 Open Access Article
Open Access ArticleIn silico description of the adsorption of cell signaling pathway proteins ovalbumin, glutathione, LC3, TLR4, ASC PYCARD, PI3K and NF-Kβ on 7.0 nm gold nanoparticles: obtaining their Lennard-Jones-like potentials through docking and molecular mechanics
Monique M. Coelho ch,
Eveline M. Bezerra
ch,
Eveline M. Bezerra *a,
Roner F. da Costa
*a,
Roner F. da Costa ab,
Érika C. de Alvarenga
ab,
Érika C. de Alvarenga c,
Valder N. Freire
c,
Valder N. Freire d,
Cláudia R. Carvalho
d,
Cláudia R. Carvalho cg,
Claudia Pessoae,
Eudenilson L. Albuquerque
cg,
Claudia Pessoae,
Eudenilson L. Albuquerque f and
Raquel A. Costac
f and
Raquel A. Costac
aPrograma de Pós-Graduação em Ciência e Engenharia de Materiais, Universidade Federal Rural do Semi-Árido (UFERSA), Mossoró, RN CEP 59625-900, Brazil. E-mail: eveline.bezerra@ufersa.edu.br
bDepartamento de Ciências, Matemática e Estatística, Universidade Federal Rural do Semi-Árido (UFERSA), Mossoró, RN CEP 59625-900, Brazil
cDepartamento de Ciências Naturais, Universidade Federal de São João del Rei (UFSJ), São João del-Rei, MG CEP 36301-160, Brazil
dDepartamento de Física, Universidade Federal do Ceará (UFC), Fortaleza, CE 60455-760, Brazil
ePrograma de Pós-Graduação em Biotecnologia, Rede Nordeste de Biotecnologia (RENORBIO, ), Universidade Federal do Ceará (UFC), Fortaleza, CE CEP 60020-181, Brazil
fDepartamento de Biofísica e Farmacologia, Universidade Federal do Rio Grande do Norte (UFRN), Natal, RN CEP 59064-741, Brazil
gDepartamento de Morfologia, Universidade Federal de Minas Gerais (UFMG), Belo Horizonte, MG CEP 31270-910, Brazil
hDepartamento de Bioquímica e Imunologia, Universidade Federal de Minas Gerais (UFMG), Belo Horizonte, MG CEP 31270-910, Brazil
First published on 5th December 2023
Abstract
The impact of vaccination on the world's population is difficult to calculate. For developing different types of vaccines, adjuvants are substances added to vaccines to increase the magnitude and durability of the immune response and the effectiveness of the vaccine. This work explores the potential use of spherical gold nanoparticles (AuNPs) as adjuvants. Thus, we employed docking techniques and molecular mechanics to describe how a AuNP 7.0 nm in diameter interacts with cell signaling pathway proteins. Initially, we used X-ray crystallization data of the proteins ovalbumin, glutathione, LC3, TLR4, ASC PYCARD, PI3K, and NF-Kβ to study the adsorption with an AuNP through molecular docking. Therefore, interaction energies were obtained for the AuNP complexes and individual proteins, as well as the AuNP and OVA complex (AuNP@OVA) with each cellular protein, respectively. Results showed that AuNPs had the highest affinity for OVA individually, followed by glutathione, ASC PYCARD domain, LC3, PI3K, NF-Kβ, and TLR4. Furthermore, when evaluating the AuNP@OVA complex, glutathione showed a greater affinity with more potent interaction energy when compared to the other studied systems.
1 Introduction
It is difficult to estimate the importance of vaccination in the health of the world's populations; in the COVID-19 pandemic, the impact of vaccination was evident after the reduction in mortality. There are several ways to protect yourself from diseases, one of which is vaccination, which remains necessary to prevent endemics and pandemics. Over time, scientific research has led to the development of many different types of vaccines, and new technologies have emerged. One of these advances was using adjuvants, compounds used to increase the intensity and duration of the protective effect caused by administering some prophylactic or therapeutic substance, such as vaccines.1 Currently, five different adjuvants constitute licensed vaccines; however, the mechanisms by which these adjuvants work remain only partially understood. Aluminum hydroxide is the most used adjuvant. However, research has directed interest to new components that can improve the efficacy and safety of vaccines.2 Especially after the 2020 pandemic, the search for new effective and viable adjuvants proved very important. In this sense, nanoparticles gained great prominence because these nanostructures can alter cellular activity, for example, by increasing the production of specific antibodies.3,4 In addition, nanomaterials with different sizes, shapes, and surface charges can have distinct effects, such as changes in bioavailability, transport, fate, uptake, and cell activation.5–7 Gold nanoparticles (AuNPs) have been used as adjuvants in some recent research and are an area of development in immunology applications.8,9 Studies suggest that AuNPs can enhance B cell responses in antibody production8,10 and cytotoxic T cell responses.11,12 For example, a formulation of AuNPs conjugated to ovalbumin (OVA) generated specific immunoglobulin G (IgG) for the antigen.8 Another study suggested that AuNPs conjugated to OVA of sizes 22 and 33 nm are more efficient in antigen delivery and induction of T cell responses than those of 10 nm.11 Additionally, cetuximab-coated AuNPs promoted tumor cell death.13 The biocompatibility and low toxicity of AuNPs make them attractive for biological applications, including as adjuvants for vaccines against pathogens.14–20 Immune signaling pathways activation can depend on the size of the AuNPs, with ultra-small size preferentially activating the NLRP3 inflammasome. At the same time, larger AuNPs are mainly absorbed by phagocytosis or endocytosis and activate inflammatory pathways.21Another pathway studied in the intracellular activation of nanoparticles is the PI3Kα pathway, which alters mechanisms of apoptosis, proliferation, and DNA damage and, therefore, is involved in the progression and development of tumors. Mahmoud and his collaborators showed that gold nanoparticles have an adjuvant effect when conjugated with a PI3K-inhibiting drug. This work showed that the conjugate administration affected transcription factors responsible for cell proliferation, apoptotic, and cell cycle arrest pathways, decreasing the expression of the PI3Kα protein.22
1.1 The potential adjuvant activity of gold nanoparticles
Zhu et al.21 evaluated the effect of administering gold nanoparticles larger and smaller than 10 nm. Their results suggest that the larger nanoparticles activated the NF-Kβ signaling pathway. The smaller nanoparticles penetrated directly into the cell cytoplasm promoted the production of ROS, and directed LC3 (microtubule-associated protein 1-light chain 3) for proteasomal degradation, a protein necessary to inhibit the activation of the NLRP3 inflammasome. Thus, the administration of these nanoparticles activated the NLRP3 inflammasome, stimulated caspase-1, and led to the production of IL-1β. Ultimately, they showed that these smaller OVA-conjugated nanoparticles could increase specific antibody production dependent on the NLRP3 inflammasome pathway.21 Another work showed that 50 nm gold nanoparticles coated with outer membrane vesicles (OMVs) from mycobacteria activate macrophages and dendritic cells through TLR2 and TLR4 receptors.23 A study by Yuan et al.24 showed that nanoparticles with approximately 25.4 nm conjugated with Pleurotus ferulae (PFPS) promoted the maturation of dendritic cells by activating MAPK and NF-Kβ through TLR4 and NLRP3 signaling pathways. Furthermore, they showed that these functionalized nanoparticles inhibited tumor growth when used as an HPV vaccine.24 An interesting study showed that a peptide conjugate with gold nanoparticles could inhibit the TLR4 signaling activated by a cigarette smoke extract, consequently inhibiting its inflammatory response; this is because the conjugate was phagocytosed by macrophages, inhibiting the endosomal acidification necessary for receptor activation and promoting the induction of autophagy and expression of antioxidant proteins.12Vyas et al.25 evaluated the uptake effect and immunomodulatory efficiency of 5, 15 and 30 nm gold nanoparticles in lung adenocarcinoma cells (A549). Further, they observed that the cells took up the 5 nm nanoparticles, decreasing inflammatory signaling molecules' expression more efficiently, and these nanoparticles also decreased TLR4 expression in A549.
Because of this, we can infer that the nanomaterial size can affect the mode of entry into the cell, but the literature is contradictory. Using AuNPs as adjuvants can be an excellent therapeutic tool, but their precise role in immune responses still needs to be investigated.
1.2 In silico study on the molecular interactions
The applications of nanotechnology in the diagnosis, monitoring, control, and treatment of diseases have gained attention in recent years.26,27 Due to its strong absorption in the UV-Vis region, gold is an exciting species in the preparation of nanoparticles.28 Simulations of systems composed of a large number of atoms, such as nanoparticles and proteins, make it impossible to use a quantum model; however, we can still model their behavior by obtaining properties of the system using a classical treatment. Thus, molecular modeling (MM) techniques have been widely applied to nanobiosystems to calculate the interaction energy between proteins and ligands.29–31 For example, the adsorption of ibuprofen,32 levodopa,33 and ascorbic acid34 on C60 fullerene was investigated using MM, proving to be an essential tool to study and understand the properties of nanoscale systems.35 Furthermore, several works study the interaction of molecules with anticancer activity and AuNPs through DFT and DM calculations, such as gold-fullerene,36 cysteamine-capped gold nanoparticles,37 narigin and AuNP38 and AuNP in the transport of molecules across the cell membrane.39 In this work, we used MM to study the interactions between a spherical gold nanoparticle (AuNP) 7.0 nm and the protein ovalbumin (OVA), as well as the interaction of the AuNP@OVA complex with other proteins related to cellular pathways. For this, we combine docking and molecular mechanics techniques using the Universal Force Field (UFF).402 Materials and methods – in silico studies
2.1 Construction of the spherical gold nanoparticle (AuNP)
We used the nanostructure building tools the Materials Studio 5.5 software41 to construct the 7.0 nm diameter spherical gold nanoparticle (AuNP), starting from the import of the gold unit cells from the program's library.2.2 Preparation of proteins
The structural details of target proteins (Table 1) were obtained from the PDB data bank.42 We used Biovia Materials Studio 5.5 package to fix the positions of heavy atoms and adjust the positions of hydrogen atoms. For all proteins, previously to the molecular docking, the hydrogen atoms were added to the structures, then an optimization procedure was executed using Universal Force Field (UFF).40 However, during optimization, hydrogen atoms were free to move, while all other atom coordinates were constrained.2.3 Molecular docking
The molecular docking was performed by a geometry based docking algorithm, the web server PatchDock Beta 1.3 (ref. 50 and 51), to the prediction of a spherical gold nanoparticle (AuNP, 7.0 nm diameter) and proteins (OVA, glutathione, LC3, TLR4, ASC PYCARD, PI3K, and NF-Kβ) interactions. Proteins were docked individually to the AuNP and AuNP@OVA. Using the energy criterion, we selected the 20 most stable poses from each simulation. Docking scores and position were used to select the proteins with the highest potential for interaction with AuNP. We refined the results with FireDock to determine the best pose configurations. Then, the best-ranked pose from AuNP in the OVA (AuNP@OVA) was used for a second docking with the other proteins under study. Subsequently, the poses of each protein in the AuNP (AuNP@OVA) were observed, and each protein's best and most energetically favorable conformations were selected.2.4 Molecular mechanics simulation conditions
The interaction energy calculations for all systems were simulated using the Forcite code using the Universal Force Field (UFF).40 The convergence limit adopted was ultra-fine with the following parameters: energy variation between two successive steps less than 2.0 × 10−5 kcal mol−1 force per atom less than 0.001 kcal mol−1 Å−1 and maximum displacement less than 1.0 × 10−5 Å with charges assignment by the force field and nonperiodic atom truncation method.2.5 Calculation of interaction energies
The final step involved calculating the single point energies. We used the Forcite module, a classical molecular mechanics tool, in Biovia Materials Studio 5.5 with the Universal Force Field (UFF) to calculate the interaction energy between AuNP and proteins. They were calculated using as inputs the best configurations of the docking. Thus, the interaction energy (ΔE) was calculated for the most favorable conformations of the molecular docking using the eqn (1) for the AuNP@proteins:| ΔE(AuNP@protein) = EAuNP@protein − [EAuNP + Eprotein] | (1) |
| ΔE(AuNP@OVA@protein) = EAuNP@OVA@protein − [EAuNP@OVA + Eprotein] | (2) |
3 Results and discussion
Many studies have shown different cellular interference caused by the application of AuNPs. Thus, they can be selected to trigger an immune reactivity to treat or prevent certain pathologies based on their properties.52–56The molecular docking of AuNP@proteins and AuNP@OVA@proteins involved was analyzed and refined using PatchDock/Firedock. The strongest interactions are those with the lowest interaction energies. The best docking poses of AuNP and AuNP@OVA with each cellular protein are illustrated in Fig. 1a–g and 2a–f, respectively.
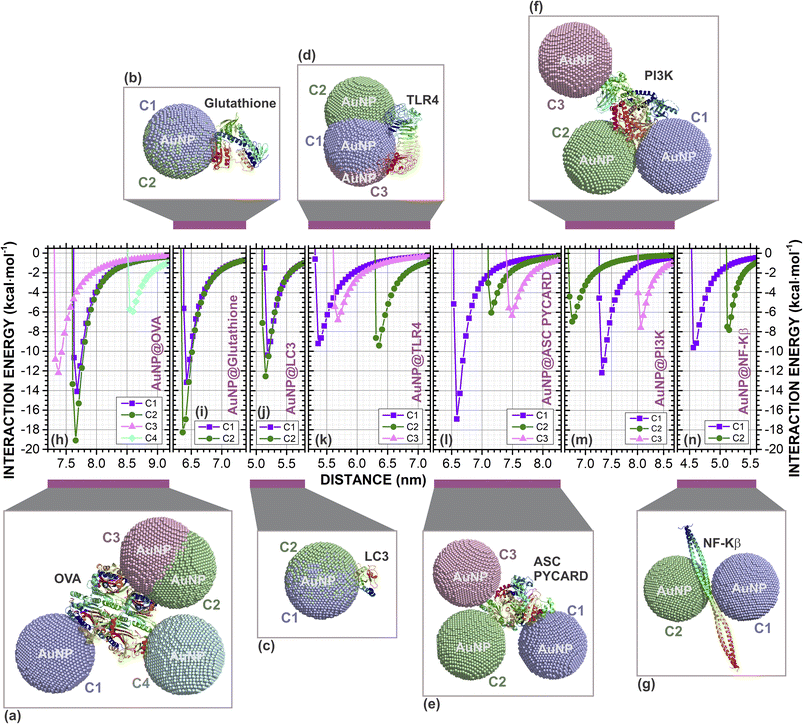 | ||
| Fig. 1 Complexes formed by molecular docking between the spherical gold nanoparticle (AuNP, 7.0 nm) and proteins: (a) OVA (four configurations C1, C2, C3 and C4); (b) glutathione (two configurations C1 and C2); (c) LC3 (two configurations C1 and C2); (d) TLR4 (three configurations C1, C2 and C3); (e) ASC PYCARD (three configurations C1, C2 and C3); (f) PI3K (three configurations C1, C2 and C3); and (g) NF-Kβ (two configurations C1 and C2). In this image we can observe the interaction site where AuNP best configuration in the docking. The classical interaction energies (in kcal mol−1) as a function of the centroids distance (in nm) between the AuNP and proteins centroids of the (h) AuNP@OVA, four configurations; (i) AuNP@glutathione, two configurations; (j) AuNP@LC3, two configurations; (k) AuNP@TLR4, three configurations; (l) AuNP@ASC PYCARD, three configurations; (m) AuNP@PIK3, three configurations; and, (n) AuNP@NF-Kβ, two configurations. The AuNPs are shown in golden dots and proteins in cartoon format. The figure was drawn using PyMOL57 (PyMOL Molecular Graphics System; https://www.pymol.org). | ||
Additionally, the variation of the interaction energy with the distance d between the centroids through classical molecular mechanics computations with Universal Force Field (UFF) of AuNP (AuNP@OVA) and proteins in kcal mol−1 of the best docking poses are presented in Fig. 1h–n (Fig. 2g–l). Our results showed that the spherical AuNP constructed in this study has a higher affinity with OVA (ΔEAuNP@OVA = −19.10), followed by glutathione (ΔEAuNP@glutathione = −18.27), ASC PYCARD domain (ΔEAuNP@ASC PYCARD = −16.87), LC3 (ΔEAuNP@LC3 = −12.53), PI3K (ΔEAuNP@PI3K = −12.16), NF-Kβ (ΔEAuNP@NF-Kβ = −9.62) and TLR4 (ΔEAuNP@TLR4 = −9.42), as shown in Fig. 1h–n. Furthermore, our results also indicate that AuNP@OVA has a higher affinity with glutathione (ΔEAuNP@OVA@glutathione = −30.26), followed by NF-Kβ (ΔEAuNP@OVA@NF-Kβ = −23.20), PI3K (ΔEAuNP@OVA@PI3K = −19.31), TLR4 (ΔEAuNP@OVA@TLR4 = −11.48), ASC PYCARD domain (ΔEAuNP@OVA@ASC PYCARD = −7.88) and LC3 (ΔEAuNP@OVA@LC3 = −5.84), as presented in Fig. 2g–l. The lower interaction energies indicate stronger interactions. The interaction between AuNP (AuNP@OVA) and proteins shows a repulsion at a very close distance and attraction at an equilibrium distance. They do not interact when at an infinite distance, as shown in Fig. 1 and 2. Thus, they behave like a Lennard-Jones-type potential.
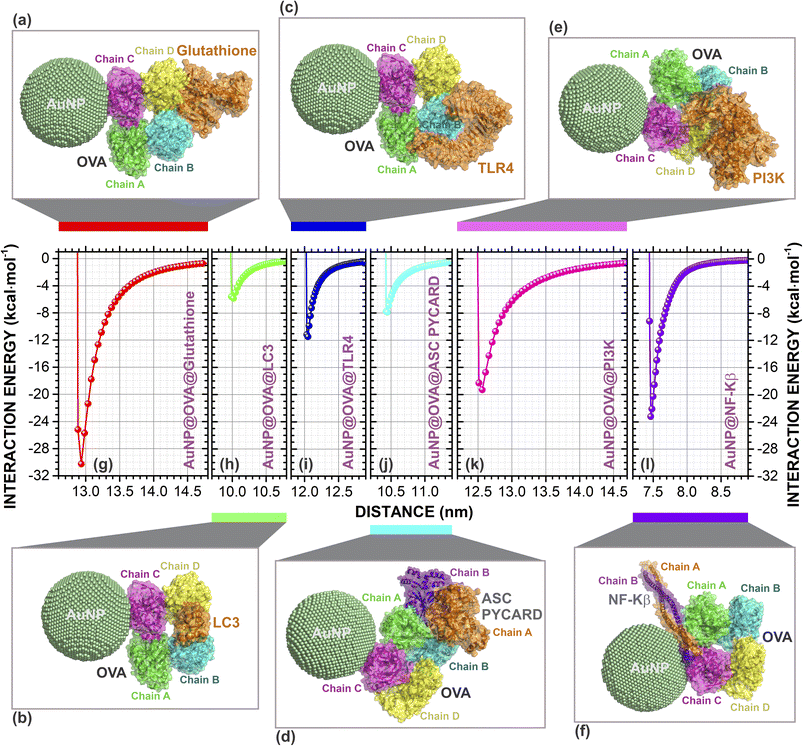 | ||
| Fig. 2 Complexes formed by molecular docking between the spherical gold nanoparticle (AuNP, 7.0 nm) with OVA (AuNP@OVA) and proteins: (a) glutathione; (b) LC3; (c) TL4; (d) ASC PYCARD; (e) PI3K; and (f) NF-Kβ. In this image we can observe the interaction site where AuNP@OVA and proteins best configuration in the docking. The classical interaction energies (in kcal mol−1) as a function of the centroids distance (in nm) between the AuNP@OVA and proteins of the (g) AuNP@OVA@glutathione; (h) AuNP@OVA@LC3; (i) AuNP@OVA@TLR4; (j) AuNP@OVA@ASC PYCARD; (k) AuNP@OVA@PIK3; and, (l) AuNP@OVA@NF-Kβ. The AuNPs are shown in golden dots and proteins in cartoon format. The figure was drawn using PyMOL57 (PyMOL Molecular Graphics System; https://www.pymol.org). | ||
We have demonstrated the in silico scenarios of seven cellular proteins (OVA, glutathione, LC3, TL4, ASC PYCARD, PI3K, and NF-Kβ) that individually interact with 7.0 nm spherical gold nanoparticle (AuNP). Similarly, we showed here that the AuNP@OVA complex could interact separately with six cellular proteins (glutathione, LC3, TL4, ASC PYCARD, PI3K, and NF-Kβ).
Glutathione is a molecule present in most cells and has a fundamental role in cellular autophagy activity. In previous studies,58 showed that treatment with AuNPs can induce autophagosome accumulation and LC3 processing, an autophagosome marker protein, resulting in the blockade of autophagy flux rather than induction.
Perhaps, AuNPs and AuNPs@OVA participate in this process by enhancing the effects of glutathione, inducing autophagosome accumulation and LC3 processing. Furthermore, the uncoated AuNP showed an affinity with ASC PYCARD domain (Fig. 1e and l) with interaction energy of −16.87 kcal mol−1 (a difference of only 2.23 kcal mol−1 for the AuNP@OVA), which contributes to signaling in inflammatory and apoptotic pathways by activating caspase, showing to be very important for NALP3 inflammasome activation. This may be further evidence of work,21 which showed that AuNPs <10.0 nm preferentially activate the NLRP3 inflammasome for caspase-1 maturation and interleukin-1β production, promoting robust ROS production and directing LC3 autophagy protein for proteasomal degradation. Furthermore, it also shows that AuNPs larger than 10.0 nm activate the NF-Kβ signaling pathway.21
In addition, Sharma et al.59 also suggested that the application of AuNPs >10.0 nm participates in NF-Kβ activation and increased antibody production. Our results showed that AuNP@OVA might have a high affinity for NF-Kβ with interaction energy of −23.20 kcal mol−1 (Fig. 2f and l). However, there is still little evidence of these considerations, and further investigations are necessary to understand the mechanisms involved in the application of these AuNPs.
We believe that these works are not contradictory but complementary, and probably the immunomodulation caused by AuNPs and AuNPs@OVA are not the same, nor are they attributed to interactions with only one protein in the cell, but with several cellular proteins, activating and/or inactivating several signaling pathways concomitantly. Furthermore, the careful and adequate selection of these adjuvants can help promote appropriate immunological changes for each treatment, as different diseases have their characteristics, requiring different body modifications to maintain systemic homeostasis, and these modifications include changes in the immune system.
4 Conclusions
The results presented in this study suggest that the spherical gold nanoparticle–ovalbumin (AuNPs@OVA) conjugate exhibits satisfactory stability to be used as an adjuvant in biomedical applications. All classical interaction energies as a function of the centroids distance between the AuNP and proteins centroids have Lennard-Jones-like potentials. Furthermore, analysis of the interaction of this conjugate with cellular signaling molecules indicates that it may be a viable option for applications in the healthcare field. The results obtained in this study point to the potential of this conjugate as an adjuvant. However, further studies are needed to investigate its safety and efficiency in biomedical applications.Author contributions
MMC, ECA, CRC, and RAC proposed the idea. MMC, EMB, RFC, ECA, CRC, and RAC contributed to the conception and design of the study. EMB, RFC, VNF, CP, RAC and ELA discussed the results, provided critical feedback, co-wrote and finalized the manuscript. All authors contributed to the manuscript revision and approved the submitted version.Conflicts of interest
There are no conflicts to declare.Acknowledgements
E. M. B. received financial support from FAPERN/CAPES. R. F. da Costa received financial support from CNPq-Universal project 434821/2018-7. E. C. A. is supported by PROPE/UFSJ, and grant from FAPEMIG [Rede Mineira de Pesquisa e Inovação para Bioengenharia de Nanosistemas (RED-00282-16), and APQ-02026-18]. This study is supported by the FAPEMIG (Fundação de Amparo à Pesquisa de Minas Gerais – the foundation for support for research in Minas Gerais) – RED-00282-16 and was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) - Finance Code 001.References
- B. Pulendran, P. S. Arunachalam and D. T. O'Hagan, Nat. Rev. Drug Discovery, 2021, 20, 454–475 CrossRef CAS PubMed.
- T. Fiyouzi and P. A. Reche, in Vaccine design: an introduction, ed. P. A. Reche, Springer US, New York, NY, 2023, pp. 1–14 Search PubMed.
- Q. Yin, W. Luo, V. Mallajosyula, Y. Bo, J. Guo, J. Xie, M. Sun, R. Verma, C. Li, C. M. Constantz, L. E. Wagar, J. Li, E. Sola, N. Gupta, C. Wang, O. Kask, X. Chen, X. Yuan, N. C. Wu, J. Rao, Y. Hsiu Chien, J. Cheng, B. Pulendran and M. M. Davis, Nat. Mater., 2023, 22, 380–390 CAS.
- S. Shrivastava, J. M. Carmen, Z. Lu, S. Basu, R. S. Sankhala, W.-H. Chen, P. Nguyen, W. C. Chang, J. King, C. Corbitt, S. Mayer, J. S. Bolton, A. Anderson, I. Swafford, G. D. Terriquez, H. V. Trinh, J. Kim, O. Jobe, D. Paquin-Proulx, R. M. Gary, G. D. Gromowski, J. R. Currier, E. Bergmann-Leitner, K. Modjarrad, N. L. Michael, M. G. Joyce, A. M. W. Malloy and M. Rao, npj Vaccines, 2023, 8, 43 CrossRef CAS PubMed.
- R. Bezbaruah, V. P. Chavda, L. Nongrang, S. Alom, K. Deka, T. Kalita, F. Ali, B. Bhattacharjee and L. Vora, Vaccines, 2022, 10, 1946 CrossRef CAS PubMed.
- V. Harish, D. Tewari, M. Gaur, A. B. Yadav, S. Swaroop, M. Bechelany and A. Barhoum, Nanomaterials, 2022, 12, 457 CrossRef CAS PubMed.
- R. Gupta and H. Xie, J. Environ. Pathol., Toxicol. Oncol., 2018, 37, 209–230 CrossRef PubMed.
- L. A. Dykman, Expert Rev. Vaccines, 2020, 19, 465–477 CrossRef CAS PubMed.
- H. Sekimukai, N. Iwata-Yoshikawa, S. Fukushi, H. Tani, M. Kataoka, T. Suzuki, H. Hasegawa, K. Niikura, K. Arai and N. Nagata, Microbiol. Immunol., 2020, 64, 33–51 CrossRef CAS PubMed.
- Y. Yang, Y. Zhang, A. Thakur, R. Li, H. Xu, Z. Wang, M. Ghavami, Z. Tu and H. Liu, Int. J. Pharm., 2019, 571, 118704 CrossRef CAS PubMed.
- K. R. Rhodes and J. J. Green, Mol. Immunol., 2018, 98, 13–18 CrossRef CAS PubMed.
- W. Gao, L. Wang, K. Wang, L. Sun, Y. Rao, A. Ma, M. Zhang, Q. Li and H. Yang, ACS Appl. Mater. Interfaces, 2019, 11, 32706–32719 CrossRef CAS PubMed.
- L. M. Andrade, E. M. N. Martins, A. F. Versiani, D. S. Reis, F. G. da Fonseca, I. P. de Souza, R. M. Paniago, E. Pereira-Maia and L. O. Ladeira, Mater. Sci. Eng., C, 2020, 107, 110203 CrossRef CAS PubMed.
- L. A. Dykman, S. A. Staroverov and A. S. Fomin, Gold Bull., 2018, 51, 197–203 CrossRef CAS.
- C. Wang, W. Zhu and B.-Z. Wang, Int. J. Nanomed., 2017, 12, 4747–4762 CrossRef CAS PubMed.
- Q. H. Quach, S. K. Ang, J.-H. J. Chu and J. C. Y. Kah, Acta Biomater., 2018, 78, 224–235 CrossRef CAS PubMed.
- G. L. Burygin, P. I. Abronina, N. M. Podvalnyy, S. A. Staroverov, L. O. Kononov and L. A. Dykman, Beilstein J. Nanotechnol., 2020, 11, 480–493 CrossRef CAS PubMed.
- R. Kumar, P. C. Ray, D. Datta, G. P. Bansal, E. Angov and N. Kumar, Vaccine, 2015, 33, 5064–5071 CrossRef CAS PubMed.
- S. Parween, P. K. Gupta and V. S. Chauhan, Vaccine, 2011, 29, 2451–2460 CrossRef CAS PubMed.
- M. G. Soliman, A. F. Mohamed, R. A. E. Sayed and A. A. A. Elqasem, Eur. J. Biomed. Pharm. Sci., 2017, 4, 529–536 CAS.
- M. Zhu, L. Du, R. Zhao, H. Y. Wang, Y. Zhao, G. Nie and R.-F. Wang, ACS Nano, 2020, 14, 3703–3717 CrossRef CAS PubMed.
- N. N. Mahmoud, D. Abuarqoub, R. Zaza, D. A. Sabbah, E. A. Khalil and R. Abu-Dahab, Int. J. Mol. Sci., 2020, 21, 3320 CrossRef CAS PubMed.
- E. George, A. Goswami, T. Lodhiya, P. Padwal, S. Iyer, I. Gauttam, L. Sethi, S. Jeyasankar, P. R. Sharma, A. A. Dravid, R. Mukherjee and R. Agarwal, Biomater. Adv., 2022, 139, 213003 CrossRef CAS PubMed.
- P. Yuan, L. Liu, A. Aipire, Y. Zhao, S. Cai, L. Wu, X. Yang, A. Aimaier, J. Lu and J. Li, Int. J. Biol. Macromol., 2023, 227, 1015–1026 CrossRef CAS PubMed.
- S. P. Vyas and R. Goswami, Nanomedicine, 2019, 14, 229–253 CrossRef PubMed.
- E. Albuquerque, U. L. Fulco, E. Caetano and V. Freire, Quantum Chemistry Simulation of Biological Molecules, Cambridge University Press, 2020, p. 416 Search PubMed.
- K. K. Cotí, M. E. Belowich, M. Liong, M. W. Ambrogio, Y. A. Lau, H. A. Khatib, J. I. Zink, N. M. Khashab and J. F. Stoddart, Nanoscale, 2009, 1, 16–39 RSC.
- H. E. Toma, V. M. Zamarion, S. H. Toma and K. Araki, J. Braz. Chem. Soc., 2010, 21, 1158–1176 CrossRef CAS.
- R. F. R. Da Costa, V. V. N. Freire, E. M. E. Bezerra, B. B. S. Cavada, E. E. W. S. Caetano, J. J. L. de Lima Filho and E. L. E. Albuquerque, Phys. Chem. Chem. Phys., 2012, 14, 1389–1398 RSC.
- K. Farias, R. F. da Costa, A. S. Meira, J. Diniz-Filho, E. M. Bezerra, V. N. Freire, P. Guest, M. Nikahd, X. Ma, M. G. Gardiner, M. G. Banwell, M. d. C. F. de Oliveira, M. O. de Moraes and C. do Ó Pessoa, ACS Med. Chem. Lett., 2020, 11, 1274–1280 CrossRef CAS PubMed.
- R. L. F. Melo, I. C. d. C. Souza, A. J. R. Carvalho, E. M. Bezerra and R. F. da Costa, Res., Soc. Dev., 2020, 9, e363974155 CrossRef.
- D. S. Dantas, J. I. N. Oliveira, J. X. Lima Neto, R. F. da Costa, E. M. Bezerra, V. N. Freire, E. W. S. Caetano, U. L. Fulco and E. L. Albuquerque, RSC Adv., 2015, 5, 49439–49450 RSC.
- N. F. Frazão, E. L. Albuquerque, U. L. Fulco, D. L. Azevedo, G. L. F. Mendonça, P. Lima-Neto, E. W. S. Caetano, J. V. Santana and V. N. Freire, RSC Adv., 2012, 2, 8306 RSC.
- S. G. Santos, J. V. Santana, F. F. Maia, V. Lemos, V. N. Freire, E. W. S. Caetano, B. S. Cavada and E. L. Albuquerque, J. Phys. Chem. B, 2008, 112, 14267–14272 CrossRef CAS PubMed.
- T. I. Adelusi, A. Q. K. Oyedele, I. D. Boyenle, A. T. Ogunlana, R. O. Adeyemi, C. D. Ukachi, M. O. Idris, O. T. Olaoba, I. O. Adedotun, O. E. Kolawole, Y. Xiaoxing and M. Abdul-Hammed, Inform. Med. Unlocked., 2022, 29, 100880 CrossRef.
- M. Sabet, S. Tanreh, A. Khosravi, M. Astaraki, M. Rezvani and M. Darvish Ganji, Diamond Relat. Mater., 2022, 126, 109142 CrossRef CAS.
- W. Phanchai, U. Srikulwong, A. Chuaephon, S. Koowattanasuchat, J. Assawakhajornsak, R. Thanan, C. Sakonsinsiri and T. Puangmali, ACS Appl. Nano Mater., 2022, 5, 9042–9052 CrossRef CAS.
- B. Singh, M. Rani, J. Singh, L. Moudgil, P. Sharma, S. Kumar, G. S. S. Saini, S. K. Tripathi, G. Singh and A. Kaura, RSC Adv., 2016, 6, 79470–79484 RSC.
- N. Farhadian, M. S. Kazemi, F. Moosavi Baigi and M. Khalaj, J. Mol. Graphics Modell., 2022, 116, 108271 CrossRef CAS PubMed.
- S. Artemova, L. Jaillet and S. Redon, J. Comput. Chem., 2016, 37, 1191–1205 CrossRef CAS PubMed.
- B. Dassault Systèmes, Biovia Materials Studio Package, 2017 Search PubMed.
- S. K. Burley, C. Bhikadiya, C. Bi, S. Bittrich, H. Chao, L. Chen, P. A. Craig, G. V. Crichlow, K. Dalenberg, J. M. Duarte, S. Dutta, M. Fayazi, Z. Feng, J. W. Flatt, S. Ganesan, S. Ghosh, D. S. Goodsell, R. K. Green, V. Guranovic, J. Henry, B. P. Hudson, I. Khokhriakov, C. L. Lawson, Y. Liang, R. Lowe, E. Peisach, I. Persikova, D. W. Piehl, Y. Rose, A. Sali, J. Segura, M. Sekharan, C. Shao, B. Vallat, M. Voigt, B. Webb, J. D. Westbrook, S. Whetstone, J. Y. Young, A. Zalevsky and C. Zardecki, Nucleic Acids Res., 2023, 51, D488–D508 CrossRef CAS PubMed.
- P. E. Stein, A. G. Leslie, J. T. Finch and R. W. Carrell, J. Mol. Biol., 1991, 221, 941–959 CrossRef CAS PubMed.
- P. Karplus and G. E. Schulz, J. Mol. Biol., 1989, 210, 163–180 CrossRef CAS PubMed.
- K. Sugawara, N. N. Suzuki, Y. Fujioka, N. Mizushima, Y. Ohsumi and F. Inagaki, Genes Cells, 2004, 9, 611–618 CrossRef CAS PubMed.
- H. M. Kim, B. S. Park, J.-I. Kim, S. E. Kim, J. Lee, S. C. Oh, P. Enkhbayar, N. Matsushima, H. Lee, O. J. Yoo and J.-O. Lee, Cell, 2007, 130, 906–917 CrossRef CAS PubMed.
- A. Hagmann, M. Hunkeler, E. Stuttfeld and T. Maier, Structure, 2016, 24, 1227–1236 CrossRef CAS PubMed.
- T. Pirali, E. Ciraolo, S. Aprile, A. Massarotti, A. Berndt, A. Griglio, M. Serafini, V. Mercalli, C. Landoni, C. C. Campa, J. P. Margaria, R. L. Silva, G. Grosa, G. Sorba, R. Williams, E. Hirsch and G. C. Tron, ChemMedChem, 2017, 12, 1542–1554 CrossRef CAS PubMed.
- A. H. Barczewski, M. J. Ragusa, D. F. Mierke and M. Pellegrini, Sci. Rep., 2019, 9, 2950 CrossRef PubMed.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2005, 33, W363–W367 CrossRef CAS PubMed.
- D. Duhovny, R. Nussinov and H. J. Wolfson, LNCS, 2002, vol. 2452, pp. 185–200 Search PubMed.
- L. Fan, W. Wang, Z. Wang and M. Zhao, Nat. Commun., 2021, 12, 6371 CrossRef CAS PubMed.
- K. Hou, J. Zhao, H. Wang, B. Li, K. Li, X. Shi, K. Wan, J. Ai, J. Lv, D. Wang, Q. Huang, H. Wang, Q. Cao, S. Liu and Z. Tang, Nat. Commun., 2020, 11, 4790 CrossRef CAS PubMed.
- H. Arami, S. Kananian, L. Khalifehzadeh, C. B. Patel, E. Chang, Y. Tanabe, Y. Zeng, S. J. Madsen, M. J. Mandella, A. Natarajan, E. E. Peterson, R. Sinclair, A. S. Y. Poon and S. S. Gambhir, Nat. Nanotechnol., 2022, 17, 1015–1022 CrossRef CAS PubMed.
- Z. Batool, G. Muhammad, M. M. Iqbal, M. S. Aslam, M. A. Raza, N. Sajjad, M. Abdullah, N. Akhtar, A. Syed, A. M. Elgorban, S. S. Al-Rejaie and Z. Shafiq, Sci. Rep., 2022, 12, 6575 CrossRef CAS PubMed.
- N. M. El-Deeb, S. M. Khattab, M. A. Abu-Youssef and A. M. A. Badr, Sci. Rep., 2022, 12, 11518 CrossRef CAS PubMed.
- PyMOL, Schrödinger LLC, 2015 Search PubMed.
- X. Ma, Y. Wu, S. Jin, Y. Tian, X. Zhang, Y. Zhao, L. Yu and X.-J. Liang, ACS Nano, 2011, 5, 8629–8639 CrossRef CAS PubMed.
- M. Sharma, R. L. Salisbury, E. I. Maurer, S. M. Hussain and C. E. W. Sulentic, Nanoscale, 2013, 5, 3747 RSC.
| This journal is © The Royal Society of Chemistry 2023 |
