Open Access Article
Open Access ArticleCurcumin I-SMA nanomicelles as promising therapeutic tool to tackle bacterial infections†
Nicola F. Virzìa,
Antonino N. Fallica a,
Giuseppe Romeoa,
Khaled Greishb,
Maha Ali Alghamdib,
Salvatore Patanèc,
Antonino Mazzagliad,
Mohammad Shahide and
Valeria Pittalà
a,
Giuseppe Romeoa,
Khaled Greishb,
Maha Ali Alghamdib,
Salvatore Patanèc,
Antonino Mazzagliad,
Mohammad Shahide and
Valeria Pittalà *ab
*ab
aDepartment of Drug and Health Science, University of Catania, Viale A. Doria 6, 95125 Catania, Italy. E-mail: valeria.pittala@unict.it
bDepartment of Molecular Medicine, Arabian Gulf University, Manama 329, Bahrain
cDepartment of Mathematical and Computer Sciences, Physical Sciences and Earth Sciences, University of Messina, V.le F. Stagno D'Alcontres, 31 98166 Messina, Italy
dNational Council of Research, Institute for the Study of Nanostructured Materials (CNR-ISMN), URT of Messina c/o Department of Chemical, Biological, Pharmaceutical and Environmental Sciences, University of Messina, V.le F. Stagno d’Alcontres 31, 98166 Messina, Italy
eDepartment of Microbiology & Immunology, Arabian Gulf University, Manama 329, Bahrain
First published on 24th October 2023
Abstract
Renewed interest towards natural substances has been pushed by the widespread diffusion of antibiotic resistance. Curcumin I is the most active and effective constituent of curcuminoids extracted from Curcuma longa and, among other beneficial effects, attracted attention for its antimicrobial potential. Since the poor pharmacokinetic profile hinders its efficient utilization, in the present paper, we report encapsulation of curcumin I in poly(styrene-co-maleic acid) (SMA-CUR) providing a nanomicellar system with improved aqueous solubility and bioavailability. SMA-CUR was characterized by means of size, zeta potential, polydispersity index, atomic force microscopy (AFM), drug release studies, spectroscopic properties and stability. SMA-CUR nanoformulation displayed exciting antimicrobial properties compared to free curcumin I towards Gram-positive and Gram-negative clinical isolates.
1 Introduction
One of the most urgent and challenging topics in modern medicine is the treatment of infections caused by multidrug-resistant (MDR) bacteria, since antibiotics classes commonly used in therapeutics have become even more inefficient in tackling the MDR-related diseases.1,2 In 2017 the World Health Organization (WHO) published a list of the most threatening bacteria resistant to the current treatments, categorizing them with different priority: critical, high, and medium.3 Among these pathogens, the Gram-negative Pseudomonas aeruginosa carbapenem-resistant and the Acinetobacter baumannii carbapenem-resistant seem to be among the most difficult bacteria to struggle with. Among Gram-positive bacteria, methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumoniae, vancomycin-resistant Enterococcus faecium (VRE), and β-lactamase-resistant strains can cause life-threatening infections.4,5 The antibacterial drug-resistance is not a recent issue but rather a natural consequence of the discovery and widespread use of antibiotics. Nowadays, there are more than 15 classes of antibiotics whose mechanisms of action target essential bacterial metabolic and physiological function; however none of them was effective in avoiding the onset of resistance phenomena.6 Bacteria can acquire antibacterial drug resistance by horizontal gene transfer of virulence factors and antibiotic resistance genes, other than improving their drug-resistance by means of biofilm formation.7 Bacterial biofilms consist in a consortium of bacteria adhered in an inert surface and enclosed in a self-produced extracellular polymeric matrix.7,8 Biofilm conditions can require 200–1000 folds higher concentrations of antibiotics than the active dose in vitro against planktonic form of bacteria. For these reasons, traditional treatment to combat microbial infections based on disrupting cell growth are failing increasingly. On the other hand, targeting microbial virulence seems to be a successful strategy, since tuning virulence factors might guide to a minor evolutionary pressure for the development of bacterial resistance. The definition of virulence factors refers to intrinsic or acquired characteristics of a pathogen that are essential to establish infection in the host environment.9In this scenario, natural compounds derived from plants seem to be a wide source that could be exploited to target and modulate virulence factors, such as the bacterial motility, quorum sensing (QS), biofilm formation, bacterial enzymes and bacterial replication.10,11 Curcumin, derived from Curcuma longa rhizome and roots (Zingiberaceae family), is widely known as the “golden spice” due to its intense yellow color and for its pleiotropic activities. Turmeric has been long used in Indian cooking and the traditional medicine; in the last decades, curcumin has attracted the attention of researchers due to its potential use as anti-cancer, anti-aging, anti-diabetics, anti-inflammatory and anti-bacterial agent.12–14 Its extract contains a mixture of bioactive curcuminoids including diferuloylmethane (curcumin I, Fig. 1), desmethoxycurcumin (curcumin II) and bisdesmethoxycurcumin (curcumin III). Curcumin I is light sensitive and nearly insoluble in water, however can be dissolved in most organic solvents.15 From a chemical point of view, curcumin I is a bis-α,β-unsaturated β-diketone showing keto–enol tautomerism as pH function.
The antibacterial activity of curcuminoids extracts is documented in literature. Curcuminoids antimicrobial activity is related to different mechanisms of action.16 One of the most important properties is the ability of curcumin to inhibit the assembly of FtsZ protein involved in cytokinesis of bacteria stopping the bacterial cell division and proliferation.17 Moreover, it has been shown that curcumin is able to bind to the cell wall of bacteria, altering its integrity causing damages and cell lysis.18 At high concentrations, curcumin induce ROS accumulation causing depolymerization of the membranes and alteration of their potential. Furthermore, ROS accumulation could similarly damage internal bacterial components as essential metabolic enzymes and DNA, leading ultimately to cell apoptosis.19,20 Lastly, it was reported that curcumin interferes with several QS mechanisms inhibiting the exopolysaccharide synthesis, diminishing clustering, motility, and swimming capability and, therefore, biofilm maturation.20 However, high variability of the Minimum Inhibitory Concentrations (MICs) towards a number of bacterial strains is reported in different studies. This is probably due to the strain types, antimicrobial resistance traits and if they are clinical isolates or not.16 Moreover, the reported MIC values vary depending whether antibacterial assays have been performed on curcumin extract mixtures or on purified curcuminoids.16 In fact, as previously reported, the antibacterial activity of curcuminoid extracts is mainly related to curcumin I.21 Another crucial issue related to curcuminoids is their adverse pharmacokinetic profile. Although the promising therapeutic prospective of curcumin I, nowadays there are still limitations in its use as therapeutic agent because of its extremely poor aqueous solubility (∼0.6 μg mL−1), bioavailability, and pharmacokinetic profile mostly responsible for its failure in clinical trials.22,23
To overcome these limitations many approaches have been implemented. It is widely recognized how nanoparticles are able to improve the pharmacokinetic profile and water solubility of bioactive compounds, increase the localization at the site of action through the enhanced permeability retention (EPR) effect, and prevent off-target side-effects ameliorating their therapeutic actions.24,25 Then the proper design of a suitable drug delivery system is of crucial importance.26 On these grounds and motivated by our expertise in the field, we selected a poly(styrene-co-maleic acid) (SMA) micellar system to encapsulate curcumin I. As a nanoplatform, SMA has several advantages, including the ability to form nanomicelles in aqueous media and to show good and active cellular uptake. This can probably be ascribed to the amphiphilic nature of SMA that favors interactions between SMA and the cells surface.27 Hence, SMA nanoparticles have been widely used to encapsulate different active principles.28–30 In the present paper, to increase the therapeutic potential of curcumin I, we encapsulated it in SMA nanoparticle (SMA-CUR). The obtained micelles have been tested on multiple drug-resistant and non-resistant bacteria and results compared to free curcumin I.
2 Results and discussion
With the aim of tackling antibacterial infections caused by multiple drug-resistant and non-resistant bacteria, we focused on curcumin I since it has been reported to be the one of the most promising curcuminoids. As first step we purified curcumin I by means of flash chromatography and characterized by means of 1H NMR and 13C NMR (Fig. S1 and S2†); hence, to overcome its poor water solubility and unfavorable pharmacokinetic profile, we encapsulated it in a nanomicellar system based on poly(styrene-co-maleic acid). The micelle formation has been achieved by using a well-established method previously reported.29 The synthesized SMA-CUR micelles had a recovery of 79.2% and loading of 14.2% of curcumin I as determined by the weight ratio of drug over SMA (Table 1). The mean diameter of SMA-CUR micelles was 102.8 ± 4.23 nm when measured by DLS. The data indicate an average particle size in the optimal range for cellular uptake in inflamed tissues.31 The polydispersity index (PDI) of SMA-CUR micelles was 0.261 ± 0.03 suggesting a homogeneous and uniform distribution of size population. The zeta (ζ) potential of the SMA-CUR micelles was −56.3 mV in deionized water (Table 1). The large negative potential is predictive of a high potential stability of the colloidal solution.| Recoveryb | Loadingc (wt/wt) | Mean diameter (nm) | PDId | ζ potential (mV) |
|---|---|---|---|---|
| a Data are shown as mean values ± standard deviation (SD). Values are the mean of triplicate experiments.b Starting from 1 g of SMA and 200 mg of free CUR, 950.8 mg of SMA-CUR have been obtained (79.2% recovery).c 135 mg of CUR encapsulated in 950.8 mg of SMA-CUR as measured by UV/vis absorbance at 433 nm (14.2% wt/wt).d PDI = polydispersity index. | ||||
| 79.2% | 14.2% | 102.8 ± 4.23 | 0.261 ± 0.03 | −56.3 mV |
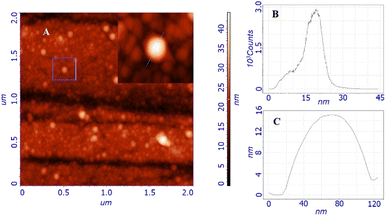
Morphological studies were carried out by atomic force microscopy (AFM). Fig. 2A shows the morphology obtained from the study of a two-by-two micron surface; the inset shows the details of a particle whose profile is shown in Fig. 2C. The topography is characterized by the presence of globular structures with an average size of around 20 nm, as stated by the size distribution shown in Fig. 2B. Larger structures are also present but not exceeding 60 nm. These globular structures resemble other similar nanomicellar systems already reported in literature.32 The observed particles uniformly cover the entire surface of the substrate, indicating that they adhere on the surface immediately without waiting for the solvent to evaporate, leading to a self-assembled homogeneous thin film. This scenario leads us to think that the observed dimensions and shapes are very similar to those of the particles in the solution, in fact, the size distribution data are in good agreement with those obtained from DLS (Dynamic Light Scattering).
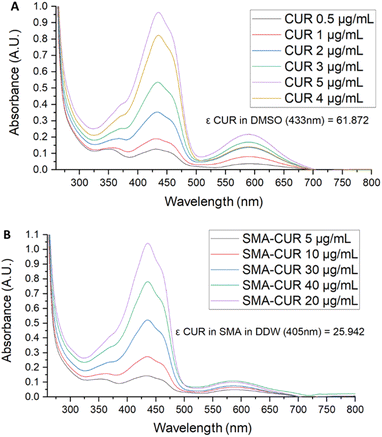
Further characterization of the micellar system has been performed by means of UV-vis and fluorescence spectra. The UV-vis spectrum of curcumin I was registered in DMSO. As displayed in Fig. 3, curcumin I showed an absorption maximum at 433 nm and an additional peak at 589 nm. Conversely, the UV-vis spectrum of SMA-CUR nanoparticles was performed in deionized water as organic solvents deteriorate the micellar nanosystem and trigger the immediate release of the encapsulated compound. As observed by the recorded spectrum (Fig. 4), SMA-CUR micelles generated one peak at 405 nm due to curcumin I, other than the absorbance band of the styrene-co-maleic acid at <280 nm.
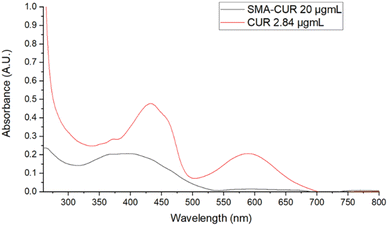 | ||
| Fig. 4 UV-vis spectra of: curcumin I in DMSO and SMA-CUR in DDW. Spectra were recorded scanning from 200 to 800 nm, using a 1 cm path length quartz cells. | ||
To further characterize the micellar nanosystem and confirm curcumin-I encapsulation, fluorescence studies have been performed. Fluorescence emission spectra of solutions of curcumin I in DMSO at different dilutions, acquired by excitation wavelength at 405 nm, showed a unique profile with three bands with a maximum centred at 495 nm (Fig. S3†). On the other hand, fluorescence emission spectra of SMA-CUR, showed only a weak fluorescence pattern, likely due to the decreased irradiation and emission of CUR incorporated into the hydrophobic core of micelle (Fig. S4†).
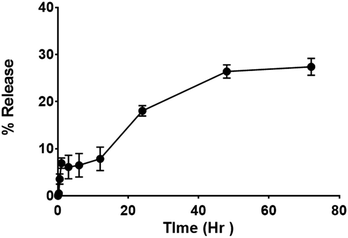
Release studies have been performed in double distilled water (DDW) to assess the efficacy and quality of the drug delivery system. The study performed by the dialysis bag method, shows an early release of ∼6% in the first hour, followed by a steady release of up to 27% at 72 h (Fig. 5). The initial release could be attributed to associate portion of curcumin I in the micelles surface; while the sustained release over 3 days can be attributed to encapsulated curcumin I in the micellar core (Fig. 4). The overall release is consistent with the sustained antibacterial effect.
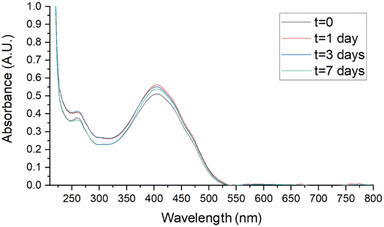
Stability studies were performed in biologically useful media to clarify the behaviour of SMA-CUR in environments simulating the antibacterial activity experiments. Samples, stored for 7 days at 4 °C, r.t., and 37 °C, were analyzed by UV-vis. Spectra were recorded at t = 0, 1, 3, and 7 days in DDW, saline (0.9 % wt NaCl), and PBS and compared to assess stability. As reported in Fig. 6 and 7, no evident changes in the UV-vis spectra were observed at T = 4 °C in saline (Fig. 6) and PBS (Fig. 7), confirming that the nanomicellar system is rather stable over the time in the utilized biological media. A mild degradation has been observed only at t = 7 in DDW at T = 4 °C, maybe due to the mild acidic pH (Fig. S5†). At r.t. no significant spectroscopic changes were observed over the first three days in saline and PBS (Fig. S8 and S10†), while a slight degradation was observed in DDW starting from t = 3 (Fig. S6†). Finally, at 37 °C mild degradation was observed starting from either t = 1 (Fig. S7 and S11†), or t = 3 (Fig. S9†).
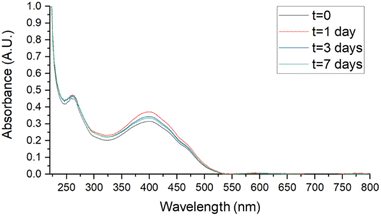 | ||
| Fig. 7 Stability studies by UV-vis of SMA-CUR at T = 4 °C in PBS aqueous solution. SMA-CUR = 60 μg mL−1. Spectra were recorded scanning from 200 to 800 nm, using a 1 cm path length quartz cells. | ||
A preliminary evaluation to assess the antibacterial activity of curcumin I and SMA-CUR has been performed. As expected, within the concentration range tested (0.5–10 mg mL−1), curcumin I did not show any antibacterial activity against the Escherichia coli isolate (Fig. S5†). Poor diffusion of curcumin I outside the wells was noticed, probably due to its hydrophobic nature and poor water solubility, hence it was seen settling down within the wells. On the other hand, within the concentration range tested (10–100 mg mL−1), screening of SMA-CUR showed antibacterial activity in concentrations 20 mg mL−1 and beyond (Fig. S12†). No antibacterial activity was noticed in the SMA-CUR concentration of 10 mg mL−1.
Hence, MICs of curcumin I in the free form or encapsulated in SMA have been evaluated in a panel of Gram-positive and Gram-negative bacteria (Table 2). Obtained MICs for free curcumin I were in the range of 25 mg mL−1 that was the highest final concentration of curcumin I used in our study; however, poor water solubility of curcumin I prevented, in some bacterial strains, its testing at this concentration. On the other hand, MICs for SMA-CUR ranged between 6.25 and 25 mg mL−1 (Table 2). Taking in consideration the loading of 14.2% of curcumin I into SMA, the effective MICs of curcumin I inside SMA ranged between 0.88 and 3.55 mg mL−1, demonstrating that SMA-CUR was more effective than curcumin I itself. In addition, SMA-CUR demonstrated to be more active on Gram-positive when compared to Gram-negative bacteria. Most interesting data have been obtained against different strains of Staphylococcus aureus both methicillin-sensitive and resistant. No specific distribution pattern in MIC values of SMA-CUR was noticed towards resistant vs. sensitive strains. Similar findings for stronger activity against Gram-positive than Gram-negative bacteria were reported by Adamczak, A et al., but with commercially purchased curcumin compound.16 On comparing the MIC values of curcumin I in SMA-CUR vs. the native form, it can be noticed that there is a substantial improvement of antibacterial activity of the active principle in the encapsulated form as opposed to that of native curcumin I (Table 2). Specifically, with respect to the methicillin-sensitive and resistant S. aureus strains tested, we observed a 4-fold reduction of MIC values for SMA-CUR micelles and a corresponding 28-fold reduction of the amount of curcumin I needed to exert its antimicrobic effect. Similarly, in the carbapenem-resistant A. baumannii strain a 2-fold reduction of MIC values was observed for SMA-CUR compared to free curcumin I and a corresponding 14-fold reduction of curcumin I required for the inhibition of bacterial growth.
| Bacterial isolates | MICs (mg mL−1) | ||
|---|---|---|---|
| Free curcumin I | SMA-CUR | Curcumin I in SMA-CURb | |
| a NT = not tested; due to poor solubility and difficulty in dissolution of curcumin I in MHB.b Data refers to a loading of 14.2%. | |||
| Escherichia coli (sensitive to all antibiotics) | 25 | 25 | 3.55 |
| Escherichia coli O157:H7 NCTC 12900 | NTa | 12.5 | 1.77 |
| Escherichia coli (carbapenem-resistant) | >25 | 25 | 3.55 |
| Methicillin-sensitive Staphylococcus aureus (isolate MSSA-1) | NTa | 6.25 | 0.88 |
| Methicillin-resistant Staphylococcus aureus (isolate MRSA-1) | NTa | 12.5 | 1.77 |
| Methicillin-sensitive Staphylococcus aureus (isolate MSSA-2) | 25 | 6.25 | 0.88 |
| Methicillin-resistant Staphylococcus aureus (isolate MRSA-2) | 25 | 6.25 | 0.88 |
| Methicillin-resistant Staphylococcus aureus (isolate MRSA-3) | 25 | 6.25 | 0.88 |
| Carbapenem-sensitive Acinetobacter baumannii (isolate CSAB-56) | NTa | 12.5 | 1.77 |
| Carbapenem-resistant Acinetobacter baumannii (isolate CRAB-57) | NTa | 12.5 | 1.77 |
| Carbapenem-sensitive Acinetobacter baumannii (isolate CSAB-59) | 25 | 25 | 3.55 |
| Carbapenem-resistant Acinetobacter baumannii (isolate CRAB-58) | 25 | 12.5 | 1.77 |
| Pseudomonas putida | 25 | 25 | 3.55 |
| Carbapenem-sensitive Klebsiella pneumoniae (isolate C16-CSKP) | NTa | 25 | 3.55 |
| Carbapenem-resistant Klebsiella pneumoniae (isolate C17-CRKP) | NTa | 12.5 | 1.77 |
Encapsulation of curcumin I in SMA micelles strongly ameliorated its aqueous solubility and allowed a more precise calculation of MIC values in clinical isolates chosen for this study. This aspect is of particular importance considering that most curcumin MIC values reported in literature largely differ on the basis of the purity of the compound used for the study, as well as on the type and/or percentage of organic solvents (DMF, DMSO, ethanol or acetone) used to dissolve curcumin.33,34 Important differences in the antimicrobial properties of curcumin have been also reported among reference strains and clinical isolates. For instance, a recent work analyzed the antimicrobial properties of curcumin in nineteen different species of bacteria.16 Almost all K. pneumoniae, A. baumannii, E. coli and MRSA S. aureus clinical isolates treated with curcumin showed MIC values from 2 up to over 5 mg mL−1, with the latter value corresponding to the highest concentration of curcumin tested. In addition, the same MIC value (5 mg mL−1) was reported in another paper for the vancomycin-resistant S. aureus (VRSA) strain.35 In agreement with these results, free curcumin I did not display any strong antimicrobic properties in our experimental conditions, whereas our SMA-CUR nanoformulation provided better MIC values compared to results obtained for non-encapsulated curcumin in the abovementioned study.
3 Experimental
3.1 Materials
Styrene-co-maleic anhydride with a number average molecular mass (Mn) ∼1,600, curcumin from Curcuma longa (turmeric), N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDAC) were obtained from Sigma-Aldrich Corp (St Louis, MO, USA). Purification of curcumin was achieved by flash column chromatography using silica gel (60, 0.040–0.063 mm, 230–400 mesh) (Merck, Kenilworth, NJ). Fractions containing the desired product were detected by thin-layer chromatography (TLC) (aluminum sheet coated with silica gel 60 F254, Merck, Kenilworth, NJ), and visualized by UV (λ = 254 and 366 nm). 1H and 13C NMR spectra were recorded on a Varian Unity Inova 200 MHz spectrometer in DMSO-d6 solution. Chemical shifts are given in δ values (ppm), using tetramethylsilane (TMS) as the internal standard; coupling constants (J) are given in Hz. Signal multiplicities are characterized as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), br (broad). UV-vis spectra were obtained on an Agilent Cary 60 spectrophotometer equipped with a xenon lamp using 1 cm path length quartz cells. Fluorescence spectra were recorded on a Cary Eclipse (Varian) fluorescence spectrophotometer using a xenon lamp as light source using 1 cm path length quartz cells. Zeta potential (ζ potential), average hydrodynamic diameter (DH) width of distribution (polydispersity index, PDI) measurements were carried out by a Zetasizer Nano ZS (Malvern Instrument, Malvern, U.K.) at 25 °C in DDW. Results are reported as the mean of three separate measurements ± the standard deviation (SD). All solvents were reagent grade and were purchased from commercial vendors (Sigma Aldrich, Merck). Following bacteria were used: E. coli O157:H7 NCTC 12900, E. coli (sensitive to all tested antibiotics), E. coli (carbapenem-resistant), carbapenem-sensitive Klebsiella pneumoniae (isolate C16-CSKP), carbapenem-resistant Klebsiella pneumoniae (isolate C17-CRKP), carbapenem-sensitive Acinetobacter baumannii (isolate CSAB-56), carbapenem-sensitive Acinetobacter baumannii (isolate CSAB-59), carbapenem-resistant Acinetobacter baumannii (isolate CRAB-57), carbapenem-resistant Acinetobacter baumannii (isolate CRAB-58), Pseudomonas putida, methicillin-sensitive Staphylococcus aureus (isolate MSSA-1), methicillin-sensitive Staphylococcus aureus (isolate MSSA-2), methicillin-resistant Staphylococcus aureus (isolate MRSA-1), methicillin-resistant Staphylococcus aureus (isolate MRSA-2), and methicillin-resistant Staphylococcus aureus (isolate MRSA-3). The bacterial isolates used in this study were taken from the culture depository of the Department of Microbiology, Immunology, & Infectious Diseases, Arabian Gulf University, Bahrain. All the isolates were maintained at −80 °C and stocked in the depository with unique identifiers. The NCTC strain (E. coli O157:H7 NCTC 12900) was procured from Mast Diagnostics, Merseyside, U.K. Other isolates, including MSSA, MRSA, CSAB, CRAB, CSKP, CRKP, were clinical isolates characterized during different research projects, and subsequently stocked in the departmental repository with unique identifiers as reflected in Table 2.3.2 Chemistry and nanomicelles characterization
Microscope glass slices were cleaned individually dipping into iso-propanol (iPr-OH) and sonicating for 5 min (ultrasonic bath Elmasonic S 30), then there were rinsed with fresh iPr-OH and dried under nitrogen flow. For the AFM characterization a 200 μg mL−1 stock dispersion of SMA-CUR in ultrapure water was prepared followed by 2 min of vortex agitation. The stock dispersion was 3-fold diluted to 25 μg mL−1 and drop-casted (150 μL) in a pre-cleaned microscope glass slice. The solvent was spontaneously evaporated in controlled atmosphere, the sample was finally dried for 2 h under mild vacuum and directly analysed.
3.3 Antibacterial studies
4 Conclusions
In this work we reported a simple and efficient method for the encapsulation of curcumin I. The obtained SMA-CUR nanomicelles greatly improved curcumin-I water solubility and good stability over storage conditions (T = 4 °C). In addition, our system demonstrated promising antibacterial activity against Gram-positive as well as Gram-negative bacteria compared to free curcumin-I. SMA-CUR was active against standard, antibiotics-sensitive, as well as antibiotics-resistant bacteria of clinical concern such as methicillin-resistant Staphylococcus aureus, carbapenem-sensitive Klebsiella pneumoniae, carbapenem-resistant Escherichia coli, carbapenem-resistant Acinetobacter baumannii. In light of the highly difficult eradication of multidrug-resistant bacterial infections, the outcomes showed in this work suggest that encapsulation of natural compounds with antibiotic properties in poly(styrene-co-maleic acid) nanoparticles represent an auspicious strategy improving their biological effects.Author contributions
Conceptualization, funding acquisition, project administration, resources, writing – review & editing: V. Pittalà. Conceptualization, data curation, supervision, writing – review & editing: K. Greish and M. Shahid. Data curation, formal analysis, investigation, methodology, software, validation, visualization, writing – original draft: N. F. Virzì, A. N. Fallica, and M. A. Alghamdi. Formal analysis, resources, writing – review & editing, supervision: G. Romeo and A. Mazzaglia.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Italian PON Project BONE++ Development of Micro and Nanotechnologies for Predictivity, Diagnosis, Therapy and Regenerative Treatments of Pathological Bone and Osteo-Articular Alterations. project number ARS01_00693.Notes and references
- S. S. Jean, D. Harnod and P. R. Hsueh, Front. Cell. Infect. Microbiol., 2022, 12, 823684 CrossRef CAS PubMed.
- Antimicrobial Resistance Collaborators, Lancet, 2022, 399, 629–655 CrossRef.
- G. Mancuso, A. Midiri, E. Gerace and C. Biondo, Pathogens, 2021, 10, 1310 CrossRef CAS.
- G. Cornaglia, Clin. Microbiol. Infect., 2009, 15, 209–211 CrossRef CAS PubMed.
- B. Jubeh, Z. Breijyeh and R. Karaman, Molecules, 2020, 25, 2888 CrossRef CAS PubMed.
- S. B. Levy and B. Marshall, Nat. Med., 2004, 10, S122–S129 CrossRef CAS.
- M. Schroeder, B. D. Brooks and A. E. Brooks, Genes, 2017, 8(1), 39 CrossRef.
- W. Yin, Y. Wang, L. Liu and J. He, Int. J. Mol. Sci., 2019, 20(14), 3423 CrossRef CAS.
- K. M. Davis, Infect. Immun., 2020, 88(7), e00911–e00919 CrossRef PubMed.
- L. N. Silva, K. R. Zimmer, A. J. Macedo and D. S. Trentin, Chem. Rev., 2016, 116, 9162–9236 CrossRef CAS PubMed.
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch, T. International Natural Product Sciences and C. T. Supuran, Nat. Rev. Drug Discovery, 2021, 20, 200–216 CrossRef CAS.
- R. R. Kotha and D. L. Luthria, Molecules, 2019, 24(16), 2930 CrossRef CAS.
- H. Li, A. Sureda, H. P. Devkota, V. Pittala, D. Barreca, A. S. Silva, D. Tewari, S. W. Xu and S. M. Nabavi, Biotechnol. Adv., 2020, 38, 107343 CrossRef CAS.
- H. Hatcher, R. Planalp, J. Cho, F. M. Tortia and S. V. Torti, Cell. Mol. Life Sci., 2008, 65, 1631–1652 CrossRef CAS PubMed.
- S. Prasad, S. C. Gupta, A. K. Tyagi and B. B. Aggarwal, Biotechnol. Adv., 2014, 32, 1053–1064 CrossRef CAS PubMed.
- A. Adamczak, M. Ożarowski and T. M. Karpiński, Pharmaceuticals, 2020, 13(7), 153 CrossRef CAS PubMed.
- D. Rai, J. K. Singh, N. Roy and D. Panda, Biochem. J., 2008, 410, 147–155 CrossRef CAS.
- S. H. Mun, S. B. Kim, R. Kong, J. G. Choi, Y. C. Kim, D. W. Shin, O. H. Kang and D. Y. Kwon, Molecules, 2014, 19, 18283–18295 CrossRef.
- D. G. Yun and D. G. Lee, Appl. Microbiol. Biotechnol., 2016, 100, 5505–5514 CrossRef CAS PubMed.
- Z. Munir, G. Banche, L. Cavallo, N. Mandras, J. Roana, R. Pertusio, E. Ficiarà, R. Cavalli and C. Guiot, Int. J. Mol. Sci., 2022, 23, 2600 CrossRef CAS PubMed.
- S. Rahbar Takrami, N. Ranji and M. Sadeghizadeh, Mol. Biol. Rep., 2019, 46, 2395–2404 CrossRef CAS.
- B. T. Kurien, A. Singh, H. Matsumoto and R. H. Scofield, Assay Drug Dev. Technol., 2007, 5, 567–576 CrossRef CAS.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharm., 2007, 4, 807–818 CrossRef CAS PubMed.
- M. A. Alghamdi, A. N. Fallica, N. Virzì, P. Kesharwani, V. Pittalà and K. Greish, J. Pers. Med., 2022, 12, 673 CrossRef PubMed.
- E. A. Azzopardi, E. L. Ferguson and D. W. Thomas, J. Antimicrob. Chemother., 2012, 68, 257–274 CrossRef.
- T. Czech, R. Lalani and M. O. Oyewumi, AAPS PharmSciTech, 2019, 20, 116 CrossRef CAS.
- H. Alimoradi, K. Greish, A. Barzegar-Fallah, L. Alshaibani and V. Pittala, Int. J. Nanomed., 2018, 13, 7771–7787 CrossRef CAS PubMed.
- G. Y. Bharate, H. Qin and J. Fang, J. Pers. Med., 2022, 12, 1650 CrossRef PubMed.
- K. Greish, A. Mathur, R. Al Zahrani, S. Elkaissi, M. Al Jishi, O. Nazzal, S. Taha, V. Pittala and S. Taurin, J. Controlled Release, 2018, 291, 184–195 CrossRef CAS PubMed.
- K. Greish, V. Pittala, S. Taurin, S. Taha, F. Bahman, A. Mathur, A. Jasim, F. Mohammed, I. M. El-Deeb, S. Fredericks and F. Rashid-Doubell, Nanomater., 2018, 8(11), 884 CrossRef PubMed.
- N. Hoshyar, S. Gray, H. Han and G. Bao, Nanomedicine, 2016, 11, 673–692 CrossRef CAS PubMed.
- P. Kesharwani, S. Banerjee, S. Padhye, F. H. Sarkar and A. K. Iyer, Colloids Surf., B, 2015, 132, 138–145 CrossRef CAS PubMed.
- H. Gunes, D. Gulen, R. Mutlu, A. Gumus, T. Tas and A. E. Topkaya, Toxicol. Ind. Health, 2016, 32, 246–250 CrossRef CAS PubMed.
- S. Y. Teow, K. Liew, S. A. Ali, A. S. Khoo and S. C. Peh, J. Trop. Med., 2016, 2016, 2853045 Search PubMed.
- F. Akhtar, A. U. Khan, L. Misba, K. Akhtar and A. Ali, Eur. J. Pharm. Biopharm., 2021, 160, 65–76 CrossRef CAS.
- M. A. Obregon-Mendoza, W. Meza-Morales, Y. Alvarez-Ricardo, M. M. Estevez-Carmona and R. G. Enriquez, Molecules, 2023, 28(1), 289 CrossRef CAS.
- N. N. Parayath, H. Nehoff, P. Müller, S. Taurin and K. Greish, Int. J. Nanomed., 2015, 10, 4653–4667 CAS.
- K. Greish, T. Sawa, J. Fang, T. Akaike and H. Maeda, J. Controlled Release, 2004, 97, 219–230 CrossRef CAS PubMed.
- R. Zagami, D. Franco, J. D. Pipkin, V. Antle, L. De Plano, S. Patane, S. Guglielmino, L. Monsu Scolaro and A. Mazzaglia, Int. J. Pharm., 2020, 585, 119487 CrossRef CAS.
- R. Zagami, A. Rubin Pedrazzo, D. Franco, F. Caldera, L. M. De Plano, M. Trapani, S. Patane, F. Trotta and A. Mazzaglia, Int. J. Pharm., 2023, 637, 122883 CrossRef CAS.
- S. M. Rahman, T. C. Telny, T. K. Ravi and S. Kuppusamy, Indian J. Pharm. Sci., 2009, 71, 139–142 CrossRef CAS PubMed.
- M. A. Khan, H. M. Khan, I. B. Ganie, S. Kumar, A. Shahzad, I. Celik and M. Shahid, Biofouling, 2022, 38, 715–728 CrossRef CAS PubMed.
- M. A. Siddiqui, R. K. Singh and A. Kumar, Int. J. Curr. Res., 2017, 9, 62114–62118 Search PubMed.
- J. N. Eloff, Planta Med., 1998, 64, 711–713 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04885c |
| This journal is © The Royal Society of Chemistry 2023 |