ray diffraction techniques. The results of spectrum techniques and powder XRD analyses deduced that the complex of bis-benzidinedioxime nickel(II) and bis-benzidinedioxime platinum(IV) have little octahedral geometry distortion. In contrast, bis-benzidinedioxime palladium(II) has square planar geometry distortion. Molecular docking studies showed that this compound had occupied both the p-aminobenzoic acid and pterin binding pockets of DHPS and DHFR. The antimicrobial activity study indicates that all complexes showed more activity than the ligand against the same microorganisms under identical experimental conditions. This provides supportive information and suggests using the complexes to control the selected microbial infection.Conflicts of interest
The authors listed immediately above certify that they did not receive support from any organization for the submitted work. No funding was received to assist with the preparation of this manuscript. No funding was received for conducting this study. No funds, grants, or other support were received.Acknowledgements
We thank professor Emadaldin Konozy, Head Department of Biotechnology, Research Center, Africa City of Technology, for his immense support during this work. Furthermore, thank Dr Babita Behera, CSIR-Indian Institute of Petroleum, Dehradun –Uttarakhand-248005, India, for the 1HNMR measurement.References
- J. Kumar, A. Rai and V. Raj, Org. Med. Chem. J, 2017, 1, 555–564 Search PubMed.
- S. N. Shariff, S. Saravu and D. Ramakrishna, in Schiff Base in Organic, Inorganic and Physical Chemistry, IntechOpen, 2022 Search PubMed.
- R. K. Mohapatra, P. K. Das, M. K. Pradhan, A. A. Maihub and M. M. El-Ajaily, J. Iran. Chem. Soc., 2018, 15, 2193–2227 CrossRef CAS.
- J. Anacona, Y. Pineda, A. Bravo and J. Camus, Med. Chem., 2016, 6, 467 Search PubMed.
- R. Nair, A. Shah, S. Baluja and S. Chanda, J. Serb. Chem. Soc., 2006, 71, 733–744 CrossRef CAS.
- M. N. Uddin, S. S. Ahmed and S. R. Alam, J. Coord. Chem., 2020, 73, 3109–3149 CrossRef CAS.
- S. A. Dalia, F. Afsan, M. S. Hossain, M. N. Khan, C. Zakaria, M.-E. Zahan and M. Ali, Int. J. Chem. Stud., 2018, 6, 2859–2867 Search PubMed.
- Y. Wang, X. Yu, B. Lu, W. Ye and S. Sheng, Chem. Abstr., 2002, 23, 257–301 CAS.
- Y. Wang, B. Lu, X. Yu, W. Ye and S. Wang, Chem. J. Internet, 2001, 3, 51 Search PubMed.
- S. Chaves-Silva, L. P. Horta, L. T. Souza, C. M. da Silva, C. S. Dohanik, G. A. Goulart, I. E. Marriel, Â. de Fátima and L. V. Modolo, Ind. Crops Prod., 2020, 145, 111995 CrossRef CAS.
- P. Ghanghas, A. Choudhary, D. Kumar and K. Poonia, Inorg. Chem. Commun., 2021, 130, 108710 CrossRef CAS.
- M. Yadav, D. Yadav, D. P. Singh and J. K. Kapoor, Inorg. Chim. Acta, 2022, 121300 Search PubMed.
- É. N. Oiye, M. F. M. Ribeiro, J. M. T. Katayama, M. C. Tadini, M. A. Balbino, I. C. Eleotério, J. Magalhães, A. S. Castro, R. S. M. Silva and J. W. da Cruz Júnior, Crit. Rev. Anal. Chem., 2019, 49, 488–509 CrossRef PubMed.
- T. Akitsu, B. Miroslaw and S. Sudarsan, Int. J. Mol. Sci., 2022, 23, 10005 CrossRef CAS PubMed.
- P. Mahadevi and S. Sumathi, Synth. Commun., 2020, 50, 2237–2249 CrossRef CAS.
- A. Campos, J. Anacona and M. Campos-Vallette, Main Group Met. Chem., 1999, 22, 283–288 CAS.
- S. Chandar, J. Indian Chem. Soc., 2004, 81, 203–206 Search PubMed.
- P. G. Cozzi, Chem. Soc. Rev., 2004, 33, 410–421 RSC.
- M. Verma, S. N. Pandeya, K. N. Singh and J. P. Stables, Acta Pharm., 2004, 54, 49–56 CAS.
- D. R. Williams, Chem. Rev., 1972, 72, 203–213 CrossRef CAS PubMed.
- S. Kumar, D. N. Dhar and P. Saxena, J. Serb. Chem. Soc., 2009, 68, 181–187 CAS.
- O. Kahn, Angew Chem. Int. Ed. Engl., 1985, 24, 834–850 CrossRef.
- L. Hua, F.-W. Zheng, H.-T. Chen, L. Wang, D.-J. Li, L. Yang, F.-J. Han, X.-Y. Duan, T.-T. Liu and W.-X. Wang, J. Solid State Chem., 2021, 122463 CrossRef CAS.
- L. Rigamonti, P. Zardi, S. Carlino, F. Demartin, C. Castellano, L. Pigani, A. Ponti, A. M. Ferretti and A. Pasini, Int. J. Mol. Sci., 2020, 21, 7882 CrossRef PubMed.
- B. Es-Sounni, A. Nakkabi, A. Bouymajane, I. Elaaraj, M. Bakhouch, F. R. Filali, M. El Yazidi, N. El Moualij and M. Fahim, Biointerface Res. Appl. Chem., 2022, 13, 1–14 Search PubMed.
- H. Kargar, A. A. Ardakani, M. N. Tahir, M. Ashfaq and K. S. Munawar, J. Mol. Struct., 2021, 1233, 130112 CrossRef CAS.
- K. Turecka, A. Chylewska, A. Kawiak and K. F. Waleron, Front. Microbiol., 2018, 9, 1594 CrossRef PubMed.
- B. Geeta, K. Shravankumar, P. M. Reddy, E. Ravikrishna, M. Sarangapani, K. K. Reddy and V. Ravinder, Spectrochim. Acta, Part A, 2010, 77, 911–915 CrossRef CAS PubMed.
- T. Y. Fonkui, M. I. Ikhile, D. T. Ndinteh and P. B. Njobeh, Trop. J. Pharm. Res., 2018, 17, 2507–2518 CrossRef CAS.
- E. Khan, ChemistrySelect, 2021, 6, 3041–3064 CrossRef CAS.
- R. A. Azzam, R. E. Elsayed and G. H. Elgemeie, ACS Omega, 2020, 5, 10401–10414 CrossRef CAS PubMed.
- H. Lade and J. S. Kim, Antibiotics, 2021, 10, 398 CrossRef CAS PubMed.
- M. K. Yun, Y. Wu, Z. Li, Y. Zhao, M. B. Waddell, A. M. Ferreira, R. E. Lee, D. Bashford and S. W. White, Science, 2012, 335, 1110–1114 CrossRef CAS PubMed.
- C. Capasso and C. T. Supuran, J. Enzyme Inhib. Med. Chem., 2014, 29, 379–387 CrossRef CAS PubMed.
- M. Graffner Nordberg, K. Kolmodin, J. Aqvist, S. F. Queener and A. Hallberg, J. Med. Chem, 2001, 44, 2391–2402 CrossRef CAS PubMed.
- A. Wrobel, K. Arciszewska, D. Maliszewski and D. Drozdowska, J. Antibiot., 2020, 73, 5–27 CrossRef CAS PubMed.
- H. Seeley and P. Van Denmark, A laboratory manual of microbiology, Taraporewala Sons and Co., Mumbai, 1975, p. 55 Search PubMed.
- Z. A. Saleh and D. K. Kafi, Phys. Chem., 2016, 6, 49–56 CAS.
- K. Buldurun, N. Turan, A. Savcı and N. Colak, J. Saudi Chem. Soc., 2019, 23, 205–214 CrossRef CAS.
- M. Mishra, K. Tiwari, A. K. Singh and V. P. Singh, Polyhedron, 2014, 77, 57–65 CrossRef CAS.
- B. Shafaatian, A. Soleymanpour, N. K. Oskouei, B. Notash and S. A. Rezvani, Spectrochim. Acta, Part A, 2014, 128, 363–369 CrossRef CAS PubMed.
- J. W. Akitt and B. E. Mann, NMR and Chemistry: An Introduction to Modern NMR Spectroscopy, CRC Press, 2017 Search PubMed.
- N. S. Gwaram, H. M. Ali, H. Khaledi, M. A. Abdulla, A. H. A. Hadi, T. K. Lin, C. L. Ching and C. L. Ooi, Molecules, 2012, 17, 5952–5971 CrossRef CAS PubMed.
- N. E. Jacobsen, NMR Spectroscopy Explained: Simplified Theory, Applications and Examples for Organic Chemistry and Structural Biology, John Wiley & Sons, 2007 Search PubMed.
- M. Babinský, K. Bouzková, M. Pipíška, L. Novosadová and R. Marek, J. Phys. Chem. A, 2013, 117, 497–503 CrossRef PubMed.
- Y. M. Dikova, D. S. Yufit and J. G. Williams, Inorg. Chem., 2023, 62, 1306–1322 CrossRef CAS PubMed.
- A. D. Aputen, M. G. Elias, J. Gilbert, J. A. Sakoff, C. P. Gordon, K. F. Scott and J. R. Aldrich-Wright, Molecules, 2022, 27, 7120 CrossRef CAS PubMed.
- M. Novák, C. Foroutan-Nejad and R. Marek, Phys. Chem. Chem. Phys., 2015, 17, 6440–6450 RSC.
- L. H. Abdel-Rahman, R. M. El-Khatib, L. A. Nassr, A. M. Abu-Dief and F. E.-D. Lashin, Spectrochim. Acta, Part A, 2013, 111, 266–276 CrossRef CAS PubMed.
- M. Neelakantan, S. Marriappan, J. Dharmaraja, T. Jeyakumar and K. Muthukumaran, Spectrochim. Acta, Part A, 2008, 71, 628–635 CrossRef CAS PubMed.
- U. El-Ayaan and A.-M. Alaa, Eur. J. Med. Chem., 2005, 40, 1214–1221 CrossRef CAS PubMed.
- Z. H. Chohan and M. Praveen, Appl. Organomet. Chem., 2001, 15, 617–625 CrossRef CAS.
- Z. H. Chohan and M. Praveen, Appl. Organomet. Chem., 2000, 14, 376–382 CrossRef CAS.
- A. Beheshti, F. Hashemi, F. Behavndi, M. Zahedi, M. Kolahi, H. Motamedi and P. Mayer, Int. J. Biol. Macromol., 2017, 104, 1107–1123 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04768g |
| This journal is © The Royal Society of Chemistry 2023 |