Open Access Article
Open Access ArticleTPN-COF@Fe-MIL-100 composite used as an electrochemical aptasensor for detection of trace tetracycline residues†
Yubo Meng a,
Yuchun Huanga,
Gailing Huangb and
Yingpan Song
a,
Yuchun Huanga,
Gailing Huangb and
Yingpan Song *b
*b
aSchool of Mechanical Engineering, Henan University of Engineering, Zhengzhou, 451191, PR China
bCollege of Material and Chemical Engineering, Zhengzhou University of Light Industry, Zhengzhou, 450002, PR China. E-mail: wangpan8624@163.com
First published on 25th September 2023
Abstract
In this work, a metal–organic framework@covalent organic framework composite (TPN-COF@Fe-MIL-100) was prepared and used as a sensing material to construct an aptasensor for trace detection of tetracycline (TET). The TPN-COF@Fe-MIL-100 integrates a large surface area, porous structure, excellent electrochemical activity, rich chemical functionality, and strong bioaffinity for aptamers, providing abundant active sites to effectively anchor aptamer strands. As a result, the TPN-COF@Fe-MIL-100-based aptasensor shows high sensitivity for detecting TET via specific recognition between aptamer and TET to form G-quadruplex. An ultralow detection limit of 1.227 fg mL−1 is deduced from the electrochemical impedance spectroscopy within a wide linear range of 0.01–10000 pg mL−1 for TET. The TPN-COF@Fe-MIL-100-based aptasensor also exhibits good selectivity, reproducibility, stability, regenerability, and applicability for a real milk sample. Therefore, the TPN-COF@Fe-MIL-100-based aptasensor will be promising for detecting trace harmful antibiotics residues for food safety.
1. Introduction
In recent years, many antibiotics, such as ampicillin (AMP), amoxicillin (AMX), cefixime (CFX), bleomycin (BLM), chlortetracycline hydrochloride (CTC), enrofloxacin (ENR), kanamycin (KAN), oxytetracycline (OTC), tetracycline (TET), and tobramycin (TOB), have been widely used in the fields of medical hygiene, food industry, and animal husbandry to treat bacterial diseases and promote animal growth.1 The dosage of antibiotics in China is astonishing, accounting for about half of the global dosage, and is massively discharged into the soil and water environment, seriously endangering human health and the ecological environment. The overuse and abuse of antibiotics will have a negative impact on human health and the environment, such as the emergence of antibiotic-resistant bacteria, the destruction of ecological balance, and the toxicity to human organs.2 Due to their low bioavailability, only a part of the antibiotics can be metabolized and absorbed in the animal body, and the remaining antibiotics will be excreted and released into the soil, surface water and groundwater, leading to serious human health risks and environmental problems. Therefore, the quantitative determination of antibiotics residues in food samples is of great significance to guarantee human health and food safety.3 Researchers have committed to the development of reliable and sensitive detection methods for antibiotics, such as high-performance liquid chromatography,4 enzyme-linked immunoassay,5 fluorescence detection,6,7 chemiluminescence,8 surface plasmon resonance,9 colorimetric assay,10 and so on. However, these detection methods often suffer from needing a skilled laborer, complex instrument operation, high cost, long analysis time, and not-sensitive enough detection, and thus cannot satisfy the requirement of the on-site assays in practical applications.11 Particularly, as for fluorescence detection, the prerequisite for the use of fluorescence sensors is that the system must contain fluorescently labeled probes, which requires either fluorescent labeling of recognition probes, or the materials used must contain fluorescent characteristics, which undoubtedly complicates the sensor construction procedure. For this reason, there is an urgent need for rapid, accurate, sensitive, and economic methods to detect antibiotics in food samples.Compared with fluorescence detection and other traditional methods, electrochemical technology shows excellent sensing performance and advantages, such as fast response, simple operation, low cost, and advanced micro-processing.12 Owing to the low cost, simple synthesis, easy to chemical modification, low immunogenicity and toxicity, high stability, high affinity and specificity with the targeted analytes, various aptamer strands (Apt) have been used as recognition probe and combined with electrochemical method to construct electrochemical aptasensors, which always exhibited high sensitivity and selectivity for detection of a variety of targeted analytes with fast response.13 In order to obtain high detection efficiency of the electrochemical aptasensors, many advanced materials with functional structures and excellent electrochemical activities were used to modify the electrode and provide more active sites for immobilization of aptamer strands and faster electron transfer between the electrode surface and the targeted analytes, including carbon nanomaterials,14 metal/metal oxides/metal hydroxide,15,16 quantum dots,17 conducting polymer,18 MXene,19 metal–organic frameworks (MOFs),20,21 covalent organic frameworks (COFs),22 etc.
Among them, MOFs and COFs have attracted more attentions because of their unique properties, such as high porosity, large specific surface area, controllable synthesis, structural diversity, modified functional groups, and thermal/chemical stability.23,24 These features and the adjustable pore sizes endow MOFs and COFs high ability to adsorb Apt by π–π* stacking and electrostatic interaction. Therefore, Apt can not only immobilize onto the surface but also penetrate into the interior of MOFs and COFs.25,26 Based on this, MOFs and COFs were chosen to fabricate electrochemical aptasensors for detection of trace antibiotics. Typically, a kind of COFs with amorphous terephthalonitrile-derived nitrogen-rich networks (denoted as TPNs), has been synthesized through trimerization reactions of terephthalonitrile monomers,27 and successfully applied in various fields.28 Therefore, one can anticipate that TPN can facilitate the aptamer strands to anchor due to its intrinsic features, such as high surface area, well-defined pore structure, and heteroatom-doped organic skeleton.
Inspired by the reported studies on MOFs as the sensitive platforms for aptasensors,29–31 a novel MOF-on-COF composite was synthesized and TET was chosen as a representative antibiotic, then the composite was employed to construct a sensitive electrochemical aptasensor for detection of TET. In the present work, Fe-MIL-100 was in situ grown on the TPN-COF surface, thus forming a novel kind of COF@MOF composite (denoted as TPN-COF@Fe-MIL-100). The synthesized TPN-COF@Fe-MIL-100 simultaneously exhibits the features of MOFs and COFs, including (i) rich nitrogen-related functional groups; (ii) excellent electrochemical activity due to the strong synergistic effect between the rich triazine rings in TPN-COF and framework containing in Fe-MIL-100; and (iii) highly strong bioaffinity interaction between the aptamer strands and the TPN-COF@Fe-MIL-100 matrix due to π–π* stacking, electrostatic interaction and hydrogen bonding. As such, the fabricated TPN-COF@Fe-MIL-100-based aptasensor exhibits extremely low detection limit (1.227 fg mL−1) within the TET concentration from 10 fg mL−1 to 10 ng mL−1. Meanwhile, it also shows high selectivity, good reproducibility, stability, regenerability, and acceptable applicability.
2. Experimental section
The additional Experimental sections, including Materials and chemicals, Synthesis of Fe-MIL-100, Synthesis of TPN-COF, Preparation of solutions, Pre-treatment of Au electrode (AE), Electrochemical measurements, and Basic characterization, were provided in the S1 Part (see the ESI†).2.1 Synthesis of TPN-COF@Fe-MIL-100
Typically, ferric chloride hexahydrate (FeCl3·6H2O, 4.9 mmol) was dissolved in 15 mL of dimethylformamide (DMF) to obtain solution A, while 1,3,5-benzenetricarboxylic acid (BTC, 2.5 mmol) was dissolved in 15 mL of DMF to obtain solution B. The two solutions were separately stirred for 30 min. Then, solution B was slowly dropped into solution A. Meanwhile, 40 mg of TPN-COF was also added into the mixture, followed by treating ultrasonically for 30 min. The following procedure was similar to the preparation of Fe-MIL-100. Finally, TPN-COF@Fe-MIL-100 powder was obtained.2.2 Fabrication of antibiotics aptasensor based on TPN-COF@Fe-MIL-100
The TPN-COF@Fe-MIL-100-based aptasensor for detection of TET was constructed as follows. First, 10 mg of TPN-COF@Fe-MIL-100 was dispersed in 10 mL of Milli-Q water and ultrasonicated for 20 min to form 1.0 mg mL−1 of TPN-COF@Fe-MIL-100 suspension. Then, about 5 μL of TPN-COF@Fe-MIL-100 dispersion was dip-coated onto the surface of pre-treated bare AE, following by natural drying for 5 h (denoted as TPN-COF@Fe-MIL-100/AE). Second, TPN-COF@Fe-MIL-100/AE was incubated in the aptamer solution (1.5 mL, 100 nM) at 4 °C for 2 h (represented by Apt/TPN-COF@Fe-MIL-100/AE) until saturation. The aptamer strands were immobilized on the electrode surface, in which the sequence of TET aptamer is 5′-TTT TTC GTA CGG AAT TCG CTA GCC CCC CGG CAG GCC ACG GCT TGG GTT GGT CCC ACT GCG CGT GGA TCC GAG CTC CAC GTG-3′.32 Third, Apt/TPN-COF@Fe-MIL-100/AE was separately immersed in different concentrations of TET solutions (0.01, 0.1, 1.0, 10, 100, 1000, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1) for electrochemical measurements (denoted as TET/Apt/TPN-COF@Fe-MIL-100/AE). For comparison, the TPN-COF-, and Fe-MIL-100-based aptasensors toward TET were constructed similarly.
000 pg mL−1) for electrochemical measurements (denoted as TET/Apt/TPN-COF@Fe-MIL-100/AE). For comparison, the TPN-COF-, and Fe-MIL-100-based aptasensors toward TET were constructed similarly.
2.3 Electrochemical measurements
The fabrication procedure of aptasensors based on the three samples was tested by electrochemical impedance spectroscopy (EIS) and cyclic voltammetry (CV). To obtain optimal conditions for detecting TET, the influences of TPN-COF@Fe-MIL-100 amount, aptamer concentration, aptamer binding time, and pH on the electrochemical performance were explored. The selectivity, regenerability, stability, and reproducibility of the developed TPN-COF@Fe-MIL-100-based aptasensor were also investigated (S1.6 in ESI†).2.4 Applicability of the proposed aptasensor
The applicability of the aptasensor was tested in real milk sample. The purchased raw milk was first treated to remove the proteins and impurities as provided in S1.6.† Then, the obtained liquid milk sample was spiked with different concentrations of TET (0.01, 0.1, 1.0, 10, 100, 1000, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1) to assess the applicability.
000 pg mL−1) to assess the applicability.
3. Results and discussion
3.1 Detection mechanism of TPN-COF@Fe-MIL-100-based aptasensor for TET
In this work, the TPN-COF@Fe-MIL-100 nanocomposite was synthesized by the hybridization of TPN-COF and Fe-MIL-100. Due to the large surface area, porous structure, excellent electrochemical activity, rich chemical functionality, and strong bioaffinity, many aptamer strands can not only anchor over the TPN-COF@Fe-MIL-100 surface, but also penetrate into the interior of porous TPN-COF@Fe-MIL-100 networks through π–π* stacking and strong electrostatic interaction between the aptamer strands and the organic frameworks (Scheme 1). All active sites of TPN-COF@Fe-MIL-100 are occupied by the aptamer strands, so non-specific adsorption between other interferents and the nanocomposite is prevented. Then, Apt/TPN-COF@Fe-MIL-100/AE is used to sensitively detect TET via specific recognition between aptamer and TET to form a G-quadruplex, leading to the variation in electrochemical signals caused by the configuration change of aptamers after TET binding. Each step of the aptasensor construction can alter the electron transfer between the electrode and electrolyte solution, which can be determined by the electrochemical techniques. As a result, the TPN-COF@Fe-MIL-100-based aptasensor could serve as biosensor for detecting TET in food sample and exhibit excellent sensitivity and selectivity.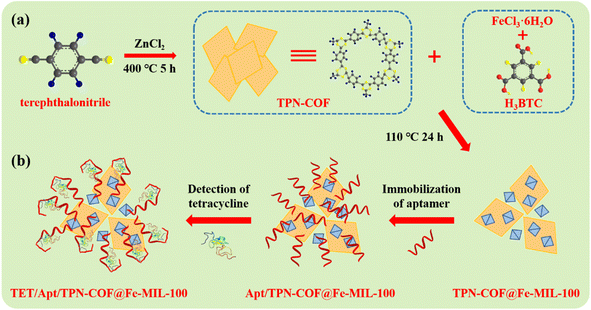 | ||
| Scheme 1 Schematic diagram of (a) the preparation of TPN-COF@Fe-MIL-100 and (b) the fabrication of TPN-COF@Fe-MIL-100-based aptasensor for detecting TET. | ||
3.2 Surface morphologies and nanostructures of TPN-COF, Fe-MIL-100, and TPN-COF@Fe-MIL-100
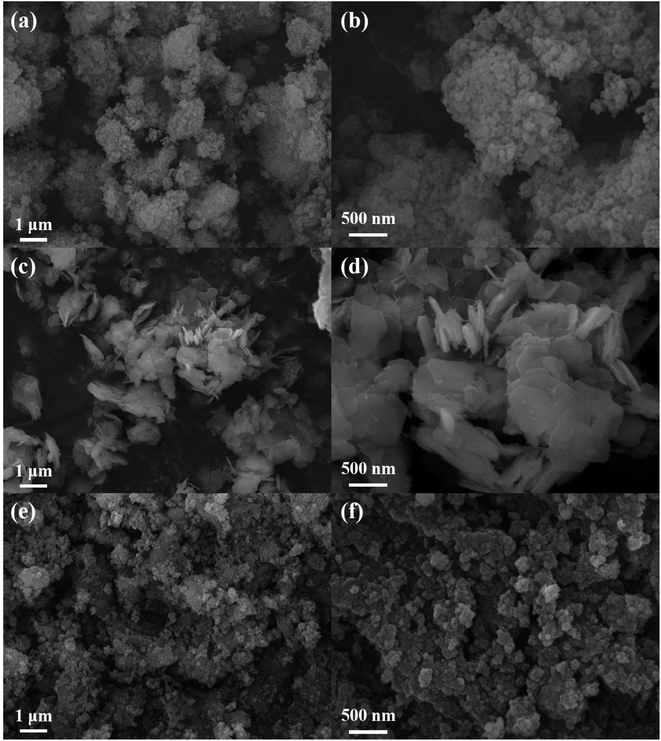
The surface morphologies and nanostructures of TPN-COF, Fe-MIL-100, and TPN-COF@Fe-MIL-100 were explored by field emission scanning electron microscopy (FE-SEM). As shown in Fig. 1a and b, Fe-MIL-100 shows a uniform, small particle size of about 100 nm. The SEM images (Fig. 1c and d) of TPN-COF exhibit two dimensional nanosheets with irregular shape and average size of about 1 μm, which are also aggregated into multilayered nanostructure. The morphologies of TPN-COF@Fe-MIL-100 composites (Fig. 1e and f) are different from those of Fe-MIL-100 and TPN-COF, indicating that the composites with different morphologies were synthesized.3.3 Chemical structures and components of TPN-COF, Fe-MIL-100, and TPN-COF@Fe-MIL-100
The chemical structure and composition of the three samples were analyzed by X-ray diffraction (XRD), Fourier transform infrared spectroscopy (FT-IR), Raman and X-ray photoelectron spectroscopy (XPS). As shown in XRD patterns (Fig. 2a), the characteristic peaks at 2θ = 10.5° and 18.9° are typical diffraction peaks of Fe-MIL-100, indicating the successful synthesis of Fe-MIL-100.33 The weak intensity of TPN-COF indicates its low crystallinity. The XRD pattern of the TPN-COF@Fe-MIL-100 composite shows that the composite retains both the diffraction peak of Fe-MIL-100 and the diffraction peak of TPN-COF at 2θ = 14.1°, suggesting that Fe-MIL-100 and TPN-COF are successfully combined to obtain the composite. As displayed in FT-IR spectra (Fig. 2b), the characteristic adsorption bands at 1560 and 1300 cm−1 indicate the formation of the triazine rings in FT-IR spectrum of TPN-COF.34 The absorption peak of terephthalonitrile disappears at 2230 cm−1, suggesting the occurrence of the trimerization reaction of terephthalonitrile to generate TPN-COF.35 As for Fe-MIL-100, the peak at 1710 cm−1 is attributed to the stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond in BTC.36 The peaks at 1629, 1573, 1452, and 1378 cm−1 represent the asymmetric vibrations of the carboxyl group.37 The peak at 1142 and 1302 cm−1 are due to the C–H vibrations,38 while the peak at 1560 cm−1 is corresponded to C
O bond in BTC.36 The peaks at 1629, 1573, 1452, and 1378 cm−1 represent the asymmetric vibrations of the carboxyl group.37 The peak at 1142 and 1302 cm−1 are due to the C–H vibrations,38 while the peak at 1560 cm−1 is corresponded to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N vibration.39 After the combination of TPN-COF and Fe-MIL-100, i.e., TPN-COF@Fe-MIL-100, since both Fe-MIL-100 and TPN-COF@Fe-MIL-100 contain trivalent metal centers and carboxylate bridging ligands, the two samples show similar spectra and same structural vibration. As shown in Fig. 2c, the Raman spectra of TPN-COF at 1340 and 1602 cm−1 correspond to the D band of carbon atoms lattice defect and the G band of in-plane stretching vibration of sp2 carbon atoms hybridization, respectively, indicating that carbon materials are formed during the calcination preparation of TPN-COF. The XPS survey scan spectra of all samples (Fig. 2d) indicate the clear signals of C 1s (284.6 eV), and O 1s (531.6 eV). Additionally, weak Fe 2p (710.3 eV) signals are also found in Fe-MIL-100, and TPN-COF@Fe-MIL-100.
N vibration.39 After the combination of TPN-COF and Fe-MIL-100, i.e., TPN-COF@Fe-MIL-100, since both Fe-MIL-100 and TPN-COF@Fe-MIL-100 contain trivalent metal centers and carboxylate bridging ligands, the two samples show similar spectra and same structural vibration. As shown in Fig. 2c, the Raman spectra of TPN-COF at 1340 and 1602 cm−1 correspond to the D band of carbon atoms lattice defect and the G band of in-plane stretching vibration of sp2 carbon atoms hybridization, respectively, indicating that carbon materials are formed during the calcination preparation of TPN-COF. The XPS survey scan spectra of all samples (Fig. 2d) indicate the clear signals of C 1s (284.6 eV), and O 1s (531.6 eV). Additionally, weak Fe 2p (710.3 eV) signals are also found in Fe-MIL-100, and TPN-COF@Fe-MIL-100.
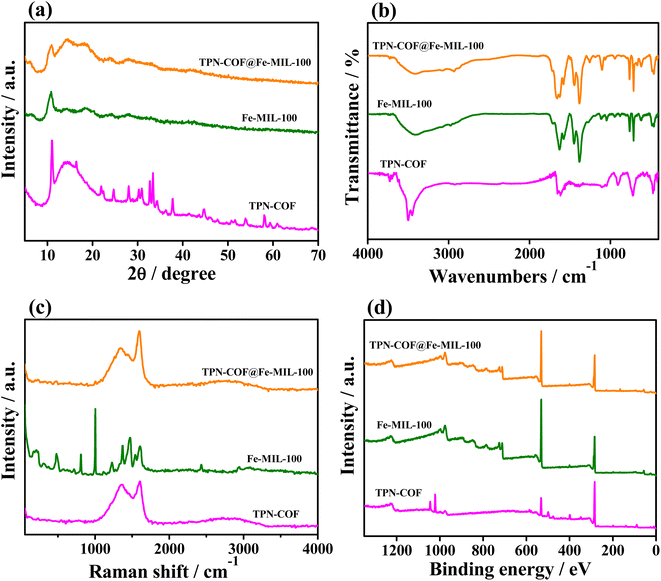 | ||
| Fig. 2 (a) XRD patterns, (b) FT-IR spectra, (c) Raman spectra, and (d) XPS survey scan spectra of TPN-COF, Fe-MIL-100 and TPN-COF@Fe-MIL-100. | ||
The C 1s and Fe 2p core-level XPS spectra of the three samples are also analyzed and illustrated in Fig. 3. Fig. 3a depicts the C 1s core-level XPS spectra of TPN-COF, Fe-MIL-100 and TPN-COF@Fe-MIL-100. The C 1s XPS measurement of the three samples reveals five peaks, including 284.6, 285.7, 286.4, 288.7, and 291.6 eV. The binding energy (BE) at 284.6 eV is attributed to the carbon atoms of the phenyl rings as well as adventitious carbon (C–C) which is used for calibration.40 The BE at around 285.7 eV is assigned to the carbon atoms in nitrile moieties (C–N). The BE at 286.4 eV is attributed to N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, whereas BE at 288.7 eV is due to the N–C
N, whereas BE at 288.7 eV is due to the N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group, indicating the partly oxidation of the carbon-related group. Besides, the weak BE at 291.6 eV corresponds to the conjugated carbon containing in benzene rings, i.e., π–π*.41 As displayed in Fig. 3b, the Fe 2p core-level XPS spectrum of Fe-MIL-100 is fitted into seven parts: the peaks at 711.2 and 712.9 eV are due to Fe2+ 2p3/2 and Fe3+ 2p3/2; the two peaks at 725.1 and 727.6 eV are assigned with Fe2+ 2p1/2 and Fe3+ 2p1/2;42 and three peaks at 715.5, 718.1, and 731.3 eV are the shake-up satellite peaks of Fe2+ and Fe3+.43 As for TPN-COF@Fe-MIL-100, the Fe 2p core-level spectrum is split into seven peaks at 711.0, 712.2, 714.4, 717.9, 724.4, 726.4, and 730.2 eV. The peaks at 712.2 and 726.4 eV are the results of Fe3+ 2p3/2 and Fe3+ 2p1/2; the peaks at 711.0 and 724.4 eV are corresponding to Fe2+ 2p3/2 and Fe2+ 2p1/2. Additionally, the satellite signals at 714.4, 717.9, and 730.2 eV are obtained from the result of the combination of Fe2+ and Fe3+. All of these results display that the TPN-COF@Fe-MIL-100 is composed of various functional groups and Fe2+/Fe3+ ions, which are helpful for the aptamer strands immobilization,44 further facilitating the sensing performance.
O group, indicating the partly oxidation of the carbon-related group. Besides, the weak BE at 291.6 eV corresponds to the conjugated carbon containing in benzene rings, i.e., π–π*.41 As displayed in Fig. 3b, the Fe 2p core-level XPS spectrum of Fe-MIL-100 is fitted into seven parts: the peaks at 711.2 and 712.9 eV are due to Fe2+ 2p3/2 and Fe3+ 2p3/2; the two peaks at 725.1 and 727.6 eV are assigned with Fe2+ 2p1/2 and Fe3+ 2p1/2;42 and three peaks at 715.5, 718.1, and 731.3 eV are the shake-up satellite peaks of Fe2+ and Fe3+.43 As for TPN-COF@Fe-MIL-100, the Fe 2p core-level spectrum is split into seven peaks at 711.0, 712.2, 714.4, 717.9, 724.4, 726.4, and 730.2 eV. The peaks at 712.2 and 726.4 eV are the results of Fe3+ 2p3/2 and Fe3+ 2p1/2; the peaks at 711.0 and 724.4 eV are corresponding to Fe2+ 2p3/2 and Fe2+ 2p1/2. Additionally, the satellite signals at 714.4, 717.9, and 730.2 eV are obtained from the result of the combination of Fe2+ and Fe3+. All of these results display that the TPN-COF@Fe-MIL-100 is composed of various functional groups and Fe2+/Fe3+ ions, which are helpful for the aptamer strands immobilization,44 further facilitating the sensing performance.
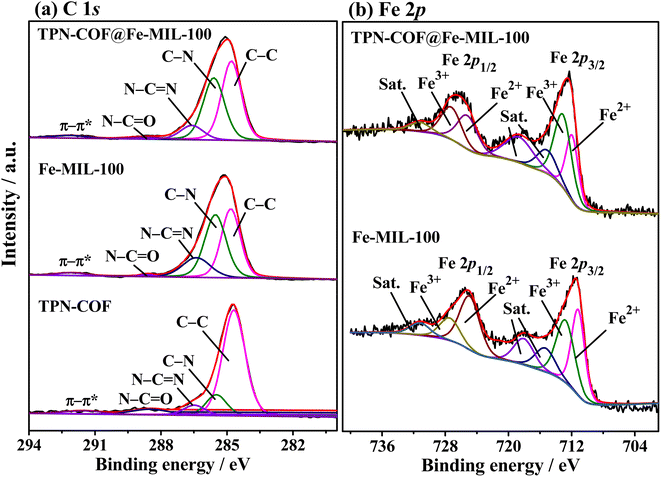 | ||
| Fig. 3 High-resolution XPS spectra of (a) C 1s, and (b) Fe 2p of TPN-COF, Fe-MIL-100 and TPN-COF@Fe-MIL-100. | ||
Additionally, Brunauer–Emmett–Teller (BET) surface area analysis was employed to investigate the porosity for the three samples (Fig. S2†). As calculated, the specific surface areas and average pore diameters of TPN-COF, Fe-MIL-100 and TPN-COF@Fe-MIL-100 are 2.74, 54.36, 68.95 m2 g−1, and 0.5, 11.7, and 12.6 nm, respectively. Results show that the TPN-COF@Fe-MIL-100 is feasible to disperse in the aqueous solution and easy to employ as the scaffold for the/immobilization of probe molecules owing to its high specific surface area and relatively larger mesopores.
3.4 Electrochemical sensing performance of TPN-COF@Fe-MIL-100-based aptasensor toward TET
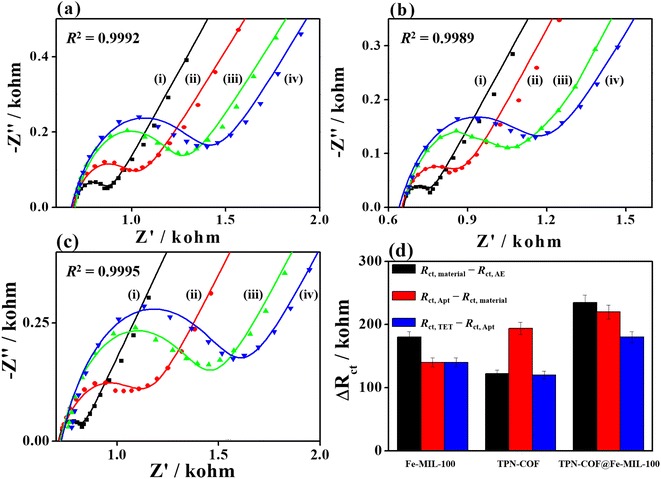
Electrochemical technique can be employed to monitor the changes during the construction of aptasensor. To obtain higher sensitivity and sensing performances of aptasensors, EIS and CV were applied to investigate the electrochemical responses for detecting TET (Fig. 4 and S3†). All EIS Nyquist plots were fitted and analyzed using Zview2 software, for which the nonlinear least-squares were fitted to determine the elemental parameters in the equivalent circuit. The goodness-of-fitting coefficients (R2) information for the fitted EIS plots are provided in Fig. 4. Fe-MIL-100 was chosen as a representative sensing material for detecting TET. In the EIS Nyquist plots (Fig. 4a), the Rct value of bare AE is 170 Ω, indicating good electrochemical activity and fast electron transfer between the electrode and electrolyte. After Fe-MIL-100 is attached to the AE, the Rct value increases to 350 Ω, because Fe-MIL-100 partly hinders electron transfer at the electrode/electrolyte interface. When the aptamer anchored over Fe-MIL-100/AE, the Rct value changes to 490 Ω due to the repulsion interaction between negatively charged phosphate of aptamer and [Fe(CN)6]3−/4− redox probe. Typically, the aptamer strands used for immobilization on the electrode surface are targeted sequences for TET, which can change their configurations to specifically recognize TET and generate the G-quadruplex, therefore realizing the specific recognition toward TET, and further impeding the electron transfer between electrolyte and TET/Apt/Fe-MIL-100/AE. Thus, the Rct value of TET/Apt/Fe-MIL-100/AE increases to 630 Ω. Besides, as for CV curves (Fig. S3a†), the peak current of modified electrode decreases continuously with the construction procedure of aptasensors due to the poorer electrochemical activity and electron transfer between the modified electrode and electrolyte. These results manifest the successful fabrication of Fe-MIL-100-based aptasensor toward the detection of TET. TPN-COF, and TPN-COF@Fe-MIL-100 were also used to construct aptasensors for the detection of TET. For TPN-COF-based aptasensor (Fig. 4b), the Rct value for bare AE is 84 Ω. When bare AE is modified with TPN-COF, incubated with aptamer strands, and used for detecting TET, the Rct value increases to 206, 400, and 520 Ω, respectively. For TPN-COF@Fe-MIL-100-based aptasensor (Fig. 4c), the Rct value for bare AE is 100 Ω, and the Rct value increases to 430, 650, and 830 Ω, when bare AE is modified with TPN-COF@Fe-MIL-100, incubated with aptamer strands, and used for detecting TET, respectively. The Rct values for each step during the fabrication procedures of aptasensors based on the three samples are summarized in Table S1.† The differences in Rct values for each step (ΔRct = Rct,i+1 − Rct,i) for different aptasensors based on TPN-COF, Fe-MIL-100, and TPN-COF@Fe-MIL-100 are summarized in Table S2† and Fig. 4d. In general, the ΔRct value can reveal the amounts of adsorbed species, such as material amount, aptamer adsorption quantity, and binding amount of TET. As shown in Table S2† and Fig. 4d, TPN-COF@Fe-MIL-100 exhibits the maximum ΔRct (Rct,material − Rct,AE) value of 330 Ω, which indicates that the TPN-COF@Fe-MIL-100 loading amounts on bare AE is the largest among the three samples, therefore showing the biggest obstruction for the electron transfer at the electrode/electrolyte interface. After aptamer adsorption, Apt/TPN-COF@Fe-MIL-100/AE shows ΔRct (Rct,Apt − Rct,material) of 220 Ω, which is supreme among the three samples. These reveal the satisfactory immobilization ability of TPN-COF@Fe-MIL-100 for aptamers and the aptamer adsorption quantities achieve the maximum value. When the aptasensors are used for TET detection, TET/Apt/TPN-COF@Fe-MIL-100/AE shows the maximum ΔRct (Rct,TET − Rct,Apt) value of 180 Ω among the three samples, proving its highest detection efficiency for TET. These results are attributed to the unique properties of TPN-COF@Fe-MIL-100, such as abundant active sites, large surface area, excellent electrochemical activity, rich chemical functionality, strong bioaffinity between the organic frameworks and aptamer strands, π–π* stacking, strong electrostatic interaction and 3D porous structure. All the factors are beneficial for immobilizing the aptamer strands and stabilizing the G-quadruplex. Based on the results of the electrochemical activity, adsorption ability, and detection efficiency of the three samples during each step, TPN-COF@Fe-MIL-100 was selected as the optimal platform for developing TET aptasensor.Additionally, the C 1s and P 2p core-level XPS spectra of the TPN-COF@Fe-MIL-100 after aptamer binding and sensing the TET analyte were conducted to investigate the structure changes during the fabrication procedure of the aptasensor (Fig. S4†). As compared with TPN-COF@Fe-MIL-100 before aptamer binding (Fig. 3a), the C 1s core-level XPS spectra of Apt/TPN-COF@Fe-MIL-100 (Fig. S4a†) were deconvoluted into different parts. Among them, the relatively high content of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/N–C
O/N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group was a result of the anchored aptamer strands. However, the relatively high content of the C
O group was a result of the anchored aptamer strands. However, the relatively high content of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/N–C
O/N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group decreased after TET was detected (Fig. S4c†). Since aptamer strands contain –PO4− in their base pairs, the successful adsorption of aptamer strands also can be proven by the presence of the P 2p signal in Apt/TPN-COF@Fe-MIL-100 (Fig. S4b†). Besides, the BEs of P 2p in TET/Apt/TPN-COF@Fe-MIL-100 (Fig. S4d†) shows no change, which suggested that the Apt/TPN-COF@Fe-MIL-100 retains its original structure when detecting TET.
O group decreased after TET was detected (Fig. S4c†). Since aptamer strands contain –PO4− in their base pairs, the successful adsorption of aptamer strands also can be proven by the presence of the P 2p signal in Apt/TPN-COF@Fe-MIL-100 (Fig. S4b†). Besides, the BEs of P 2p in TET/Apt/TPN-COF@Fe-MIL-100 (Fig. S4d†) shows no change, which suggested that the Apt/TPN-COF@Fe-MIL-100 retains its original structure when detecting TET.
After optimization of experimental parameters (S4 Part, Fig. S5†), 1.0 mg mL−1 TPN-COF@Fe-MIL-100 dispersion, 100 nM aptamer solution, 80 min of binding time with aptamers, and pH 7.4 of phosphate buffer were chosen for TET detection.
3.5 Sensitivity of the TPN-COF@Fe-MIL-100-based aptasensor toward TET
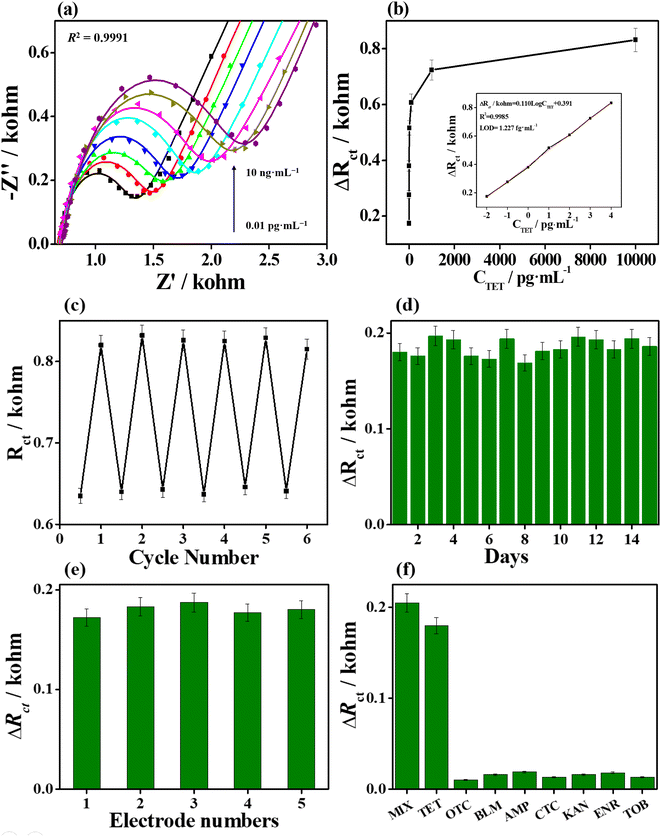
Sensitivity is an important index to evaluate the sensing performance for the proposed aptasensor. High sensitivity can provide more accurate information in the fast and trace detection of antibiotics. A series of TET concentrations were applied to examine the analytical performance of the TPN-COF@Fe-MIL-100-based aptasensor by EIS in 0.1 M phosphate buffer containing [Fe(CN6)]3−/4− redox probe (Fig. 5a). The Rct values increases with increasing TET concentrations in 0.01–10000 pg mL−1, which is ascribed to the specific recognition between aptamer and TET to generate configurational changes in aptamers and form the G-quadruplex, also hindering the electron transfer at the electrode/electrolyte interface. The ΔRct value is deduced from the difference in Rct values before and after TET detection, representing the binding amount of TET. After linear fitting, the ΔRct value acts as a function of the logarithm value of TET concentration, with a linear regression equation ΔRct (kΩ) = 0.110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CTET (pg mL−1) + 0.391 (Fig. 5b), and the correlation coefficient R2 is 0.9985. The limit of detection (LOD) was calculated to be 1.227 fg mL−1 at a signal-to-noise (S/N) ratio of 3. The superior sensitivity of the proposed aptasensor toward TET detection is due to the high porosity and biocompatibility of TPN-COF@Fe-MIL-100, rich chemical functionality and diverse binding forms of TPN-COF@Fe-MIL-100 with aptamers, and high stability of TPN-COF@Fe-MIL-100 to stabilize the G-quadruplex.
CTET (pg mL−1) + 0.391 (Fig. 5b), and the correlation coefficient R2 is 0.9985. The limit of detection (LOD) was calculated to be 1.227 fg mL−1 at a signal-to-noise (S/N) ratio of 3. The superior sensitivity of the proposed aptasensor toward TET detection is due to the high porosity and biocompatibility of TPN-COF@Fe-MIL-100, rich chemical functionality and diverse binding forms of TPN-COF@Fe-MIL-100 with aptamers, and high stability of TPN-COF@Fe-MIL-100 to stabilize the G-quadruplex.
3.6 Regenerability, stability, reproducibility, and selectivity of the TPN-COF@Fe-MIL-100-based aptasensor toward TET
The regenerability and stability of the TPN-COF@Fe-MIL-100-based aptasensor were determined via EIS measurements to evaluate the long-term preservation performance. The regenerability of aptasensor was performed by soaking TET/Apt/TPN-COF@Fe-MIL-100/AE in 0.5 M H2SO4 for 5 min, and then rinsed with phosphate buffer. Afterward, the electrode was incubated with the TET solution (10 fg mL−1) again until the Rct value returned to the origin level. The same procedure was repeated by several cycles. Fig. 5c indicates that the aptasensor can recover an intense EIS value after detecting TET (10 fg mL−1). Moreover, the aptasensor can preserve 98.6% of the original detection efficiency after five cycles, revealing the good regenerability. The stability of aptasensor was evaluated by measuring the ΔRct values at regular intervals (Fig. 5d), with no clear change in response to TET (10 fg mL−1) after storage for 15 days. The aptasensor retains almost 100.7% of the initial electrochemical signal, and the relative standard deviation (RSD) for detecting TET is 4.93%, revealing its superior stability. Five Apt/TPN-COF@Fe-MIL-100/AEs prepared under the same conditions were used to study the reproducibility of the TPN-COF@Fe-MIL-100-based aptasensor toward TET by EIS measurements (Fig. 5e). The ΔRct values show no substantial changes and the RSD for detecting TET (10 fg mL−1) is 3.17%, indicating the good reproducibility of the proposed aptasensor. The selectivity of the TPN-COF@Fe-MIL-100-based electrochemical aptasensor was measured by examining the response of the proposed aptasensor to other interferents, which possibly coexist with TET, including AMP, BLM, CTC, ENR, KAN, OTC, and TOB, as well as the mixture of TET with other antibiotics. As shown in Fig. 5f, the ΔRct values of the TPN-COF@Fe-MIL-100-based aptasensor for detecting TET and a mixture of interferents and TET exhibit significant signal responses, whereas the ΔRct values for other individual interferents show weak signal responses. Notably, the TET concentration (10 fg mL−1) was only 1% of other interferents (1 pg mL−1). These results suggest the superior selectivity of the TPN-COF@Fe-MIL-100-based sensor due to the specific recognition between aptamers and TET.During the regenerability test, XRD patterns were also used to investigate the stability of Apt/TPN-COF@Fe-MIL-100 when detecting TET (Fig. S6†). Obviously, after separately detecting TET for 3, 6, and 9 cycles, the XRD patterns of Apt/TPN-COF@Fe-MIL-100 (i.e., after rinsed TET of TET/Apt/TPN-COF@Fe-MIL-100 with phosphate buffer) are similar with that of pristine Apt/TPN-COF@Fe-MIL-100, indicating that Apt/TPN-COF@Fe-MIL-100 can retain its crystalline structure and keep its stability for detecting TET during a long period and after several cycles.
3.7 Real sample analysis
The practical use of the TPN-COF@Fe-MIL-100 aptasensor was investigated in the pretreated milk sample using spiking and recovery method. Different concentrations of TET were spiked into milk, and further detected using the TPN-COF@Fe-MIL-100-based aptasensor. The TET concentrations in milk sample are calculated and listed in Table 1 according to the calibration curve (Fig. 5b inset) and the deduced equation. As listed in Table 1, the TPN-COF@Fe-MIL-100-based aptasensor shows good recovery in the range of 97.3–107.8%, and the RSDs are in the range of 1.19–3.67%. These results suggest the high sensitivity, reproducibility, regenerability, and feasibility of the proposed aptasensor for TET detection.| Added amount (pg mL−1) | Found amount (pg mL−1) | Recovery (%) | RSD% |
|---|---|---|---|
| 0.01 | 0.0104 | 104.0 | 2.81 |
| 0.1 | 0.0989 | 98.9 | 3.42 |
| 1 | 0.973 | 97.3 | 1.19 |
| 10 | 10.648 | 106.5 | 3.67 |
| 100 | 105.81 | 105.8 | 2.94 |
| 1000 | 1067.04 | 106.7 | 1.97 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 782.69 782.69 |
107.8 | 2.66 |
4. Conclusion
In this work, a highly sensitive platform for detecting TET was developed using the TPN-COF@Fe-MIL-100 nanocomposite as biosensing material. TPN-COF@Fe-MIL-100 possesses advantages such as porous network, large surface area, excellent electrochemical activity, rich chemical functionality, and strong bioaffinity, all of which contributes to largely immobilize aptamer strands and further efficiently determine TET. The TPN-COF@Fe-MIL-100-based aptasensor shows good sensitivity, satisfactory selectivity, stability, reproducibility, regenerability, and practicability for real milk sample. Therefore, TPN-COF@Fe-MIL-100 can be applied in detecting trace amounts of harmful antibiotics residues in food safety.Author contributions
Yubo Meng: investigation, writing – original draft. Yuchun Huang: data curation, methodology. Gailing Huang: formal analysis, writing – review & editing. Yingpan Song: supervision, visualization, project administration, funding acquisition, writing – review & editing.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was supported by Henan Provincial Science and Technology Research Project (No. 222102310493 and 232102320293).References
- C. A. Nadgir and D. A. Biswas, Cureus, 2023, 15, e38251 Search PubMed.
- Y. Sun, J. Zhao and L. Liang, Microchim. Acta, 2021, 188, 21 CrossRef CAS PubMed.
- J. Hong, M. Su, K. Zhao, Y. Zhou, J. Wang, S. Zhou and X. Lin, Biosensors, 2023, 13, 327 CrossRef CAS.
- T. Yao, H. Zhang, C. Feng and Y. He, J. Dairy Sci., 2023, 106, 5916–5929 CrossRef CAS PubMed.
- E. V. Ezenduka, O. J. Okorie-Kanu and J. A. Nwanta, J. Immunoassay Immunochem., 2019, 40, 617–629 CrossRef CAS.
- M. Zhang, S. Zhang, Z. Xu, T. Lv, X. Liu, L. Wang and B. Liu, Talanta, 2024, 266, 124982 CrossRef CAS PubMed.
- F. Hu, Q. Fu, Y. Li, C. Yan, D. Xiao, P. Ju, Z. Hu, H. Li and S. Ai, Food Chem., 2024, 431, 137097 CrossRef CAS PubMed.
- S. Quan, K. Ji, F. Liu, T. H. Barkae, M. I. Halawa, S. Hanif, B. Lou, J. Li and G. Xu, J. Food Drug Anal., 2022, 30, 293–302 CrossRef.
- F. Gao, D. Fan, M. Xiao, H. Liu, Y. Liu, J. Zhang, M. Fang, X. Tan and M. Kong, Environ. Sci. Pollut. Res., 2023, 30, 66696–66704 CrossRef CAS.
- K. Yang, R. Zhu, Z. Li, S. Shuang, Y. Zhai and C. Dong, Talanta, 2023, 266, 125077 CrossRef PubMed.
- L. Haiping, W. Jiangyue, M. Fanping and L. Aifeng, Food Control, 2021, 130, 108356 CrossRef.
- Y. Pang, Int. J. Electrochem. Sci., 2020, 15, 5232–5244 CrossRef CAS.
- G. Liang, L. Song, Y. Gao, K. Wu, R. Guo, R. Chen, J. Zhen and L. Pan, Toxics, 2023, 11, 513 CrossRef CAS.
- K. L. Lin, T. Yang, H. Y. Zou, Y. F. Li and C. Z. Huang, Talanta, 2019, 192, 400–406 CrossRef CAS.
- J. Li, L. Liu, Y. Ai, Y. Liu, H. Sun and Q. Liang, ACS Appl. Mater. Interfaces, 2020, 12, 5500–5510 CrossRef CAS.
- K. Xiang, D. Wu, X. Deng, M. Li, S. Chen, P. Hao, X. Guo, J. Luo and X. Fu, Adv. Funct. Mater., 2020, 30, 1909610 CrossRef CAS.
- F. Duan, C. Guo, M. Hu, Y. Song, M. Wang, L. He, Z. Zhang, R. Pettinari and L. Zhou, Sens. Actuators, B, 2020, 310, 127844 CrossRef CAS.
- T. Xu, H. Dai and Y. Jin, Microchim. Acta, 2020, 187, 1–7 CrossRef PubMed.
- J. Jiang, F. Li, S. Bai, Y. Wang, K. Xiang, H. Wang, J. Zou and J. Hsu, Nano Res., 2023, 16, 4656–4663 CrossRef CAS.
- C. Gu, C. Guo, Z. Li, M. Wang, N. Zhou, L. He, Z. Zhang and M. Du, Biosens. Bioelectron., 2019, 134, 8–15 CrossRef CAS.
- J. Li, J. Zhao, S. Li, Y. Chen, W. Lv, J. Zhang, L. Zhang, Z. Zhang and X. Lu, Nano Res., 2021, 14, 4689–4695 CrossRef CAS.
- X. Ma, C. Pang, S. Li, Y. Xiong, J. Li, J. Luo and Y. Yang, Biosens. Bioelectron., 2019, 146, 111734 CrossRef CAS.
- Y. Deng, Y. Wang, Z. Di, M. Xie, F. Dai, S. Zhan and Z. Zhang, Small Methods, 2022, 6, 2200265 CrossRef CAS PubMed.
- Y. Deng, Y. Wang, X. Xiao, B. J. Saucedo, Z. Zhu, M. Xie, X. Xu, K. Yao, Y. Zhai, Z. Zhang and J. Chen, Small, 2022, 18, 2202928 CrossRef CAS PubMed.
- R. Liu, K. T. Tan, Y. Gong, Y. Chen, Z. Li, S. Xie, T. He, Z. Lu, H. Yang and D. Jiang, Chem. Soc. Rev., 2021, 50, 120–242 RSC.
- Y. Xue, S. Zheng, H. Xue and H. Pang, J. Mater. Chem. A, 2019, 7, 7301–7327 RSC.
- P. Kuhn, M. Antonietti and A. Thomas, Angew. Chem., Int. Ed., 2008, 47, 3450–3453 CrossRef CAS PubMed.
- C. E. Chan-Thaw, A. Villa, P. Katekomol, D. Su, A. Thomas and L. Prati, Nano Lett., 2010, 10, 537–541 CrossRef CAS.
- Y. Meng, Z. Ma, Y. Huang and Y. Song, Anal. Sci., 2023, 39, 901–909 CrossRef CAS PubMed.
- Y. Song, K. Chen, S. Li, L. He, M. Wang, N. Zhou and M. Du, Microchim. Acta, 2022, 189, 338 CrossRef CAS PubMed.
- Y. Song, L. He, S. Zhang, X. Liu, K. Chen, Q. Jia, Z. Zhang and M. Du, Food Chem., 2021, 351, 129248 CrossRef CAS.
- Y. Kim, Y. S. Kim, J. H. Niazi and M. B. Gu, Bioprocess Biosyst. Eng., 2010, 33, 31–37 CrossRef CAS.
- M. Tong, D. Liu, Q. Yang, S. Devautour-Vinot, G. Maurin and C. Zhong, J. Mater. Chem. A, 2013, 1, 8534–8537 RSC.
- M. J. Bojdys, J. Jeromenok, A. Thomas and M. Antonietti, Adv. Mater., 2010, 22, 2202–2205 CrossRef CAS.
- X. Liu, M. Hu, M. Wang, Y. Song, N. Zhou, L. He and Z. Zhang, Biosens. Bioelectron., 2019, 123, 59–68 CrossRef CAS PubMed.
- C. Zhao, J. Wang, X. Chen, Z. Wang, H. Ji, L. Chen, W. Liu and C. Wang, Sci. Total Environ., 2021, 752, 141901 CrossRef CAS.
- R. A. S. Dias, E. R. Sousa, G. S. Silva, L. K. Silva, A. S. Freitas, D. L. D. Lima and É. M. L. Sousa, Microchem. J., 2021, 160, 105633 CrossRef CAS.
- L. Ai, C. Zhang, L. Li and J. Jiang, Appl. Catal., B, 2014, 148–149, 191–200 CrossRef CAS.
- Q. Yang, R. Lu, S. Ren, C. Chen, Z. Chen and X. Yang, Chem. Eng. J., 2018, 348, 202–211 CrossRef CAS.
- W. Li, J. Cao, W. Xiong, Z. Yang, S. Sun, M. Jia and Z. Xu, Chem. Eng. J., 2020, 392, 124844 CrossRef CAS.
- X. Liu, M. Hu, M. Wang, Y. Song, N. Zhou, L. He and Z. Zhang, Biosens. Bioelectron., 2019, 123, 59–68 CrossRef CAS.
- Y. Luo, M. Balogun, W. Qiu, R. Zhao, P. Liu and Y. Tong, Chem. Commun., 2015, 51, 13016–13019 RSC.
- Z. Ma, L. Ren, S. Xing, Y. Wu and Y. Gao, J. Phys. Chem. C, 2015, 119, 23068–23074 CrossRef CAS.
- G. Mayer, Angew. Chem., Int. Ed., 2009, 48, 2672–2689 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra05452g |
| This journal is © The Royal Society of Chemistry 2023 |