Open Access Article
Open Access ArticleControllable mitochondrial aggregation and fusion by a programmable DNA binder†
Longyi
Zhu
 a,
Yiting
Shen
a,
Shengyuan
Deng
a,
Yiting
Shen
a,
Shengyuan
Deng
 c,
Ying
Wan
c,
Ying
Wan
 *b,
Jun
Luo
*b,
Jun
Luo
 a,
Yan
Su
b,
Mingxu
You
a,
Yan
Su
b,
Mingxu
You
 d,
Chunhai
Fan
d,
Chunhai
Fan
 e and
Kewei
Ren
e and
Kewei
Ren
 *a
*a
aSchool of Chemistry and Chemical Engineering, Nanjing University of Science and Technology, Nanjing 210094, China. E-mail: kwren@njust.edu.cn
bIntelligent Microsystem Technology and Engineering Center, School of Mechanical Engineering, Nanjing University of Science and Technology, Nanjing 210094, China. E-mail: wanying@njust.edu.cn
cSchool of Environmental and Biological Engineering, Nanjing University of Science and Technology, Nanjing 210094, China
dDepartment of Chemistry, University of Massachusetts, Amherst, MA 01003, USA
eSchool of Chemistry and Chemical Engineering, Shanghai Jiao Tong University, Shanghai 200127, China
First published on 6th July 2023
Abstract
DNA nanodevices have been feasibly applied for various chemo-biological applications, but their functions as precise regulators of intracellular organelles are still limited. Here, we report a synthetic DNA binder that can artificially induce mitochondrial aggregation and fusion in living cells. The rationally designed DNA binder consists of a long DNA chain, which is grafted with multiple mitochondria-targeting modules. Our results indicated that the DNA binder-induced in situ self-assembly of mitochondria can be used to successfully repair ROS-stressed neuron cells. Meanwhile, this DNA binder design is highly programmable. Customized molecular switches can be easily implanted to further achieve stimuli-triggered mitochondrial aggregation and fusion inside living cells. We believe this new type of DNA regulator system will become a powerful chemo-biological tool for subcellular manipulation and precision therapy.
Introduction
DNA nanotechnology has been widely used to accomplish intelligent processes in many fields including synthetic soft materials, bio-computing, sensing and bioimaging, due to their outstanding programmability, high flexibility and diverse structures.1–3 Recently, DNA nanodevices have become a hotspot in exploiting manipulation strategies in bio-processes, such as substrate channeling, cell migration, immune activation and cell identification.4–7 The specificity and well predictability of Watson–Crick–Franklin base pairing provides the fidelity of constructing the responsively artificial DNA nano-machines by catalytic hairpin assembly, proximity-based ligation, rolling circle amplification (RCA), hybridization chain reaction (HCR).8–11 Furthermore, convenient modification on DNA strands with various functional organic molecules greatly expanded their applications to manipulate cascade enzymatic reactions, cell surface receptors assembly and cell–cell interactions.4–6 For example, cholesterol-functionalized DNA nanostructures have been applied for cell surface engineering to realize adaptive cellular interactions.12–14 DNA nanomachines demonstrate precise controllability and robust operation with effective biological outputs in extracellular environment.15,16 However, a large number of biological processes occurs inside cells, the development of DNA-based manipulation of intracellular network still remains a big challenge.Mitochondria are intracellular organelles playing a critical role in energy production and proliferation.17 Under oxidative stress, the dynamic fusion–fission process of mitochondria can be used to regulate metabolism: fission facilitates cell apoptosis, whereas, fusion can mitigate oxidative stress. Specifically, when abnormal level of reactive oxygen species (ROS) prompts the mutation of mitochondria genomes,18 fusion occurs between mitochondria with wild-type DNA and ones with mutant DNA to compensate for the defects.19 As an important communication process among mitochondria in the cytosol, mitochondrial fusion (mito-fusion) is regulated by membrane-anchored dynamics, of which Fzo1 heptads from two adjacent mitochondria form coiled-coil structures to tether the membrane.20 Abnormal mito-fusion can cause neurodegenerative diseases, such as Charcot–Marie–Tooth 2A and dominant optic atrophy.21 Excessive fission are closely related to tumors.22,23 Traditional mito-therapeutic strategy is normally based on modulating the expression of fusion proteins, which requires either delicate gene engineering process or screening of organic molecules.24,25 Recently, Wang and coworkers reported another approach for the direct induction of mitochondrial fusion based on host-guest supramolecular assembly, which coins the artificial manipulation of mito-fusion as an alternative therapeutic strategy.26 Yang and coworkers developed a DNA tetrahedron machine as polyanionic barriers to successfully modulate mitochondria communications.27 However, designing molecular tools that can achieve controllable assemblies of mitochondria and precise manipulation of the fusion processes remains largely underdeveloped. In this work, a new DNA nanomachine will be introduced to fill this gap.
We designed herein a DNA binder that contains multiple mitochondrial targeting modules for the controllable manipulation of mitochondrial aggregation and fusion in living cells. Such DNA binder-mediated artificial handing of mitochondria can also result in mitigated reactive oxygen species (ROS) levels and cell repairs (Fig. 1A and B). By further taking advantages of the highly programmable DNA architectures, molecular switches could be integrated into the DNA binder to achieve precise 2′-deoxyadenosine-5′-triphosphate (ATP)-controlled mitochondrial fusion. In addition, a variety of stimuli-triggered process could also be rationally activated using this approach. We believe the DNA binder described in this study opens a new avenue for effective and easy manipulation of intracellular organelles and related metabolism.
Results and discussions
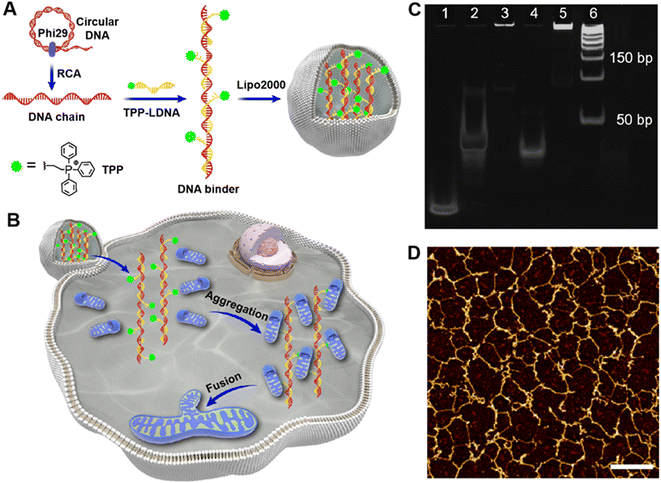
Fabrication and in vitro performance of DNA binders
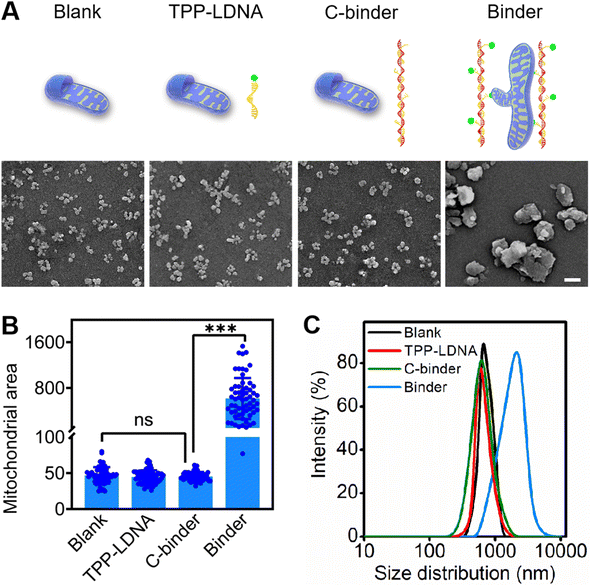
To develop mitochondria-targetable DNA binder, a long single-stranded DNA chain was first prepared by a phi29 polymerase-catalyzed RCA reaction, which consists of 48 nt-long repeated sequences containing 24 nt (Table S1†) that can be recognized by multiple triphenylphosphonium (TPP)-modified linkage DNAs (TPP-LDNAs) (Fig. 1A and Scheme S1†). Here, the TPP cation is used to target mitochondrial and induce mito-fusion.28,29 TPP-LDNA was synthesized based on a copper-catalyzed azide/alkyne cycloaddition reaction30 between an azide-modified linkage DNA and an alkynyl-functionalized TPP cation (Fig. S1–S4†). Native PAGE (15 wt%) showed complete formation of TPP-LDNA (Fig. S5†), indicating the high yield of the cycloaddition reaction (nearly 100%).31 The amounts of phi29 polymerase for DNA chain synthesis were optimized. The agarose gel electrophoresis revealed that changing amounts of phi29 polymerase led to no obvious variation on the length of DNA chains.32 Due to the low yield, there is no obvious DNA chain bands were observed at 0.05 and 0.1 U μL−1 of phi29 polymerase. The yield of DNA chains plateaued at 0.2 U μL−1 of phi29 polymerase by comparing the band intensities of the RCA products. Thus, 0.2 U μL−1 of phi29 polymerase was chosen as the optimized amount for RCA, and the DNA chain was obtained with major length ranging from 1500 to 2000 bases (Fig. S6A†). After hybridization between the DNA chain and TPP-LDNA, and purification by ultrafiltration, only the band of DNA binder that shifted toward higher base of band ruler was observed, indicating the excess TPP-LDNA was removed and the DNA binder was prepared (Fig. S6B†). The successful preparation of the DNA binder was also confirmed by 10% denature polyacrylamide gel electrophoresis and atomic force microscopy (AFM) (Fig. 1C and D). The average length of the DNA binder was measured by observing thread features in the AFM images,33 and found to be 500 ± 150 nm (Fig. S7†). Given that 48 nt-long repeated sequences were about 16.3 nm,34 31 ± 9 TPP-LDNA repeating units could be counted for each DNA binder. A 93% grafting ratio of TPP-LDNA on the DNA chain was further estimated by fluorescence measurements (Fig. S8†), demonstrating sufficient mito-targeting modules in the DNA binder.We first wanted to test if the designed DNA binder can be used to induce mitochondrial aggregation and fusion in an extracellular experiment. As shown in Fig. 2A, the addition of either TPP-LDNA alone or a non-TPP-modified DNA chain/LDNA complex (C-binder) did not change the size or aggregation status of mitochondria. While in contrast, when mitochondria were incubated with the DNA binder (binder), particle stacking with an integrated morphology could be clearly observed, suggesting a DNA binder-induced aggregation and fusion of the mitochondria. A 15-fold enlargement in the area of mitochondrial aggregates was displayed upon the treatment of DNA binder (Fig. 2B), which was in accordance with the dynamic light scattering results (Fig. 2C). The length of DNA binder was optimized35 to 500 nm, and the number of interspersed bases in-between TPP-LDNAs of DNA binder was optimized to 24 nt for the efficient aggregation and fusion of mitochondria (Fig. S9†), which 31 of mito-targeting modules is require as the minimum number. The manipulation of extracellular mitochondrial behavior can be further visualized in solution under confocal fluorescence microscopy. The TPP-LDNA and DNA binder were hybridized with FAM-labeled oligonucleotides (FAM-O1 and FAM-O2) respectively. The fluorescence signals of these two complexes localized well with extracted mitochondria in the bright field channel of confocal images, which proved their successfully attachments on the mitochondria (Fig. S10†). To investigate the fusion behaviors of mitochondria, Förster resonance energy transfer (FRET) study was carried out by using fluorescent probes N-(7-nitro-2-1,3-benzoxadiazol-4-yl)-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (NBD-PE) and N-(lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (Rhod-PE) as the donor and acceptor.36 Two sets of mitochondria were respectively stained with NBD-PE and Rhod-PE, followed washing with PBS and mixing them together. Upon addition of 1.0 μM DNA binder into the above mixture, increased particle size was observed in the confocal images, together with FRET signals (red) under the excitation of 488 nm (Fig. S11 and S12A†). This result was also verified by the enhanced emission intensity at 580 nm in the fluorescence spectra (Fig. S12B†).37,38 Further disassembly of DNA binder and aggregated mitochondria via toehold-mediated strand displacement did not change the FRET results, indicating the FRET signal was generated by mitochondrial fusion. On the other hand, reduced FRET efficiency could be achieved by mixing one set of FRET pairs-labeled mitochondria with the unlabeled set39,40 under the treatment of DNA binders (Fig. S13†). Besides, colocalization study showed overlapping of the two sets of mitochondria signals41 were observed with increased particle size (Fig. S14†), demonstrating the fusion of mitochondria by DNA binders. All these above results confirmed that the DNA binder could successfully induce mitochondrial fusion in vitro. Both mito-targeting TPP moieties and the long DNA chain are essential for the function of DNA binder.
The performance and therapeutic results of DNA binder in living cells
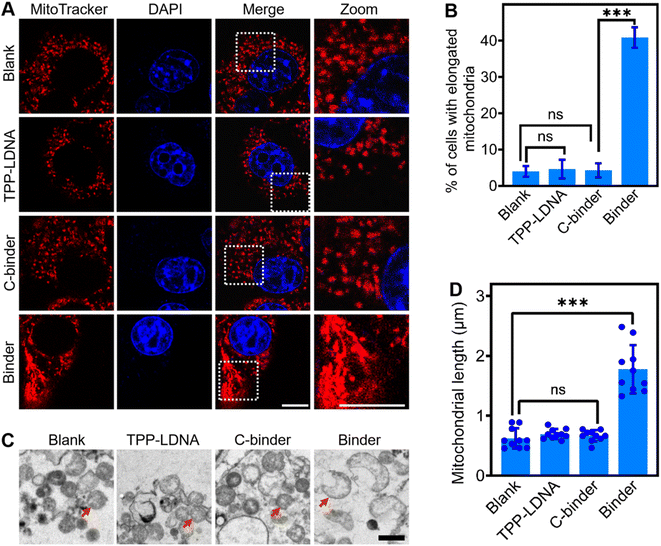
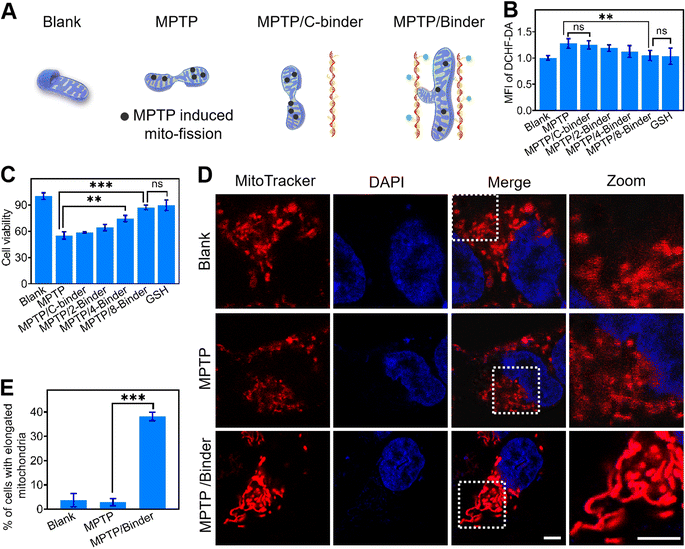
Next, we studied if the DNA binder can be further used to regulate mitochondrial aggregations in living cells. After demonstrating the minimal cell toxicity of the DNA binder (Fig. S15†), these DNA nanodevices were transfected into 4T1 cells by means of liposomes (Fig. S16†). As shown in Fig. 3A, upon the introduction of 1 μM DNA binders, the mitochondria in the cytosol of 4T1 cells (as stained by the MitoTracker) clearly changed from scattered small spheres (∼0.5 μm diameter) into elongated structures (≥2 μm length). The percentage of cells bearing these elongated mitochondria increased from 5% to 40% (Fig. 3B), the efficacy of mitochondria elongation might be attributed to the combined results of transfection, mitochondria targeting and aggregating processes. As a control, the addition of either TPP-LDNA or C-binder did not change the cellular mitochondrial size or shape (Fig. 3A and B). Transmission electron microscopy images have further evidenced the DNA binder-induced formation of fused mitochondria with increased lengths (Fig. 3C). The mitochondria in 4T1 cells remained individual form before and after treated with TPP-LDNA and C-binder. Upon addition of DNA binder, elongation of mitochondria was observed with average length of 1.8 μm (Fig. 3D). Thus, the DNA binder can indeed be used to effectively manipulate mito-aggregation and fusion in living cells.We next asked whether the DNA binder could be potentially used as a therapeutic tool in neurodegenerative diseases, which are known to be closely associated with the abnormal mitochondrial fusion process. Human neuroblastoma SH-SY5Y cells were selected as a model disease cell line. 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) was used to interrupt the mitochondrial metabolism of SH-SY5Y cells by increasing the intracellular ROS and Ca2+ levels (Fig. 4A, B and S17†).42,43 While after the addition of 2–8 μM DNA binders, the cellular generation of ROS was gradually decreased to that of the blank sample. Meanwhile, an obvious increase in the SH-SY5Y cell viability (from 55 to 87%) was shown after adding the DNA binders (Fig. 4C). However, addition of C-binder construct (non-TPP-modified DNA chain/LDNA complex) in MPTP treated cells showed negligible change on the ROS level and cell viability, which indicated the DNA binder construct (without mito-targeting modules) did not contribute to the oxidative stress mitigation. Similar therapeutical results could be achieved by using glutathione (GSH), an antioxidant, as the positive control. The above therapeutic process was also verified by the intracellular Ca2+ levels measurements and propidium iodide staining44 experiments (Fig. S17 and S18†).
Confocal fluorescence microscopy was also utilized for the visual evaluation of subcellular status upon different treatment. As shown in Fig. 4D, after adding MPTP, stress-induced mito-fission resulted in obviously more mitochondrial fragmentations as compared to the blank sample.26 While the introduction of DNA binders in these MPTP-pretreated cells can instead result in distinct mitochondrial aggregations and elongations. The percentage of cells bearing these elongated mitochondria increased from 2.86% to 38.1% upon addition of DNA binders (Fig. 4E). Therefore, the DNA binder could be considered as an artificial cell repair tool for mitochondria-related therapy. The easy modification of oligonucleotides enables the DNA binder system to be functionalized with cell-targeting and stimuli-recognition modules for improved performance.
The controlled manipulation of mitochondria aggregation and fusion by DNA binder in living cells
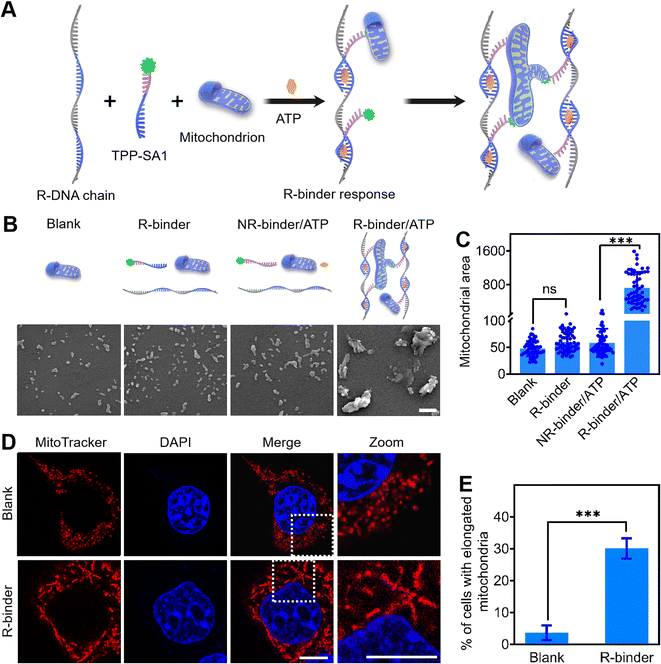
The DNA binder system was further studied on engineering molecular switch to achieve stimuli-triggered mitochondrial fusion. Since ATP is overexpressed in cancer cells and can be used as a trigger for controlling anticancer therapy,45,46 we chose an ATP-recognizing aptamer as a proof-of-concept (Fig. 5A). This 27 nt-long ATP aptamer was first divided into two parts, named as SA1 and SA2,47,48 and labelled with a TAMRA dye and a BHQ2 quencher, respectively (Table S1†). Due to the fluorescence resonance energy transfer,49 the fluorescence of TAMRA-SA1 could be efficiently quenched by the nearby BHQ2 upon the formation of an ATP/SA1/SA2 complex (Fig. S19A†). By integrating repeated SA2 sequences into the long RCA chain, a responsive DNA chain (R-DNA chain) was generated (Table S1†). Further fluorescence experiment verified that ATP could be used to selectively induce the self-assembly of R-DNA chain and TAMRA-SA1, and as a result, to develop an ATP-triggered responsive DNA binder (R-binder) (Fig. S19B†).As shown in Fig. 5B, C, and S20,† in the presence of ATP, R-binder can induce an efficient in vitro aggregation of mitochondria with increased particle sizes and integrated morphology. As a control, R-binder itself (no ATP) resulted in no morphological changes. These results have been further validated in living 4T1 cells, as the formation of elongated mitochondria can be triggered by intracellular ATP (Fig. 5D and E). The efficacy (30.1%) of mitochondria elongation by R-binder might be attributed to the combined results of transfection, in situ self-assembly, mitochondria targeting and aggregating processes. To further confirm that the ATP-triggered mitochondrial aggregation and fusion, we treated the cells with oligomycin, a commonly chemical inhibitor that is known to suppress the expression of ATP in cells.48 As expected, after adding oligomycin, no obviously morphological changes were observed in cells (Fig. S21†). As another control, by replacing the SA1 with a non-ATP aptamer sequence (CSA1), no mitochondrial aggregation was observed both in vitro and in living cells (Fig. 5B, C, and S21†). Taken together, these results demonstrated that aggregation and fusion of mitochondria were triggered by the ATP. This proof-of-concept could be extended to further control the mitochondria metabolism by programmable DNA binder, such as fusion triggered by damage-related marker cytochrome c50 and terminated by stand displacement.51
Conclusion
In sum, we report here a programmable DNA-based molecular tool for the manipulation of mitochondrial aggregation and fusion in living cells. The designed DNA binder could efficiently gather mitochondria together based on multivalent mito-targeting modules. The DNA binder may potentially even be used as an alternative strategy for cell therapy. By redesigning a stimuli-responsive DNA binder, we demonstrated that ATP could be used as a switch to achieve regulatable mitochondrial aggregation. Further conjugation of cell targeting moieties, such as folate and aptamers, could reduce the nonspecific therapeutical effects of the system.52 The highly editable and predictable nature of Watson–Crick–Franklin base pairing endows the DNA binder as a new versatile platform for regulating intracellular metabolism, which also further extended the functions of existing DNA nanodevices.Methods
Reagents
All the chemicals were purchased from Sigma unless otherwise noted. Commercial reagents were used without further purification. 4′,6-Diamidino-2-phenylindole (DAPI) and SYBR gold were purchased from invitrogen (Carlsbad, CA, USA). N-(7-Nitro-2-1,3-benzoxadiazol-4-yl)-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (NBD-PE) and N-(lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (Rhod-PE) were obtained from Macklin (Shanghai, China). Mitochondrial extraction kit was obtained from Abbkine Scientific Co., Ltd (USA). Mitochondrial tracker red (MitoRed) purchased from KeyGEN Biotech Co., Ltd (Nanjing, China). Phi29 DNA polymerase, T4 DNA ligase, exonuclease I, exonuclease III and dNTPs were purchased from New England Biolabs Ltd (USA). DNA oligonucleotides were synthesized and purified by Sangon Biotech Co., Ltd (Shanghai, China). The detailed sequences have been listed in Table S1.† All aqueous solutions were prepared using ultrapure water (18.2 MΩ cm, Milli-Q, Millipore).Apparatus
The concentrations of nucleic acids were all measured using a NanoDrop one UV-vis spectrophotometer. The gel electrophoresis was performed on a Tanon EPS-300 electrophoresis analyser (Tanon Science & Technology Company, China) and imaged on Bio-rad ChemDoc XRS (Bio-Rad, USA). All the in vitro fluorescence measurements were measured on an F-7000 spectrometer (HITACHI, Japan). All the intracellular images were taken by a Nikon A1& SIM-S&STORM super resolution microscope (Tokyo, Japan). ROS levels and cell viabilities were measured with a Safire microplate analyzer (Molecular Devices, America). AFM imaging was performed under ambient conditions with a dimension icon atomic force microscopy (Bruker, Germany). Dynamic lights scattering (DLS) was observed on a 90 plus/BI-MAS equipment (Brookhaven, USA). Scanning electron microscopy (SEM) images were recorded using a JEOL JSM-7800F field emission scanning electron microscope. MALDI-TOF MS was carried out by ABSCIEX MALDI TOF–TOF 4800 plus.Synthesis of alkynyl TPP cation (but-3-yn-1-yltriphenylphosphonium)
Triphenylphosphine (520 mg, 2 mmol) and 4-bromobutyne (400 mg, 3 mmol) was dissolved in acetonitrile (20 mL). The reaction mixture was heated to 80 °C for 72 h under nitrogen. The solvent was removed at room temperature followed by addition of benzene (80 mL). The resulting mixture was cooled to −20 °C for 0.5 h and the product was filtered off as white solid (512 mg, 82% yield). 1H NMR (500 MHz, DMSO-d6) δ: 8.27–7.24 (m, 15H), 3.93 (dt, J = 13.1, 7.5 Hz, 2H), 3.02 (d, J = 2.7 Hz, 1H), 2.57 (dtd, J = 12.9, 7.6, 2.7 Hz, 2H). 13C NMR (126 MHz, DMSO-d6) δ: 135.55, 135.52, 134.28, 134.20, 130.75, 130.65, 118.72, 118.04, 81.60, 81.46, 74.47, 20.50, 20.09, 12.51, 12.49. ESI-MS: C22H20P+ [M]+ calculated, 315.12; found, 315.03. The spectroscopic data agreed with previous reports.53Synthesis of TPP-LDNA
The TPP-LDNA was synthesized by adding 5 mM (5 μL) alkynyl TPP cation to 100 μM (200 μL) of linkage DNA (or SA1). The mixture was diluted with 250 μL DMSO before addition of 40 μL (50 mM) ascorbic acid. The cycloaddition reaction between azide group of linkage DNA (or SA1) and alkynyl group of TPP cation was initiated by adding 10 mM (50 μL) CuSO4 stock solution. The reaction was incubated for 24 hours and the resulting TPP-LDNA was then purified by gel filtration.54 MALDI-TOF MS: LDNA [M + K]+ calculated, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944.3; found, 10
944.3; found, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 951.7. TPP-LDNA [M + K]+ calculated, 11
951.7. TPP-LDNA [M + K]+ calculated, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259.3; found, 11
259.3; found, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 266.8. MALDI-TOF MS results confirmed the successful synthesis of TPP-LDNA (Fig. S4†).
266.8. MALDI-TOF MS results confirmed the successful synthesis of TPP-LDNA (Fig. S4†).
Preparation of circular DNA (circ-DNA)
For the synthesis of circ-DNA1, 4.2 μL of 100 μM phosphorylated linear DNA1 (linear DNA-12 or linear DNA-36) and ligation DNA1 were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After the annealing procedure, 1 μL of T4 DNA ligase (400 U μL−1), 2 μL of 10× T4 DNA buffer and 8.6 μL of ultrapure water were added for 16 h incubation at 25 °C. Then, the mixture was heated at 65 °C for 10 min to inactivate T4 DNA ligase. After reaction, 4 μL of exonuclease I (20 U μL−1) and 4 μL of exonuclease III (100 U μL−1) were added, and the mixture was incubated at 37 °C for 8 h to degrade ligation DNA1. Afterward, the mixture was incubated at 80 °C for 15 min to denature the exonuclease I and exonuclease III. The circ-DNA1 (circ-DNA12 or circ-DNA36) was generated and stored at 4 °C before use. The circ-DNA2 was obtained by using phosphorylated linear DNA2 and ligation DNA2 following the same approach.
1. After the annealing procedure, 1 μL of T4 DNA ligase (400 U μL−1), 2 μL of 10× T4 DNA buffer and 8.6 μL of ultrapure water were added for 16 h incubation at 25 °C. Then, the mixture was heated at 65 °C for 10 min to inactivate T4 DNA ligase. After reaction, 4 μL of exonuclease I (20 U μL−1) and 4 μL of exonuclease III (100 U μL−1) were added, and the mixture was incubated at 37 °C for 8 h to degrade ligation DNA1. Afterward, the mixture was incubated at 80 °C for 15 min to denature the exonuclease I and exonuclease III. The circ-DNA1 (circ-DNA12 or circ-DNA36) was generated and stored at 4 °C before use. The circ-DNA2 was obtained by using phosphorylated linear DNA2 and ligation DNA2 following the same approach.
Preparation of DNA chain
The DNA chain was synthesized by rolling circle amplification (RCA).55 10 μL of 3 μM circ-DNA1 (or circ-DNA2) and 0.5 μL of 100 μM DNA primer1 (or DNA primer2) were mixed and annealed at 95 °C for 4 min, then gradually cooled to 25 °C at a rate of 1 °C min. The mixture was incubated with phi29 DNA polymerase (0.2 U μL−1), bovine serum albumin (BSA) (0.4 μg μL−1) and deoxynucleotide triphosphates (dNTPs) (0.1 mM) at 37 °C for 5 min in 150 μL of 1× Phi29 reaction buffer. The RCA reaction was terminated through heated at 65 °C for 10 min, then purified by 30 kDa MWCO ultrafiltration device (Millipore) to obtain the long DNA chain (R-DNA chain). DNA chain has repeating strand segments with sequences complementary to circ-DNA. The 36 nt and 12 nt interspersed bases DNA chains were respectively obtained by using circ-DNA36 and DNA primer1, and circ-DNA12 and DNA primer12 following the same approach. The concentration of the repeat unit in DNA chain was obtained by measuring ultraviolet absorbance at 260 nm and used as the concentration of DNA chain.56 Briefly, DNA chain with n repeats had an approximate extinction coefficient of 471500n, where 471![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 L (mole−1 cm−1) is the ε260 of linear RCA repeats unit. Therefore, the molar concentration of RCA repeats could be calculated using the absorbance data and 1 cm length, which was used as the concentration of DNA chain for a given experiment.
500 L (mole−1 cm−1) is the ε260 of linear RCA repeats unit. Therefore, the molar concentration of RCA repeats could be calculated using the absorbance data and 1 cm length, which was used as the concentration of DNA chain for a given experiment.
Preparation of DNA binder
25 μL of 10 μM TPP modified linkage DNA (TPP-LDNA) and 300 μL of DNA chain were mixed together for 2 h at 37 °C to synthesize the DNA binder. The prepared DNA binder was purified by 30 kDa MWCO ultrafiltration device (Millipore) for three times. The concentration of TPP-LDNA on the DNA chain was used as the concentration of DNA binder. Given that the excess amount of TPP-LDNAs were used, all the binding sites of DNA chain were considered to be occupied by TPP-LDNA. Therefore, the concentration of DNA binder is considered equal to the concentration of RCA repeats. As controls, the C-binder was obtained by using linkage DNA and DNA chain following the same approach.To measure the amounts of linkage DNA in each DNA binder, 5-carboxyfluorescein (FAM)-labelled linkage DNA (FAM-LDNA) was used as components to synthesize a fluorescent DNA binder (F-binder). From the fluorescence intensities of FAM in DNA binder and the standard calibration curves for FAM-LDNA, its amounts could be obtained.
The responsive DNA binder (R-binder) that was able to activate with ATP was constructed by using TPP modified SA1 (TPP-SA1) and R-DNA chain. As controls, no responsive DNA binder (NR-binder) that was unable to activate with ATP was constructed by using TPP modified negative control SA1 (TPP-CSA1) and R-DNA chain.
To demonstrate the response of the split ATP aptamer to ATP, 2 μM tetramethylrhodamine labeled SA1 (TAMRA-SA1) or tetramethylrhodamine labeled CSA1 (TAMRA-CSA1), and 2 μM black hole quencher (BHQ2) labeled SA2 (BHQ-SA2) with various concentrations of ATP (CTP, GTP or UTP) were used and incubated for 1 hour at 37 °C. Fluorescence measurements were performed with excitation at 550 nm and emission spectra were collected in the range of 570–650 nm. The slit width was set to be 2 nm for both excitation and emission.
To characterize the ATP activated R-binder response, 2 μM TAMRA-SA1 and 2 μM R-DNA chain with 4 mM ATP were used and incubated at 37 °C for 1 hour. Afterward, the solution was purified by ultrafiltration (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MW cut off membrane, Millipore) for three times. The fluorescence measurement of concentrate and filtrate at the third ultrafiltration were performed with excitation at 550 nm and emission spectra were collected in the range of 570–650 nm. All the data were plotted using the Origin software.
000 MW cut off membrane, Millipore) for three times. The fluorescence measurement of concentrate and filtrate at the third ultrafiltration were performed with excitation at 550 nm and emission spectra were collected in the range of 570–650 nm. All the data were plotted using the Origin software.
Polyacrylamide gel electrophoresis analysis
10% native polyacrylamide gel was prepared using 1× TBE buffer. The loading samples were prepared by mixing 7.5 μL DNA samples and 1.5 μL 6× loading buffer, and placed for 3 min before injected into polyacrylamide gel. The gel electrophoresis was run at 100 V for 60 min in 1× TBE buffer, stained with 1× SYBR gold, and scanned with a molecular imager Gel Doc XR.AFM imaging
6 μL DNA binder was deposited on freshly cleaved mica surface and incubated for 10 min. After the solvent water was absorbed with filter paper, 20 μL ultrapure water was added to wash the mica for three times. The mica was then dried with a nitrogen flow and scanned in tapping mode.Cell culture
The SH-SY5Y and 4T1 cell lines (Procell Life Science & Technology, Wuhan, China) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 μg mL−1 streptomycin and 100 U per mL penicillin-streptomycin at 37 °C in a humidified incubator containing 5% CO2 and 95% air. Short tandem repeats (STR) profiling and mycoplasma testing were conducted for each cell line before use. Cell numbers were determined with a Petroff-Hausser cell counter (USA).3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
MTT assays were performed to investigate DNA binder biocompatibility. SH-SY5Y and 4T1 cell lines were respectively seeded within replicate 96-well plates at a density of 1 × 106 cells per well. Plates were maintained at 37 °C for 24 h. After discarding the medium, the cells were washed twice with PBS and incubated with serial concentrations of the DNA binder transfected with Lipofectamine® 2000 transfection reagent. Cells incubated with only the PBS was served as control. After 24 hour-incubation, the cells were washed twice with PBS buffer and 100 μL of a fresh medium containing 10 μL of MTT (5 mg mL−1) was added and incubated for 4 h. After removing the remaining MTT solution, 100 μL of dimethylsulphoxide was added to dissolve the formazan crystals precipitates. After shaking the plate for 10 min, the optical density at a wavelength of 490 nm was measured with a Safire microplate analyzer.Measurement of the ROS generation
Human neuroblastoma SH-SY5Y cells were seeded in a 6-well plate at a density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well for 24 h at 37 °C. Then, incubated with 1 mM MPTP in the absence and in the presence of 8 μM C-binder, 2, 4 and 8 μM of DNA binders transfected with Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific), as well as 5 mM GSH for another 24 h. After washing with PBS for three times, the cells were stained with 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), or 10 μg mL−1 propidium iodide and 5 mg mL−1 of DAPI, and measured by a Safire microplate Analyzer or Nikon A1& SIM-S&STORM super resolution microscope, respectively.
000 cells per well for 24 h at 37 °C. Then, incubated with 1 mM MPTP in the absence and in the presence of 8 μM C-binder, 2, 4 and 8 μM of DNA binders transfected with Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific), as well as 5 mM GSH for another 24 h. After washing with PBS for three times, the cells were stained with 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), or 10 μg mL−1 propidium iodide and 5 mg mL−1 of DAPI, and measured by a Safire microplate Analyzer or Nikon A1& SIM-S&STORM super resolution microscope, respectively.
Measurement of Ca2+ levels
Intracellular Ca2+ levels were measured according to the instruction of Fluo-4AM calcium kit from Beyotime (Jiangsu, China). SH-SY5Y cell lines were seeded within replicate 96-well plates at a density of 1 × 106 cells per well. Plates were maintained at 37 °C for 24 h. After discarding the medium, the cells were washed twice with PBS and incubated with 1 mM MPTP in the absence and in the presence of 8 μM C-binder, 2 μM, 4 μM or 8 μM of DNA binders transfected with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific), or 5 mM GSH for another 24 h. The cells were washed three times and stained with 5 μM Fluo-4AM at 37 °C in the dark for 30 min. Then, the cells in the wells were washed three times with PBS and the level of fluorescence was acquired at λex/em = 488/516 nm with Safire microplate analyzer (Molecular Devices, America).57Cellular imaging and data analysis
1 × 104 4T1 cells were seeded in a confocal dish for 24 h at 37 °C, then transfected, respectively, with 1 μM binder, TPP-LDNA, C-binder, or F-binder using a Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific). 24 h after transfection, the cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS) and stained with 200 nM of mitochondrial tracker red and 5 mg mL−1 of DAPI for 15 min for imaging.To demonstrate the potential capability of DNA binder for molecular mitochondrial aggregation/fusion, 1 × 104 human neuroblastoma SH-SY5Ycells were seeded in a confocal dish for 24 h at 37 °C, then incubated with 1 mM MPTP in the absence and in the presence of 1 μM DNA binder transfected with Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific) for another 24 h. Afterward, the cells were washed twice with DPBS and stained with 200 nM of mitochondrial tracker red, as well as 5 mg mL−1 of DAPI for 15 min for imaging.
For the ATP controlled mitochondrial aggregation/fusion experiment, 1 × 104 4T1 cells were seeded in a confocal dish for 24 h at 37 °C, then transfected, respectively, with 1 μM TPP-SA1 and R-DNA chain, TPP-CSA1 and R-DNA chain, or TPP-SA1, R-DNA chain and 10 μM oligomycin using a Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific). After incubation for 24 h, the cells were washed with DPBS and stained with 200 nM of mitochondrial tracker red and 5 mg mL−1 of DAPI for imaging.
All the fluorescence images were collected with Nikon A1& SIM-S&STORM super resolution microscope. FAM, mitochondrial tracker red and DAPI were excited with 488 nm, 640 nm and 405 nm lasers, respectively. A 100× oil immersion objective was used for imaging cells. Image analysis was performed with a NiS-elements AR analysis software. Data analysis was done using the Origin and GraphPad Prism software.
Author contributions
Longyi Zhu: conceptualization, investigation, writing – original draft. Yiting Shen: investigation. Shengyuan Deng: data curation. Ying Wan: conceptualization, resources. Jun Luo: resources. Yan Su: data curation. Mingxu You: methodology. Chunhai Fan: resources, supervision. Kewei Ren: conceptualization, writing – original draft, supervision.Conflicts of interest
The authors report no declarations of interest.Acknowledgements
The authors gratefully acknowledge the National Key Research and Development Program of China for International Science & Innovation Cooperation Major Project between Governments (Grant No. 2018YFE0113200), National Natural Science Foundation of China (21775072, 21874071, 22104058, 22174066), the Natural Science Foundation of Jiangsu Province (BK20200459), and the Program of Jiangsu Specially-Appointed Professor.References
- A. Saminathan, J. Devany, A. T. Veetil, B. Suresh, K. S. Pillai, M. Schwake and Y. Krishnan, A DNA-based voltmeter for organelles, Nat. Nanotechnol., 2021, 16, 96–103 CrossRef CAS PubMed.
- P. S. Kwon, S. K. Ren, S. J. Kwon, M. E. Kizer, L. L. Kuo, M. Xie, D. Zhu, F. Zhou, F. M. Zhang, D. Kim, K. Fraser, L. D. Kramer, N. C. Seeman, J. S. Dordick, R. J. Linhardt, J. Chao and X. Wang, Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition, Nat. Chem., 2020, 12, 26–35 CrossRef CAS PubMed.
- J. Chao, J. B. Wang, F. Wang, X. Y. Ouyang, E. Kopperger, H. J. Liu, Q. Li, J. Y. Shi, L. H. Wang, J. Hu, L. H. Wang, W. Huang, F. C. Simmel and C. H. Fan, Solving mazes with single-molecule DNA navigators, Nat. Mater., 2019, 18, 273–279 CrossRef CAS PubMed.
- Y. H. Chen, G. L. Ke, Y. L. Ma, Z. Zhu, M. H. Liu, Y. Liu, H. Yan and C. Y. J. Yang, A synthetic light-driven substrate channeling system for precise regulation of enzyme cascade activity based on DNA origami, J. Am. Chem. Soc., 2018, 140, 8990–8996 CrossRef CAS PubMed.
- M. X. You, Y. F. Lyu, D. Han, L. P. Qiu, Q. L. Liu, T. Chen, C. S. Wu, L. Peng, L. Q. Zhang, G. Bao and W. H. Tan, DNA probes for monitoring dynamic and transient molecular encounters on live cell membranes, Nat. Nanotechnol., 2017, 12, 453–459 CrossRef CAS PubMed.
- L. L. Sun, F. Y. Shen, J. Xu, X. Han, C. H. Fan and Z. Liu, DNA-edited ligand positioning on red blood cells to enable optimized T cell activation for adoptive immunotherapy, Angew. Chem., Int. Ed., 2020, 59, 14842–14853 CrossRef CAS PubMed.
- H. Li, J. Gao, L. Cao, X. Xie, J. H. Fan, H. D. Wang, H. H. Wang and Z. Nie, A DNA molecular robot that autonomously walks on the cell membrane to drive cell motility, Angew. Chem., Int. Ed., 2021, 60, 26087–26095 CrossRef CAS PubMed.
- C. Yao, C. X. Zhu, J. P. Tang, J. H. Ou, R. Zhang and D. Y. Yang, T lymphocyte-captured DNA network for localized immunotherapy, J. Am. Chem. Soc., 2021, 143, 19330–19340 CrossRef CAS PubMed.
- C. Jung, P. B. Allen and A. D. Ellington, A stochastic DNA walker that traverses a microparticle surface, Nat. Nanotechnol., 2016, 11, 157–163 CrossRef CAS PubMed.
- Y. N. Tang, Z. X. Wang, X. L. Yang, J. B. Chen, L. A. Liu, W. A. Zhao, X. C. Le and F. Li, Constructing real-time, wash-free, and reiterative sensors for cell surface proteins using binding-induced dynamic DNA assembly, Chem. Sci., 2015, 6, 5729–5733 RSC.
- K. W. Ren, R. Wu, A. P. K. K. Mudiyanselage, Q. K. Yu, B. Zhao, Y. W. Xie, Y. Bagheri, Q. Tian and M. X. You, In situ genetically cascaded amplification for imaging RNA subcellular locations, J. Am. Chem. Soc., 2020, 142, 2968–2974 CrossRef CAS PubMed.
- J. Li, K. Y. Xun, K. Pei, X. J. Liu, X. Y. Peng, Y. L. Du, L. P. Qiu and W. H. Tan, Cell-membrane-anchored DNA nanoplatform for programming cellular interactions, J. Am. Chem. Soc., 2019, 141, 18013–18020 CrossRef CAS PubMed.
- L. P. Wang, H. Liang, J. Sun, Y. C. Liu, J. Y. Li, J. Y. Li, J. Li and H. H. Yang, Bispecific aptamer induced artificial protein-pairing: a strategy for selective inhibition of receptor function, J. Am. Chem. Soc., 2019, 141, 12673–12681 CrossRef CAS PubMed.
- J. Li, K. Y. Xun, L. Y. Zheng, X. Y. Peng, L. P. Qiu and W. H. Tan, DNA-based dynamic mimicry of membrane proteins for programming adaptive cellular interactions, J. Am. Chem. Soc., 2021, 143, 4585–4592 CrossRef CAS PubMed.
- E. Akbari, M. Y. Mollica, C. R. Lucas, S. M. Bushman, R. A. Patton, M. Shahhosseini, J. W. Song and C. E. Castro, Engineering cell surface function with DNA origami, Adv. Mater., 2017, 29, 1703632 CrossRef PubMed.
- P. Keshri, B. Zhao, T. F. Xie, Y. Bagheri, J. Chambers, Y. B. Sun and M. X. You, Quantitative and multiplexed fluorescence lifetime imaging of intercellular tensile forces, Angew. Chem., Int. Ed., 2021, 60, 15548–15555 CrossRef CAS PubMed.
- Y. M. Qin, X. Jiang, Q. Yang, J. Q. Zhao, Q. Zhou and Y. H. Zhou, The functions, methods, and mobility of mitochondrial transfer between cells, Front. Oncol., 2021, 11, 672781 CrossRef CAS PubMed.
- D. C. Chan, Mitochondria: dynamic organelles in disease, aging, and development, Cell, 2006, 125, 1241–1252 CrossRef CAS PubMed.
- R. J. Youle and A. M. van der Bliek, Mitochondrial fission, fusion, and stress, Science, 2012, 337, 1062–1065 CrossRef CAS PubMed.
- S. Hoppins, L. Lackner and J. Nunnari, The machines that divide and fuse mitochondria, Annu. Rev. Biochem., 2007, 76, 751–780 CrossRef CAS PubMed.
- B. N. Whitley, E. A. Engelhart and S. Hoppins, Mitochondrial dynamics and their potential as a therapeutic target, Mitochondrion, 2019, 49, 269–283 CrossRef CAS PubMed.
- S. Srinivasan, M. Guha, A. Kashina and N. G. Avadhani, Mitochondrial dysfunction and mitochondrial dynamics-the cancer connection, Biochim. Biophys. Acta, 2017, 1858, 602–614 CrossRef CAS PubMed.
- X. Y. Lei, H. Lin, J. Q. Wang, Z. P. Ou, Y. Ruan, A. Sadagopan, W. X. Chen, S. L. Xie, B. S. Chen, Q. X. Li, J. Wang, H. Y. Lin, X. F. Zhu, X. Q. Yuan, T. Tian, X. B. Lv, S. Fu, X. R. Zhu, J. Zhou, G. K. Pan, X. Xia, B. A. Tannous, S. Ferrone, S. Fan and J. S. Li, Mitochondrial fission induces immunoescape in solid tumors through decreasing MHC-I surface expression, Nat. Commun., 2022, 13, 3882 CrossRef CAS PubMed.
- D. L. Wang, J. N. Wang, G. M. C. Bonamy, S. Meeusen, R. G. Brusch, C. Turk, P. Y. Yang and P. G. Schultz, A small molecule promotes mitochondrial fusion in mammalian cells, Angew. Chem., Int. Ed., 2012, 51, 9302–9305 CrossRef CAS PubMed.
- H. M. Wang, P. J. Lim, M. Karbowski and M. J. Monteiro, Effects of overexpression of huntingtin proteins on mitochondrial integrity, Hum. Mol. Genet., 2009, 18, 737–752 CrossRef CAS PubMed.
- C. Sun, Z. Y. Wang, L. D. Yue, Q. X. Huang, Q. Cheng and R. B. Wang, Supramolecular induction of mitochondrial aggregation and fusion, J. Am. Chem. Soc., 2020, 142, 16523–16527 CrossRef CAS PubMed.
- F. Li, Y. J. Liu, Y. H. Dong, Y. W. Chu, N. C. Song and D. Y. Yang, Dynamic assembly of DNA nanostructures in living cells for mitochondrial interference, J. Am. Chem. Soc., 2022, 144, 4667–4677 CrossRef CAS PubMed.
- Y. L. Shao, J. Zhao, J. Y. Yuan, Y. L. Zhao and L. L. Li, Organelle-specific photoactivation of DNA nanosensors for precise profiling of subcellular enzymatic activity, Angew. Chem., Int. Ed., 2021, 60, 8923–8931 CrossRef CAS PubMed.
- M. P. Murphy, Selective targeting of bioactive compounds to mitochondria, Trends Biotechnol., 1997, 15, 326–330 CrossRef CAS PubMed.
- V. K. Tiwari, B. B. Mishra, K. B. Mishra, N. Mishra, A. S. Singh and X. Chen, Cu-catalyzed click reaction in carbohydrate chemistry, Chem. Rev., 2016, 116, 3086–3240 CrossRef CAS PubMed.
- K. Dan, A. T. Veetil, K. Chakraborty and Y. Krishnan, DNA nanodevices map enzymatic activity in organelles, Nat. Nanotechnol., 2019, 14, 252–259 CrossRef CAS PubMed.
- F. B. Dean, J. R. Nelson, T. L. Giesler and R. S. Lasken, Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification, Genome Res., 2001, 11, 1095–1099 CrossRef CAS PubMed.
- G. D. Hamblin, J. F. Rahbani and H. F. Sleiman, Sequential growth of long DNA strands with user-defined patterns for nanostructures and scaffolds, Nat. Commun., 2015, 6, 7065 CrossRef CAS PubMed.
- K. L. Lau, G. D. Hamblin and H. F. Sleiman, Gold nanoparticle 3D-DNA building blocks: high purity preparation and use for modular access to nanoparticle assemblies, Small, 2014, 10, 660–666 CrossRef CAS PubMed.
- Y. W. Xu, Z. Y. Lv, C. Yao and D. Y. Yang, Construction of rolling circle amplification-based DNA nanostructures for biomedical applications, Biomater. Sci., 2022, 10, 3054–3061 RSC.
- M. M. Rahman, M. Ueda, T. Hirose and Y. Ito, Spontaneous formation of gating lipid domain in uniform-size peptide vesicles for controlled release, J. Am. Chem. Soc., 2018, 140, 17956–17961 CrossRef CAS PubMed.
- D. K. Struck, D. Hoekstra and R. E. Pagano, Use of resonance energy transfer to monitor membrane fusion, Biochemistry, 1981, 14, 4093–4099 CrossRef PubMed.
- L. Oshry, P. Meers, T. Mealy and A. I. Tauber, Annexin-mediated membrane fusion of human neutrophil plasma membranes and phospholipid vesicles, Biochim. Biophys. Acta, 1991, 1066, 239–244 CrossRef CAS PubMed.
- M. Piffoux, A. K. A. Silva, C. Wihelm, F. Gazeau and D. Tareste, Modification of extracellular vesicles by fusion with liposomes for the design of personalized biogenic drug delivery systems, ACS Nano, 2018, 12, 6830–6842 CrossRef CAS PubMed.
- K. Thorsteinsson, E. Olsén, E. Schmidt, H. Pace and M. Bally, FRET-based assay for the quantification of extracellular vesicles and other vesicles of complex composition, Anal. Chem., 2020, 92, 15336–15343 CrossRef CAS PubMed.
- N. B. Samanas and S. Hoppins, Cell-free analysis of mitochondrial fusion by fluorescence microscopy, Methods Mol. Biol., 2020, 2159, 129–140 CrossRef CAS PubMed.
- A. Tomac, E. Lindqvist, L.-F. H. Lin, S. O. Ögren, D. Young, B. J. Hoffer and L. Olson, Protection and repair of the nigrostriatal dopaminergic system by GDNF in vivo, Nature, 1995, 373, 335–339 CrossRef CAS PubMed.
- J. Liu, F. F. Dou and Z. H. Yu, The potassium channel KCa3.1 represents a valid pharmacological target for microgliosis-induced neuronal impairment in a mouse model of Parkinson's disease, J. Neuroinflammation, 2019, 16, 273 CrossRef PubMed.
- L. Zhang, K. Mizumoto, N. Sato, T. Ogawa, M. Kusumoto, H. Niiyama and M. Tanaka, Quantitative determination of apoptotic death in cultured human pancreatic cancer cells by propidium iodide and digitonin, Cancer Lett., 1999, 142, 129–137 CrossRef CAS PubMed.
- H. Choi, G. Park, E. Shin, S. W. Shin, B. Jana, S. Jin, S. Kim, H. M. Wang, S. K. Kwak, B. Xu and J. H. Ryu, Intramitochondrial co-assembly between ATP and nucleopeptides induces cancer cell apoptosis, Chem. Sci., 2022, 13, 6197–6204 RSC.
- R. Mo, T. Y. Jiang, R. DiSanto, W. Y. Tai and Z. Gu, ATP-triggered anticancer drug delivery, Nat. Commun., 2014, 5, 3364 CrossRef PubMed.
- X. L. Zuo, Y. Xiao and K. W. Plaxco, High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices, J. Am. Chem. Soc., 2009, 131, 6944–6945 CrossRef CAS PubMed.
- P. Peng, Y. Du, J. Zheng, H. H. Wang and T. Li, Reconfigurable bioinspired framework nucleic acid nanoplatform dynamically manipulated in living cells for subcellular imaging, Angew. Chem., Int. Ed., 2019, 58, 1648–1653 CrossRef CAS PubMed.
- A. J. Shuendler, K. Y. Pu, L. N. Cui, J. P. Uetrecht and J. H. Rao, Real-time imaging of oxidative and nitrosative stress in the liver of live animals for drug-toxicity testing, Nat. Biotechnol., 2014, 32, 373–380 CrossRef PubMed.
- T. T. Chen, X. Tian, C. L. Liu, J. Ge, X. Chu and Y. F. Li, Fluorescence activation imaging of cytochrome c released from mitochondria using aptameric nanosensor, J. Am. Chem. Soc., 2015, 137, 982–989 CrossRef CAS PubMed.
- T. Mayer, L. Oesinghaus and F. C. Simmel, Toehold-mediated strand displacement in random sequence pools, J. Am. Chem. Soc., 2023, 145, 634–644 CrossRef CAS PubMed.
- Z. R. Goddard, M. J. Marín, D. A. Russell and M. Searcey, Active targeting of gold nanoparticles as cancer therapeutics, Chem. Soc. Rev., 2020, 49, 8774–8789 RSC.
- N. R. Perl, N. D. Ide, S. Prajapati, H. H. Perfect, S. G. Durón and D. Y. Gin, Annulation of thioimidates and vinyl carbodiimides to prepare 2-aminopyrimidines, competent nucleophiles for intramolecular alkyne hydroamination. Synthesis of (−)-Crambidine, J. Am. Chem. Soc., 2010, 132, 1802–1803 CrossRef CAS PubMed.
- H. Qian and L. He, Surface-initiated activators generated by electron transfer for atom transfer radical polymerization in detection of DNA point mutation, Anal. Chem., 2009, 81, 4536–4542 CrossRef CAS PubMed.
- D. Han, C. C. Wu, M. X. You, T. Zhang, S. Wan, T. Chen, L. P. Qiu, Z. Zheng, H. Liang and W. H. Tan, A cascade reaction network mimicking the basic functional steps of adaptive immune response, Nat. Chem., 2015, 7, 835–841 CrossRef CAS PubMed.
- G. D. Hamblin, K. M. M. Carneiro, J. F. Fakhoury, K. E. Bujold and H. F. Sleiman, Rolling circle amplification-templated DNA nanotubes show increased stability and cell penetration ability, J. Am. Chem. Soc., 2012, 134, 2888–2891 CrossRef CAS PubMed.
- H. Chen, C. L. Li, H. Y. Hu and B. Zhang, Activated TRPA1 plays a therapeutic role in TMZ resistance in glioblastoma by altering mitochondrial dynamics, BMC Mol. Cell Biol., 2022, 23, 38 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc07095b |
| This journal is © The Royal Society of Chemistry 2023 |