Open Access Article
Open Access ArticleEngineering oncogene-targeted anisamide-functionalized pBAE nanoparticles as efficient lung cancer antisense therapies†
Cristina Fornaguera *,
Antoni Torres-Coll,
Laura Olmo,
Coral Garcia-Fernandez,
Marta Guerra-Rebollo* and
Salvador Borrós
*,
Antoni Torres-Coll,
Laura Olmo,
Coral Garcia-Fernandez,
Marta Guerra-Rebollo* and
Salvador Borrós
Grup d'Enginyeria de Materials (Gemat), Institut Químic de Sarrià (IQS), Universitat Ramon Llull (URL), Spain. E-mail: cristina.fornaguera@iqs.url.edu
First published on 13th October 2023
Abstract
Non-small cell lung cancer (NSCLC) is one of the leading causes of worldwide death, mainly due to the lack of efficient and safe therapies. Currently, NSCLC standard of care for consist on the use of traditional chemotherapeutics, non-selectively distributed through the whole body, thus causing severe side effects while not achieving high efficacy outcomes. Consequently, the need of novel therapies, targeted to modify specific subcellular routes aberrantly expressed only in tumor cells is still urgent. In this context, the delivery of siRNAs that can know-down overexpressed oncogenes, such as mTOR, could become the promised targeted therapy. However, siRNA effective delivery remains a challenge due to its compromised stability in biological fluids and its inability to cross biological and plasmatic membranes. Therefore, polymeric nanoparticles that efficiently encapsulate siRNAs and are selectively targeted to tumor cells could play a pivotal role. Accordingly, we demonstrate in this work that oligopeptide end-modified poly(beta aminoester) (OM-pBAE) polymers can efficiently complex siRNA in small nanometric particles using very low polymer amounts, protecting siRNA from nucleases attack. These nanoparticles are stable in the presence of serum, advantageous fact in terms of in vivo use. We also demonstrated that they efficiently transfect cells in vitro, in the presence of serum and are able to knock down target gene expression. Moreover, we demonstrated their antitumor efficacy by encapsulating mTOR siRNA, as a model antisense therapy, which showed specific lung tumor cell growth inhibition in vitro and in vivo. Finally, through the addition of anisamide functionalization to the surface of the nanoparticles, we proved that they become selective to lung tumor cells, while not affecting healthy cells. Therefore, our results are a first step in the discovery of a tumor cell-targeted efficient silencing nanotherapy for NSCLC patients survival improvement.
Introduction
Lung cancer is one of the most common malignancies and leading causes of worldwide cancer-related deaths, being non-small cell lung cancer (NSCLC) the most common type and thus a major cause of death over the world.1 The European Society for Medical Oncology (ESMO) has rated an age-standardized incidence of around 50 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants, with an increase death rate in women in Europe for the years to come, mainly due to the growing of smoking habits in the last decades.2
000 inhabitants, with an increase death rate in women in Europe for the years to come, mainly due to the growing of smoking habits in the last decades.2
Currently, various treatment options are available, including surgery, radiotherapy, chemotherapy and immunotherapy. Although, chemotherapy remains the standard of care for most NSCLC subtypes and disease stages.3 Chemotherapeutic agents (i.e. cisplatin) are intravenously administered, so they are rapidly spread throughout the whole body via the bloodstream circulation, affecting tumor malignant cells but also all other rapid dividing cells in the body. Consequently, adverse side effects such as gastrointestinal problems, infection, fatigue, and hair loss are common. Therefore, the need for novel targeted therapies is urgent.
It has been known for a long time that tumor cells present aberrant gene expression, being some genes, named as oncogenes, overexpressed in tumor cells. In this context, gene therapy devoted to modifying oncogene expression aroused as a promising alternative to produce localized effects in the tumor microenvironment.3 It has the potential to treat a huge variety of congenic and acquired diseases, such as cancer, in which traditional pharmacological therapies consisting of small molecule drugs have failed.4
Oncogenes usually correspond to apoptosis and cell growth metabolic routes, such as those involved in the Akt/mTOR pathway3,5 (see Graphical abstract). The mechanistic mammal target of rapamycin (mTOR) pathway is abnormally activated in many NSCLC patients and it has been attributed to NSCLC development and maintenance. Specifically, NSCLC patients with overexpressed mTOR experience shorter overall and progression-free survival, thus requiring higher doses of current treatments to achieve tumor regression.6 In addition, many other common alterations in oncogenic pathways found in NSCLC, such as EGFR, TP53 and KRAS genes converge in alterations of the mTOR gene. Therefore, mTOR has a key role in the control of cell proliferation and metabolism, through its function negatively regulating apoptosis for promoting cell growth and replication.3,6,7 Consequently, inhibition of the mTOR overexpression has been described as a promising strategy to treat NSCLC patients showing different mutational patterns thanks to the decrease of tumor cells growth.6–9 mTOR has been also related with metastasis development from lung tumors,10,11 so therapies inhibiting mTOR overexpression are envisaged in any stage of NSCLC development. To achieve this inhibition, small interference RNA (siRNA) can be used as a gene knockdown strategy.
siRNAs, short non-coding RNA duplexes that target mRNA to produce gene silencing, have broadened the realm of nucleic acids. They have played an increasingly important laboratory tool in the last twenty years since their discovery by Fire, Mello and coworkers in the late nineties.12 They are naturally occurring macromolecules present in mammal cells, that target specific sequences of mRNA transcripts to avoid their expression (gene silencing) through cleavage in a sequence-specific manner. They are advantageous among other silencing nucleic acids in terms of single mRNA target specificity, which enhances safety of resulting therapies by reducing off-target effects.4,13–19 Therefore, their use has spurred cancer research over the last years to silence genes of interest, including those impossible to target by using small molecules or proteins.20 Using a siRNA complementary to the mTOR mRNA transcript, it was possible to knockdown the expression of this overexpressed oncogene and thus, reduce the replicating capacity of tumor cells.3,16 However, siRNAs are labile molecules, prone to degradation by nucleases, which are present in the whole-body fluids and tissues. In addition, they can enhance immune system activation and their ability to penetrate cells to be located in the cytoplasm, their subcellular site of action, is really limited.3,4,17,21 Therefore, although they hold great promise for overexpressed tumor oncogenes silencing, their therapeutic potential is hampered when they are used naked. In this context, polymeric nanoparticles appeared as a promising solution for the selective delivery of labile nucleic acids. Although the numerous studies reporting the design of novel polymers for biomedical applications, with good performance in vitro and even in preclinical tests, their translation to clinics is markedly limited.22–26 This is due to the fact that although nanoparticles present many advantages in a theoretical field, such as specific vectorization to the target organ that potentiates therapeutic effects and reduces side effects, protection of the active principle inside the nanosystem and spatial and time-controlled release of the actives they bring; these advantages are difficult to accomplish in practice.22–26
In our group, we designed a family of cationic oligopeptide end-modified poly(beta aminoester) (OM-pBAE) biocompatible polymers to electrostatic interact with anionic nucleic acids and form nanometric polyplexes. These polymers are easily biodegraded thanks to the presence of ester groups along the polymer backbone, thus avoiding accumulation-related toxicities. Moreover, thanks to the presence of cationic amino acids, they promote proton sponge effect when they are endocytosed, thus releasing their cargo directly in the cytoplasm, only a few hours after their in vivo administration.27,28 Additionally, they can be functionalized to include targeting moieties on their surface, thus promoting a selective delivery to target cells.29 Thus, they could be selectively directed to tumor cells.
In this context, the main objective of the present study is to demonstrate that mTOR-encoding siRNA-loaded OM-pBAE nanoparticles can be easily obtained and that these nanoparticles can promote efficient gene knockdown thus resulting in a selective tumor cells death without toxic effects in other cell types. To achieve a selective accumulation of the nanoparticles only on tumor cells, they have been functionalized, for the first time, with anisamide chemical group. We selected anisamide as the active targeting moiety. Anisamide is a ligand that binds to Sigma receptor, a receptor known to be overexpressed in a variety of tumors like non-small cell lung cancer.30,31 Although its mechanism of action to determine interaction with the receptor has not been clearly elucidated yet32,33 its use for tumor cells selective targeting has demonstrated clear, namely for prostate cancer, where numerous references exists.30,31,34 Thus, the presence of anisamide on the surface of pBAE nanoparticles was hypothesized to promote their selective accumulation on tumor cells. To achieve our objective, we started with the synthesis of this new polymer, including anisamide as the targeting moiety. Following, once nanoparticles synthesized, they were physicochemically characterized. Then, nucleic acid binding, entrapment efficiency and stability in serum to achieve cytoplasmatic delivery of the siRNA was demonstrated, together with their hemo- and biocompatibility. Next in vitro and in vivo studies demonstrated their capacity to produce a selective mTOR gene knockdown that ends up with tumor cells death, together with the selective lung tumor cell targeting.
Results
OM-pBAE polymers efficiently encapsulate siRNA forming small cationic nanoparticles
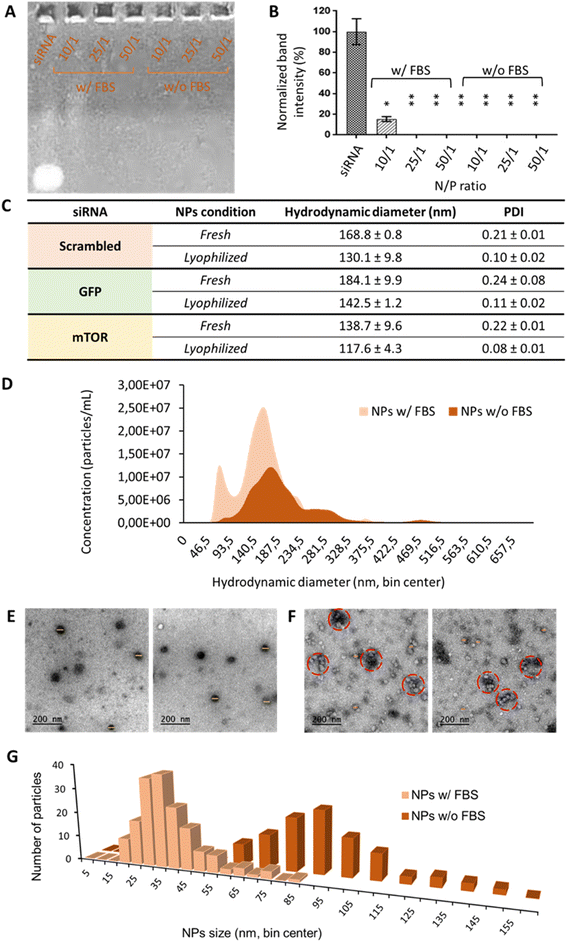
Oligopeptide end-modified poly(beta aminoester) (OM-pBAE) nanoparticles were formulated, based on our previous experience,35–37 using the so-called C6 polymer, which includes hexylamine as a monomer to achieve the half of its lateral chains with a higher hydrophobicity, previously demonstrated to result in more stable nanoparticles, able to be lyophilized without losing their properties.36,38 Concerning the end-oligopeptide modifications, 60/40 w/w mixture of arginine/histidine (R/H)-modified pBAEs was used, since they were previously reported to form discrete nanometric particles. The addition of a lyoprotectant buffer enabled the freeze-drying of the nanoparticles without losing their quality control attributes. In the current work, since it was the first time that this specific combination of OM-pBAE was used for the encapsulation of small interfering RNA (siRNA, codifying either from GFP, mTOR or non-codifying – scrambled), we first confirmed their capacity to encapsulate this kind of nucleic acids. As it can be observed in Fig. 1, and as expected, naked siRNA penetrated and run though agarose gel. When it was encapsulated in OM-pBAE nanoparticles, the signal disappeared, which indicated that siRNA had been retained in the well by the polymer, thus, all tested ratios were able to allow the electrostatic interaction between the cationic pBAE polymers and the anionic siRNA for the complexation of siRNA in discrete nanoparticles. To go deep into the study of complexation capacity, the same experiment was performed with a previous incubation of NPs with FBS, to simulate the physiological environment that nanoparticles will find when intravenously administered. The destabilization under the presence of proteins has been described previously, thus not enabling systemic delivery and transforming therapies to local deliveries, as performed before in our group.16 Different polymer/siRNA mass ratios were tested. In this present case, as quantified in Fig. 1B, 10/1 ratio was not sufficient to enable the complete encapsulation of siRNA. Therefore, blood proteins, as described in previous studies,37 have a key role in the biological identity of nanoparticles, which is required to study if in vivo systemic administration is envisaged; thus, it was decided to continue the experiments with 25/1 N/P ratio, since it was the minimum polymer amount required to complex siRNA. It must be remarked that 10/1 and even 25/1 N/P ratios represents a very low amount of polymer, as compared to previous pBAE nanoparticles, required to encapsulate siRNA and protect it from destabilization by protein attachment, advantageously from our previous studies in terms of toxicity reduction,16 effect that can be attributed to the possibility to lyophilize nanoparticles. Although there are a few previous studies dealing with nucleic acids encapsulation that report so low N/P ratios,3 they did not achieve a complete siRNA encapsulation and, in fact, most previous reports required higher amounts of polymer,16–18,39–42 reaching to N/P ratios around 100/1 in most cases and even requiring 450/1 to achieve a complete gene silencing;42 which is detrimental in terms of nanoparticles efficiency and biocompatibility, due to the above-mentioned low N/P ratio and consequent reduced polymer amount, which only plays the carrier function. Therefore, our polymers are efficient in complexing high amounts of siRNA.Once selected the most appropriate N/P ratio to complex siRNA, resulting nanoparticles were physicochemically characterized. The three siRNAs used along this work were encapsulated since they have different molecular weight (scrambled = 13.3 kDa; GFP = 12 kDa; mTOR = 9.5 kDa), which could affect properties of nanoparticles. Results, observed in Fig. 1C, show that independently on the siRNA molecular weight, we were able to achieve small nanometric particles (diameters lower than 200 nm in all formulations) and a very low polydispersity, which enables to define most of them as monodisperse systems. It is worth remarking the small sizes with lower PDI achieved as compared to other pBAE nanoparticles,39–41 which are advantageous in terms of our final aim: the intravenous administration. In addition, and as we demonstrated before,36 we have a robust methodology for the efficient lyophilization of our nanoparticles that does not affect their physicochemical properties. To remark, as already discussed in our previous studies,43,44 the hydrodynamic diameters of the nanoparticles slightly shrinked due to freeze-drying, which can be attributed either to the elimination of possible solvents/buffers form the inner structure of the nanoparticle or to the interactions of the sucrose, added as lyoprotectant, with the polymer/nucleic acid, contributing to the stabilization of the polyplexes.
Small nanometric sizes below 200 nm were confirmed by cryoTEM analysis (Fig. 1E). In fact, by cryoTEM, mean diameter sizes of serum-free nanoparticles were found to be around 80 nm (Fig. 1G), markedly lower than those found by DLS/NTA, difference already expected since the first technique measures the hard sphere size of the nanoparticles, while light scattering techniques measure the hydrodynamic diameter. When nanoparticles were incubated with FBS (Fig. 1F and G), their size was found even smaller, of around 35 nm of mean diameter. This effect, previously reported in bibliography to affect nucleic acid packaging by cationic polymers,45,46 could be attributed to a nanoparticle shrinkage produced by their interaction with FBS proteins: it seems that serum proteins produce some kind of protective effect by coating nanoparticles surface to look for the most thermodynamically stable conformation. As it can be seen in micrographs of Fig. 1F, nanoparticles, although without losing their integrity and sphere morphology, interacted with FBS producing nanometric aggregates of sizes around 160 nm.
siRNA encapsulation in pBAE nanoparticles allows its in vivo use thanks to protecting it from nucleases and ensuring safe formulations
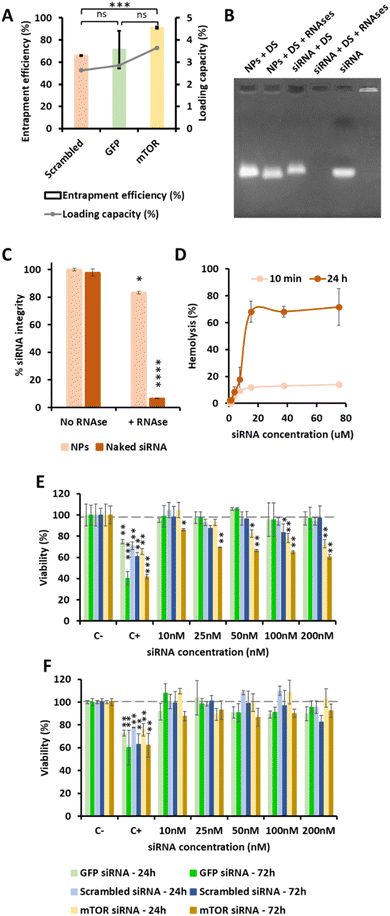
OM-pBAE showed an entrapment efficiency higher than 60% in all cases (Fig. 2A). Although no significant differences were found, it is clear that mTOR siRNA was the one demonstrating a higher encapsulation (around 90%), which was attributed to the smaller size of this siRNA as compared to the other two. The smaller the nucleic acid length to encapsulate, the easier its encapsulation when using pBAE polymers, as we expected from our experience encapsulating other types of nucleic acids (see Table S1, from ESI†). These entrapment efficiencies are high as compared to other nanoparticle types, and taking into account that the N/P ratios used include less polymer than most previously reported nanoparticles.39–41,47 In addition, the therapeutic formulation, that one encapsulating siRNA mTOR, shows an almost complete encapsulation, thus the results are promising. Nevertheless, if required, polymer ratio could be increased to increase entrapment efficiency. In close relationship, the loading capacity was around 3%, showing an increase when mTOR siRNA was used. Although the notable amount of polymer that contributes to the low drug loading percentage, these values are higher than those obtained by other authors using similar polymers.41,48 This means that the formulation is balanced through the siRNA: using a same amount of polymer, we are able to encapsulate a higher amount of siRNA; which added to the lower N/P ratios as compared to those found in previous bibliography (see Fig. 1) makes our nanoparticles more efficient in terms of using a reduced amount of the carrier as compared to the active principle (the siRNA). In fact, the number of siRNA molecules per nanoparticle can be calculated knowing the siRNA concentration added and the number of nanoparticles per volume unit (found from NTA measurements, Fig. 1D). Thanks to this low N/P ratio, we are able to encapsulate more than 4000 siRNA molecules/NP, which is two orders of magnitude higher than values reported previously.49 This is a clear competitive advantage in terms of costs and process efficiency that should facilitate the arrival of nanoparticles to marketing.To enable the in vivo use of these nanosystems, it is required to protect the encapsulated siRNA from degradation by nucleases, which are common in biological systems. A first indication that pBAEs are able to protect encapsulated siRNAs was found when testing the complexation stability. As reported above, when nanoparticles were incubated with FBS, to simulate physiological proteins, we found siRNA encapsulation from 25/1 ratios (Fig. 1). Further, to confirm nucleases protection, nanoparticles were incubated with ribonucleases, which specifically degrade naked RNA before it arrives to its biological fate. OM-pBAE nanoparticles were able to circumvent RNAses attack, thus, achieving more than 80% of the RNA integrity protection (Fig. 2B and C), slightly higher than data from previous studies with similar particles.18,41 Therefore, our OM-pBAE nanoparticles are able to protect RNA from nucleases, being this another competitive advantage in terms of direct in vivo administration of nanoparticles.
Concerning cellular compatibility, since we aim a parenteral use of our formulation, both hemocompatibility and cell compatibility were studied in parallel (Fig. 2D to F). As for hemocompatibility, as expected, the increase on the nanoparticle concentration and the incubation time, produced an increase in hemolysis from 0 to 20 μM siRNA concentration, where a plateau of hemolysis was reached (Fig. 2D). After 10 min incubation, 10% hemolysis was produced from 5 μM siRNA concentration. Nevertheless, increasing the incubation time up to 24 h, around 70% hemolysis was produced at siRNA concentrations equal or higher than 10 μM. Hemotoxicity was also found in previous studies using other kinds of polymeric nanoparticles,50 although hemolysis has not been routinely studied in most studies that have already performed in vivo tests. Our nanoparticles are intended to be administered through intravenous route, thus, their blood compatibility is a must. Although after 24 h, high hemolysis percentages were found at high siRNA concentrations, this is not an issue concerning in vivo use. It must be taken into account that, once in circulation, nanoparticles are distributed to organs and extravasated in some minutes;51 therefore, they are not expected to circulate through bloodstream more than this time. In addition, concentrations tested could be in the range or even higher than those required for in vivo use,10 so decreasing the concentration has been demonstrated to reduce hemolysis. Consequently, since at 10 min hemolysis percentages are very low, these nanoparticles are considered appropriate for their intravenous use.
Cytotoxicity studies, performed in two model cell lines: A549 cells as tumor model to treat, and BEAS-2B as healthy epithelial non-target cells, showed that only using mTOR siRNA, a concentration-dependent cytotoxicity was produced, and only in target tumor cells (Fig. 2E and F). As expected, in cultures of model healthy cells, no cytotoxicity was observed in any condition, except for lipofectamine (positive control) used, as widely reported previously.35 In addition, OM-pBAE polymers neither siRNA macromolecules are toxic by themselves since the use of a non-codifying siRNA as well as that codifying for a reporter protein (GFP) did not produce any toxicity (Fig. 2E and F). It is important to remark the lung tumor cells death produced by mTOR siRNA encapsulated nanoparticles. These results will be further discussed in the next section, where the efficacy of engineered nanoparticles is detailed.
pBAE NPs promoted enhanced uptake and transfection of encapsulated reporter siRNA
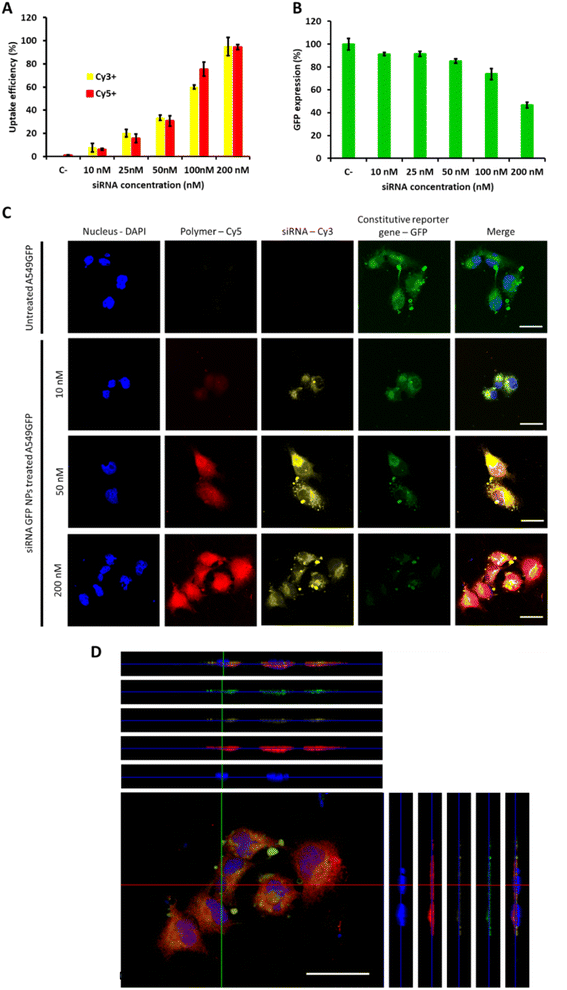
Once confirmed the safety of the proposed mTOR siRNA-loaded OM-pBAE nanoparticles treatment, their cellular interaction was studied. Firstly, NPs uptake was studied in model lung cancer cells, by labelling the polymer and the siRNA simultaneously (Fig. 3A). As expected, we observed a concentration dependent uptake, both for the polymer and for the siRNA, arriving to the whole uptake at the highest dose tested (200 nM) after 24 h nanoparticle incubation with cells, according to previous studies which found the maximum uptake after this time using similar or even higher concentrations.41,52 Following, silencing efficacy was tested using a siRNA codifying for a reporter gene, GFP, to silence the expression of A549-GFP cells, previously labelled to constitutively express this reporter gene. As shown in Fig. 3B, the dose of 200 nm was required to achieve 50% of GFP silencing, which could be attributed to its higher uptake efficacy. Therefore, our nanoparticles are efficient in silencing genes in vitro. Free siRNA cannot diffuse through plasmatic membranes, due to their high molecular weight and overall negative charge,41 therefore, with these data, we re-confirm our previous study in which we demonstrated the ability of cationic OM-pBAE polymer to facilitate association with anionic cell membranes and subsequent internalize nucleic acids.27 The correlation between polymer and siRNA uptake also confirms that even using this reduced N/P ratio, the polymer amount is sufficient to enable the complete internalization of the whole siRNA amount and reinforce the idea of the nanoparticle stability until they penetrate cells (see Fig. 2). At this point, it is important to remark that all in vitro experiments were performed in the presence of FBS, since we previously confirmed particles stability in serum conditions. This is advantageous as compared to other particles, such as the commonly used polyethyleneimine (PEI) polyplexes, which require a serum-free media to enable nucleic acids transfection and thus, they cannot be used for in vivo applications.53,54 These silencing efficiencies are like those found in previous studies using similar polymers to encapsulate siRNAs codifying by different genes.42,55 Nevertheless, taking into account that this dose was far from being toxic and in our previous studies using other nucleic acid types we could use higher particle concentrations,35 if a higher silencing percentage would be necessary, we could test higher siRNA doses, which is not possible in previous studies found in literature, in which they achieved very high silencing efficiencies but together with high mortality ratios, probably caused by the use of high doses of polymer and high N/P ratios.18,42,56 Accordingly, longer nanoparticle incubation times could also result in higher silencing efficiencies.55Going deep into particles uptake and transfection studies, we further performed confocal microscopy to localize particles inside cells (Fig. 3C and D). We selected three siRNA doses, 10, 50 and 200 nM; corresponding to 10, 40 and 100% uptake and 10, 15 and 60% silencing efficiencies respectively. Concerning uptake labelling, both fluorophores can be found in the whole cell cytoplasm (Fig. 3C and D), and not only on cell surface, thus confirming that both, polymer and siRNA penetrate cells. Although Cy5 and Cy3 signals are both higher when increasing the concentration, the overall fluorescence of Cy5 is much higher than that from Cy3, as expected, since even using this low N/P ratios, polymer concentration is in an excess in the formulation, but this excess is required to maintain nanoparticles formed, as we demonstrated in our previous study,27 due to the dynamic character of the formulation. In addition, it is worth noting that both signals do not colocalize completely, which means that, once inside the cell, particles decomplex and each component follows its fate. This result was already expected, thanks to the ability of pBAE polymers to escape from endosomes due to the proton sponge effect,39,40 but this is the first time that we can visualize this effect under the microscope.
pBAE NPs are able to silence mTOR and produce a tumor cell specific growth inhibition in vitro
mTOR silencing and specific capacity to inhibit tumor cells growth was studied by comparing the effect of siRNA codifying for mTOR with that of a non-codifying siRNA (scrambled), to discard the possible interference of the polymer on the result. mTOR protein expression after 72 h nanoparticle incubation, quantified by western blot, was normalized using β-actin, a constitutively expressed protein (Fig. 4A–C). As expected, the use of a scrambled siRNA did not produce any effect on the mTOR protein expression; while the use of anti mTOR siRNA was able to decrease mTOR expression in a concentration dependent manner, thus achieving the highest mTOR protein expression inhibition when higher siRNA mTOR concentrations were used. Our OM-pBAE nanoparticles were able to silence up to 60% of mTOR expression, when siRNA mTOR was encapsulated at 100 nM. As discussed by GFP silencing, this silencing efficiency is within the order of magnitude of previous reports, but in our case, our nanoparticles can be considered more efficient since we achieved these high silencing after shorter times of particles incubation (72 h vs. >5 days in previous studies) and with lower N/P ratios and concentrations that are completely biocompatible with cells and could be increased safely in case of higher silencing required.35,42,55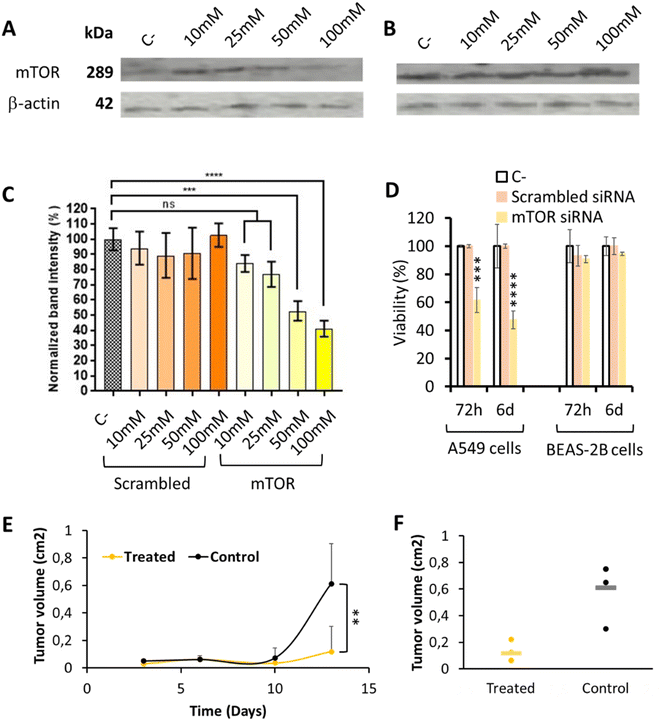 | ||
| Fig. 4 mTOR silencing efficiency in lung tumor model cells. (A)–(C) Expression of mTOR protein determined by western blot analysis in A549 cells, after 72 h nanoparticle incubation, by using increasing concentrations of pBAE nanoparticles loaded with scrambled or mTOR siRNAs (see whole western blots in Fig. S9†). (A) Western blot image showing a marked reduction of mTOR protein when cells were transfected with increasing amounts of mTOR siRNA encapsulated in OM-pBAE nanoparticles. (B) Western blot image showing a similar concentration of mTOR protein when cells were transfected with increasing amounts of scrambled siRNA encapsulated. (C) Quantification of the band intensity signal corresponding to the mTOR protein concentration, normalized by β-actin expression. (D) Percentage of viability, as a function of incubation time and type of siRNA encapsulated, for two different cell lines incubated with 200 nM siRNA. (E) Tumor volume growth as a function of time, for the treated and control groups. (F) Individual values of the tumor volume quantification at the final time point. Results correspond to the mean ± standard deviation calculated from at least, a triplicate. | ||
Additionally, tumor cells specific growth inhibition was studied by quantifying their viability in vitro, as represented in Fig. 4D. As already indicated, only those nanoparticles loading the mTOR siRNA produced a dose-dependent cytotoxic effect only in A549 lung tumor model cells, in a time-dependent manner while healthy cells and those treated with scrambled siRNA growth normally. This toxicity, higher after 6 days mTOR siRNA incubation, can be attributed to the inhibition of the mTOR gene, overexpressed in lung tumor cells, as for bibliographic data,3,52 since nanoparticles loaded with other siRNAs did not produced any cytotoxicity. In addition, the incubation of these nanoparticles with healthy cells did not produce toxic effects. Therefore, this experiment represents the confirmation of the safety character of our nanoparticles in healthy, non-targeted tissues, as well as their efficacy in lung tumor cells, which make them specific against the target cells.
Additionally, in vivo local administration of the mTOR siRNA-loaded nanoparticles confirmed their capacity to significantly retard tumor growth (Fig. 4E and F), which is another promising result referring to the efficacy of our system. Nevertheless, when thinking on translating these preclinical results to clinics, for lung tumor patients, the intratumoral administration is only feasible for resected patients during the surgical procedure. However, in most cases, they are diagnosed in late metastatic disease stages, where tumors are spread and surgery is not recommended, and this is why the addition of an active targeting moiety is encouraged.57–62
The addition of a targeting moiety is required to achieve a selective effect on tumor cells: anisamide functionalization as the solution
Although the previously promising results in terms of selective mTOR silencing of tumor cells, it is fair to comment that this selectivity can be observed in a non-competitive in vitro environment, where no other cells are present. Therefore, for the success of a systemic parenteral in vivo therapy, the addition of a selective targeting moiety was required.In our case, we covalently attached the anisamide chemical group to our OM-pBAE polymers (Fig. 5A). Specifically, this new targeted polymer was oligo-peptide end modified with arginine and the new targeted nanoparticles (named in the further as AA-pBAE) were a mixture of 30% of R polymer + 30% of R polymer functionalized with the anisamide (AA) targeting moiety + 40% of H polymer (30/30/40). To remark, 40% of histidine-modified polymer was maintained to enable the correct lyophilization, as we described previously.63 Since it was the first time that this specific mixture of polymers was used for the encapsulation of genetic material, we first confirmed their capacity to encapsulate siRNA mTOR. Different ratios of OM-pBAE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA were tested from 25
siRNA were tested from 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 100
1 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. EMSA gel shows that while naked siRNA has penetrated and run thought agarose gel, when siRNA was encapsulated in OM-pBAE nanoparticles, the signal disappeared, indicating that de genetic materials is encapsulated into discrete nanoparticles (Fig. 5B). The ratio chosen was the 75
1. EMSA gel shows that while naked siRNA has penetrated and run thought agarose gel, when siRNA was encapsulated in OM-pBAE nanoparticles, the signal disappeared, indicating that de genetic materials is encapsulated into discrete nanoparticles (Fig. 5B). The ratio chosen was the 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which demonstrated a full encapsulation for siRNA mTOR (Fig. 5B and C). To note, more polymer amount is required when anisamide targeting was added, which could be attributed to the addition of the targeting moiety using a poly(ethylene glycol) side chain, which decreases the overall cationic charge of the polymer chain, making it necessary a higher amount of polymer to encapsulate a same quantity of nucleic acid. The hydrodynamic diameter of the AA-pBAE NPs was slightly higher than those found for siRNA mTOR encapsulating non-targeted nanoparticles, also attributed to the presence of the anisamide-functionalized lateral chains, maintaining a monodisperse and cationic population (Fig. 5D).
1, which demonstrated a full encapsulation for siRNA mTOR (Fig. 5B and C). To note, more polymer amount is required when anisamide targeting was added, which could be attributed to the addition of the targeting moiety using a poly(ethylene glycol) side chain, which decreases the overall cationic charge of the polymer chain, making it necessary a higher amount of polymer to encapsulate a same quantity of nucleic acid. The hydrodynamic diameter of the AA-pBAE NPs was slightly higher than those found for siRNA mTOR encapsulating non-targeted nanoparticles, also attributed to the presence of the anisamide-functionalized lateral chains, maintaining a monodisperse and cationic population (Fig. 5D).
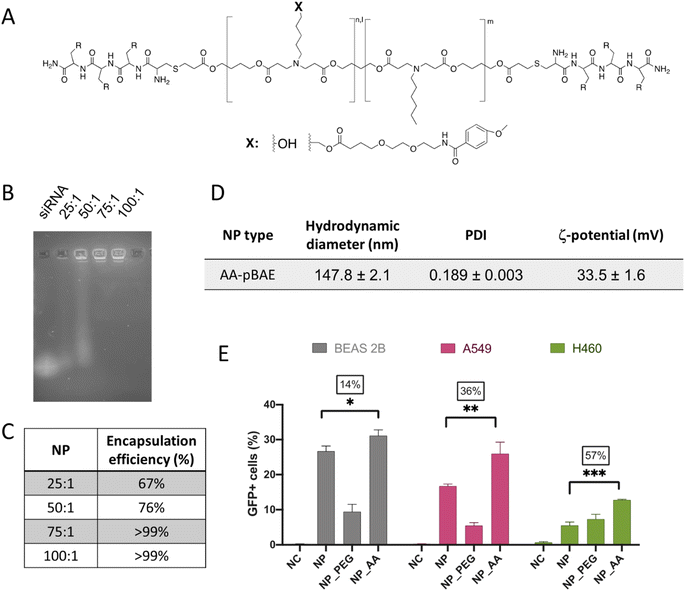 | ||
| Fig. 5 Development of the anisamide-targeted nanoparticles that selectively act on tumor cells. (A) Chemical structure of AA-pBAE (see synthetic procedure in Fig. S7, from ESI†). (B) Encapsulation efficacy of pBAE nanoparticles by EMSA gel. (C) Quantification of the EMSA gel. (D) Table summarizing the NPs physicochemical characteristics. (E) Transfection efficiency of pGFP-loaded NPs, as a function of the NP type and cell line. Results correspond to the mean values ± standard deviation calculated from at least, a triplicate. | ||
Regarding the effect of the anisamide targeting, it was studied in vitro, first by a selectivity test of transfection efficacy in non-target (BEAS-2B) as compared to target (A549 and H460) lung tumor cell models. As clearly shown in Fig. 5E, while in non-target cells, transfection was enhanced only 14%, the increase on the transfection raised up to 36% and to 57% in A549 and H460 target tumor cells. Thus, it is clear the selective targeting effect of the anisamide functionalization in tumor cells, attributed to the interaction of NPs with the plasmatic membrane, not only by electrostatic charges but also to the sigma receptor.
Discussion
mTOR gene has been widely related with different types of tumors development, being nowadays levels of mTOR a biomarker for predicting lung tumor responses. Specifically, in NSCLC, its overexpression has been demonstrated as a predictor of shorter overall and progression-free survival in patients under current treatments.6,7 Therefore, any strategy that aims to the downregulation or silencing mTOR gene could represent a promising treatment, that, administered before current standard of care therapies, could sensitize patients and prolong their progression-free survival. Small-interfering RNA (siRNA) are molecules of interest for gene silencing, since they are non-coding short duplexes of RNA that interact specifically with only one mRNA target, thus achieving its silencing in a post-transcriptional level avoiding off-target effects. Nevertheless, as small nucleic acids, siRNA are very labile molecules, which easily degrade when in contact with physiological tissues and fluids, so for their in vivo use, a carrier that enables its stability until delivery site is a must.21,48,64,65 Therefore, the ultimate objective of our present work was the development of a polymeric delivery system capable of mTOR silencing through a siRNA selective delivery to non-small cell lung cancer cells, using oligopeptide-end modified poly(beta aminoester) nanoparticles (OM-pBAE). OM-pBAE nanoparticles are polymers of quite low molecular weight, advantageous fact in terms of siRNA encapsulation thanks to the polymer flexibility and adaptability to the nucleic acid conformation to form small nanometric particles.56 Non-specific delivery strategies that widespread active principles through the whole body have a reduced efficacy and, consequently, increased toxic side effects, due to accumulation in non-target organs. Therefore, the use of specifically targeted nanosystems could represent a competitive advantage among traditional strategies. To achieve this objective, in here, we used AA-pBAE nanoparticles, specifically modified with the anisamide moiety on their surface, since it was previously demonstrated the capacity of this benzyl derivative to selectively interact with sigma receptors, overexpressed in tumor cells.30,31,33,34In here, we achieved small nanometric particles (Fig. 1), with sizes appropriate to facilitate cellular uptake.3 These nanoparticles have a high encapsulation efficiency for the protection of siRNA from RNAses attack (Fig. 2), and were stable in the presence of serum, which usually hampers nanoformulations due to the formation of protein corona surrounding nanoparticles that interferes with cationic polymers packaging capacity and does not enable or modifies their biological fate.3,47,53,66,67 This was demonstrated not only by incubating nanoparticles with FBS (Fig. 2), but also by the possibility to perform transfections under serum presence (Fig. 3), only achieved by a reduced number of nanoparticle types before.53,54,56 Additionally, our nanoparticles were formed using low N/P ratios (maximum 75/1 when adding the targeting moiety), which is advantageous in terms of safety and economic issues, as siRNAs are the active principles to maximize and polymers could produce undesired toxicity, being them usually the most toxic component of the formulations. Although the higher stability in the presence of FBS is an advantage, it can be also a drawback, since the decomplexation of the polyplexes is required to achieve further cell transfection.53 This double edge sword was not problematic for our nanoparticles, since serum-stable pBAE nanoparticles were able to efficiently transfect cell cultures in the presence of serum proteins, since, as previously discussed, FBS did not compromise their integrity and stability (Fig. 3); advantageous fact as compared to previous studies, in which the formation of protein coronas surrounding nanoparticles destabilized them and did not enable cell transfection.53,54 It is worth remarking this effect, since the addition of polyanions, coming from serum proteins, did not hamper the stability of our pBAE nanoparticles, formed by electrostatic interactions, as previously reported in other electrostatic polyplexes, which can be attributed to the use of the so-called C6 polymer, as previously referred to be more stable than the first C32 polymer version, thanks to its higher hydrophobicity in lateral chains.35,36,54,68 In this sense, our nanoparticles are advantageous for their further in vivo use, since after their intravenous administration, they are expected to retain their capability to transfect target cells. Even having this high stability in presence of serum, pBAE were also able to deliver siRNA in the cytosol thanks to the demonstrated buffering capacity of their primary and secondary amine groups (proton sponge effect), required to escape from endosomes,39,40 thus making siRNA accessible to promote efficient gene silencing in vitro (Fig. 3 and 4). The cytosol localization of the gene material was demonstrated in our previous article27 without the need of adapting polymers for a cytoplasmic targeting, required in other studies.15 At the same time, during endosomal scape, pBAEs are degraded thanks to their ester, amine and sulfide bonds in their structure, easily hydrolysable in aqueous conditions, in reducing environments and due to the presence of specific enzyme, as it happens in late endosomes.52 This was demonstrated here by confocal microscopy studies (Fig. 3), in which the decomplexation of the particles is demonstrated by the differential subcellular localization of polymer and siRNA. Additionally, the in vivo demonstration of the efficacy of our nanoparticles in silencing mTOR gene and, consequently, reducing tumor growth in mice models became a promising milestone that encouraged further use of this formulation for lung cancer antitumor therapy. However, this study required a local intratumor administration, not feasible for patients, usually diagnosed at late stages of the disease, involving a metastatic, non-localized tumor development.58,69
Being aware that a possible limitation of our study was the lack of a specific targeting moiety; we continued it by adding anisamide in the lateral chains of the polymer (AA-pBAE), since this benzyl derivative was previously reported to selectively bind to sigma receptors, overexpressed in many tumor cells, and, among them, lung cancer.30,31,33,34 Through an array of in vitro studies (Fig. 5), we were able to demonstrate that the addition of this active targeting did really produced a selective action of the mTOR siRNA on lung tumor model cells. In comparison with numerous previous biocompatibility studies in which they only demonstrated the cytotoxicity expected in lung tumor models,3,15,52 we went a step further and successfully demonstrated the safety of our proposed novel therapy in healthy cells (i.e. epithelial cells), which is a must even including a targeting moiety, since receptors are overexpressed, but not solely expressed, in tumor cells. In addition, we demonstrated that this toxicity is produced by the specific inhibition of mTOR overexpression in tumor cells, since lactate dehydrogenase (LDH) viability studies, which measure membrane integrity, did not show any differences between negative controls and samples tested (Fig. S8†), thus confirming that even membrane integrity is not lost, the specific inhibition of the mTOR gene is the cause of the specific tumor cells mortality. It is fair to recognize that cytotoxic effects were present in tumor cells because they are continuously growing cells and mTOR is implicated in the cell growth and proliferation metabolic pathway by regulating protein synthesis in NSCLC cells,3 not so activated in other healthy cell types. However, in organisms, there exist continuously growing tissues, which could also be affected by side effects of our therapy. This is the reason why we selected two epithelial cell models as non-target cells, to demonstrate the safety of our nanoparticles expected in vivo in continuously growing cells (Fig. 4 and 5).
Therefore, taking all results together, we are convinced that we have found a promising candidate, appropriate to be intravenously administered, for the treatment of NSCLC patients in a near future.
Experimental
Materials
Reagents and solvents used for synthesis were purchased from Sigma Aldrich and Panreac. CK3 (NH2-Cys-Lys-Lys-Lys-COOH) and CH3 (NH2-Cys-His-His-His-COOH) peptides were obtained from GL Biochem (Shanghai) Ltd with a purity of at least 98%. pGFP used was amplified in the research group, using the PureLink™ HiPure Plasmid Filter Maxiprep Kit. siRNAs were acquired from Dharmacon™ (GE Dharmacon USA). Three different siRNAs were used, that codified for: (1) nothing (scrambled, as a control; MW = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1); (2) GFP protein (GFP duplex I; MW = 13
000 g mol−1); (2) GFP protein (GFP duplex I; MW = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 g mol−1); and (3) mTOR protein (SMARTpool: ON-TARGETplus mTOR siRNA with the following sequences: GGCCAUAGCUAGCCUCAUA, CAAAGGACUUCGCCCAUAA, GCAGAAUUGUCAAGGGAUA, and CCAAAGCACUACACUACAA MW = 9500 g mol−1). In addition, cyanin-3 labelled siRNA anti-GFP (Silencer™ Cy™ 3-labeled siRNA) was also used for uptake studies. They were used at an initial concentration of 50 μM (0.266 μg μL−1) in DEPC water. siRNA working concentration was set up at 0.03 μg μL−1. Anti-mTOR antibody mouse IgG1 clone 6H9B10, anti-β-actin antibody mouse IgG2 clone 2F1-1 and goat anti-IgG mouse antibody clone Poly4053, conjugated to HRP were acquired from BioLegend. Immobilon-FL PVDF, 0.45 μm membranes were obtained from EMD Millipore USA. All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Cyanine dyes were purchased from Lumiprobe. Paraformaldehyde (PFA), bovine serum albumin (BSA), glucose, sucrose, AcONa, Hepes, DEPC treated water, PBS, DMSO were purchase from Sigma-Aldrich®. Potassium phosphate, EDTA and sodium chloride were from PanReac.
300 g mol−1); and (3) mTOR protein (SMARTpool: ON-TARGETplus mTOR siRNA with the following sequences: GGCCAUAGCUAGCCUCAUA, CAAAGGACUUCGCCCAUAA, GCAGAAUUGUCAAGGGAUA, and CCAAAGCACUACACUACAA MW = 9500 g mol−1). In addition, cyanin-3 labelled siRNA anti-GFP (Silencer™ Cy™ 3-labeled siRNA) was also used for uptake studies. They were used at an initial concentration of 50 μM (0.266 μg μL−1) in DEPC water. siRNA working concentration was set up at 0.03 μg μL−1. Anti-mTOR antibody mouse IgG1 clone 6H9B10, anti-β-actin antibody mouse IgG2 clone 2F1-1 and goat anti-IgG mouse antibody clone Poly4053, conjugated to HRP were acquired from BioLegend. Immobilon-FL PVDF, 0.45 μm membranes were obtained from EMD Millipore USA. All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Cyanine dyes were purchased from Lumiprobe. Paraformaldehyde (PFA), bovine serum albumin (BSA), glucose, sucrose, AcONa, Hepes, DEPC treated water, PBS, DMSO were purchase from Sigma-Aldrich®. Potassium phosphate, EDTA and sodium chloride were from PanReac.
Cell lines
A549 (ATCC CCL-185), H460 (ATCC HTB-177), A549GFP (previously engineered to constitutively express GFP), healthy bronchial epithelium cell line, BEAS 2B (ATCC® CCL-9609™), and ARPE-19 (ATCC CRL-2302) cell lines were obtained from ATCC. They were maintained in DMEM supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS, research grade, HyCloneTM), 100 units per mL penicillin G, 100 μg per mL streptomycin, and 2 mmol per L L-glutamine. All cells were cultured at 37 °C, under a 5% CO2/95% air atmosphere until 90% confluence before starting transfections.Animals
C57BL/6 mice of 6–8 weeks were obtained from Envigo and stabled under pathogen-free conditions in laminar flow boxes. Maintenance and experiments were performed according to corresponding guidelines of the Catalan Government (protocol 9938, approved by the “Direcció General del Medi Natural” from the “Generalitat de Catalunya”).Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5
0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 molar ratio), as we described previously.28,39,70 Oligopeptide end-modified pBAE were obtained by end-modification of acrylate-terminated C6 polymer with thiol-terminated oligopeptide at a molar ration of 1
1.2 molar ratio), as we described previously.28,39,70 Oligopeptide end-modified pBAE were obtained by end-modification of acrylate-terminated C6 polymer with thiol-terminated oligopeptide at a molar ration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 in dimethyl sulfoxide (DMSO). The mixture was stirred overnight at room temperature. The resulting polymer was precipitated in a mixture 7
2.5 in dimethyl sulfoxide (DMSO). The mixture was stirred overnight at room temperature. The resulting polymer was precipitated in a mixture 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v of diethyl ether/acetone. For the synthesis of anisamide-functionalized polymers (AA-pBAE in the following), a Schotten–Baumann reaction was first performed. The 4-methoxybenzoyl chloride (0.0125 mol), dissolved in 1.25 mL of THF, was added to a stirred solution of 6-aminocaproic acid (0.0125 mol) in 25 mL of NaOH 1 M. The pH was kept constant between 9 and 11 adding more NaOH if it was necessary. After 2 h, the final product was precipitated by acidification (until pH 1–2) of the medium adding concentrated hydrochloric acid. Then was filtered, washed with water and air-dried. Following, a Steglish esterification between this intermediate and the C6 polymer was performed. The C6 pBAE (300 mg), without oligo-peptide end modified, was dissolved in 2 mL of dichloromethane anhydrous (solution 1). The resulting acid from the Schotten–Bauman Reaction (94.75 mg), previously dissolved in 2 mL of dichloromethane anhydrous, and DMAP (small quantity) was added to the solution 1 and mixed for 5′ until dissolution. DCC (90.5 mg) was dissolved in 2 mL of dichloromethane anhydrous. Both solutions were put in the fridge for 15–30′. After the required time, the DCC solution was added to the other solution and were stirred overnight at 25 °C in N2 atmosphere. The resulting product was filtered and dried. All the polymers and intermediate products were characterized by 1H-NMR (see spectra on Fig. S1–S6, from ESI†). Final polymers were dried overnight and dissolved in DMSO in order to obtain a solution of 100 mg mL−1.
3 v/v of diethyl ether/acetone. For the synthesis of anisamide-functionalized polymers (AA-pBAE in the following), a Schotten–Baumann reaction was first performed. The 4-methoxybenzoyl chloride (0.0125 mol), dissolved in 1.25 mL of THF, was added to a stirred solution of 6-aminocaproic acid (0.0125 mol) in 25 mL of NaOH 1 M. The pH was kept constant between 9 and 11 adding more NaOH if it was necessary. After 2 h, the final product was precipitated by acidification (until pH 1–2) of the medium adding concentrated hydrochloric acid. Then was filtered, washed with water and air-dried. Following, a Steglish esterification between this intermediate and the C6 polymer was performed. The C6 pBAE (300 mg), without oligo-peptide end modified, was dissolved in 2 mL of dichloromethane anhydrous (solution 1). The resulting acid from the Schotten–Bauman Reaction (94.75 mg), previously dissolved in 2 mL of dichloromethane anhydrous, and DMAP (small quantity) was added to the solution 1 and mixed for 5′ until dissolution. DCC (90.5 mg) was dissolved in 2 mL of dichloromethane anhydrous. Both solutions were put in the fridge for 15–30′. After the required time, the DCC solution was added to the other solution and were stirred overnight at 25 °C in N2 atmosphere. The resulting product was filtered and dried. All the polymers and intermediate products were characterized by 1H-NMR (see spectra on Fig. S1–S6, from ESI†). Final polymers were dried overnight and dissolved in DMSO in order to obtain a solution of 100 mg mL−1.
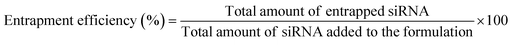 | (1) |
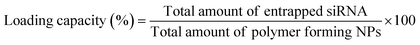 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution for 1 h under agitation to further detect the chemoluminescence (ImageQuant LAS 400 mini), after the incubation for 5 min of luminol substrate (Thermo Scientific Pierce). ImageJ was used to quantify mTOR silencing by measuring the bands intensity.
000 dilution for 1 h under agitation to further detect the chemoluminescence (ImageQuant LAS 400 mini), after the incubation for 5 min of luminol substrate (Thermo Scientific Pierce). ImageJ was used to quantify mTOR silencing by measuring the bands intensity.Conclusions
In conclusion, in a context where safe and efficient intracellular siRNA delivery remains still a challenging obstacle, the design of novel delivery vectors is of key importance. In this work, we have demonstrated, for the first time, the advantages of adding anisamide moiety to pBAE nanoparticles for the selective targeting of tumor cells in oncogene silencing targeted therapies. Using siRNA codifying for mTOR as a proof-of-concept, we have demonstrated the potential of our targeted pBAE nanoparticles in producing selective lung tumor cells death. Therefore, our nanoparticles represent a promising novel therapy for NSCLC patients.Author contributions
Conceptualization: CF, MGR, SB. Data curation: CF, MGR. Formal analysis: CGF, SB. Funding acquisition: CFP, SB. Investigation: ATC, LO, CGF. Methodology: CF, CGF, SB. Project administration: CFP, SB. Resources: CF, SB. Software: CGF, ATC, LO, MGR. Supervision: MGR, CF, SB. Validation: SB. Visualization: CGF, ATC, LO. Writing – original draft: CF, ATC, LO. Writing – review & editing: CF, CGF, MGR, SB.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from MINECO/FEDER (grants RTC-2015-3751-1, SAF2015-64927-C2-1-R and SAF2015-64927-C2-2-R) is acknowledged. The support of Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) from Generalitat de Catalunya for their support trough 2021 SGR 00357 grant, as well as that from COST Actions CA21154, CA CIG 17104 is also acknowledged. Tomàs Moliner Usón and Joan Balibrea Rull, are acknowledged for their contribution in the experimental work.References
- N. S. Gandhi, et al., Bioreducible Poly(Amino Ethers) Based mTOR siRNA Delivery for Lung Cancer, Pharm. Res., 2018, 35, 1–20 CrossRef PubMed.
- European Society for Medical Oncology (ESMO), 2019.
- N. S. Gandhi, et al., Bioreducible Poly(Amino Ethers) Based mTOR siRNA Delivery for Lung Cancer, Pharm. Res., 2018, 35, 1–20 CrossRef PubMed.
- A. M. Portis, G. Carballo, G. L. Baker, C. Chan and S. P. Walton, Confocal microscopy for the analysis of siRNA delivery by polymeric nanoparticles, Microsc. Res. Tech., 2010, 73, 878–885 CrossRef CAS PubMed.
- N. Karachaliou, et al., BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer, Sci. Rep., 2015, 5, 17499 CrossRef CAS PubMed.
- N. Karachaliou, et al., BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer, Sci. Rep., 2015, 5, 17499 CrossRef CAS PubMed.
- D. Mossmann, S. Park and M. N. Hall, mTOR signalling and cellular metabolism are mutual determinants in cancer, Nat. Rev. Cancer, 2018, 18, 744–757 CrossRef CAS PubMed.
- R. S. Herbst, D. Morgensztern and C. Boshoff, The biology and management of non-small cell lung cancer, Nature, 2018, 553, 446–454 CrossRef CAS PubMed.
- W. Jiang, et al., Designing nanomedicine for immuno-oncology, Nat. Biomed. Eng., 2017, 1, 0029 CrossRef CAS.
- K. Seip, et al., Fibroblast-induced switching to the mesenchymal-like phenotype and PI3K/mTOR signaling protects melanoma cells from BRAF inhibitors, Oncotarget, 2016, 7(15), 19997–20015 CrossRef PubMed.
- S. Viel, et al., TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway, Sci. Signaling, 2016, 9, ra19 CrossRef PubMed.
- A. Fire, et al., Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- K. L. Kozielski, S. Y. Tzeng and J. J. Green, A bioreducible linear poly(b-amino ester) for siRNA delivery, Chem. Commun., 2013, 49, 5319–5321 RSC.
- K. L. Kozielski, S. Y. Tzeng and J. J. Green, Bioengineered nanoparticles for siRNA delivery, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 449–468 CAS.
- K. L. Kozielski, S. Y. Tzeng, B. A. Hurtado De Mendoza and J. J. Green, Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells, ACS Nano, 2014, 8, 3232–3241 CrossRef CAS PubMed.
- N. Segovia, et al., Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer, Adv. Healthcare Mater., 2015, 4, 271–280 CrossRef CAS PubMed.
- Y. Zhou, et al., Schwann cells augment cell spreading and metastasis of lung cancer, Cancer Res., 2018, 121973, canres.1702.2018 Search PubMed.
- L. Zhou, et al., Branched Glycerol-Based Copolymer with Ultrahigh p65 siRNA Delivery Efficiency for Enhanced Cancer Therapy, ACS Appl. Mater. Interfaces, 2018, 10, 4471–4480 CrossRef CAS PubMed.
- J. K. W. Lam, M. Y. T. Chow, Y. Zhang and S. W. S. Leung, siRNA Versus miRNA as Therapeutics for Gene Silencing, Mol. Ther., 2015, 4, e252 CAS.
- T. Tieu, et al., Maximizing RNA Loading for Gene Silencing Using Porous Silicon Nanoparticles, ACS Appl. Mater. Interfaces, 2019, 11, 22993–23005 CrossRef CAS PubMed.
- J. K. W. Lam, M. Y. T. Chow, Y. Zhang and S. W. S. Leung, siRNA versus miRNA as Therapeutics for Gene Silencing, Mol. Ther., 2015, 4, e252 CAS.
- L. I. Selby, C. M. Cortez-Jugo, G. K. Such and A. P. R. Johnston, Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9(5), e1452 Search PubMed.
- F. Danhier, et al., PLGA-based nanoparticles: an overview of biomedical applications, J. Controlled Release, 2012, 161, 505–522 CrossRef CAS PubMed.
- S. Svenson, What nanomedicine in the clinic right now really forms nanoparticles?, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6, 125–135 CAS.
- C. Vauthier and K. Bouchemal, Methods for the Preparation and Manufacture of Polymeric Nanoparticles, Pharm. Res., 2009, 26, 1025–1058 CrossRef CAS PubMed.
- C. Pinto Reis, R. J. Neufeld, A. J. Ribeiro and F. Veiga, Nanoencapsulation I. Methods for preparation of drug-loaded polymeric nanoparticles, Nanomedicine, 2006, 2, 8–21 CrossRef PubMed.
- R. Riera, et al., Tracking DNA complexation state of pBAE polyplexes in cells with super resolution microscopy, Nanoscale, 2019, 11, 17869–17877 RSC.
- C. Fornaguera, et al., mRNA Delivery System for Targeting Antigen-Presenting Cells in Vivo, Adv. Healthcare Mater., 2018, 7(17), e1800335 CrossRef PubMed.
- C. Fornaguera, et al., In Vivo Retargeting of Poly(beta aminoester) (OM-PBAE) Nanoparticles is Influenced by Protein Corona, Adv. Healthcare Mater., 2019, 8(19), e1900849 CrossRef PubMed.
- K. A. Fitzgerald, et al., A novel, anisamide-targeted cyclodextrin nanoformulation for siRNA delivery to prostate cancer cells expressing the sigma-1 receptor, Int. J. Pharm., 2016, 499, 131–145 CrossRef CAS PubMed.
- D. Qu, et al., Anisamide-functionalized pH-responsive amphiphilic chitosan-based paclitaxel micelles for sigma-1 receptor targeted prostate cancer treatment, Carbohydr. Polym., 2020, 229, 115498 CrossRef CAS PubMed.
- A. Dasargyri, C. D. Kümin and J. C. Leroux, Targeting Nanocarriers with Anisamide: Fact or Artifact?, Adv. Mater., 2017, 29(7), 1603451 CrossRef PubMed.
- N. K. Garg, P. Dwivedi, C. Campbell and R. K. Tyagi, Site specific/targeted delivery of gemcitabine through anisamide anchored chitosan/poly ethylene glycol nanoparticles: an improved understanding of lung cancer therapeutic intervention, Eur. J. Pharm. Sci., 2012, 47, 1006–1014 CrossRef CAS PubMed.
- X. Luan, et al., Anisamide-targeted PEGylated gold nanoparticles designed to target prostate cancer mediate: enhanced systemic exposure of siRNA, tumour growth suppression and a synergistic therapeutic response in combination with paclitaxel in mice, Eur. J. Pharm. Biopharm., 2019, 137, 56–67 CrossRef CAS PubMed.
- C. Fornaguera, et al., mRNA Delivery System for Targeting Antigen-Presenting Cells in Vivo, Adv. Healthcare Mater., 2018, 1800335, 1–11 Search PubMed.
- C. Fornaguera, C. Castells-sala, M. Á. Lázaro, A. Cascante and S. Borrós, Development of an optimized freeze-drying protocol for OM-PBAE nucleic acid polyplexes, Int. J. Pharm., 2019, 569, 118612 CrossRef CAS PubMed.
- C. Fornaguera, et al., In Vivo Retargeting of Poly(beta aminoester) (OM-PBAE) Nanoparticles is Influenced by Protein Corona, Adv. Healthcare Mater., 2019, 1900849, DOI:10.1002/adhm.201900849.
- P. Dosta, V. Ramos and S. Borrós, Stable and efficient generation of poly(β-amino ester)s for RNAi delivery, Mol. Syst. Des. Eng., 2018, 3, 677–689 RSC.
- N. Segovia, P. Dosta, A. Cascante, V. Ramos and S. Borrós, Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization, Acta Biomater., 2014, 10, 2147–2158 CrossRef CAS PubMed.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide-modified poly(beta-amino ester)s (PBAEs), Acta Biomater., 2015, 20, 82–93 CrossRef CAS PubMed.
- N. S. Gandhi, et al., Bioreducible Poly(Amino Ethers) Based mTOR siRNA Delivery for Lung Cancer, Pharm. Res., 2018, 35(10), 188 CrossRef PubMed.
- K. L. Kozielski, S. Y. Tzeng and J. J. Green, A bioreducible linear poly(b-amino ester) for siRNA delivery, Chem. Commun., 2013, 49, 5319–5321 RSC.
- N. González-Ríos, et al., Novel α-mannose-functionalized poly(β-amino ester) nanoparticles as mRNA vaccines with increased antigen presenting cell selectivity in the spleen, J. Mater. Chem. B, 2023, 11, 6412–6427 RSC.
- C. Bofill-Bonet, et al., Fine-tuning formulation and biological interaction of doxorubicin-loaded polymeric nanoparticles via electrolyte concentration modulation, J. Mol. Liq., 2023, 390(Part A), 122986 CrossRef CAS.
- D. Zhu, et al., Detailed Investigation on How the Protein Corona Modulates the Physicochemical Properties and Gene Delivery of Polyethylenimine (PEI) Polyplexes, Biomater. Sci., 2018, 6, 1800–1817 RSC.
- R. S. Burke and S. H. Pun, Extracellular Barriers to in Vivo PEI and PEGylated PEI Polyplex-Mediated Gene Delivery to the Liver, Bioconjugate Chem., 2008, 19, 693–704 CrossRef CAS PubMed.
- T. Tieu, et al., Maximizing RNA Loading for Gene Silencing Using Porous Silicon Nanoparticles, ACS Appl. Mater. Interfaces, 2019, 11, 22993–23005 CrossRef CAS PubMed.
- Y. Tang, Y. Liu, Y. Xie, J. Chen and Y. Dou, Apoptosis of A549 cells by small interfering RNA targeting survivin delivery using poly-β-amino ester/guanidinylated O-carboxymethyl chitosan nanoparticles, Asian J. Pharm. Sci., 2020, 15, 121–128 CrossRef PubMed.
- J. Lee, et al., Gold, Poly(beta-amino ester) Nanoparticles for Small Interfering RNA Delivery, Nano Lett., 2009, 9, 2402–2406 CrossRef CAS PubMed.
- M. Alameh, et al., siRNA Delivery with Chitosan: Influence of Chitosan Molecular Weight, Degree of Deacetylation, and Amine to Phosphate Ratio on in Vitro Silencing Efficiency, Hemocompatibility, Biodistribution, and in Vivo Efficacy, Biomacromolecules, 2018, 19, 112–131 CrossRef CAS PubMed.
- C. Fornaguera, et al., Interactions of PLGA nanoparticles with blood components: protein adsorption, coagulation, activation of the complement system and hemolysis studies, Nanoscale, 2015, 12–14, 10.1039/C5NR00733J.
- K. L. Kozielski, et al., Cancer-selective nanoparticles for combinatorial siRNA delivery to primary human GBM in vitro and in vivo, Biomaterials, 2019, 209, 79–87 CrossRef CAS PubMed.
- D. Zhu, et al., Detailed Investigation on How the Protein Corona Modulates the Physicochemical Properties and Gene Delivery of Polyethylenimine (PEI) Polyplexes, Biomater. Sci., 2018, 6, 1800–1817 RSC.
- R. S. Burke and S. H. Pun, Extracellular Barriers to in Vivo PEI and PEGylated PEI Polyplex-Mediated Gene Delivery to the Liver, Bioconjugate Chem., 2008, 19, 693–704 CrossRef CAS PubMed.
- C. J. Bishop, S. Y. Tzeng and J. J. Green, Degradable polymer-coated gold nanoparticles for co-delivery of DNA and siRNA, Acta Biomater., 2015, 11, 393–403 CrossRef CAS PubMed.
- M. Alameh, et al., siRNA Delivery with Chitosan: Influence of Chitosan Molecular Weight, Degree of Deacetylation, and Amine to Phosphate Ratio on in Vitro Silencing Efficiency, Hemocompatibility, Biodistribution, and in Vivo Efficacy, Biomacromolecules, 2018, 19, 112–131 CrossRef CAS PubMed.
- C. Y. X. Chua, J. Ho, S. Demaria, M. Ferrari and A. Grattoni, Emerging Technologies for Local Cancer Treatment, Adv. Ther., 2020, 3(9), 2000027 CrossRef PubMed.
- G. A. Silvestri, et al., Methods for staging non-small cell lung cancer: diagnosis and management of lung cancer, 3rd ed: American college of Chest Physicians evidence-based clinical practice guidelines, Chest, 2013, 143, e211S–e250S CrossRef PubMed.
- C. Zappa and S. A. Mousa, Non-small cell lung cancer: current treatment and future advances, Transl. Lung Cancer Res., 2016, 5, 288–300 CrossRef CAS PubMed.
- M. Majem, et al., SEOM clinical guidelines for the treatment of non-small cell lung cancer (2018), Clin. Transl. Oncol., 2019, 21, 3–17 CrossRef CAS PubMed.
- A. M. C. Dingemans, et al., Small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up, Ann. Oncol., 2021, 32, 839–853 CrossRef PubMed.
- L. E. Hendriks, et al., Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up, Ann. Oncol., 2023, 34(4), 358–376 CrossRef CAS PubMed.
- C. Fornaguera, C. Castells-Sala, M. A. Lázaro, A. Cascante and S. Borrós, Development of an optimized freeze-drying protocol for OM-PBAE nucleic acid polyplexes, Int. J. Pharm., 2019, 569, 118612 CrossRef CAS PubMed.
- J. McCaskill, et al., Efficient Biodistribution and Gene Silencing in the Lung Epithelium via Intravenous Liposomal Delivery of siRNA, Mol. Ther.--Nucleic Acids, 2013, 2, e96 CrossRef PubMed.
- K. L. Kozielski, S. Y. Tzeng, B. A. Hurtado De Mendoza and J. J. Green, Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells, ACS Nano, 2014, 8, 3232–3241 CrossRef CAS PubMed.
- S. Ritz, et al., Protein Corona of Nanoparticles: Distinct Proteins Regulate the Cellular Uptake, Biomacromolecules, 2015, 16, 1311–1321 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Protein-nanoparticle interactions, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- M. Navalón-López, A. Dols-Perez, S. Grijalvo, C. Fornaguera and S. Borrós, Unravelling the role of individual components in pBAE/polynucleotide polyplexes in the synthesis of tailored carriers for specific applications: on the road to rational formulations, Nanoscale Adv., 2023, 5, 1611–1623 RSC.
- S. Peters, et al., Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up, Ann. Oncol., 2008, 3, 1–2 Search PubMed.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide-modified poly(beta-amino ester)s (PBAEs), Acta Biomater., 2015, 20, 82–93 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra05830a |
| This journal is © The Royal Society of Chemistry 2023 |