Open Access Article
Open Access ArticleHidden hydrophobicity impacts polymer immunogenicity†
Zhefan
Yuan
 a,
Patrick
McMullen
a,
Sijin
Luozhong
a,
Pranab
Sarker
b,
Chenjue
Tang
a,
Tao
Wei
a,
Patrick
McMullen
a,
Sijin
Luozhong
a,
Pranab
Sarker
b,
Chenjue
Tang
a,
Tao
Wei
 *b and
Shaoyi
Jiang
*b and
Shaoyi
Jiang
 *a
*a
aMeinig School of Biomedical Engineering, Cornell University, Ithaca, NY 14853, USA. E-mail: sj19@cornell.edu
bDepartment of Chemical Engineering, Howard University, Washington, D.C. 20059, USA. E-mail: tao.wei@howard.edu
First published on 30th January 2023
Abstract
Antibodies against poly(ethylene glycol) (PEG) have been found to be the culprit of side reactions and efficacy loss of a number of PEGylated drugs. Fundamental mechanisms of PEG immunogenicity and design principles for PEG alternatives still have not been fully explored. By using hydrophobic interaction chromatography (HIC) under varied salt conditions, we reveal the “hidden” hydrophobicity of those polymers which are generally considered as hydrophilic. A correlation between the hidden hydrophobicity of a polymer and its polymer immunogenicity is observed when this polymer is conjugated with an immunogenic protein. Such a correlation of hidden hydrophobicity vs. immunogenicity for a polymer also applies to corresponding polymer–protein conjugates. Atomistic molecular dynamics (MD) simulation results show a similar trend. Based on polyzwitterion modification and with this HIC technique, we are able to produce extremely low-immunogenic protein conjugates as their hydrophilicity is pushed to the limit and their hydrophobicity is eliminated, breaking the current barriers of eliminating anti-drug and anti-polymer antibodies.
Introduction
Poly(ethylene glycol) (PEG) immunogenicity has been attracting attention in the pharmaceutical and biomedical areas, where several incidents associated with the use of PEGylated drugs have been attributed to pre-existing or induced anti-PEG antibodies (Abs).1,2 Conjugating PEG to proteins could amplify the generation of anti-PEG antibodies to a degree depending on protein immunogenicity,3i.e., the ‘haptenic’ characteristics of PEG. Our previous studies revealed a strong correlation of the titers of anti-PEG Abs to the immunogenicity of conjugated proteins, whereas the generation of anti-polymer Abs against poly(N-(3-acrylamidopropyl)carboxybetaine) (PCB) was minimal and insensitive to protein immunogenicity.3 Although the existence of anti-PEG antibodies is now widely recognized, fundamental mechanisms of PEG immunogenicity are still not clear. Answers to these questions such as why PEG could induce a high level of antibodies and how the chemical and physical characteristics of a polymer are related to its potential immunogenicity when conjugated to proteins will provide not only a fundamental understanding of PEG immunogenicity but also molecular principles for the design of PEG alternatives for better pharmaceutical efficacy and safety.PEG has a quite simple molecular structure. The repetitive unit consists of one ethylene group plus one oxygen atom. Hydrogen bonding between oxygen and surrounding water molecules makes PEG readily soluble in water. On the other hand, PEG can also easily dissolve in many water-immiscible organic solvents, indicating its amphiphilic characteristics. While PEG reduces protein adsorptions on many material surfaces,4,5 it can induce the formation of protein corona and capsule via non-specific interactions.6,7 All of these imply that PEG, which is generally recognized as “inert”, can still trigger a certain level of biological interactions through its intrinsic hydrophobicity. A recent study on the crystal structure of anti-PEG Ab further strengthens this hypothesis. Anti-PEG Ab has an open ring-like sub-structure in its Fab paratope, wherein the PEG backbone is captured via van der Waals interactions.8 It suggests that hydrophobic contact rather than hydrogen bonding dominates PEG/anti-PEG Ab interactions. Similar hydrophobic interactions were also observed in two other PEG/anti-PEG Ab complexes, where the main part of PEG binds with both monoclonal Abs at their aromatic residues.9 Given that hydrophobicity is believed to play an important role in protein immunogenicity10 and many modeling studies use it as an important index to predict epitopes and neoantigens,11,12 it is hypothesized that the immunogenicity of PEG is strongly related to its ‘hidden’ hydrophobicity, which can be applied to many polymers which are generally considered as “hydrophilic”. To verify the hypothesis, we would like to first set up the ranking in the ‘hidden’ hydrophobicity of common hydrophilic polymers. Currently, there is no standard definition of the hydrophobicity of hydrophilic polymers or a method to test the ‘hidden’ hydrophobicity. While the degree of protein hydrophobicity has been extensively explored using different theoretical and experimental approaches,13 here, we employ hydrophobic interaction chromatography (HIC) to evaluate polymer hydrophobicity. HIC, as an analytical technique, is widely used as a scalable process for protein purification.13 By incubating proteins with a high concentration of salts, protein surfaces are de-solvated, resulting in amplified interactions between the hydrophobic portions of the proteins and the hydrophobic ligands on a stationary phase. Employing high-salt-induced hydrophobic interaction, we could reveal the ‘hidden’ hydrophobicity of many water-soluble polymers and polymer–protein conjugates. Moreover, HIC also provides certain advantages over other techniques, such as reverse-phase liquid chromatography system (HPLC)14 as it reveals the hydrophobicity of analytes in an aqueous solution under relatively low pressure. Most analytes, like proteins or protein conjugates, are not denatured under HIC conditions and can be used in downstream studies after a simple desalting process.
Results and discussion
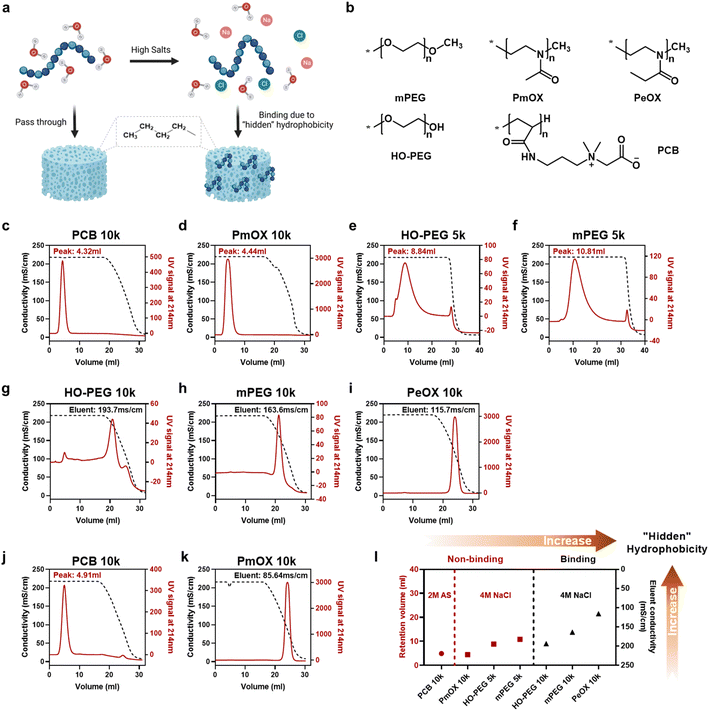
In this study, we selected four synthetic polymers (Fig. 1a), PEG, PCB, poly(2-methyl-2-oxazoline) (PmOX), and poly(2-ethyl-2-oxazoline) (PeOX), as typical examples to establish the ranking of the ‘hidden’ hydrophobicity of the polymers. The main reason to choose these polymers is that they are all hydrophilic, electrically neutral, and have been reported with ‘stealth’ properties.15 Furthermore, all of them have been used as protein modifiers to prepare protein–drug conjugates in academic research or clinical trials.16–18 Before on-column tests, we first examined polymer solubilities in two types of salted buffers, 0–4 M sodium chloride (NaCl) and 0–2 M ammonium sulfate (AS), which are also typical ‘salting out’ substances used in HIC. As a high salt buffer leads to polymer aggregation or liquid–liquid phase separation, polymer solubility could be monitored by the change of O.D. values at 350nm.19 The results are summarized in Fig. S1.† Generally, all polymers steadily dissolved in the buffer containing up to 4 M of NaCl while only PeOX had a slight OD increase at the highest salt buffer. The difference in solubility was revealed in AS solutions, a stronger ‘salting out’ substance according to the Hofmeister series.20 As expected, PeOX showed the lowest solubility limit, followed by MPEG and PmOX, which formed phase separation at 1.2 M AS. Only the PCB solution remained clear even in 2 M AS buffer. Thus, the ranking based on solubility-based polymer hydrophobicity could be roughly written as PCB < mPEG ∼ PmOX < PeOX.Column-based HIC tests were performed to establish the detailed ranking in ‘hidden’ hydrophobicity. Typical HIC columns are functionalized with butyl, octyl, or phenyl groups. Based on the ‘like dissolves like’ rule, we chose a butyl group functionalized HIC medium to reveal potential hydrophobicity originating from polymer backbone or pendant groups. An on-column HIC experiment was performed: polymers were first loaded and isocratically eluted at 4 M NaCl until all non-binding or weak binding portions passed through, followed by gradient elution from 4 M to 0 M NaCl to free trapped sample portions. The eluent profiles of polymers were monitored using UV signals at 214 nm, reflecting the hydrophobicity of the backbone of polyoxazoline or the amide group in the side chain of PCB. As the PEG backbone lacks spectral signals in the UV-vis range, we used mono-maleimide-modified derivatives to evaluate PEG hydrophobicity in all HIC tests. The conductivity of the eluent buffer was monitored simultaneously and values corresponding to eluting peaks were recorded to describe the strength of hydrophobic interactions between the polymer and the column. The curves of HIC experiments performed at 4 M NaCl are shown in Fig. 1c–i. Generally, seven tested polymers could be divided into two groups: 1. Polymers eluting at 4 M NaCl (Fig. 1c–f), where larger retention volumes indicate stronger hydrophobic column interactions; 2. Polymers eluting at lowered NaCl concentrations (Fig. 1g–i), where lower solution conductivities correspond to eluent peaks, indicating stronger hydrophobic column interactions. For example, no column binding occurred in HIC tests for PCB 10k and PmOX 10k as they eluted along with 4 M NaCl immediately, leaving sharp peaks and indicating low ‘hidden’ hydrophobicity. Most of HO-PEG 5k passed through the butyl column without decreasing salt concentration, but a broadened and delayed eluent peak profile implied the existence of weak interactions. Changing the terminal group from hydroxyl to methyl did not significantly alter curve patterns, but a delay of eluent volume was observed, suggesting reduced hydrophobicity. PeOX 10k, mPEG 10k, and HO-PEG 10k showed much higher hydrophobicity as they completely bound to the column at 4 M NaCl. Eluting bound PeOX 10k required a much lower salt concentration (eluent peak, 115.7 mS cm−1) than mPEG 10k (163.6 mS cm−1) and HO-PEG 10k (193.7 mS cm−1), indicating its strongest column interactions. The impact of molecular weight on the polymer hydrophobicity could be also interpreted by the different HIC curves of the four PEGs. Both 10k PEGs were completely bound to the column media while 5k PEGs could elute out under the same eluent conditions, implying a higher molecular weight led to a higher ‘hidden’ hydrophobicity. To further compare PCB 10k with PmOX 10, additional HIC experiments were carried out by changing the incubation buffer from 4 M NaCl to 2 M AS, which has a stronger ‘salting out’ capacity. A large difference is observed in Fig. 1j and k. While PmOX 10k (Fig. 1k) completely bound to the column in 2 M AS, a very low AS concentration (85.6 mS cm−1, equals 0.82 M) is required to elute out these bound PmOX molecules. The peak of PCB 10k (Fig. 1j) was slightly right-shifted compared to the one in 4 M NaCl test, but no broadened peak occurred. This phenomenon was likely due to the shrinkage of the hydrodynamic size of PCB induced by a stronger ‘salting out’ effect rather than the consequence of hydrophobic interactions. The size exclusion effect of the porous column medium played the main role here. To summarize, a more detailed ‘hidden’ hydrophobicity ranking based on HIC studies is shown as follows (Fig. 1i): PCB (10k) ≪ PmOX (10k) < HO-PEG (5k) ∼ mPEG (5k) < HO-PEG (10k) ∼ mPEG (10k) < PeOX (10k). The ranking could be further divided into three regions. Region 1 (low), non-binding polymers at 2 M AS; region 2 (medium), non-binding polymers at 4 M NaCl; region 3 (high), polymers bound to columns at 4 M NaCl.
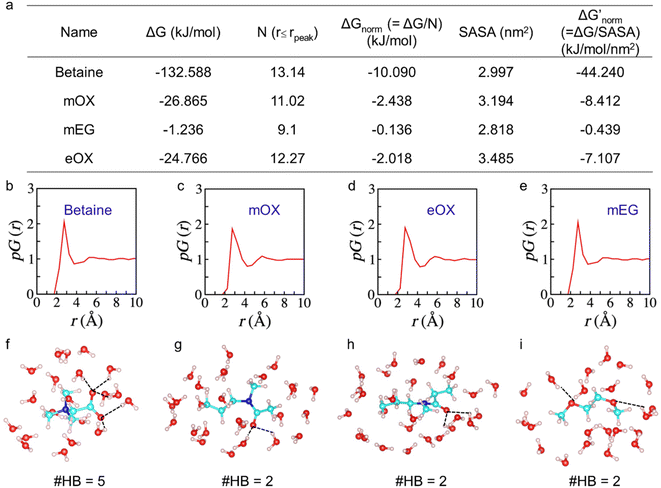
To further investigate the interactions of different compounds with water, we performed atomistic molecular dynamics (MD) simulations and estimated their solvation free energies ΔG using the free energy perturbation method.21 To eliminate the molecular size effect on ΔG and compare their affinity with water, we only computed ΔG of the monomers and then normalized them with the number of hydration water molecules close to the surface of the monomers i.e., water molecules within the first hydration shell of the monomers up to the peak position of the proximal radial distribution pG(r) (Fig. 2).22,23 We also normalized ΔG with the solvent-accessible surface area (SASA) to provide an additional evaluation. As one –OH group could significantly change water binding to one EG monomer, we did not calculate the monomer of OH-terminated PEG here. As shown in Fig. 2a, results by both ways of normalization show the same trends of normalized solvation free energies of the monomers. The zwitterionic betaine monomer has an absolute value of the normalized solvation free energy |ΔGnorm| one order of magnitude greater than that of all other compounds, due to its strongest electrostatic interactions with water. Moreover, our simulation results show that a betaine can form five hydration bonds with the surrounding hydration water molecules, whereas for other monomers (mOX, mEG and eOX), there are only two hydrogen bonds detected (Fig. 2f–i). The ranking of |ΔGnorm| is betaine > mOX > eOX > mEG (see Fig. 2a). Although the general trend of hydration ranking is consistent with that obtained from HIC experiments, a different order of eOX and mEG has been observed. This is because the hydration of molecules in simulations is not completely equal to their hydrophobicity in HIC experiments where there are additional interactions between polymers and hydrophobic moieties (butyl groups) on the column matrix. The latter represents the interactions between polymers and antibodies better.
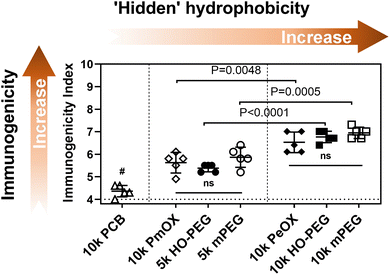
Next, we conjugated each polymer to a high immunogenic carrier protein, keyhole limpet hemocyanin (KLH), to induce anti-polymer Abs.3 Similar polymer density was achieved to make a fair comparison (Table S1†). Each group of C57bl/6 mice (n = 5) was then treated with two doses of polymer-KLH conjugate via biweekly subcutaneous (s.c.) injections. Serum samples for the Ab test were collected one week later after the 2nd injection and Ab titers were measured by indirect ELISA. For the quantitative comparison of polymer immunogenicity, we employed a parameter, called ‘immunogenicity index’,3 which was defined as an arithmetic sum of the log values of IgG and IgM titers. It should be noted that the theoretical minimum of immunogenicity index in the current study is 4 because serum dilution for IgG and IgM ELISA tests started from 100-fold. The relationship between polymer immunogenicity and their intrinsic hydrophobicity is summarized in Fig. 3. Seven polymers can be divided into three groups based on the level of immunogenicity indexes. PCB (10k) with the most hydrophilicity possessed the lowest immunogenicity. PEGs (5k) and PmOX (10k), which all passed through the butyl column at 4 M NaCl, showed similar and modest immunogenicity index values. PEGs (10k) and PeOX (10k), which completely bound to the HIC column, triggered the highest level of anti-polymer Abs. The order of polymer immunogenicity indexes is estimated as: PCB (10k) < PmOX (10k) ∼ HO-PEG (5k) ∼ mPEG (5k) < PeOX (10k) ∼ HO-PEG (10k) ∼ mPEG (10k), which matches with their hydrophobicity ranking. It should be noted that the tested immunogenicity indexes of both PeOX (10k) and mPEG (10k) are equally high and have no statistical difference. To better understand structural effects, we compared polymers sharing similar structures for their immunogenicity indexes. The additional methylene moiety in every repeating unit makes PeOX more hydrophobic than PmOX. As a result, PeOX induced higher antibody levels than PmOX did (P = 0.0048). PEGs (10k) showed significantly higher immunogenicity indexes than PEGs (5k) as increased molecular weight enhanced overall hydrophobicity interactions. However, turning the methoxy terminal group into a more hydrophilic hydroxyl group did not provide a significant improvement over PEG immunogenicity. Previous reports showed that the terminal hydroxyl group of PEG-induced stronger complement activation than the methoxy group terminated PEG.24 While OH is more hydrophilic, its complement activation should be also taken into account when it comes to polymer immunogenicity.
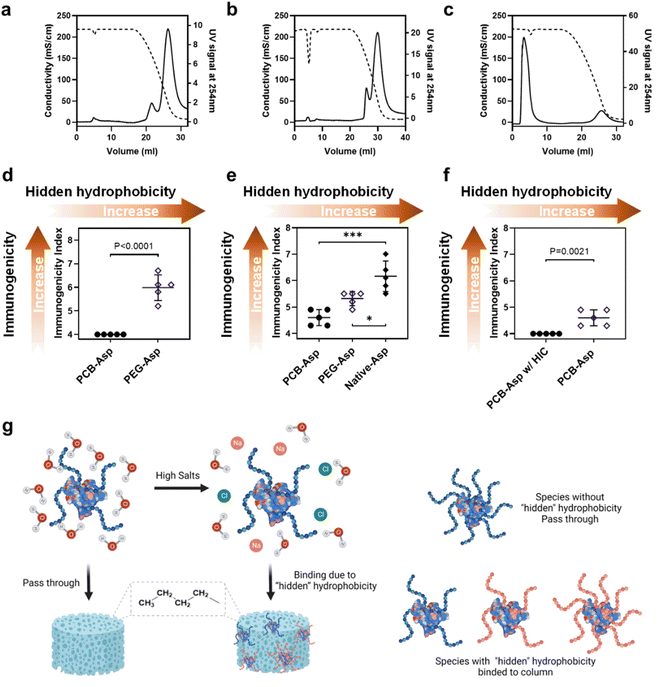
The correlation between polymer hydrophobicity and immunogenicity suggested a new way to mitigate anti-polymer Abs by reducing the hydrophobic feature of a polymer. In clinical practice, anti-drug Ab (ADA) for the whole polymer–protein conjugate is more meaningful because it is a combined result contributed by the immunogenicity of both the protein and polymer. The measurement of ADA is also on the must-do checklist both in preclinical and clinical trials. Thus, we would like to explore if the ‘‘hidden’ hydrophobicity–immunogenicity’ correlation can be applied to the relationship between the hydrophobicity of a polymer–protein conjugate and its ADA. MPEG (5k)-modified Asparaginase (Asp) with a similar structure to Oncaspar® (FDA-approved Pegylated therapeutic protein) was prepared and studied here. 10k PCB-Asp conjugate was also prepared with similar polymer density and particle size for a fair comparison. SDS-PAGE (Fig. S5a†) and SEC (Fig. S5b and Table S2†) data confirm their compositions. Asp and Asp conjugates were then loaded on the butyl HIC column and eluted with 4 M NaCl by the same method used in polymer tests. Native Asp (Fig. 4a) showed a similar curve pattern to10k PEGs and PeOX did but needed a lower NaCl concentration to be released from the column, indicating its high level of hydrophobicity. After modification with 5k mPEG, entire PEG-Asp conjugates (Fig. 4b) still tightly bind to the HIC column at 4 M NaCl but the main eluent peak advanced from 43.6 to 51.9 mS cm−1 compared to Asp protein. Although 5k mPEG showed very weak column binding at 4 M NaCl, it only slightly reduced the apparent hydrophobicity of the Asp conjugate. Modification of the Asp with multiple PEG chains made the protein conjugate more like a huge star-shaped mPEG nanoparticle; thus, the HIC profile of mPEG-Asp conjugate was supposed to behave like high molecular weight PEGs. A huge contrast was observed in the PCB-Asp conjugate (Fig. 4c). Most of PCB-Asp freely passed through the butyl column in the same manner as the free PCB polymer did although the apparent molecular weight of PCB increased tens of times. Antibodies were induced in C57BL/6 mice by three weekly s.c. injections of Asp or Asp conjugates. The calculated immunogenicity index of the whole PEG-Asp conjugate reached 5.99 while PCB-Asp did not induce any positive signals (Fig. 4d). This result verified our assumption that the apparent hydrophobicity of polymer–protein conjugates may also be used to predict their potential immunogenicity. With a similar purpose, we further compared anti-Asp immunogenicity indexes for Asp, PEG-Asp, and PCB-Asp (Fig. 4e), which correlate well with their hydrophobicity ranking as shown in Fig. 4a–c. PEG reduced Asp immunogenicity by shielding surface epitopes, which was reflected as improved hydrophilicity. It is noted that the anti-protein index was lower than the anti-conjugate index detected in those mice treated with PEG-Asp, indicating the existence of PEG-specific Abs. PCB significantly mitigated Asp immunogenicity, but anti-Asp Abs were still detected in PCB-Asp treated mice although small.
Thus, we further tested the non-binding portion of PCB-Asp from HIC purification (or further purified PCB-ASP conjugate) in mice by the same three-injection route. No anti-Asp Abs could be detected in this experiment (Fig. 4f). It is interesting that SEC curves and SDS-PAGE (Fig. S5†) did not reveal any significant difference caused by HIC treatment. A slight increase in average PCB density from 9.1 to 9.3 chains per Asp monomer was found by SEC-MALS measurement (Table S2†). There are many factors that can affect the surface chemistry and hydrophilicity of polymer–protein conjugates such as the hydrophilicity, molecular weight, chain number, and distribution of a polymer to shield the hydrophobic regions of a protein. The HIC method in this work picks up the overall most hydrophilic polymer–protein conjugates. The conventional purification of polymer–protein conjugates highly relies on liquid chromatography technologies like SEC, ion exchange, hydrophobic interaction, or affinity chromatography. These techniques are effective to separate mono-, di-, or even tri-polymer conjugated proteins from the reaction mixture but the process becomes tedious and inefficient,25 particularly when completely shielding polymers over the polymer–protein conjugate is required. By combining super-hydrophilic PCB modification and the HIC purification technique, the best PCB–protein conjugates could be selectively collected in a scalable process, benefiting both drug production and quality control.
Conclusions
In this study, we revealed and established a ranking order in the hidden hydrophobicity of hydrophilic polymers and their polymer–protein conjugates by using a HIC technique under varied salt conditions. The immunogenicity reflected by the generation of Abs was evaluated for these polymers and their polymer–protein conjugates. Experimental and atomistic molecular dynamics simulation results supported our hypothesis on the relationship between the hidden hydrophobicity and immunogenicity of a polymer. Higher hidden polymer hydrophobicity induced a higher level of anti-polymer Abs when the polymer is conjugated to an immunogenic protein. The correlation could be seen more clearly when those polymers sharing a similar structure were compared. This work mainly focuses on the immunogenic risk of a polymer used for protein modification. Issues such as compromise in efficacy and complement activation that commonly exist in PEG, POX, or other amphiphilic polymer-modified nanoparticles are often associated with their hidden polymer hydrophobicity.26–28 Although the reduction in polymer hydrophobicity through altering polymer structures or reducing molecular weights may mitigate polymer immunogenicity issues to a certain extent, using a super-hydrophilic alternative with the aid of HIC purification under varied salt conditions could be a fundamental solution both in principle and practice.Data availability
We have provided all the data in the manuscript and ESI.†Author contributions
Z. Yuan prepared the samples and carried out data analysis. Z. Yuan and P. McMullen performed the HIC tests. Z. Yuan and S. Luozhong performed the animal study. P. Sarker and T. Wei did the MD simulations. C. Tang provided suggestions for the project. Z. Yuan and S. Jiang wrote the manuscript. S. Jiang and T. Wei directed the experimental and computational components of this project, respectively. All the authors provided suggestions and comments on the manuscript.Conflicts of interest
S. J. is a co-founder of ZWI therapeutics. S. J. and Z. Y. are listed as inventors on a provisional patent application filed by Cornell University.Acknowledgements
S. J. acknowledges financial support from the National Science Foundation (CBET- 1911478), the Office of Naval Research (N00014-20-1-2731), and start-up support from Cornell University, including Robert Langer ‘70 Family and Friends Professorship and Cornell NEXT Nano Initiative. T. W. is grateful for financial support from the Office of Naval Research (N00014-21-1-2215) and the computational resources from the program of Extreme Science and Engineering Discovery Environment (XSEDE) and the Texas Advanced Computing Center (TACC).Notes and references
- N. J. Ganson, T. J. Povsic, B. A. Sullenger, J. H. Alexander, S. L. Zelenkofske, J. M. Sailstad, C. P. Rusconi and M. S. Hershfield, J. Allergy Clin. Immunol., 2016, 137, 1610–1613 CrossRef CAS PubMed.
- P. Zhang, F. Sun, S. Liu and S. Jiang, J. Controlled Release, 2016, 244, 184–193 CrossRef CAS PubMed.
- B. Li, Z. Yuan, H. C. Hung, J. Ma, P. Jain, C. Tsao, J. Xie, P. Zhang, X. Lin and K. Wu, Angew. Chem., Int. Ed., 2018, 57, 13873–13876 CrossRef CAS PubMed.
- N.-P. Huang, R. Michel, J. Voros, M. Textor, R. Hofer, A. Rossi, D. L. Elbert, J. A. Hubbell and N. D. Spencer, Langmuir, 2001, 17, 489–498 CrossRef CAS.
- V. C. F. Mosqueira, P. Legrand, A. Gulik, O. Bourdon, R. Gref, D. Labarre and G. Barratt, Biomaterials, 2001, 22, 2967–2979 CrossRef CAS PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Nat. Nanotechnol., 2016, 11, 372–377 CrossRef PubMed.
- L. Zhang, Z. Cao, T. Bai, L. Carr, J.-R. Ella-Menye, C. Irvin, B. D. Ratner and S. Jiang, Nat. Biotechnol., 2013, 31, 553–556 CrossRef CAS PubMed.
- J. T. Huckaby, T. M. Jacobs, Z. Li, R. J. Perna, A. Wang, N. I. Nicely and S. K. Lai, Commun. Chem., 2020, 3, 124 CrossRef CAS PubMed.
- C.-C. Lee, Y.-C. Su, T.-P. Ko, L.-L. Lin, C.-Y. Yang, S. S.-C. Chang, S. R. Roffler and A. H. J. Wang, J. Biomed. Sci., 2020, 27, 12 CrossRef CAS PubMed.
- D. Chowell, S. Krishna, P. D. Becker, C. Cocita, J. Shu, X. Tan, P. D. Greenberg, L. S. Klavinskis, J. N. Blattman and K. S. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E1754–E1762 CrossRef CAS PubMed.
- L. P. Richman, R. H. Vonderheide and A. J. Rech, Cell Syst., 2019, 9, 375–382 CrossRef CAS PubMed.
- T. P. Riley, G. L. J. Keller, A. R. Smith, L. M. Davancaze, A. G. Arbuiso, J. R. Devlin and B. M. Baker, Front. Immunol., 2019, 10 Search PubMed.
- A. Mahn, M. E. Lienqueo and J. C. Salgado, J. Chromatogr. A, 2009, 1216, 1838–1844 CrossRef CAS PubMed.
- K. J. Wilson, A. Honegger, R. P. Stötzel and G. J. Hughes, Biochem. J., 1981, 199, 31–41 CrossRef CAS PubMed.
- Z. Amoozgar and Y. Yeo, WIREs Nanomed. Nanobiotechnol., 2012, 4, 219–233 CrossRef CAS PubMed.
- S. Liu and S. Jiang, Nano Today, 2016, 11, 285–291 CrossRef CAS.
- R. W. Moreadith, T. X. Viegas, M. D. Bentley, J. M. Harris, Z. Fang, K. Yoon, B. Dizman, R. Weimer, B. P. Rae, X. Li, C. Rader, D. Standaert and W. Olanow, Eur. Polym. J., 2017, 88, 524–552 CrossRef CAS.
- M. J. Vicent and R. Duncan, Trends Biotechnol., 2006, 24, 39–47 CrossRef CAS PubMed.
- GE Healthcare (now Cytiva), Application note 28-9964-9949 AA, High-throughput screening of HIC media in PreDictor™ plates for capturing recombinant Green Fluorescent Protein from E. coli, GE Healthcare Bio-Sciences AB, 2011 Search PubMed.
- A. M. Hyde, S. L. Zultanski, J. H. Waldman, Y.-L. Zhong, M. Shevlin and F. Peng, Org. Process Res. Dev., 2017, 21, 1355–1370 CrossRef CAS.
- M. R. Shirts, D. L. Mobley and J. D. Chodera, in Annu. Rep. Comput. Chem., ed. D. C. Spellmeyer and R. Wheeler, Elsevier, 2007, vol. 3, pp. 41–59 Search PubMed.
- H. Huang, C. Zhang, R. Crisci, T. Lu, H.-C. Hung, M. S. J. Sajib, P. Sarker, J. Ma, T. Wei, S. Jiang and Z. Chen, J. Am. Chem. Soc., 2021, 143, 16786–16795 CrossRef CAS PubMed.
- P. Sarker, G. T. Chen, M. S. J. Sajib, N. W. Jones and T. Wei, Colloids Surf., A, 2022, 653, 129943 CrossRef CAS.
- Y. Arima, M. Toda and H. Iwata, Biomaterials, 2008, 29, 551–560 CrossRef CAS PubMed.
- V. Gaberc-Porekar, I. Zore, B. Podobnik and V. Menart, Curr. Opin. Drug Discovery Dev., 2008, 11, 242–250 CAS.
- M. J. Grace, S. Lee, S. Bradshaw, J. Chapman, J. Spond, S. Cox, M. DeLorenzo, D. Brassard, D. Wylie, S. Cannon-Carlson, C. Cullen, S. Indelicato, M. Voloch and R. Bordens, J. Biol. Chem., 2005, 280, 6327–6336 CrossRef CAS PubMed.
- P. H. Kierstead, H. Okochi, V. J. Venditto, T. C. Chuong, S. Kivimae, J. M. J. Fréchet and F. C. Szoka, J. Controlled Release, 2015, 213, 1–9 CrossRef CAS PubMed.
- R. Tavano, L. Gabrielli, E. Lubian, C. Fedeli, S. Visentin, P. Polverino De Laureto, G. Arrigoni, A. Geffner-Smith, F. Chen, D. Simberg, G. Morgese, E. M. Benetti, L. Wu, S. M. Moghimi, F. Mancin and E. Papini, ACS Nano, 2018, 12, 5834–5847 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and supplementary figures. See DOI: https://doi.org/10.1039/d2sc07047b |
| This journal is © The Royal Society of Chemistry 2023 |