Open Access Article
Open Access ArticleTarget-mediated self-assembly of DNA networks for sensitive detection and intracellular imaging of APE1 in living cells†
Xue-Mei
Zhou
,
Ying
Zhuo
 ,
Ruo
Yuan
,
Ruo
Yuan
 * and
Ya-Qin
Chai
*
* and
Ya-Qin
Chai
*
Key Laboratory of Luminescence Analysis and Molecular Sensing, Ministry of Education, College of Chemistry and Chemical Engineering, Southwest University, Chongqing 400715, PR China. E-mail: yuanruo@swu.edu.cn; yqchai@swu.edu.cn
First published on 10th February 2023
Abstract
Herein, giant DNA networks were assembled from two kinds of functionalized tetrahedral DNA nanostructures (f-TDNs) for sensitive detection and intracellular imaging of apurinic/apyrimidinic endonuclease 1 (APE1) as well as gene therapy in tumor cells. Impressively, the reaction rate of the catalytic hairpin assembly (CHA) reaction on f-TDNs was much faster than that of the conventional free CHA reaction owing to the high local concentration of hairpins, spatial confinement effect and production of giant DNA networks, which significantly enhanced the fluorescence signal to achieve sensitive detection of APE1 with a limit of 3.34 × 10−8 U μL−1. More importantly, the aptamer Sgc8 assembled on f-TDNs could enhance the targeting activity of the DNA structure to tumor cells, allowing it to endocytose into cells without any transfection reagents, which could achieve selective imaging of intracellular APE1 in living cells. Meanwhile, the siRNA carried by f-TDN1 could be accurately released to promote tumor cell apoptosis in the presence of endogenous target APE1, realizing effective and precise tumor therapy. Benefiting from the high specificity and sensitivity, the developed DNA nanostructures provide an excellent nanoplatform for precise cancer diagnosis and therapy.
Introduction
Human apurinic/apyrimidinic endonuclease 1 (APE1) is a multifunctional enzyme in human cells, which plays a critical role in the base excision repair (BER) pathway by recognizing apurinic/apyrimidinic (AP) sites in double-stranded DNA (dsDNA).1–3 It has been confirmed that the occurrences of various tumors are accompanied by changes in the expression level or intracellular distribution of APE1;4–6 thus effective monitoring of intracellular APE1 expression is beneficial for tumor diagnosis and further treatment. Recently, several methods have been developed to assess the activity of APE1,7–9 however, these methods only utilized the cleavage function of APE1 to output signals without cyclic amplification of the target, resulting in low detection sensitivity and unifunctional application of APE1, which still hindered the accurate diagnosis and treatment of diseases. Therefore, it is necessary to introduce efficient amplification methods for sensitive detection and multifunctional application of APE1. Recently, RNA interference (RNAi) has been proposed as a novel multifunctional gene-silencing tool for tumor treatment, in which small interfering RNAs (siRNAs) are used as effector molecules to guide efficient gene therapy.10–13 However, the lack of efficient and non-toxic siRNA delivery vectors and the precise release mechanism seriously hinder the clinical application of RNAi technology.14–18 Therefore, with the help of APE1, designing efficient nanocarriers of siRNA with a precise release mechanism may be an effective therapy approach, and how to achieve sensitive detection of APE1 and accurately release siRNA remains to be explored.As a classical enzyme-free nucleic acid circuit amplification method, catalytic hairpin assembly (CHA)19–21 achieves cyclic amplification of the target through toehold-mediated strand displacement22–24 with simplicity and versatility,25–29 which is one of the best candidates to achieve sensitive detection and versatility of APE1. Nevertheless, traditional CHA reactions relied on random collisions of reactants in a homogeneous solution with low collision efficiency and long reaction time, resulting in a low reaction rate and nonspecific background leakage, which limit further exploration and application of CHA.30–33 Thus, how to improve the CHA reaction kinetics is the direction of our continuous efforts. For the past few years, tetrahedral DNA nanostructures (TDNs), as the classical frame nucleic acid, have attracted great attention33–36 owing to their pre-engineered sizes, structural stability, excellent biocompatibility and cell permeability.37–40 Based on precise programmability, TDNs can not only serve as efficient nanocarriers for probes and siRNAs, but also provide a spatial-confinement effect to aggregate reactants together in a compact space, which could increase the local concentration of DNA reactants accelerating the reaction rate and improving the dynamic efficiency. Therefore, it is worthwhile to explore the use of TDNs as siRNA vectors and amplification scaffolds to improve the kinetics and rate of CHA reactions.
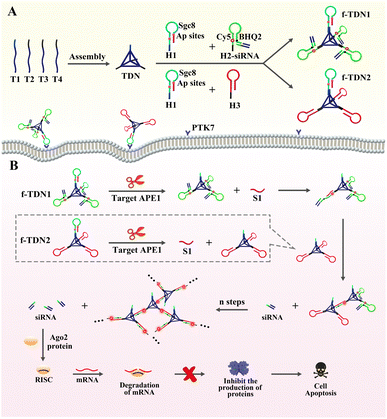
Herein, giant DNA networks were assembled from two kinds of functionalized tetrahedral DNA nanostructures f-TDNs (f-TDN1 and f-TDN2) triggered by a target for sensitive detection and intracellular imaging of apurinic/apyrimidinic endonuclease 1 (APE1) as well as delivery of siRNA into cells for tumor therapy. The mechanical principle is displayed in Scheme 1. First, tetrahedral DNA nanostructures (TDNs) were assembled in one step41 from four single DNA strands (T1, T2, T3 and T4), which were designed with four anchors (three of them containing the same DNA sequence). The hairpin H1 was marked with two AP sites in the stem, and the loop was the aptamer Sgc8 of protein tyrosine kinase 7 (PTK7), which is expressed on cell membranes associated with tumors. H2 was labeled with fluorophore (Cy5) and quencher (BHQ2), and the fluorescent signal was in a quenched state because of Förster resonance energy transfer. siRNA was assembled at the end of H2 by complementary base pairing (H2-siRNA). Then, H1 and H2-siRNA were assembled on the TDNs at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 via base complementary pairing to form functionalized tetrahedral DNA nanostructure 1 (f-TDN1). Among them, H1 hybridized with the 5′ end of T1, and H2 hybridized with the 5′ end of T2–T4. Similarly, H1 and H3 were assembled on the TDNs at a ratio of 1
3 via base complementary pairing to form functionalized tetrahedral DNA nanostructure 1 (f-TDN1). Among them, H1 hybridized with the 5′ end of T1, and H2 hybridized with the 5′ end of T2–T4. Similarly, H1 and H3 were assembled on the TDNs at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 to form functionalized tetrahedral DNA nanostructure 2 (f-TDN2). After that, the aptamer Sgc8 modified on f-TNDs targeted and delivered probes into tumor cells directly. In the presence of target APE1, the AP sites of H1 were cleaved to release single-stranded S1 for triggering CHA reactions on f-TND1 and f-TND2 to form giant network nanostructures with significant generation of fluorescent signals, which achieved sensitive detection and intracellular imaging of APE1. Meanwhile, siRNA released during the f-TDNs CHA reaction could promote cell apoptosis to achieve the purpose of precise tumor treatment. Hence, the proposed target-mediated self-assembly of DNA networks provides an excellent and effective nanoplatform for sensitive detection of biomarkers, specific diagnosis of diseases and precise gene therapy of tumors.
3 to form functionalized tetrahedral DNA nanostructure 2 (f-TDN2). After that, the aptamer Sgc8 modified on f-TNDs targeted and delivered probes into tumor cells directly. In the presence of target APE1, the AP sites of H1 were cleaved to release single-stranded S1 for triggering CHA reactions on f-TND1 and f-TND2 to form giant network nanostructures with significant generation of fluorescent signals, which achieved sensitive detection and intracellular imaging of APE1. Meanwhile, siRNA released during the f-TDNs CHA reaction could promote cell apoptosis to achieve the purpose of precise tumor treatment. Hence, the proposed target-mediated self-assembly of DNA networks provides an excellent and effective nanoplatform for sensitive detection of biomarkers, specific diagnosis of diseases and precise gene therapy of tumors.
Results and discussion
PAGE analysis of the proposed strategy
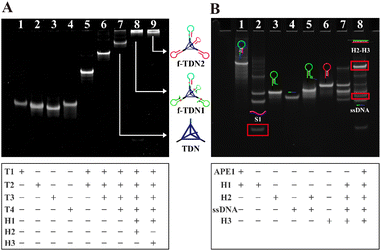
In order to validate the successful assembly of the DNA nanostructures and the feasibility of the proposed analysis strategy, polyacrylamide gel electrophoresis analysis was adopted. As displayed in Fig. 1A, samples in lanes 1–4 were four single-stranded DNA T1, T2, T3 and T4 forming tetrahedral DNA nanostructures (TDNs), which corresponded to a band with a faster migration rate, respectively. The lanes 5–7 were respectively loaded with T1 + T2, T1 + T2 + T3 and T1 + T2 + T3 + T4, and the mobility of bands was observed to be lower and lower with the addition of single-stranded DNA, indicating the formation of TDNs. After incubating the mixture of TDNs, H1 and H2, a bright band was observed at the top of the lane 8, demonstrating that TNDs, H1 and H2 successfully formed into f-TDN1 via base complementary pairing. Similarly, the reaction products of TDNs, H1 and H3 showed a bright band at the top of the lane 9, which proved the formation of f-TDN2. The above results showed that the proposed DNA structures could be successfully assembled.In Fig. 1B, the cleavage ability of the target APE1 and the catalytic hairpin assembly (CHA) reaction between the hairpins were verified by PAGE analysis. Lane 1 was loaded with hairpin H1 with two AP sites, and lane 2 was loaded with the products of H1 cleavage by APE1. It can be seen in lane 2 that H1 almost disappeared and several new bands appeared, indicating that substrate H1 could be efficiently cleaved by APE1 to release S1. The samples of lane 3 and lane 4 were H2 and ssDNA (ssDNA was used instead of siRNA), respectively, showing a clear band. Lane 5 was loaded with a mixture of H2 and ssDNA, which displayed a slower migration band in comparison with H2, indicating that the ssDNA could be efficiently tethered to the end of H2 through base pairing. Lane 6 was loaded with H3 and shows a clear band. Lane 7 was loaded with a mixture of H2 and H3, and a shallow and almost negligible band appeared due to the spontaneous reaction of H2 and H3, indicating that H2 and H3 hardly reacted with each other in the absence of APE1. In contrast, after incubating the sample of H1, H2, H3 and APE1, an obvious band corresponding to dsDNA of H2 and H3 was observed at the top of lane 8, and a band corresponding to ssDNA was also observed. These observations demonstrated that S1 produced by APE1 cleavage of H1 triggered the CHA between H2 and H3. Furthermore, it was also demonstrated that siRNA could be released in the case of the CHA reaction.
Feasibility and reaction rate analysis of different CHA systems
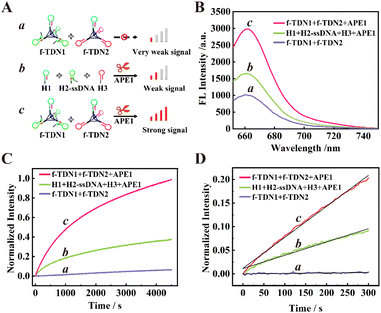
The feasibility of target-mediated self-assembly of DNA networks for sensitive detection of APE1 was evaluated in vitro (Fig. 2B). In the reaction system of 125 nM f-TDNs CHA without the target APE1 (schematic illustration in Fig. 2A, part a), a low background signal was observed (Fig. 2B, column a). However, with the addition of 0.1 U μL−1 APE1 (schematic illustration in Fig. 2A, part c), the fluorescence signal was significantly enhanced (Fig. 2B, column c), indicating that APE1 was the key to triggering the CHA reaction and the recovery of the fluorescence signal. Besides, it was worth noting that in the presence of 0.1 U μL−1 APE1, the fluorescence signal of the conventional CHA reaction with the same concentrations of hairpins as in the f-TDNs system (schematic illustration in Fig. 2A, part b) was weaker (Fig. 2B, column b) than that of the f-TDNs CHA reaction system under the same conditions. The results implied that the assembly of f-TDNs could promote the CHA reaction and produce a stronger fluorescence signal.The real-time fluorescence monitoring further confirmed the necessity for the presence of APE1 to achieve recovery of the fluorescence signal and compared the reaction rate of different reaction systems. As shown in Fig. 2C, in the absence of APE1, the fluorescence signal barely increased with time in the 125 nM f-TDNs system (Fig. 2C, curve a). In contrast, the fluorescence signal recovered rapidly over time in the presence of 0.1 U μL−1 APE1 (Fig. 2C, curve c). In addition, the real-time fluorescence of the conventional CHA reaction with free hairpins was also monitored (Fig. 2C, curve b), showing that the fluorescence signal generated by f-TDNs CHA was significantly higher than that of traditional CHA with the same concentration at the same time. The initial rates of different CHA systems were calculated using the slope of the time-dependent FL intensity in the first 5 minutes of the reactions,42,43 and the linear regression equations of the proposed f-TDNs CHA and the traditional CHA were Ic = 6.57 × 10−4t + 1.16 × 10−2 (Fig. 2D, curve c) and Ib = 2.82 × 10−4t + 1.11 × 10−2 (Fig. 2D, curve b), respectively. Therefore, the normalized initial reaction rate of f-TDN CHA was 6.57 × 10−4 s−1, which was 2.33-fold faster than that of conventional CHA (2.82 × 10−4 s−1). The results demonstrated that the assembly of hairpins on TDNs could accelerate the reaction between the hairpins due to the enrichment effect of TDNs.
AFM characterization of the DNA nanostructures
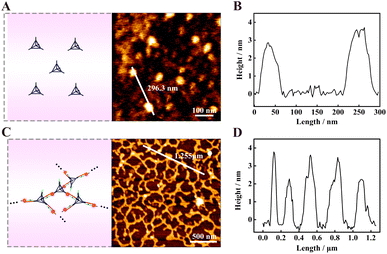
Atomic force microscopy (AFM) was used to characterize the assembled DNA nanostructures. All the samples were prepared by the typical dehydration of solvents on mica method. Fig. 3A shows the AFM image of TDNs, and there were some uniformly dispersed particles with a height of about 3–4 nm (Fig. 3B), which was close to the theoretical height of 4.62 nm, further indicating the successful assembly of the TDNs. The characterization of products yielded by f-TDNs CHA is displayed in Fig. 3C, which showed huge and uniformly distributed DNA network nanostructures. The height of the DNA network nanostructures (Fig. 3D) almost corresponded to that of TDNs. These results further revealed that f-TDN1 and f-TDN2 could assemble into huge DNA networks triggered by APE1.Analysis capability of the proposed biosensor
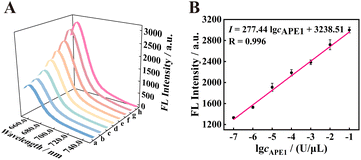
To study the analysis ability of the proposed strategy for APE1, the fluorescence intensity was recorded at different concentrations of APE1 in vitro. As shown in Fig. 4 A, with the increase in concentration of APE1 from 0 to 1 × 10−1 U μL−1, the fluorescence signal of the 125 nM f-TDN system was obviously enhanced. The fluorescence intensity showed a good linear relationship with different concentrations of APE1 with the correlation equation of I = 277.44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cAPE1 + 3238.51, and the detection limit of the proposed biosensor was 3.34 × 10−8 U μL−1 (Fig. 4B). More importantly, the proposed strategy has a significantly wider linear range and an extremely low detection limit compared to other APE1 assays (Table S2†), suggesting the potential application of the proposed CHA on f-TDNs for ultrasensitive biomarker detection.
cAPE1 + 3238.51, and the detection limit of the proposed biosensor was 3.34 × 10−8 U μL−1 (Fig. 4B). More importantly, the proposed strategy has a significantly wider linear range and an extremely low detection limit compared to other APE1 assays (Table S2†), suggesting the potential application of the proposed CHA on f-TDNs for ultrasensitive biomarker detection.
APE1 is an endonuclease, therefore, several different nucleases of hOGG1, Nt.BbvcI, EcoRI and Exonuclease I were selected as interferences to evaluate the specificity of the proposed strategy. As displayed in Fig. S1,† with the addition of 0.1 U μL−1 hOGG1, Nt.BbvcI, EcoRI and Exonuclease I, respectively, the fluorescence signal barely recovered. However, the fluorescence signal of the mixture of 0.01 U μL−1 APE1 and 0.1 U μL−1 interference nucleases was consistent with that of only adding target APE1, clearly indicating that the proposed method could distinguish APE1 from other interference nucleases with high selectivity.
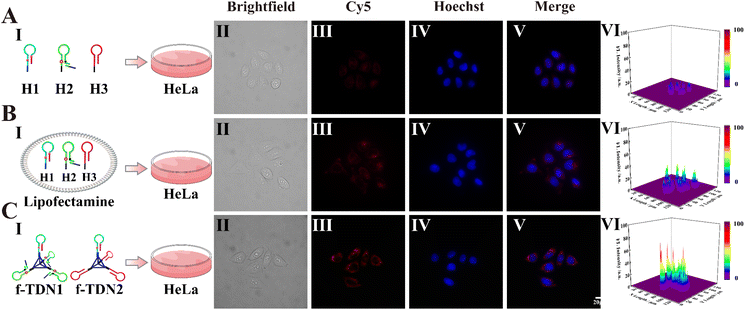
The cellular uptake of f-TDNs
After validating the excellent detection ability of f-TDNs in vitro, the imaging capability of intracellular APE1 and gene therapy were investigated. First, the uptake mechanism of f-TDNs was verified in HeLa cells, which have high expression of the PTK7 receptor on the cell membrane. Brightfield (Panel II) and Hoechst staining images (Panel IV) showed the healthy state of the HeLa cells in the experiments. As shown in Fig. 5A, after incubating with the free hairpins, almost no fluorescent signal was observed in HeLa cells, indicating that the free hairpins were hardly taken up into cells without the help of any transfection reagents. This is mainly due to the permeability resistance of the cell membranes to prevent hairpins from entering the cells. Only with the help of the transfection agent lipofectamine, the fluorescent signal was observed obviously (Fig. 5B). In addition, after incubation of f-TDNs and HeLa cells, a more obvious fluorescent signal was observed (Fig. 5C), indicating that f-TDNs could be directly taken up by cells through the recognition of aptamer Sgc8 with the protein PTK7 on the cell membrane of HeLa cells. Moreover, the fluorescence signal of the f-TDNs CHA reaction triggered by APE1 was brighter than that of the free hairpin CHA reaction in HeLa cells, further emphasizing that f-TDNs could promote CHA reaction occurrence. These results indicated that the proposed f-TDNs CHA system could be used to evaluate the activity of APE1 in tumor cells.Activity assessment of intracellular APE1
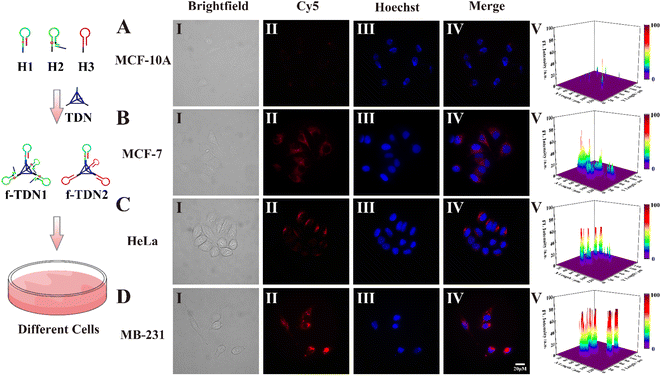
After verifying how f-TDNs were taken up by cells, we investigated the imaging capability of f-TDNs for APE1 in different cells with different expressions.1,44,45 Both brightfield (Panel I) and Hoechst staining images (Panel III) showed that all cells maintained high activity in the experiments. The f-TDNs were incubated with different cells for 4 hours under the same conditions. When normal cells MCF-10A were incubated with f-TDNs (Fig. 6A), almost no fluorescent signal was observed because of low expression of PTK7 and APE1. Although APE1 is highly expressed in MCF-7 cells, a small amount of f-TDNs were endocytosed into MCF-7 cells via sgc8 targeting PTK7 due to the low expression of PTK7, thus only a relatively obvious fluorescence signal was observed in MCF-7 cells (Fig. 6B). Similarly, HeLa cells with high expression of PTK7 and low expression of APE1 showed only a relatively obvious fluorescence signal compared with MCF-10A cells. Notably, only the double positive MDA-MB-231 cells displayed a significant fluorescent signal, which was attributed to the synergistic effect of highly expressed PTK7 and APE1. The above results indicated that the constructed f-TDNs CHA system could not only achieve sensitive intracellular APE1 imaging, but also distinguish tumor cells with different PTK7 and APE1 expression levels.Cell proliferation assays
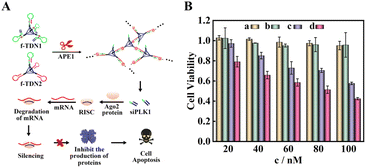
According to the results of fluorescence experiments in vitro and intracellular imaging experiments, we concluded that APE1 could trigger the CHA reaction between f-TDN1 and f-TDN2 with simultaneous release of siRNA. As a proof of concept, small interfering RNA of polo-like kinase 1 (siPLK1) was chosen to demonstrate the feasibility of gene therapy (Fig. 7A), which was a key regulator of cellular proliferation that over-expressed in many malignant tumor cells. We studied the cell viability activities by MTT assays, and the experimental results are shown in Fig. 7B. The proliferation activity of HeLa cells cultured with different concentrations of f-TDN1 without siPLK1 and f-TDN2 (Fig. 7B, column a) was not significantly inhibited, indicating that the constructed DNA nanostructures did not affect the viability of cells. Similarly, after culturing with different concentrations of free siPLK1 (Fig. 7B, column b), the proliferation activity of HeLa cells was also not inhibited because free siPLK1 would not be taken up into cells without the help of transfection reagents. However, after incubation with different concentrations of the mixture of f-TDN1 with siPLK1 and f-TDN2 (Fig. 7B, column c), the viability of HeLa cells decreased with increasing concentrations, indicating that f-TDNs with siPLK1 could indeed achieve gene silencing to inhibit tumor growth. Notably, the proliferation of MAD-MB-231 cells was more effectively inhibited by the same concentration of f-TDNs with siPLK1 (Fig. 7B, column d) than that of HeLa cells, possibly due to the release of more siPLK1 triggered by the high expression of APE1. Overall, these results demonstrated that the prepared f-TDNs with siPLK1 could effectively silence the PLK1 gene to realize tumor treatment with the aid of the endogenous marker APE1.Conclusions
In conclusion, giant DNA networks were assembled from two kinds of functionalized tetrahedral DNA nanostructures (f-TDN1 and f-TDN2) in the presence of APE1, which achieved sensitive detection and intracellular imaging of APE1 as well as gene therapy by siRNA delivery of f-TDN1. Specifically, the constructed f-TDNs efficiently targeted and entered tumor cells by the accurate recognition of aptamer Sgc8 and PTK7. After entering into tumor cells, the AP sites in the stem of H1 could be efficiently cleaved in the presence of APE1 and release S1, which triggered the CHA reaction between f-TDN1 and f-TDN2. Notably, compared with the traditional free CHA reaction, f-TDNs CHA had a faster reaction rate and stronger fluorescence signal due to the spatial confinement effect, which realized the sensitive detection and intracellular imaging of APE1. In addition, the endogenous tumor biomarker APE1 triggered the release of siRNA loaded on f-TDN1 precisely, which promoted tumor cell apoptosis to achieve gene therapy. The proposed f-TDNs achieved functions of diagnosis and treatment integration, and provided a potential method for accurate tumor diagnosis and treatment in clinical practice.Data availability
All the data supporting the findings of this study are available within the article and its ESI files† and from the corresponding author upon reasonable request.Author contributions
Xue-Mei Zhou and Ying Zhuo conceived the project. Xue-Mei Zhou performed the experiments. All authors contributed to the analysis and the interpretation of the results. The manuscript was written by Xue-Mei Zhou and revised by Ruo Yuan and Ya-Qin Chai. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the NNSF of China (21974108, 22174113 and 22176153).References
- M. M. Lv, J. W. Liu, R. Q. Yu and J. H. Jiang, Chem. Sci., 2020, 11, 10361–10366 RSC.
- C. C. Li, H. Y. Chen, J. Hu and C. Y. Zhang, Chem. Sci., 2020, 11, 5724–5734 RSC.
- T. C. Liu, C. T. Lin, K. C. Chang, K. W. Guo, S. Wang, J. W. Chu and Y. Y. Hsiao, Nat. Commun., 2021, 12, 601 CrossRef CAS PubMed.
- Z. Xiang, J. Zhao, J. Qu, J. Song and L. Li, Angew. Chem., Int. Ed., 2022, 61, e202111836 CAS.
- F. Yu, Y. Shao, X. Chai, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2022, 61, e202203238 CAS.
- Y. L. Shao, J. Zhao, J. Y. Yuan, Y. L. Zhao and L. L. Li, Angew. Chem., Int. Ed., 2021, 60, 8923–8931 CrossRef CAS PubMed.
- X. M. Zhou, Y. Zhuo, T. T. Tu, R. Yuan and Y. Q. Chai, Anal. Chem., 2022, 94, 8732–8739 CrossRef CAS PubMed.
- Z. Fan, J. Zhao, X. Chai and L. Li, Angew. Chem., Int. Ed., 2021, 60, 14887–14891 CrossRef CAS PubMed.
- Y. Shao, J. Zhao, J. Yuan, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2021, 60, 8923–8931 CrossRef CAS PubMed.
- L.-L. Zhou, Q. Guan, W. Zhou, J.-L. Kan and Y.-B. Dong, Chem. Sci., 2022, 13, 7846–7854 RSC.
- X. Gong, H. Wang, R. Li, K. Tan, J. Wei, J. Wang, C. Hong, J. Shang, X. Liu, J. Liu and F. Wang, Nat. Commun., 2021, 12, 3953 CrossRef CAS PubMed.
- F. Li, W. Yu, J. Zhang, Y. Dong, X. Ding, X. Ruan, Z. Gu and D. Yang, Nat. Commun., 2021, 12, 1138 CrossRef CAS PubMed.
- J. Li, Y. Zhang, J. Sun, J. Ouyang and N. Na, Chem. Sci., 2021, 12, 15353–15361 RSC.
- F. Ding, Q. Mou, Y. Ma, G. Pan, Y. Guo, G. Tong, C. H. J. Choi, X. Zhu and C. Zhang, Angew. Chem., Int. Ed., 2018, 57, 3064–3068 CrossRef CAS PubMed.
- X. Guan, F. Meng, H. Tan, X. Wang, J. Li, J. Wei, J. Ouyang and N. Na, Chem. Sci., 2022, 13, 8657–8666 RSC.
- F. Li, Z. Lv, X. Zhang, Y. Dong, X. Ding, Z. Li, S. Li, C. Yao and D. Yang, Angew. Chem., Int. Ed., 2021, 60, 25557–25566 CrossRef CAS PubMed.
- H. Hu, N. Yang, J. Sun, F. Zhou, R. Gu, Y. Liu, L. Wang, X. Song, R. Yun, X. Dong and G. Wang, Chem. Sci., 2022, 13, 7355–7364 RSC.
- H. Qiao, L. Zhang, D. Fang, Z. Zhu, W. He, L. Hu, L. Di, Z. Guo and X. Wang, Chem. Sci., 2021, 12, 4547–4556 RSC.
- B. Li, X. Chen and A. D. Ellington, Anal. Chem., 2012, 84, 8371–8377 CrossRef CAS PubMed.
- Y. Tang, Z. Zhu, B. Lu and B. Li, Chem. Commun., 2016, 52, 13043–13046 RSC.
- Y. Tang, B. Lu, Z. Zhu and B. Li, Chem. Sci., 2018, 9, 760–769 RSC.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- M. Mohammadniaei, M. Zhang, J. Ashley, U. B. Christensen, L. J. Friis-Hansen, R. Gregersen, J. G. Lisby, T. L. Benfield, F. E. Nielsen, J. Henning Rasmussen, E. B. Pedersen, A. C. R. Olinger, L. T. Kolding, M. Naseri, T. Zheng, W. Wang, J. Gorodkin and Y. Sun, Nat. Commun., 2021, 12, 5089 CrossRef CAS PubMed.
- Y. Wang, Y. Fang, Y. Zhu, S. Bi, Y. Liu and H. Ju, Chem. Sci., 2022, 13, 2062–2070 RSC.
- F. Ma, Q. Zhang and C. Y. Zhang, Nano Lett., 2019, 19, 6370–6376 CrossRef CAS PubMed.
- Y. Qiao, J. Du, R. Ge, H. Lu, C. Wu, J. Li, S. Yang, S. Zada, H. Dong and X. Zhang, Anal. Chem., 2022, 94, 5538–5545 CrossRef CAS PubMed.
- F. Li, Y. Chen, J. Shang, Q. Wang, S. He, X. Xing and F. Wang, Anal. Chem., 2022, 94, 4495–4503 CrossRef CAS PubMed.
- H. Wang, Y. He, J. Wei, H. Wang, K. Ma, Y. Zhou, X. Liu, X. Zhou and F. Wang, Angew. Chem., Int. Ed., 2022, 61, e202115489 CAS.
- Q. Liu, Y. Huang, Z. Li, L. Li, Y. Zhao and M. Li, Angew. Chem., Int. Ed., 2022, e202214230 CAS.
- X. L. Zhang, Y. Yin, S. M. Du, L. Q. Kong, Y. Q. Chai, Z. H. Li and R. Yuan, Anal. Chem., 2021, 93, 13952–13959 CrossRef CAS PubMed.
- X. L. Zhang, Y. Yin, S. M. Du, L. Q. Kong, Z. H. Yang, Y. Y. Chang, Y. Q. Chai and R. Yuan, Adv. Sci., 2022, 9, e2104084 CrossRef PubMed.
- Y. Feng, Q. Liu, X. Zhao, M. Chen, X. Sun, H. Li and X. Chen, Anal. Chem., 2022, 94, 2934–2941 CrossRef CAS PubMed.
- Z. Qing, J. Hu, J. Xu, Z. Zou, Y. Lei, T. Qing and R. Yang, Chem. Sci., 2020, 11, 1985–1990 RSC.
- J. Su, F. Wu, H. Xia, Y. Wu and S. Liu, Chem. Sci., 2020, 11, 80–86 RSC.
- N. Liao, M. C. Pan, L. Wang, F. Yang, R. Yuan and Y. Zhuo, Anal. Chem., 2021, 93, 4051–4058 CrossRef CAS PubMed.
- D. Zhu, Y. Wei, T. Sun, C. Zhang, L. Ang, S. Su, X. Mao, Q. Li, C. Fan, X. Zuo, J. Chao and L. Wang, Anal. Chem., 2021, 93, 2226–2234 CrossRef CAS PubMed.
- B. Zhang, T. Tian, D. Xiao, S. Gao, X. Cai and Y. Lin, Adv. Funct. Mater., 2022, 32, 2109728 CrossRef CAS.
- F. Li, Y. Liu, Y. Dong, Y. Chu, N. Song and D. Yang, J. Am. Chem. Soc., 2022, 144, 4667–4677 CrossRef CAS PubMed.
- D. Wang, S. Li, Z. Zhao, X. Zhang and W. Tan, Angew. Chem., Int. Ed., 2021, 60, 15816–15820 CrossRef CAS PubMed.
- D. X. Wang, J. Wang, Y. X. Wang, Y. C. Du, Y. Huang, A. N. Tang, Y. X. Cui and D. M. Kong, Chem. Sci., 2021, 12, 7602–7622 RSC.
- R. P. Goodman, R. M. Berry and A. J. Turberfield, Chem. Commun., 2004, 1372–1373 RSC.
- J. W. Hill, T. K. Hazra, T. Izumi and S. Mitra, Nucleic Acids Res., 2001, 29, 430–438 CrossRef CAS PubMed.
- L. J. Wang, L. Liang, B. J. Liu, B. Jiang and C. Y. Zhang, Chem. Sci., 2021, 12, 5544–5554 RSC.
- F. Chen, M. Bai, K. Cao, Y. Zhao, J. Wei and Y. X. Zhao, Adv. Funct. Mater., 2017, 27, 1702748 CrossRef.
- R. Abbotts and S. Madhusudan, Cancer Treat. Rev., 2010, 36, 425–435 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc06968g |
| This journal is © The Royal Society of Chemistry 2023 |