Open Access Article
Open Access ArticleA microbiological system for screening the interference of XNA monomers with DNA and RNA metabolism†
Aude Blancharda,
Mikhail Abramovb,
Camille Hassana,
Philippe Marlièrecd,
Piet Herdewijn b and
Valérie Pezo
b and
Valérie Pezo *a
*a
aGénomique Métabolique, Genoscope, Institut François Jacob, CEA, CNRS, Univ Evry, Université Paris-Saclay, 2 Rue Gaston Crémieux, 91057 Evry, France. E-mail: vpezo@genoscope.cns.fr
bLaboratory for Medicinal Chemistry, Rega Institute, Herestraat 49, KU Leuven, Leuven, Belgium
cTheraxen SA, 296 route de Longwy, L-1940, Luxembourg
dTESSSI, 81 Rue Réaumur, Paris 75002, France
First published on 12th October 2023
Abstract
We explored the toxicity and mutagenicity of a wide range of xenobiotic nucleoside triphosphates to an Escherichia coli strain equipped with a nucleoside triphosphate transporter. This bacterial test provides a tool to evaluate and guide the synthesis of nucleotides for applications such as the propagation of non-natural genetic information or the selection of potential drugs.
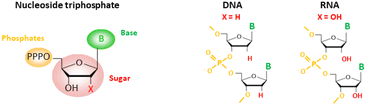
Genetic information is stored and conveyed by only two types of nucleic acids DNA and RNA (Scheme 1), in all living species. Over the past few decades, different systems have revealed that genetic information can also be carried and propagated by non-natural nucleic acids.1,2 These chemically modified molecules, called XNA (for Xenobiotic Nucleic Acids),3,4 exhibit modified nucleobases, sugars or backbones or a combination of these modifications and even novel bases or base pairs. XNAs have mainly been considered to improve the physicochemical or pharmacological properties of nucleic acids. These characteristics make them potentially useful for numerous biotechnological, therapeutic and diagnostic applications,5 but also for the understanding of molecular mechanisms and the origins of life.6
In the natural world, bacteriophage genomes exhibit the widest chemical diversity of non-canonical nucleobases.7 We have recently elucidated the biosynthetic pathway of a non-canonical nucleotide: aminoadenine (Z) which completely replaces adenine of siphoviruses genomes8 and characterized a specific DNA polymerase responsible for its incorporation.9 These natural variations mostly prevent cleavage by prokaryotic host restriction endonucleases. The expression of genetic information by non-natural nucleic acids to increase the spectrum of coding possibilities is a major objective of biotechnology and synthetic biology. Another important outcome of this approach is also to limit genetic pollution by minimizing the possible interactions of these non-canonical molecules with the rest of the living world.10 In this sense, considerable efforts have been made in recent years to produce non-natural base pairs to expand the genetic alphabet and produce new functionalities.11–13 In a previous study, we constructed an E. coli strain equipped with a dNTP transporter from an algal plastid that allows the transport of nucleoside triphosphates into the bacterium.14 Using this E. coli strain, we here tested the in vivo toxicity and the mutagenic effect of our chemically modified nucleoside triphosphates containing modifications in the sugar, nucleobase or backbone and also in different combinations (Fig. 1 and 2). The ability of modified nucleosides to interact with the nucleic acids of the cell is an important prerequisite to be tested for their use to efficiently carry genetic information in vivo.
 | ||
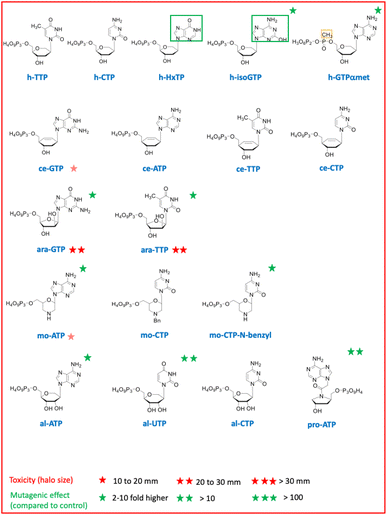
| Fig. 2 List of chemically synthetized deoxyribonucleotide triphosphates with non-canonical nucleobases. | ||
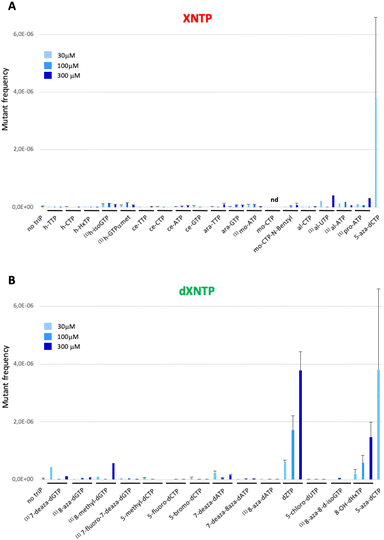
To test the toxicity of chemically modified nucleoside triphosphates in vivo, a dilution of an overnight pre-culture of the E. coli strain equipped with a dNTPs transporter (XE858) was spread on a rich medium plate in the presence of a 100 mM solution of nucleoside triphosphates placed in the central well of the plate. Toxicity was measured by the size of the inhibition halo observed after the overnight growth of the strain on plate (Fig. 3A). A positive control of toxicity was performed with the dideoxyCTP (ddCTP), a replication chain terminator. As a negative control, the strain devoid of dNTP transporter was used under the same conditions (Fig. 3B). To test the mutagenic effect of these XNA triphosphates transported into bacterial cells, the XE858 strain was incubated overnight with different concentrations of nucleoside triphosphates and spread on LB rifampicin plates. Rifampicin resistance, most frequently results from a mutation in the rpoB gene encoding for the beta subunit of bacterial RNA polymerase. This selection provides a direct assay of the mutagenicity of nucleoside triphosphate by interfering with DNA replication. 5-Aza-dCTP (decitabine triphosphate) known to generate G- > C transversions when incorporated into DNA was used as positive control of mutagenesis. It was previously hypothesized that decitabine could undergo a ring-opening step, followed by hydrolysis to yield a structure capable of pairing with cytosine. Thus, decitabine triphosphate incorporated as dCTP can pair with a cytosine inducing G to C mutations.15 Mutation frequencies were determined by the ratio of rifampicin-resistant clones to the number of viable clones counted on LB plates at each concentration (Fig. 4).
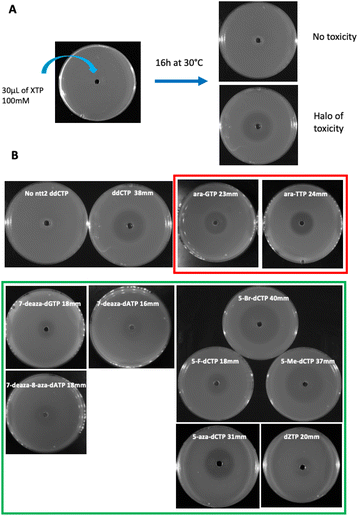 | ||
| Fig. 3 Toxicity on plates (A) toxicity test description (B) pictures of the toxic compounds revealed in our selection strain and their relative halo diameters. | ||
Chemically synthesized nucleotides with different sugar moieties (hexitol, cyclohexene or arabinose) (Fig. 1) were initially chosen for their chemical and enzymatic stability. As XNA polymerases able to copy DNA in XNA and back to DNA were successfully developed, these XNA were established to be capable of evolution.2,3 HNA were also selected for the development of efficient aptamers and catalysts. Using an E. coli selection, we have also previously shown that genetic information can be conveyed to DNA from these three sugar-modified oligonucleotides, in vivo, with up to twelve consecutive HNA residues converted into DNA by cellular DNA polymerases.16 The import of these sugar-modified nucleoside triphosphates revealed no toxicity for any hexitol triphosphates even when coupled to a non-canonical nucleobase or a backbone modification and low toxicity for only one of the four cyclohexene triphosphates.
The toxicity effect observed for the nucleoside triphosphates assayed may result from their incorporation either into DNA or RNA. The mutagenic effect most likely results from random incorporation of a nucleoside triphosphate in DNA followed by faulty pairing of the incorporated nucleoside during subsequent rounds of replication. The assessment of mutagenicity amounts to measure the pairing ambiguity of XNA nucleoside triphosphates, and therefore reflects their efficiency of incorporation into bacterial DNA.
Hexitol triphosphate and cyclohexene triphosphate did not induce a mutagenic effect in vivo when coupled to a canonical base (Fig. 4A). However, a slight mutagenic effect was observed when hexitol triphosphates were coupled to a non-canonical and ambiguous base with the h-isoGTP or to a phosphate modification with the h-GTPαmet. This result suggests that these sugar modified nucleoside triphosphates can be weakly incorporated by cellular DNA polymerases during DNA synthesis as they can serve as templates in oligonucleotides for canonical DNA synthesis in an vivo selection.16 The toxicity of arabinose nucleoside triphosphates is consistent with their known capacity to inhibit both DNA synthesis and repair (Fig. 2B).
Prolinol oligonucleotides widely studied for their ability to hybridize in antisense applications showed no toxicity as nucleoside triphosphates but one log of mutagenic effect probably due to their destabilizing effect when incorporated in DNA, leading to mutations17 (Fig. 4A). Altritol triphosphates, studied for their stabilizing characteristics in SiRNA, led to a mutagenic effect comparable to that of prolinol when added to the cell culture (Fig. 4A), suggesting possible incorporation into DNA by cellular DNA polymerases. Hexitol and cyclohexene nucleoside triphosphates appear to be the best candidates to consider for a propagation of genetic information that would weakly interfere with cellular DNA and RNA biosynthesis.
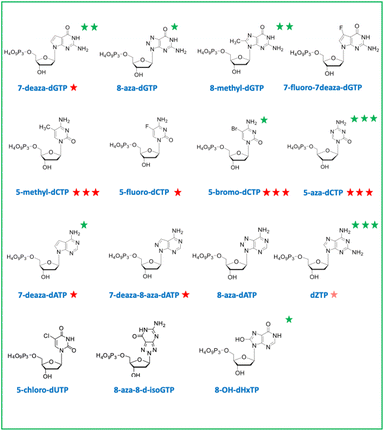
In a second part, we selected and tested base-modified XNA triphosphates derived from the 4 canonical deoxyribose nucleotides with the ambitious objective to fully morph DNA by substituting the G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C and A
C and A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) T base pairs. Among the four modified deoxyguanosine triphosphates tested, only 7-deaza-dGTP showed in vivo toxicity (Fig. 3B). The mutagenic effect was 15 to 20 fold higher than the negative control with the 7-deaza-dGTP and the 8-methyl-dGTP respectively, with the weakest mutagenic effect of dGTP-derived nucleotide triphosphates being 7-fluoro-7-deaza-dGTP, comparable to E. coli spontaneous mutation rate (Fig. 4B). Interestingly, all three dCTP derivatives exhibited a strong toxicity on plates without mutagenic effect (Fig. 3B and 4B). This result may be due to an efficient incorporation of these compounds into RNA, previously reported with canonical dCTP using a related dNTPs transporter.18 The best candidate to replace dCTP would be the 5-fluoro-dCTP which has the lowest toxicity and no mutagenic effect. 7-Deaza-dGTP and 7-fluoro-deaza-dGTP have previously been shown to be an effective alternative to replacing dG with any of the modified dCTPs tested in in vitro DNA polymerisation assays.19 Our in vivo tests point to a preference for the 7-fluoro-7-deaza-dGTP over the 7-deaza-dGTP which could be coupled to 5-fluoro-dCTP.
T base pairs. Among the four modified deoxyguanosine triphosphates tested, only 7-deaza-dGTP showed in vivo toxicity (Fig. 3B). The mutagenic effect was 15 to 20 fold higher than the negative control with the 7-deaza-dGTP and the 8-methyl-dGTP respectively, with the weakest mutagenic effect of dGTP-derived nucleotide triphosphates being 7-fluoro-7-deaza-dGTP, comparable to E. coli spontaneous mutation rate (Fig. 4B). Interestingly, all three dCTP derivatives exhibited a strong toxicity on plates without mutagenic effect (Fig. 3B and 4B). This result may be due to an efficient incorporation of these compounds into RNA, previously reported with canonical dCTP using a related dNTPs transporter.18 The best candidate to replace dCTP would be the 5-fluoro-dCTP which has the lowest toxicity and no mutagenic effect. 7-Deaza-dGTP and 7-fluoro-deaza-dGTP have previously been shown to be an effective alternative to replacing dG with any of the modified dCTPs tested in in vitro DNA polymerisation assays.19 Our in vivo tests point to a preference for the 7-fluoro-7-deaza-dGTP over the 7-deaza-dGTP which could be coupled to 5-fluoro-dCTP.
For a thymine alternative, an almost complete substitution of this base (98.4%) with chlorouracil was achieved in E. coli genomic DNA by metabolic engineering and evolution.20 As expected, the corresponding deoxyribochlorouracil triphosphate showed no toxicity or mutagenic effect confirming its potential for synthetic genome construction and other applications such as aptamers.21 Since 7-deaza-dATP, 8-aza-dATP and 7deaza-8aza-dATP can be polymerized with comparable efficiency by cellular DNA polymerases, using a polyT template in vitro,22 the 8-aza-dA would be the best candidate, regarding its characteristics as a triphosphate in vivo (no toxicity or mutagenic effect), to pair with chlorouracil. The mutagenic effect of 8-OH-dHxTP, 12 to 71 fold higher than the spontaneous mutation rate and closely related to the concentration used, was expected because this nucleobase can base pair with an A or a C after a 180° rotation around the glycosidic bond (Fig. 2 and 4B).
Aminoadenine deoxyribonucleotide triphosphate, which totally replaces adenine in genomes of some siphoviruses exhibited an unexpectedly high mutagenic effect and low toxicity suggesting weak interaction with RNA and efficient incorporation into DNA. The high mutagenic effect probably results from the spontaneous deamination of aminoadenine to guanine after incorporation of the dZTP into DNA.23 However, it remains to be determined whether an E. coli enzyme would catalyze this in situ deamination reaction on DNA.
Conclusions
In this in vivo system, the fact that at least one compound of each type of modification is toxic or mutagenic strongly indicates that the synthetic compounds cross the bacterial membrane and reach replication and transcription machineries. In general, we observed that the most mutagenic nucleoside triphosphates are little toxic and the most toxic are little mutagenic. A prominent exception to this pattern is 5-aza-dCTP, which is known to be highly mutagenic and which we found to be highly toxic. Altogether, our bacterial screening system provides a simple and effective way for assessing both the toxicity and mutagenicity of synthetic nucleotides. In the future, noncanonical nucleoside triphosphates inducing either of these two effects should be avoided as building blocks of XNA carriers of genetic information in vivo. The interactions of certain nucleoside triphosphates with cellular components, would certainly require further investigation to understand the underlying mechanisms. The combination of organic synthesis of non-canonical DNA and RNA monomers and the construction of reporter strains should allow the screening of numerous drug candidates and new types of nucleic acids propagated in vivo.Author contributions
VP, PM and PH conceived the project. VP and PM designed experiments. AB, MA, and CH performed experiments. VP wrote the manuscript. All authors reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement No 965135.Notes and references
- P. Herdewijn and P. Marlière, Chem. Biodiversity, 2009, 6, 791–808 CrossRef CAS PubMed.
- V. B. Pinheiro, A. I. Taylor, C. Cozens, M. Abramov, M. Renders, S. Zhang, J. C. Chaput, J. Wengel, S.-Y. Peak-Chew, S. H. McLaughlin, P. Herdewijn and P. Holliger, Science, 2012, 336, 341–344 CrossRef CAS PubMed.
- V. B. Pinheiro and P. Holliger, Curr. Opin. Chem. Biol., 2012, 16, 245–252 CrossRef CAS PubMed.
- J. C. Chaput and P. Herdewijn, Angew. Chem., Int. Ed., 2019, 58, 11570–11572 CrossRef CAS PubMed.
- K. Duffy, S. Arangundy-Franklin and P. Holliger, BMC Biol., 2020, 18, 112 CrossRef PubMed.
- A. I. Taylor, V. B. Pinheiro, M. J. Smola, A. S. Morgunov, S. Peak-Chew, C. Cozens, K. M. Weeks, P. Herdewijn and P. Holliger, Nature, 2015, 518, 427–430 CrossRef CAS PubMed.
- P. Weigele and E. A. Raleigh, Chem. Rev., 2016, 116, 12655–12687 CrossRef CAS PubMed.
- D. Sleiman, P. S. Garcia, M. Lagune, J. Loc’h, A. Haouz, N. Taib, P. Röthlisberger, S. Gribaldo, P. Marlière and P. A. Kaminski, Science, 2021, 372, 516–520 CrossRef CAS PubMed.
- V. Pezo, F. Jaziri, P.-Y. Bourguignon, D. Louis, D. Jacobs-Sera, J. Rozenski, S. Pochet, P. Herdewijn, G. F. Hatfull, P.-A. Kaminski and P. Marliere, Science, 2021, 372, 520–524 CrossRef CAS PubMed.
- M. Schmidt, Bioessays, 2010, 32, 322–331 CrossRef CAS PubMed.
- S. A. Benner, N. B. Karalkar, S. Hoshika, R. Laos, R. W. Shaw, M. Matsuura, D. Fajardo and P. Moussatche, Cold Spring Harbor Perspect. Biol., 2016, 8, a023770 CrossRef PubMed.
- K. Hamashima, M. Kimoto and I. Hirao, Curr. Opin. Chem. Biol., 2018, 46, 108–114 CrossRef CAS PubMed.
- V. T. Dien, S. E. Morris, R. J. Karadeema and F. E. Romesberg, Curr. Opin. Chem. Biol., 2018, 46, 196–202 CrossRef CAS PubMed.
- V. Pezo, C. Hassan, D. Louis, B. Sargueil, P. Herdewijn and P. Marlière, ACS Synth. Biol., 2018, 7, 1565–1572 CrossRef CAS PubMed.
- L. Jackson-Grusby, P. W. Laird, S. N. Magge, B. J. Moeller and R. Jaenisch, Proc. Natl. Acad. Sci. U.S.A., 1997, 94, 4681–4685 CrossRef CAS PubMed.
- V. Pezo, F. W. Liu, M. Abramov, M. Froeyen, P. Herdewijn and P. Marlière, Angew. Chem., Int. Ed., 2013, 52, 8139–8143 CrossRef CAS PubMed.
- G. Ceulemans, A. Van Aerschot, B. Wroblowski, J. Rozenski, C. Hendrix and P. Herdewijn, Chem.–Eur. J., 1997, 3, 1997–2010 CrossRef CAS.
- A. W. Feldman, E. C. Fischer, M. P. Ledbetter, J.-Y. Liao, J. C. Chaput and F. E. Romesberg, J. Am. Chem. Soc., 2018, 140, 1447–1454 CrossRef CAS PubMed.
- E. Eremeeva, M. Abramov, L. Margamuljana and P. Herdewijn, Chem.–Eur. J., 2017, 23, 9560–9576 CrossRef CAS PubMed.
- P. Marlière, J. Patrouix, V. Döring, P. Herdewijn, S. Tricot, S. Cruveiller, M. Bouzon and R. Mutzel, Angew Chem. Int. Ed. Engl., 2011, 50, 7109–7114 CrossRef PubMed.
- C. Gasse, M. Zaarour, S. Noppen, M. Abramov, P. Marlière, S. Liekens, B. De Strooper and P. Herdewijn, Chembiochem, 2018, 19, 754–763 CrossRef CAS PubMed.
- E. Eremeeva, M. Abramov, P. Marlière and P. Herdewijn, Org. Biomol. Chem., 2017, 15, 168–176 RSC.
- M. Levy and S. L. Miller, Proc. Natl. Acad. Sci., 1998, 95, 7933–7938 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra06172h |
| This journal is © The Royal Society of Chemistry 2023 |