Open Access Article
Open Access ArticleComprehensive analysis of crystal structure, spectroscopic properties, quantum chemical insights, and molecular docking studies of two pyrazolopyridine compounds: potential anticancer agents†
Efraín Polo-Cuadrado a,
Lorena López-Cuellarab,
Karen Acosta-Quirogac,
Cristian Rojas-Peñac,
Iván Brito
a,
Lorena López-Cuellarab,
Karen Acosta-Quirogac,
Cristian Rojas-Peñac,
Iván Brito d,
Jonathan Cisternae,
Jorge Trilleras
d,
Jonathan Cisternae,
Jorge Trilleras f,
Joel B. Aldereteg,
Yorley Duarte*hi and
Margarita Gutiérrez
f,
Joel B. Aldereteg,
Yorley Duarte*hi and
Margarita Gutiérrez *a
*a
aLaboratorio Síntesis Orgánica y Actividad Biológica (LSO-Act-Bio), Instituto de Química de Recursos Naturales, Universidad de Talca, Casilla 747, Talca 3460000, Chile. E-mail: mgutierrez@utalca.cl
bUniversidad de la Amazonia, Programa de Química, Cl. 17 Diagonal 17 con, Cra. 3F, Florencia 180001, Colombia
cDoctorado en Química, Departamento de Química Inorgánica y Analítica, Universidad de Chile, Santiago, Chile
dDepartamento de Química, Facultad de Ciencias Básicas, Universidad de Antofagasta, Avenida. Universidad de Antofagasta, Campus Coloso, Antofagasta 02800, Chile
eDepartamento de Química, Facultad de Ciencias, Universidad Católica del Norte, Sede Casa Central, Av. Angamos 0610, Antofagasta, Chile
fGrupo de Investigación en Compuestos Heterocíclicos, Universidad del Atlántico, Puerto Colombia 081007, Colombia
gInstituto de Química de Recursos Naturales (IQRN), Universidad de Talca, Avenida Lircay S/N, Casilla 747, Talca, Chile
hCenter for Bioinformatics and Integrative Biology, Facultad de Ciencias de la Vida, Universidad, Andrés Bello, Av. Republica 330, Santiago 8370146, Chile. E-mail: yorley.duarte@unab.cl
iInterdisciplinary Centre for Neuroscience of Valparaíso, Facultad de Ciencias, Universidad de Valparaíso, Valparaíso 2381850, Chile
First published on 16th October 2023
Abstract
In this study, two pyrazolo[3,4-b]pyridine derivatives (4a and 4b) were grown using a slow evaporation solution growth technique and characterized by FT-IR, HRMS, 1H/13C NMR spectroscopy, and X-ray crystallography. The 4a and 4b structures crystallized in monoclinic and triclinic systems with space groups P21/n and P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , respectively. Theoretical calculations were performed at the DFT/B3LYP level for the optimized geometries. The results were in excellent agreement with the experimental data (spectroscopic and XRD). This investigation encompasses molecular modeling studies including Hirshfeld surface analysis, energy framework calculations, and frontier molecular orbital analysis. Intermolecular interactions within the crystal structures of the compounds were explored through Hirshfeld surface analysis, which revealed the notable presence of hydrogen bonding and hydrophobic interactions. This insight provides valuable information on the structural stability and potential solubility characteristics of these compounds. The research was extended to docking analysis with eight distinct kinases (BRAF, HER2, CSF1R, MEK2, PDGFRA, JAK, AKT1, and AKT2). The results of this analysis demonstrate that both 4a and 4b interact effectively with the kinase-binding sites through a combination of hydrophobic interactions and hydrogen bonding. Compound 4a had the best affinity for proteins; this is related to the fact that the compound is not rigid and has a small size, allowing it to sit well at any binding site. This study contributes to the advancement of kinase inhibitor research and offers potential avenues for the development of new therapeutic agents for cancer treatment.
, respectively. Theoretical calculations were performed at the DFT/B3LYP level for the optimized geometries. The results were in excellent agreement with the experimental data (spectroscopic and XRD). This investigation encompasses molecular modeling studies including Hirshfeld surface analysis, energy framework calculations, and frontier molecular orbital analysis. Intermolecular interactions within the crystal structures of the compounds were explored through Hirshfeld surface analysis, which revealed the notable presence of hydrogen bonding and hydrophobic interactions. This insight provides valuable information on the structural stability and potential solubility characteristics of these compounds. The research was extended to docking analysis with eight distinct kinases (BRAF, HER2, CSF1R, MEK2, PDGFRA, JAK, AKT1, and AKT2). The results of this analysis demonstrate that both 4a and 4b interact effectively with the kinase-binding sites through a combination of hydrophobic interactions and hydrogen bonding. Compound 4a had the best affinity for proteins; this is related to the fact that the compound is not rigid and has a small size, allowing it to sit well at any binding site. This study contributes to the advancement of kinase inhibitor research and offers potential avenues for the development of new therapeutic agents for cancer treatment.
1. Introduction
Kinases are naturally occurring enzymes that play an important role in the regulation of cellular and physiological processes through phosphorylation of proteins, lipids, and carbohydrates. More than 500 different types of kinases have been identified in the human genome, which are classified based on their structure, substrate specificity, and the location of phosphorylated residues on the substrate.1,2 Kinases regulate cell growth and division, cell differentiation, apoptosis, and immune response.3 However, abnormal activation of these enzymes can contribute to the development of various diseases, such as cancer and autoimmune diseases. Kinases are important biological targets in the treatment of diseases, such as cancer and inflammatory diseases, and kinase inhibitors are a therapeutic option to block the activity of abnormal kinases, thereby reducing uncontrolled cell growth, destruction, and inflammation.4Human protein kinases use adenosine triphosphate (ATP) for phosphorylate serine, threonine, or tyrosine residues in their target proteins. This ATP-binding site is located between the two lobes of the protein and is connected by a hinge region with a short chain of hydrogen-bonded clusters in an acceptor–donor–acceptor arrangement (see Fig. 1). Both ATP and most ATP-competing kinase inhibitors use hydrogen-bond interactions with the hinge region. Generally, kinase inhibitors are designed around a heterocyclic scaffold that forms hydrogen bonds with the hinge region and interacts with an ATP-binding pocket.5
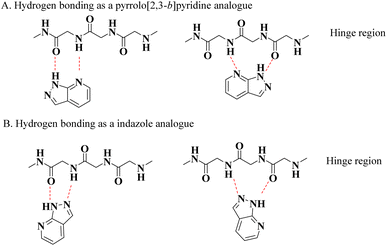 | ||
| Fig. 1 Interaction of the pyrazolo[3,4-b]pyridine core with the hinge region of kinases. (A) Hydrogen bonding as a pyrrolo[2,3-b]pyridine analogue. (B) Hydrogen bonding as an indazole analogue. | ||
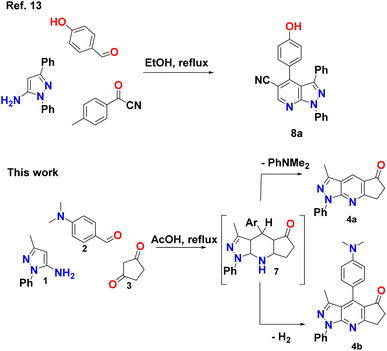
Different heterocyclic structures serve as the basis for the generation of kinase inhibitors, among which pyrazolo[3,4-b]pyridine derivatives stand out, in addition to demonstrating interesting inhibitory properties in the many main families of kinases. It has biological properties such as antitumor, antioxidant, anti-inflammatory, antimicrobial, and therapeutic effects in autoimmune diseases.2,6–11 Recently, Barghash et al., reported the synthesis and evaluation of novel pyrazolo[3,4-b]pyridine derivatives as potential anticancer agents. Screening of pyrazolo[3,4-b]pyridine derivatives for antitumor activity revealed that several compounds exhibited potent anticancer effects. Among the tested compounds, pyrazolo[3,4-b]pyridine 8a showed the most efficient antiproliferative activity, with broad-spectrum activity against almost all examined cancer cell lines (Scheme 1).12 This study suggests that these derivatives have the potential to be used as lead molecules for the development of potent anticancer candidates.12–15
However, it has been reported that this heterocyclic system possesses two key structural features for kinase inhibition: (a) its ability to form hydrogen bonds and (b) its combination of pyrrolo[2,3-b]pyridine and an indazole moiety, which allows it to achieve multiple modes of binding both at the hinge region and at different active kinase-binding sites (see Fig. 1).5 Likewise, it has been observed that this scaffold provides various advantages in terms of intellectual property, biological activities, physical properties, and synthetic flexibility, which has aroused great interest among researchers because of the fact that of the total number of references included in SciFinder, around pyrazolo[3,4-b] pyridines since 1908, more than 50% corresponds to the period from 2012 to 2022, showing an almost exponential increase, half of which are patents, clearly indicating that this type of structure currently plays an important role as a scaffold for the development of drug candidates.15
Computational docking is a potent method for understanding and forecasting the molecular interactions of ligands with various biological receptors such as protein active sites. This fascinating protein–ligand interaction can be used to direct the design of compounds and experiments, offering a vast pool of possibilities for therapeutic use.
The understanding of kinase–ligand interactions and selectivity has advanced significantly over the past few years. Experimentally established structures of more than 2800 catalytic kinase domains from mice and humans have shed significant light on fundamental structural factors.16 Using this knowledge, we selected seven exemplary kinase structures for comparison. With the two most promising ligands, the emphasis was on examining the key structural characteristics in relation to their binding affinities. The structure–activity interactions of these kinases and the possible effects of our ligands were better understood through this preliminary analysis.
Taking this into account, in the present work, we synthesized and crystallized two nuclei derived from the pyrazolo[3,4-b]pyridine system, which were fully characterized using the experimental techniques XRD, FT-IR, HRMS, and NMR (Scheme 1). Likewise, confirmation of the stable crystal structure has been based on quantum chemistry results, such as geometry optimization, Hirshfeld surface analysis, and energy frame calculations of frontier molecular orbitals additionally, we conducted a molecular docking study of these two molecules against eight types of kinases linked to cancer cell lines, B-Raf proto-oncogene, serine/threonine kinase (BRAF), human epidermal growth factor receptor 2 (HER2), Colony Stimulating Factor 1 Receptor (CSF1R), Mitogen-Activated Protein Kinase 2 (MEK2), Platelet-Derived Growth Factor Receptor Alpha (PDGFRA), Janus kinase (JAK), Protein Kinase B alpha (AKT1), and Protein Kinase B beta (AKT2). This study was driven by the vast potential of pyrazolo[3,4-b]pyridine derivatives as kinase inhibitors. We were particularly interested in predicting the modes of action of the most active compounds.
2. Experimental
2.1. Materials and methods
All chemical reagents and organic solvents were obtained from commercial suppliers and were used without further purification. The experiments were performed in a Discover microwave apparatus (CEM Corporation, USA) and Branson 1510 ultrasonic cleaning bath with a mechanical timer and heater switch at 47 kHz. Thin-layer chromatography (TLC) was performed on silica gel 60 HF254 plates (Merck, Germany) to determine the purity of the compounds. The melting point ranges (m.p.) were recorded on an Electrothermal IA9100 apparatus (Stone, UK) using the one-end open capillary method and were uncorrected. IR spectra (KBr discs, 500–4000 cm−1) were recorded on a NEXUS 670 FT-IR spectrophotometer (Thermo Nicolet, USA). 1H and 13C NMR spectra were recorded using DMSO-d6 and CDCl3 as solvents, and tetramethylsilane (TMS) as an internal reference on an AM-400 spectrometer (Bruker, Germany) at 400 and 100 MHz, respectively. High-resolution mass spectrometry (HRMS) analyses were carried out using a Bruker “Compact” quadrupole time-of-flight mass spectrometry (qTOF-MS, Germany) coupled with an Apollo II ion funnel electrospray ionization (ESI) source.2.2. Reaction conditions to obtain pyrazolopyridine 4a and 4b
A mixture of aminopyrazole 1 (2 mmol), 4-(dimethylamino)benzaldehyde 2 (2 mmol), and cyclopentane-1,3-dione 3 (2 mmol) in glacial acetic acid (8 mL) was maintained at 120 °C for 3 h. The boiling reaction mixture was diluted with water (4 mL) and stirred. The cooled mass was filtered and the solid was washed with water. The reaction mixture was purified by column chromatography using a mixture of ethyl acetate and petroleum ether (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) to obtain compounds 4a and 4b Compounds 4a and 4b were crystallized by redissolving them in a mixture of DCM
3) to obtain compounds 4a and 4b Compounds 4a and 4b were crystallized by redissolving them in a mixture of DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOH (1
EtOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), leaving them to stand until the crystals formed.
1), leaving them to stand until the crystals formed.
2.3. X-ray crystallography
Diffraction data were collected in a range of 295–296 K on a Bruker D8 Venture diffractometer equipped with a bidimensional CMOS Photon 100 detector, using graphite monochromated Mo-Kα (λ = 0.71073 Å) radiation. The diffraction frames were integrated using the APEX3 package17 and corrected for absorption using SADABS.18 The structure of (1) was solved by intrinsic phasing19 using the OLEX2 software20 and refined with full-matrix least-squares methods based on F2 (SHELXL).21 Non-hydrogen atoms were refined using anisotropic displacement parameters. The hydrogen atoms were included in their calculated positions and assigned the isotropic and shift-limited thermal parameters of their parent atoms as constants. All geometric calculations were performed using Platon software.223. Computational methods
3.1. Theoretical calculations
Density functional theory (DFT) was employed to optimize the ground-state geometries and compute the vibrational frequencies of 4a and 4b. These calculations were performed using the Gaussian 16 computational package with the B3LYP functional] and 6-31G* basis set.23–28 The calculated vibrational frequencies were scaled by 0.9627.The ionization potential (IP) and electronic affinity (EA) were estimated from the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) energies as IP = −εHOMO and EA = −εLUMO, respectively. These estimates were employed to compute the electronegativity (χ), chemical hardness (η) and softness (S) parameters as χ = (IP + EA)/2, η = (IP − EA)/2 and S = 1/η.29
3.2. Hirshfeld surface analysis
CrystalExplorer 21.3 software30 was used to calculate the Hirshfeld surface31 and associated 2D-fingerprint plots32 of the title compound using the crystallographic information file (CIF) as input for the analysis. The normalized contact distance dnorm, defined in terms of the de, di, and vdW radii of the atoms, was calculated using eqn (1), where the distance from the Hirshfeld isosurface to the nearest external (de) or internal (di) nucleus, and vdW is the van der Waals radii of atoms taken from the literature.33,34
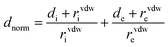 | (1) |
Hirshfeld surface analysis was performed using the 6-31G(d, p) basis set at the B3LYP level of theory over a range of ±0.002 au (ref. 35) using the TONTO computational package, which was integrated into the program CrystalExplorer.35 The bond lengths of the hydrogen atoms involved in the interactions were normalized to standard values from neutron diffraction measurements (C–H = 1.083 Å, N–H = 1.009 Å, O–H = 0.983 Å).36 The intermolecular energies of the molecular pairs in the crystal packing were calculated, at B3LYP/6-31G(d, p) level of theory, in a cluster of radius 3.8 Å around the molecule.23,37
3.3. Molecular docking
Molecular docking was used to investigate the molecular basis and potential biological activity of the two pyrazolopyridine compounds as anticancer agents by targeting eight specific kinases.The Schrödinger's Small-Molecule Drug Discovery Suite facilitated the execution of molecular docking calculations.38 Using the Protein Preparation Wizard of Schrödinger, the basic setup of the kinase enzyme structures (BRAF – (Pdb:4MNF), HER2-(Pdb:3PP0), CSF1R-(Pdb:7MFC), MEK2 (Pdb:1S9I), PDGFRA (Pdb:6JOJ), JAK (Pdb:2B7A), AKT1 (Pdb:4GV1), and AKT2 (Pdb:2X39)) was created by adding hydrogen atoms, assigning bond ordering, generating rotamers, and protonation states. Lig-Prep software was used to construct compound structures, and ChemDraw (PerkinElmer, Waltham, Massachusetts, USA) was used to design the compound structures. Epik was used to forecast the ionization and tautomeric states. Using the impact module, the protein was subjected to molecular minimization. Furthermore, the protein was subjected to molecular minimization using the impact module. Docking calculations were carried out with Glide in the Single Precision (SP) mode, considering rigid receptors and flexible ligands. The co-crystallized ligands of the kinases served as a reference point for the grid box, and the docking grid box was oriented accordingly. Finally, Emodel was used to examine the docking poses for each molecule.
4. Results and discussion
4.1. Chemistry and characterization
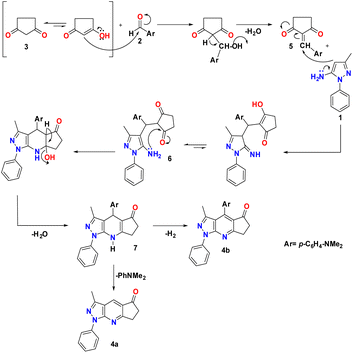
A reasonable mechanism for the formation of 4a and 4b is presented in Scheme 2. The formation proceeds via initial condensation of aldehyde (2) with cyclopentane-1,3-dione (3) to give an intermediate [5], which further undergoes Michael addition with (1) to give an open-chain intermediate [6], which is subsequently cyclized, dehydrated and dehydrogenated to afford the aromatized intermediate [7] with the splitting off N,N-dimethylaniline and the formation of a polycondensed heterocyclic system with a γ-unsubstituted pyridine ring (pyrazolo[3,4-b]quinoline, 4a), and in a separate process, dehydrogenated and elimination of N,N-dimethylaniline to afford the aromatized product 4b.The FT-IR spectra of the synthesized pyrazolopyridine derivatives 4a and 4b showed bands at the stretching frequencies of 1727–1709 cm−1 and 1589–1598 cm−1, respectively, which are characteristic of (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and (–C
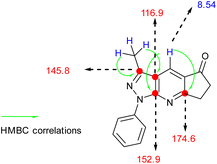
O) and (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) groups (see Table 1). 1H-NMR was characterized by the presence of three groups of signals (aromatic protons, protons near heteroatoms, and aliphatic protons). In compound 4a, a signal at approximately 8 ppm was found, which is typical of a γ-unsubstituted pyridine ring, as shown in Fig. 2.
C) groups (see Table 1). 1H-NMR was characterized by the presence of three groups of signals (aromatic protons, protons near heteroatoms, and aliphatic protons). In compound 4a, a signal at approximately 8 ppm was found, which is typical of a γ-unsubstituted pyridine ring, as shown in Fig. 2.
| Normal mode | B3LYP/6-31G(d) | Experimental in this study | Approximate assignments | |
|---|---|---|---|---|
| aFreq (cm−1) | Intensity (km mol−1) | Freq (cm−1) | ||
| 4a | ||||
| 1 | 3083 | 0.09 | 3043 | Symmetric stretching C–H sp2 (phenyl) |
| 2 | 3029 | 0.15 | 3033 | Asymmetric stretching C–H sp2 (phenyl) |
| 3 | 2952 | 0.18 | 2924 | Symmetric stretching C–H sp3 (N,N-dimethyl) |
| 4 | 2891 | 0.33 | 2854 | Asymmetric stretching C–H sp3 (N,N-dimethyl) |
| 5 | 1724 | 0.49 | 1709 | Stretch C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
| 6 | 1601 | 0.55 | 1622 | Stretch C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
| 7 | 1547 | 1 | 1589 | Stretch C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
| 8 | 1486 | 0.8 | 1495 | Balanceo en el plano C–H |
| 9 | 1340 | 0.9 | 1383 | Stretch C–N (aryl) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| 4b | ||||
| 1 | 3068 | 0.08 | 3075 | Asymmetric stretching C–H sp2 (phenyl) |
| 2 | 2976 | 0.04 | 2964 | Asymmetric stretching C–H sp3 (methyl-pyrazole) |
| 3 | 2929 | 0.07 | 2852 | Symmetric stretching C–H sp3 (methyl-pyrazole) |
| 4 | 1739 | 1 | 1727 | Stretch C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
| 5 | 1578 | 0.80 | 1598 | Stretch C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
| 6 | 1378 | 0.42 | 1382 | Out-of-plane torsion C–H |
The 13C NMR spectrum of 4b compound was measured in deuterated chloroform and revealed the presence of nineteen carbon atoms, which was consistent with the target compound (see Fig. ESI 5, 6, and Table ESI 1†). 13C NMR showed a signal at δC = 202.8 ppm assigned to the carbonyl carbon of cyclopentanone and the signals at δC = 15.6 and 40.2 ppm were attributed to methyl groups, pyrazole ring and N,N-methyl respectively.
Electrospray ionization-mass spectrometry (ESI-MS) of compounds 4a and 4b exhibited [M + H]+ peaks at 264.1134 and 383.1867, respectively, corresponding to the molecular formula C16H14N3O+ and C24H23N4O+ thus, based on the above spectral data, the synthesized structures were confirmed (Fig. ESI 7 and 8†).
4.2. FT-IR spectra
The experimental and simulated (B3LYP/6-31G*) infrared (IR) spectra of 4a and 4b are shown in Fig. 3. The resulting vibrational frequencies of the optimized geometries, proposed vibrational assignments, and IR intensities are listed in Table 1. The modes are numbered from the lowest to the highest frequency. Comparison of the theoretical and experimental IR spectra showed t that the strong vibrations in the experimental spectrum were also strong in the theoretical spectrum.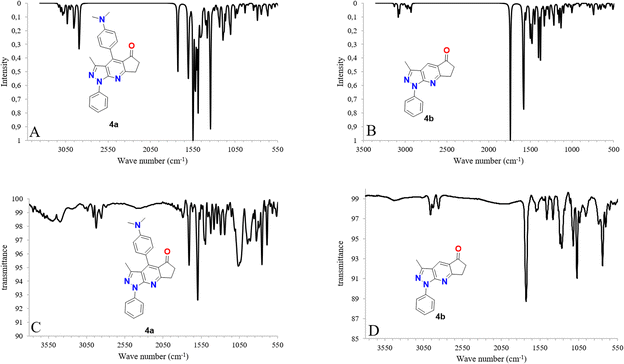 | ||
| Fig. 3 IR spectra calculated with DFT (B3LYP/6-31G*) (A and B) along with experimental (C and D) IR spectra for molecules 4a and 4b. | ||
4.3. Frontier molecular orbitals (FMOs) analysis and molecular reactivity
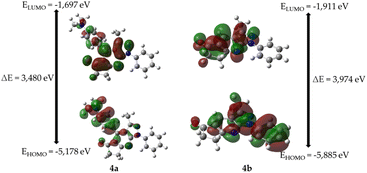
As shown in Table 2, the frequencies calculated using the B3LYP/6-31G* method were in good agreement with the experimentally obtained results. Therefore, this computational method is valuable for identifying important functional groups for characterizing the molecules studied here.39To obtain information on the reactivity and stability of compounds 4a and 4b, descriptors obtained from density functional theory were studied: HOMO and LUMO energies, HOMO–LUMO gap, hardness, softness, electronegativity, and electron affinity (see Table 2). It was found that for molecule 4a the values were −5.178, −1.697, 3.481, 1.740, 0.574, 3.438, and 1.697 eV, respectively, while those of 4b were −5.885, −1.911, 3.975, 1.987, 0.503, 3.898, and 1.911 eV, respectively.
The highest energy occupied molecular orbital (HOMO) characterizes the ability of a compound to donate electrons and undergo electrophilic additions. In contrast, the energy of the lowest unoccupied molecular orbital (LUMO) supplies information about a compound's readiness to accept electrons and its susceptibility to nucleophilic attack and is related to properties such as electronegativity (tendency to attract electron density) and electron affinity (ability to accept electrons). In addition, the difference in energy between the HOMO and LUMO orbitals (HOMO–LUMO gap) provides information on chemical reactivity and kinetic stability. A molecule with a high energy gap is associated with low chemical reactivity but high kinetic stability and vice versa (see Fig. 4). Finally, chemical hardness and softness are related to the polarizability of a molecule. In other words, higher hardness implies lower polarizability, whereas higher softness is associated with higher polarizability.
Fig. 4 shows the molecular frontier orbitals of compounds 4a and 4b. For molecule 4a, it is evident that the HOMO orbital has a high electronic density located in the 4-(dimethylamino)phenyl ring; in the case of the LUMO orbital, the region of highest probability is in the ring of the pentanone cycle; on the other hand, for the 4b molecule, the HOMO orbital shows an electron density in the 3-methyl-1-phenylpyrazolo[3,4-b]pyridine nucleus, while the LUMO orbital shows that the region of highest probability lies both on the 3-methylpyrazolo[3,4-b]pyridine ring and on the cyclopentanone ring in the molecule.
In general, 4a has a smaller HOMO–LUMO gap, less chemical hardness, greater chemical softness, lower electronegativity, and lower electron affinity than 4b. Therefore, 4a exhibits greater reactivity, less kinetic stability, greater polarizability, and less ability to attract electron density and accept electrons.
4.4. Molecular docking
We used the two most promising ligands to understand crucial structural traits in connection with their binding affinities to some kinases implicated in carcinogenesis, taking into account kinase–ligand interactions and the number of kinase structures. To accomplish this, we used molecular docking, a method that simulates interactions between protein ligands. This discovery lays the groundwork for understanding the processes governing kinase–pyrazolopyridine interactions, possibly positioning these compounds as crucial structurally modifiable scaffolds for targeted cancer therapy.The synthesized novel compounds were successfully docked within the active site of the kinase enzyme, demonstrating a favorable binding affinity with the active site amino acids based on several intermolecular interactions.
Eight kinases were used in the docking analysis, and the results showed that compounds 4a and 4b fit well into the binding site, stabilizing it mostly through hydrophobic interactions and hydrogen bonds. The docking score for compound 4a was more negative than that of compound 4b for the six kinases, suggesting superior affinity for these proteins (Table 3).
| Kinase | Compound 4a dock score (kcal mol−1) | Compound 4b dock score (kcal mol−1) | Ref. ligand dock score (kcal mol−1) | Compound 4a interactions | Compound 4b interactions | Ref. ligand interactions |
|---|---|---|---|---|---|---|
| CSFR1 | −7.60 | −8.27 | −11.18 | Hydro. I: Met637, Cys774, Asp796, Ile646, Phe797, Val647, Leu640 | Hydro. I: Met637, Ile636, Thr663, Asp796, Ile646, Ile794, Val647 | Hydro. I: Phe797, Glu664, Val596, Val647, Cys666, Tyr665 |
| HB: Glu633, Gly795 | HB: Asp796 | HB: Asp796, Cys666 | ||||
| AKT2 | −6.05 | −4.27 | −10.42 | Hydro. I: Met229, Asp293, Glu236, Met282, Gly161, Phe439, Thr213 | Hydro. I: Lys160, Val166, Thr292, lys181, Gly161 | Hydro. I: lys181, Val166, Tyr231, Gly164, Met229, Met282, Ala232, Thr292, Phe439 |
| HB: Tyr231, Arg6, Glu230 | HB: Asp293, Glu236, Arg6, π-cation Arg6 | HB: Glu230, Ala232, Asp293 | ||||
| HER2 | −8.81 | −5.52 | −10.74 | Hydro. I: Met801, Leu800, Lys753, Asp863, Met801, Leu852, Thr733 | Hydro. I: Met801, Lys753, Thr862, Cys805, Gly727, Leu852 | Hydro. I: Leu796, Met774, Phe864, Gln799, Leu852 |
| HB: Asp863, Ile752 | HB: Phe864, Arg849 | HB: Leu796, Asp863, Lys753, Met801, Cys805 | ||||
| PDGFRA | −6.94 | −6.10 | −9.37 | Hydro. I: Cys677, Leu599, Ala625, Ala840,Val607, Arg841 | Hydro. I: Phe678, Tyr679, Leu599, Ala840, Cys677 | Hydro. I: Tyr679, Asn684, Phe679, Leu599, Ala840, R841, K627 |
| HB: Aromatic HB Glu675 | HB: Asp681, Arg841, Cys677 | HB: Asp681, Cys677, salt bridge Asp681 | ||||
| BRAF | −7.70 | −7.83 | −11.0 | Hydro. I: Trp531, Asp594, Gly466, Thr526 | Hydro. I: Gly466, Leu514, Asp594, Ile527, Asn581, Gly593 | Hydro. I: Trp531, Ile527, Phe583, Gly466, Lys483 |
| HB: Phe583 (π–π), Cys532, Ser536 (WM) Trp531(WM) | HB: Cys532, Asp594 | HB: Glu501, Cys532, Asn580 (WM) | ||||
| MEK2 | −6.24 | −4.70 | −9.60 | Hydro. I: Arg193, Lys196, Leu119, Leu219, Cys211, Phe213, Lys101 | Hydro. I: Met234, Arg193, Lys196, Leu119, Leu219, Cys211 | Hydro. I: Lys196, Arg193, Leu119, Leu219, Met141 |
| HB: Ser216 | HB: Lys101 | HB: Asp194, Lys101, Phe213 (π–π), Val131 | ||||
| JAK | −9.25 | <3.0 | −10.9 | Hydro. I: Met929, Tyr931, Gly993, Leu855 | Hydro. I: — | Hydro. I: Met929, Tyr931, Gly993, Leu855, Arg980, Asn981, Gly935 |
| HB: Glu930, Leu932, Leu983 (WM) | HB: — | HB: Glu930, Leu932 | ||||
| AKT1 | −6.90 | <3.0 | −11.2 | Hydro. I: Asp292, Lys158, Lys276, Asn279, Thr291 | Hydro. I: — | Hydro. I: Asn279, Phe438, Lys279, Asp292, Ala230, Met281 |
| HB: Ala230 | HB: — | HB: Ala230,Glu228, Met281, Glu234, Asn279 |
However, it should be noted that this affinity remained inferior to that exhibited by the corresponding reference compounds, as detailed in Table 3.
Compound 4a has lower rigidity and smaller size, allowing it to fit better into each binding site, leading to increased binding affinity. These characteristics are particularly beneficial for kinases with restricted binding sites, such as AKT1 and JAK, which possess smaller cavities incapable of accommodating larger molecules. Notably, compound 4a demonstrated the highest affinity against JAK, with less than a 1.0 kcal mol−1 difference compared to the reference compound.
While all kinases have the same basic functions as enzymes that enable the transfer of a phosphate group from ATP to another molecule, are essential in cell signaling pathways, and may serve as targets for cancer treatment, there are noticeable distinctions in their active sites.40 These variations control the molecules to which each kinase can bind. For instance, the binding sites of HER2 and CSF1R are predominantly negatively charged, which influences the types of molecules with which they can interact.41
In contrast, the active sites of the kinases BRAF, MEK2, and PDGFRA have neutral electrostatic characteristics.42–44 JAK, AKT1, and AKT2 have positively charged binding sites, making them attractive to negatively charged ligands.45–47 These electrostatic properties have significant implications for the affinities of compounds 4a and 4b. According to Table 3, compounds 4a and 4b, which are primarily positively charged owing to the presence of nitrogen heteroatoms, demonstrate strong affinities to the binding sites of the kinases HER2 and CSF1R, respectively. Here, the interactions primarily involved charge-positive residues.
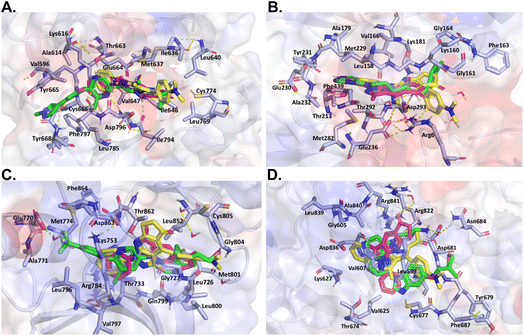
On the other hand, despite its divergent electrostatic characteristics, compound 4a can attach to JAK with a high degree of affinity because of its smaller size. According to the results of the molecular docking study, these pyrazole derivatives inhibited the activity of HER2, BRAF, JAK, PDGFRA, and AKT1. Despite having lower docking score values than the reference ligand, the docked molecules had significant score values. Compound 4a showed a better binding affinity to the afore-mentioned targets than the other two docked molecules. The theoretical foundation for the rational design of novel pyrazolopyridine compounds as cancer inhibitors was provided by these docking results. It also is essential to note that despite the existence of various drug-targeting kinases, these proteins continue to represent a significant class of drug targets. Thus, the development of specific drugs targeting these targets remains a challenge. This is where pyrazolopyridines come into play, serving as a fundamental fragment capable of binding to specific kinase pockets, and hence, constituting a critical scaffold for kinase drugs, as evidenced in several approved pharmaceuticals.48 Moreover, the structural differences between the binding sites of kinases can be used to dock more suitable fragments, thereby aiding in the achievement of high selectivity for new molecules interacting with these proteins (Fig. 6).
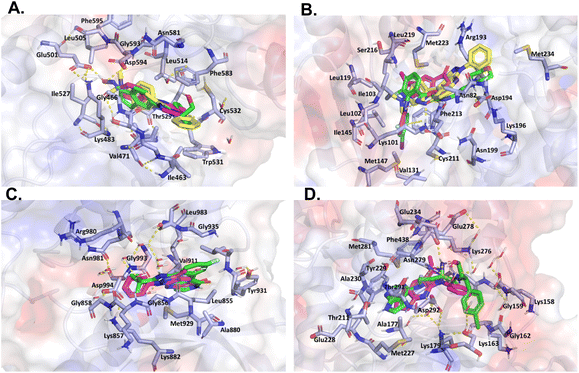 | ||
| Fig. 6 Like Fig. 5, this figure presents a comparative in silico analysis of the binding interactions among four additional kinases and their corresponding reference ligands, as well as the interactions with compounds 4a and 4b. Panel (A) binding interactions of BRAF. Panel (B) binding interactions of MEK2. Panel (C) binding interactions of JAK. Panel (D) binding interactions of AKT1. In all panels, each compound demonstrates a precise orientation in the protein's binding site that aligns seamlessly with the reference ligand. However, in some instances, the reference ligand is of a larger size. | ||
4.5. X-ray structure
The molecular structures of the synthesized compounds 4a and 4b crystallized in monoclinic and triclinic unit cells with space groups P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (Z = 2) and P21/n (Z = 4), respectively. The molecular structures of the compounds agreed with the spectroscopic characterization and the proposed structures, and both showed a centrosymmetric setting with normal bond distances and angles36 (see Fig. ESI 9†). The dihedral angles between the mean planes of the phenyl and pyrazolo[3,4-b]pyridyl rings were 7.93(4) and 17.93(6)°, respectively, with the phenyl ring being more coplanar in compound 4a than in 4b.
(Z = 2) and P21/n (Z = 4), respectively. The molecular structures of the compounds agreed with the spectroscopic characterization and the proposed structures, and both showed a centrosymmetric setting with normal bond distances and angles36 (see Fig. ESI 9†). The dihedral angles between the mean planes of the phenyl and pyrazolo[3,4-b]pyridyl rings were 7.93(4) and 17.93(6)°, respectively, with the phenyl ring being more coplanar in compound 4a than in 4b.
In contrast, in the case of 4b, the dihedral angle between the pyrazolo[3,4-b]pyridyl and diethylaminophenyl rings was 65.47(6)°. A summary of the details of the crystal data and collection is presented in Table 4, and additional crystallographic details are provided in the CIF file. ORTEP views were drawn using OLEX2 software.20
| 4a | 4b | 4a | 4b | ||
|---|---|---|---|---|---|
| Empirical formula | C16H13N3O | C24H22N4O | μ mm−1 | 0.088 | 0.082 |
| Formula mass (g mol−1) | 263.29 | 382.45 | F(000) | 552.0 | 404.0 |
| Collection T (K) | 296.19 | 295.2 | Crystal size (mm−3) | 0.273 × 0.166 × 0.145 | 0.718 × 0.67 × 0.504 |
| Crystal system | Monoclinic | Triclinic | 2Θ range for data collection (°) | 5.874–52.818 | 7.188–61.29 |
| Space group | P21/n | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
Index ranges (hkl) | −10/10, −6/6, −34/34 | −11/11, −14/14, −19/18 |
| a (Å) | 8.7898(16) | 8.0100(9) | Reflections collected | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 284 284 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 838 838 |
| b (Å) | 5.2594(9) | 9.8032(11) | Independent reflections | 2623 [Rint = 0.0830, Rsigma = 0.0737] | 5989 [Rint = 0.0406, Rsigma = 0.0293] |
| c (Å) | 27.871(5) | 13.6657(15) | Comp. qmax (%) | 99.7 | 99.1 |
| α (°) | 90 | 71.974(2) | Max/min transmission | 0.735, 0.677 | 0.746, 0.683 |
| β (°) | 95.483(5) | 74.329(3) | Data/restraints/parameters | 2623/0/199 | 5989/0/282 |
| γ (°) | 90 | 89.987(3) | Goodness-of-fit on F2 | 1.032 | 1.060 |
| V (Å3) | 1282.6(4) | 978.40(19) | Final R indexes [I ≥ 2σ (I)] | R1 = 0.0523, wR2 = 0.1289 | R1 = 0.0595, wR2 = 0.1349 |
| Z | 4 | 2 | Final R indexes [all data] | R1 = 0.0767, wR2 = 0.1425 | R1 = 0.1031, wR2 = 0.1733 |
| ρcalcd (g cm−3) | 1.364 | 1.298 | Largest diff. peak/hole/e Å−3 | 0.23/−0.15 | 0.30/−0.24 |
Additionally, the crystal packing of 4a and 4b does not present geometrical parameters corresponding to classical hydrogen bonding49 and is stabilized by intra- and intermolecular non-conventional hydrogen bond-like interactions C–H⋯N and C–H⋯O. In 4a, the intramolecular C12–H12⋯N1 hydrogen bond can be described using the graph set motifs S(6). Likewise, intermolecular hydrogen bond interactions generate a ring motif that can be described with R22 (10) graph set motifs (1−x, 2−y, 1−z).50 In compound 4b, C14–H14⋯O1 and C3–H3B⋯N3 interactions (+x, 1+y, +z) form extended chains running along the [111] direction, forming C11 (n) (n = 8 and 11) graph set motif (Fig. ESI 10†).
4.6. Hirshfeld surface analysis and 2D fingerprint plots
To see other intermolecular contacts across the crystal structure, Hirshfeld surface analysis was performed to complement XRD analysis. Intermolecular interactions are constituted by C–H⋯O and, to a lesser extent, by C–H⋯N contacts, which are shown as red (dnorm < vdW radii), white (dnorm = vdW radii), and blue (dnorm > vdW radii) spots on the dnorm surfaces for all compounds. Moreover, there is evidence of another interesting weak contact in the crystal structures of all compounds. The reciprocal contacts, their respective contributions, and all fingerprint plots with dnorm (where dnorm = di + de) surfaces for their intermolecular contacts are shown in Fig. ESI 11.†Additionally, the H⋯H contacts in each compound, generate a significant effect on the molecular packing in the crystal structure stabilization because their contacts are di + de < 2.4 Å, in other words, these contacts are slightly shorter than the sum of the vdW radii for these atoms,33 which can support the crystal packing of each compound as dihydrogen bond interactions. These are shown as sharp needles in 4a and diffuse spots in compound 4b. This last feature can be attributed to the force of these interactions, with di + de ≈ 2.2 (Fig. ESI 12†).
In addition, another type of weak interaction was observed in Hirshfeld surface analysis. For example, the ditetrel bond was verified for compound 4a (see Fig. ESI 13†). Only a few examples of this type of interaction, which works as an electron donor in a σ-hole noncovalent bond,51,52 with a contribution of around 5.1% with de + di of > 3.6 Å, are shown as an arrow tip pattern in the fingerprint plot.
In the case of compound 4b, π⋯π stacking was also observed, which was verified over the heterocycles in the title compound (see Fig. ESI 14†), with a contribution of approximately 1.3% with de + di ≈ 3.5 Å. This was verified using the shape index surface, which allowed us to determine the presence of these weak interactions. The yellow–orange spots show surface subsidence owing to the proximity of the neighboring moieties, and the blue–green spots show the reciprocal contacts of the moieties that generate the subsidence.
4.7. Energy frameworks
Finally, the energy framework53 was analyzed to better understand the packing and topology of the crystal structure and supramolecular rearrangement. This method allows the calculation and comparison of different energy components, that is, repulsion (E_rep), electrostatic (E_ele), dispersion (E_dis), polarization (E_pol), and total (E_tot) energies, based on the anisotropy of the topology of pairwise intermolecular interaction energies (see Fig. ESI 15† and Table 5). The thickness of the cylinder radius indicates the grade of interactions, is directly related to the energy magnitude, and offers information about the stabilization of the crystal packing.54 Depending on the direction of the pipe, it can be concluded that the framing is driven by translation or centrosymmetric elements. However, this rearrangement leads to the formation of another weak interaction in the crystal structure.| N | Symop | R | E_ele | E_pol | E_dis | E_rep | E_tot |
|---|---|---|---|---|---|---|---|
| 4a | |||||||
| 1 | −x, −y, −z | 7.64 | −8.7 | −1.1 | −44.6 | 25.8 | −33.0 |
| 2 | x, y, z | 9.80 | −7.0 | −2.1 | −14.9 | 9.4 | −16.1 |
| 1 | −x, −y, −z | 8.24 | −5.1 | −1.8 | −46.8 | 28.1 | −30.1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| 4b | |||||||
| 0 | −x, −y, −z | 7.22 | 1.0 | −1.9 | −28.7 | 14.5 | −16.4 |
| 0 | −x + 1/2, y + 1/2, −z + 1/2 | 9.14 | −3.3 | −0.6 | −15.5 | 10.4 | −11.4 |
| 0 | x + 1/2, −y + 1/2, −z + 1/2 | 15.05 | 0.0 | −0.0 | −0.1 | 0.0 | −0.1 |
| 0 | −x + 1/2, y + 1/2, −z + 1/2 | 8.01 | −4.8 | −0.4 | −15.7 | 10.2 | −12.7 |
| 0 | x + 1/2, −y + 1/2, z + 1/2 | 14.25 | −0.1 | −0.0 | −0.2 | 0.0 | −0.3 |
The results of the calculations revealed that dispersion interactions exhibit approximately honeycomb-shaped energy topologies in compound 4a, whereas in compound 4b, this topology zig-zag a ladder-shaped topology.
5. Conclusions
Compounds 4a and 4b were characterized by FT-IR, HRMS, 1H NMR, 13C NMR, and X-ray crystallography. The X-ray findings showed that 4a crystallized in a monoclinic system with a P21/n space group, Z = 4, and unit cell parameters a = 8.7898(16) Å, b = 5.2594(9) Å, c = 27.871(5) Å, β = 95.483(5)°, and V = 1282.6(4) Å3, whereas compound 4b crystallized in the triclinic system with a P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group, Z = 2, and unit cell parameters a = 8.0100(9) Å, b = 9.8032(11) Å, c = 13.6657(15) Å, β = 74.329(3)°, and V = 978.40(19) Å3. In general, good agreement was found between all the investigated theoretical properties (structural, electronic, and spectroscopic) and the experimental results. Hirshfeld surface analysis shows that intermolecular interactions are constituted mainly by C–H⋯O contacts, and to a lesser extent by C–H⋯N contacts, for both compounds. In contrast, FMO analysis and chemical reactivity descriptors revealed that 4a was more reactive and less stable than 4b.
space group, Z = 2, and unit cell parameters a = 8.0100(9) Å, b = 9.8032(11) Å, c = 13.6657(15) Å, β = 74.329(3)°, and V = 978.40(19) Å3. In general, good agreement was found between all the investigated theoretical properties (structural, electronic, and spectroscopic) and the experimental results. Hirshfeld surface analysis shows that intermolecular interactions are constituted mainly by C–H⋯O contacts, and to a lesser extent by C–H⋯N contacts, for both compounds. In contrast, FMO analysis and chemical reactivity descriptors revealed that 4a was more reactive and less stable than 4b.
The docking results provide a theoretical basis for the rational design of novel pyrazolo[3,4-b]pyridine compounds as inhibitors for cancer treatment. This study highlighted the potential of these compounds as essential fragments capable of binding to specific kinase pockets, making them a critical scaffold for the development of kinase-targeted drugs. This observation aligns with the use of similar scaffolds in approved pharmaceutical products.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the Research Group of the Laboratory of Organic Synthesis and Biological Activity of the University of Talca. E. P.-C. Thanks FONDECYT Post-Doctoral Fellowship No. 3220681. Fondecyt Project 1200531 the authors also acknowledge FONDEQUIP program (EQM 130021, 160063 and 180024).References
- R. Roskoski, Pharmacol. Res., 2015, 100, 1–23 CrossRef CAS PubMed.
- G. Manning, D. B. Whyte, R. Martinez, T. Hunter and S. Sudarsanam, Science, 2002, 298, 1912–1934 CrossRef CAS PubMed.
- Y. Keshet and R. Seger, Methods Mol. Biol., 2010, 661, 3–38 CrossRef CAS PubMed.
- J. Zhang, P. L. Yang and N. S. Gray, Nat. Rev. Cancer, 2009, 9, 28–39 CrossRef CAS PubMed.
- S. Wenglowsky, Expert Opin. Ther. Pat., 2013, 23, 281–298 CrossRef CAS PubMed.
- C. Chen, P. Pan, Z. Deng, D. Wang, Q. Wu, L. Xu, T. Hou and S. Cui, Bioorg. Med. Chem. Lett., 2019, 29, 912–916 CrossRef CAS PubMed.
- L. Jing, Y. Tang and Z. Xiao, Bioorg. Med. Chem. Lett., 2018, 28, 1386–1391 CrossRef CAS PubMed.
- A. E. M. Mekky and S. M. H. Sanad, Polycyclic Aromat. Compd., 2021, 41, 936–949 CrossRef CAS.
- S. B. Bharate, T. R. Mahajan, Y. R. Gole, M. Nambiar, T. T. Matan, A. Kulkarni-Almeida, S. Balachandran, H. Junjappa, A. Balakrishnan and R. A. Vishwakarma, Bioorg. Med. Chem., 2008, 16, 7167–7176 CrossRef CAS PubMed.
- Y. K. Abdel-Monem, S. A. Abou El-Enein and M. M. El-Sheikh-Amer, J. Mol. Struct., 2017, 1127, 386–396 CrossRef CAS.
- Y. Huang, Y. Li, G. Dong, W. Zhang, N. Liu and C. Sheng, Arch. Pharm., 2018, 351, 1–8 Search PubMed.
- R. F. Barghash, W. M. Eldehna, M. Kovalová, V. Vojáčková, V. Kryštof and H. A. Abdel-Aziz, Eur. J. Med. Chem., 2022, 227, 113952 CrossRef CAS PubMed.
- B. Bhukya, R. Korra and H. Guguloth, J. Heterocycl. Chem., 2023, 60, 872–878 CrossRef CAS.
- A. A. Farahat, E. M. Samir, M. Y. Zaki, R. A. T. Serya and H. A. Abdel-Aziz, Arch. Pharm., 2022, 355, 2100302 CrossRef CAS PubMed.
- A. Donaire-Arias, A. M. Montagut, R. P. de la Bellacasa, R. Estrada-Tejedor, J. Teixidó and J. I. Borrell, Molecules, 2022, 27, 2237 CrossRef CAS PubMed.
- A. J. Kooistra, G. K. Kanev, O. P. J. Van Linden, R. Leurs, I. J. P. De Esch and C. De Graaf, Nucleic Acids Res., 2016, 44, D371 CrossRef PubMed.
- APEX3 SAINT and SADABS Bruker, AXS Inc., Madison, Wisconsin, USA, 2015.
- G. M. Sheldrick, SADABS Version 2.03, University of Göttingen, Germany, 2002 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- O. V Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- A. L. Spek, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 148–155 CrossRef CAS PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- W. J. Hehre, K. Ditchfield and J. A. Pople, J. Chem. Phys., 2003, 56, 2257 CrossRef.
- P. Geerlings, F. De Proft and W. Langenaeker, Chem. Rev., 2003, 103, 1793–1874 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. V. Marenich, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Streatmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Danneberg, S. Dapperich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision A.08, Gaussian, Inc., Wallingford CT, 2009 Search PubMed.
- P. J. Stephens, F. J. Devlin, C. F. Chabalowski and M. J. Frisch, J. Phys. Chem., 1994, 98, 11623–11627 CrossRef CAS.
- F. Sonmez, Z. Gunesli, B. Z. Kurt, I. Gazioglu, D. Avci and M. Kucukislamoglu, Mol. Diversity, 2019, 23, 829–844 CrossRef CAS PubMed.
- P. R. Spackman, M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, D. Jayatilaka and M. A. Spackman, J. Appl. Crystallogr., 2021, 54, 1006–1011 CrossRef CAS PubMed.
- M. A. Spackman and D. Jayatilaka, CrystEngComm, 2009, 11, 19–32 RSC.
- M. A. Spackman and J. J. McKinnon, CrystEngComm, 2002, 4, 378–392 RSC.
- A. Bondi, J. Phys. Chem., 1964, 68, 441–451 CrossRef CAS.
- S. S. Batsanov, Inorg. Mater., 2001, 37, 871–885 CrossRef CAS.
- M. A. Spackman, J. J. McKinnon and D. Jayatilaka, CrystEngComm, 2008, 10, 377–388 CAS.
- F. H. Allen, O. Kennard, D. G. Watson, L. Brammer, A. G. Orpen and R. Taylor, J. Chem. Soc., 1987, 2, S1–S19 Search PubMed.
- J. Tirado-Rives and W. L. Jorgensen, J. Chem. Theory Comput., 2008, 4, 297–306 CrossRef CAS PubMed.
- S. M. Anjum, K. S. Kumar, A. Umamaheswari, D. Lakhanpal, S. Swargam, K. Riazunnisa and T. Chandrasekhar, Results Chem., 2023, 5, 100970 CrossRef CAS.
- E. Polo-Cuadrado, C. Rojas-Peña, K. Acosta-Quiroga, L. Camargo-Ayala, I. Brito, J. Cisterna, F. Moncada, J. Trilleras, Y. A. Rodríguez-Núñez and M. Gutierrez, RSC Adv., 2022, 12, 33032–33048 RSC.
- J. A. Adams, Chem. Rev., 2001, 101, 2271–2290 CrossRef CAS PubMed.
- Z. Hartman, W. J. Geldenhuys and Y. M. Agazie, J. Biol. Chem., 2020, 295, 3563–3575 CrossRef CAS PubMed.
- D. Nguyen, L. Y. Lin, J. O. Zhou, E. Kibby, T. W. Sia, T. D. Tillis, N. Vapuryan, M. R. Xu, R. Potluri, Y. Shin, E. A. Erler, N. Bronkema, D. J. Boehlmer, C. D. Chung, C. Burkhard, S. H. Zeng, M. Grasso, L. A. Acevedo, R. Marmorstein and D. Fera, Biochemistry, 2020, 59, 4755 CrossRef CAS PubMed.
- S. Keretsu, S. Ghosh and S. J. Cho, Int. J. Mol. Sci., 2020, 21, 8232 CrossRef CAS PubMed.
- J. A. Martinez Fiesco, D. E. Durrant, D. K. Morrison and P. Zhang, Nat. Commun., 2022, 13, 1–14 Search PubMed.
- K. Sanachai, P. Mahalapbutr, K. Choowongkomon, R. P. Poo-Arporn, P. Wolschann and T. Rungrotmongkol, ACS Omega, 2020, 5, 369–377 CrossRef CAS PubMed.
- S. Lu, R. Deng, H. Jiang, H. Song, S. Li, Q. Shen, W. Huang, R. Nussinov, J. Yu and J. Zhang, Structure, 2015, 23, 1725–1734 CrossRef CAS PubMed.
- I. S. Lucet, E. Fantino, M. Styles, R. Bamert, O. Patel, S. E. Broughton, M. Walter, C. J. Burns, H. Treutlein, A. F. Wilks and J. Rossjohn, Blood, 2006, 107, 176–183 CrossRef CAS PubMed.
- Z. Z. Wang, X. X. Shi, G. Y. Huang, G. F. Hao and G. F. Yang, Trends Pharmacol. Sci., 2021, 42, 551–565 CrossRef CAS PubMed.
- T. Steiner, Angew. Chem., Int. Ed., 2002, 41, 48–76 CrossRef CAS.
- J. Bernstein, R. E. Davis, L. Shimoni and N.-L. Chang, Angew. Chem., Int. Ed., 1995, 34, 1555–1573 CrossRef CAS.
- W. Dong, Q. Li and S. Scheiner, Molecules, 2018, 23, 1681 CrossRef PubMed.
- S. Scheiner, Phys. Chem. Chem. Phys., 2020, 22, 16606–16614 RSC.
- C. F. Mackenzie, P. R. Spackman, D. Jayatilaka and M. A. Spackman, IUCrJ, 2017, 4, 575–587 CrossRef CAS PubMed.
- H. A. Khamees, K. Chaluvaiah, N. A. El-Khatatneh, A. Swamynayaka, K. H. Chong, J. P. Dasappa and M. Madegowda, Acta Crystallogr., Sect. E: Crystallogr. Commun., 2019, 75, 1620–1626 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2278999 and 2279000. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3ra04874h |
| This journal is © The Royal Society of Chemistry 2023 |