Open Access Article
Open Access ArticleQuantifying cell viability through organelle ratiometric probing†
Rui
Chen
 a,
Kangqiang
Qiu
b,
Guanqun
Han
a,
Bidyut Kumar
Kundu
a,
Kangqiang
Qiu
b,
Guanqun
Han
a,
Bidyut Kumar
Kundu
 a,
Guodong
Ding
a,
Yujie
Sun
a,
Guodong
Ding
a,
Yujie
Sun
 *a and
Jiajie
Diao
*a and
Jiajie
Diao
 *b
*b
aDepartment of Chemistry, University of Cincinnati, Cincinnati, OH 45221, USA. E-mail: yujie.sun@uc.edu
bDepartment of Cancer Biology, College of Medicine, University of Cincinnati, Cincinnati, OH 45267, USA. E-mail: jiajie.diao@uc.edu
First published on 7th September 2023
Abstract
Detecting cell viability is crucial in research involving the precancerous discovery of abnormal cells, the evaluation of treatments, and drug toxicity testing. Although conventional methods afford cumulative results regarding cell viability based on a great number of cells, they do not permit investigating cell viability at the single-cell level. In response, we rationally designed and synthesized a fluorescent probe, PCV-1, to visualize cell viability under the super-resolution technology of structured illumination microscopy. Given its sensitivity to mitochondrial membrane potential and affinity to DNA, PCV-1's ability to stain mitochondria and nucleoli was observed in live and dead cells, respectively. During cell injury induced by drug treatment, PCV-1's migration from mitochondria to the nucleolus was dynamically visualized at the single-cell level. By extension, harnessing PCV-1's excellent photostability and signal-to-noise ratio and by comparing the fluorescence intensity of the two organelles, mitochondria and nucleoli, we developed a powerful analytical assay named organelle ratiometric probing (ORP) that we applied to quantitatively analyze and efficiently assess the viability of individual cells, thereby enabling deeper insights into the potential mechanisms of cell death. In ORP analysis with PCV-1, we identified 0.3 as the cutoff point for assessing whether adding a given drug will cause apparent cytotoxicity, which greatly expands the probe's applicability. To the best of our knowledge, PCV-1 is the first probe to allow visualizing cell death and cell injury under super-resolution imaging, and our proposed analytical assay using it paves the way for quantifying cell viability at the single-cell level.
Introduction
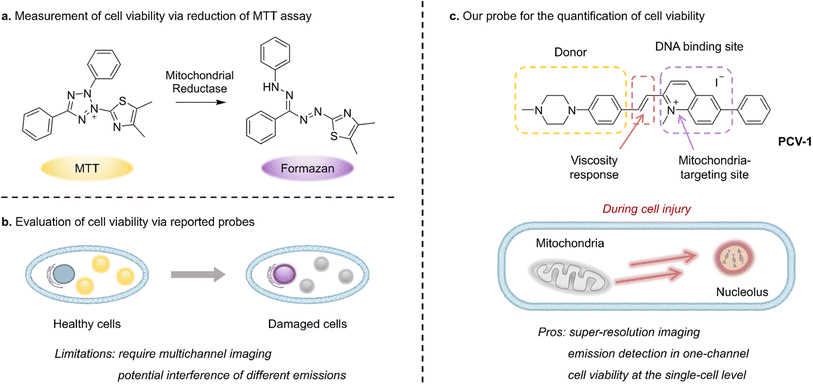
Cells, as the essential unit of organisms, have been broadly studied in relation to biological processes, medical treatments, and nerve conduction. Processes that alter cell viability, including apoptosis, autophagy, and necrosis, are fundamental to eliminating damaged cells and maintaining homeostasis.1,2 In particular, modulating the specific cellular response of dysregulated cell death, as is prevalent in diseases such as cancer, autoimmune disease, and neurodegeneration,3,4 has shown success as a therapeutic strategy. Therefore, detecting cell viability is crucial in the discovery of precancerous cells, the evaluation of treatments, and drug toxicity testing.4,5Whereas earlier methods applied to detect cell viability include cellular morphology studies, flow cytometric analysis, and DNA degradation,6,7 in the past few decades colorimetric assays have been broadly used to monitor cell viability. Among such materials, the well-known yellow 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) is absorbed by live cells and reduced by mitochondrial reductase into purple formazan, which can be quantified by measuring its absorbance using a microplate reader (Fig. 1a).8 However, because this method can involve factors that may hinder mitochondrial reductase (e.g., amount of cells and incubation time), definite information about cell viability is difficult to obtain.9 Moreover, owing to its high susceptibility to metabolic interference, MTT-based assay can easily provide false-positive results.4 Such assays also report the cumulative results of cell viability based on a great number of cells and do not permit investigating cell viability at the single-cell level. Consequently, next-generation biological tools are urgently needed to monitor cell viability on individual cells and thereby enable the rapid screening of drug treatment, minimize experimental error due to external interference, and reduce medical costs.
While fluorescence microscopy allows the in situ, real-time observation of subcellular organelles and biological processes, fluorescence imaging, as a non-invasive technique that causes relatively little damage, readily accommodates the study of cell viability. Therefore, several fluorescent probes have been developed to distinguish live from dead cells based on various tracking mechanisms and are thus versatile tools for biological research and applications in medicine.5 As one such fluorescent dye, 4′,6-diamidino-2-phenylindole (DAPI) is widely used to visualize nuclei in dead cells due to its weak ability to penetrate intact plasma membranes in live cells.
In past research, mitochondrial membrane potential (ΔΨm), as a vital feature representing mitochondrial function, has been used as an indicator to discriminate the states of cells.10,11 A decrease in mitochondrial membrane potential signifies a disturbance in mitochondrial homeostasis that can lead to cell dysfunction or even death. Tian et al., for instance, reported a dual-color fluorescent probe that migrated from mitochondria to the nucleus when the mitochondrial membrane potential decreased during apoptosis; in that way, cell viability could be monitored by identifying the localization and fluorescent intensity of the probe (Fig. 1b).12 However, the dual-color probe demonstrated several limitations, including the need for a multichannel fluorescence microscope, the potential overlap and interference of different emissions, and the incomparability of fluorescence images from various channels. Meanwhile, Li et al. have developed a red fluorescent probe that can reflect different situations of mitochondrial membrane potential (i.e., normal, decreasing, and vanishing) according to various fluorescence morphologies of the nucleus and mitochondria and thereby indicate the cell's state.13 However, the probe's main limitation is that cell viability cannot be quantitatively assessed by analyzing images owing to the high degree of fluorescence interference between the two organelles and the poor image quality available with conventional confocal microscopy. Thus, clearly observing the interaction of organelles and efficiently quantifying cell viability using fluorescence microscopy remains challenging, especially at the single-cell level.
Recently, super-resolution imaging, as a cutting-edge technology able to overcome the Abbe's diffraction limit (<200 nm) of conventional confocal microscopy has been greatly applied to observe subcellular organelles and biological interactions.14–19 Therefore, fluorescence microscopy with super-resolution imaging might be ideal for synchronously visualizing the dynamics of organelles and consequently quantifying cell viability.20 In view of the size of eukaryotic organelles, including lysosomes (i.e., 100–1200 nm), mitochondria (i.e., 500–5000 nm), and nuclei (i.e., approx. 10 μm),21 structured illumination microscopy (SIM) may be a promising alternative, especially given its super spatial resolution (i.e., approx. 100 nm), short acquisition time, and only slight photodamage.22–25
Against this backdrop, we rationally designed and synthesized a fluorescent probe PCV-1 to visualize cell viability under SIM. As shown in Fig. 1c, PCV-1 consists of a donor–π–acceptor structure, in which the positively charged π-conjugated system endows PCV-1 with the ability to target mitochondria given the highly negative mitochondrial membrane potential in healthy cells.26,27 The cationic methylquinolinium moiety gives PCV-1 a high affinity to DNA due to their electrostatic interaction.28,29 PCV-1 exhibited extended π-conjugation but decreased π–π stacking compared to the cell viability probe previously reported, based on its molecular design.12 Moreover, owing to its alkene bridge, PCV-1 can readily respond to viscosity.30,31 In an environment of high viscosity, PCV-1's molecular rotation is likely restricted, thereby rendering a more planar configuration that reduces its non-radiative decay and hence increases the emission intensity. By performing the fluorescence imaging of PCV-1-stained HeLa cells using SIM, we observed mitochondria and nucleoli as distinct organelles in live and dead cells in single-channel mode, respectively. Moreover, owing to the excellent super-resolution imaging and viscosity-enhanced emission, PCV-1 is able to provide more subcellular details that cannot be achieved by conventional confocal microscopy. Thus, during cell injury induced by drug treatment, PCV-1's migration from mitochondria to the nucleolus was dynamically visualized at the single-cell level. Beyond that, based on PCV-1's translocation from mitochondria to the nucleolus following damage and by comparing the two organelles' fluorescence intensity at the subcellular level, we developed a new analytical assay, organelle ratiometric probing (ORP), that can be used to quantitatively analyze and efficiently assess the viability of individual cells. In the ORP analysis with PCV-1, we systematically investigated the relationship between the various fluorescence intensity ratios of organelles and their corresponding cell viability values at different stages, as a result, identified 0.3 as the cutoff point for assessing whether adding a drug will cause apparent cytotoxicity. Furthermore, regardless of various routes to cell death, including apoptosis, autophagy, and necrosis, a ratio significantly higher than 0.3 for the ORP analysis with PCV-1 indicated nearly dead cells. To our knowledge, PCV-1 is the first probe to allow visualizing cell death and cell injury under super-resolution imaging, and our proposed ORP assay using PCV-1 enables the quantification of cell viability at the single-cell level.
Results and discussion
Synthesis, photophysical properties, and computational calculations of PCV-1
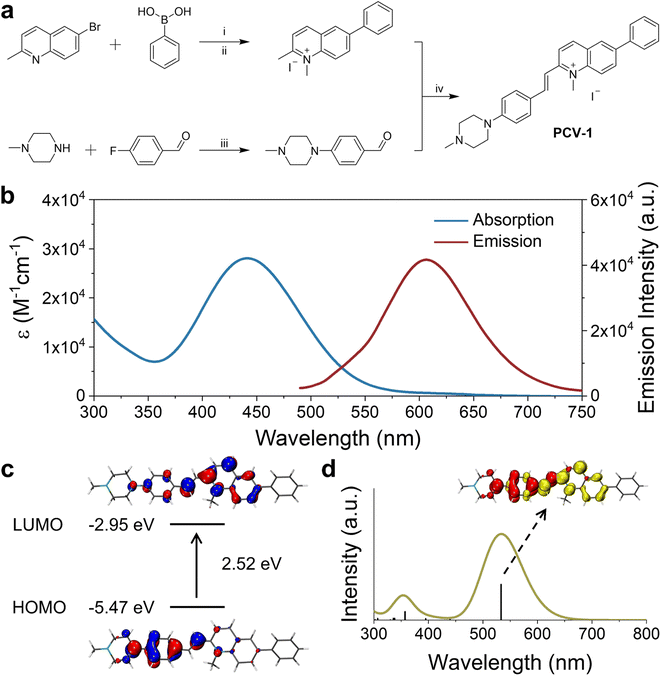
As shown in Fig. 2a, the synthesis of PCV-1 began with commercially available 6-bromo-2-methylquinoline. The intermediate 2-methyl-6-phenylquinoline was generated by a typical Suzuki coupling reaction between 6-bromo-2-methylquinoline and phenylboronic acid. Next, methylation at the nitrogen position of 2-methyl-6-phenylquinoline using methyl iodide resulted in 1,2-dimethyl-6-phenylquinolin-1-ium. Meanwhile, 1-methylpiperazine reacting with p-fluorobenzaldehyde under an alkaline conditions formed another intermediate 4-(4-methylpiperazin-1-yl) benzaldehyde with a nearly quantitative yield. Finally, the Knoevenagel condensation reaction between 1,2-dimethyl-6-phenylquinolin-1-ium and 4-(4-methylpiperazin-1-yl) benzaldehyde in anhydrous ethanol generated our designed product, PCV-1. Details of the synthesis procedure and characterizations of 1H, 13C NMR, and MS for intermediates and products are included in the ESI (Fig. S1–S5).†With the designed probe PCV-1 in hand, we first investigated its photophysical properties in aqueous solution. As shown in Fig. 2b, PCV-1 exhibited a broad absorption spectrum with a tail exceeding 570 nm due to its large π–π conjugation. The maximum absorption of PCV-1 was located at 442 nm with a high molar extinction coefficient of 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060 M−1 cm−1. Under 450 nm excitation, PCV-1 showed a strong red emission with a maximum peak at 606 nm (Φ = 0.08%), which is favorable for bioimaging applications. The Stokes shift of PCV-1 was beyond 160 nm due to its charge-enhanced intramolecular charge transfer system and extended conjugation length.32 Moreover, when PCV-1's emission spectra were investigated in different pH buffer solutions, no apparent change in emission intensity was observed in the pH range of 4.0–8.0, as shown in Fig. S6.† Next, the potential influence of biochemical species on PCV-1's emission intensity was investigated. As illustrated in Fig. S7,† various sources of interference in the cytoplasm and microenvironment were added to the aqueous solution of PCV-1, including the cations of Na+, K+, Zn2+, Fe2+, Fe3+, Mg2+, Ag+, and Ni2+, the anions of NO3−, SO42−, AcO−, HCO3−, HSO3−, SCN−, F−, Cl−, Br−, and I−, the biomolecules of GSH, Hcy, and Cys, and the reactive substance H2O2. No evident change in PCV-1's emission intensity was observed in the presence of any aforementioned species.
060 M−1 cm−1. Under 450 nm excitation, PCV-1 showed a strong red emission with a maximum peak at 606 nm (Φ = 0.08%), which is favorable for bioimaging applications. The Stokes shift of PCV-1 was beyond 160 nm due to its charge-enhanced intramolecular charge transfer system and extended conjugation length.32 Moreover, when PCV-1's emission spectra were investigated in different pH buffer solutions, no apparent change in emission intensity was observed in the pH range of 4.0–8.0, as shown in Fig. S6.† Next, the potential influence of biochemical species on PCV-1's emission intensity was investigated. As illustrated in Fig. S7,† various sources of interference in the cytoplasm and microenvironment were added to the aqueous solution of PCV-1, including the cations of Na+, K+, Zn2+, Fe2+, Fe3+, Mg2+, Ag+, and Ni2+, the anions of NO3−, SO42−, AcO−, HCO3−, HSO3−, SCN−, F−, Cl−, Br−, and I−, the biomolecules of GSH, Hcy, and Cys, and the reactive substance H2O2. No evident change in PCV-1's emission intensity was observed in the presence of any aforementioned species.
To shed light on PCV-1's electronic transitions, density functional theory (DFT) calculations were performed using the Gaussian 16W program package (Tables S1 and S2†). As shown in Fig. 2c, PCV-1's highest occupied molecular orbital (HOMO) was located in the electron-donating phenyl piperazine group. With the introduction of the positive charge via methylation, the central methylquinolinium unit showed greater electronegativity. As expected, PCV-1's lowest unoccupied molecular orbital (LUMO) was centered on the electron-accepting methylquinolinium moiety. The calculated energies of HOMO and LUMO were −5.47 eV and −2.95 eV, respectively, resulting in an energy gap of 2.52 eV. Moreover, time-dependent DFT calculations indicated that PCV-1's lowest singlet excited state S1 was mainly HOMO to LUMO transition (see the electron density difference map (EDDM) image in Fig. 2d) and located at 533 nm (Fig. 2d), which generally matched the experimental absorption spectrum (Fig. 2b).
Viscosity-enhanced optical properties of PCV-1
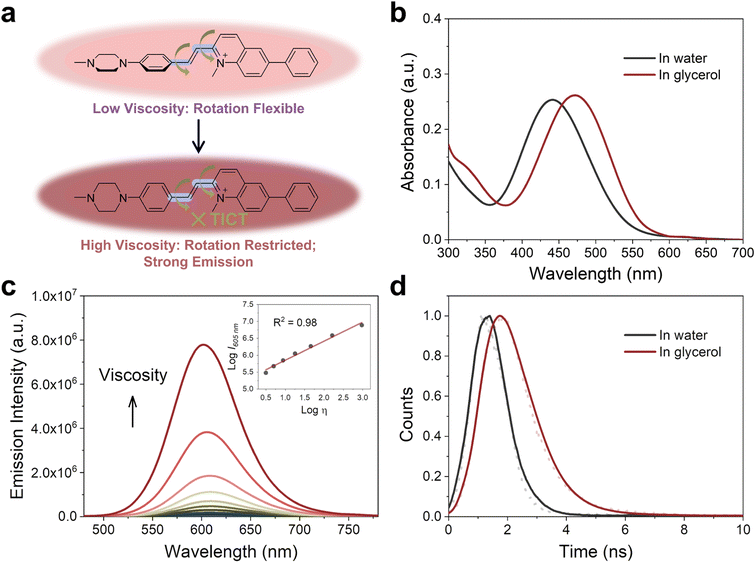
The viscosity of subcellular organelles is far higher than cytosol's.33–35 Lysosomes in particular are known to contain an internal environment with viscosity ranging from 50 to 90 cP.36 The average viscosity of mitochondria, by comparison, is estimated to be approximately 62 cP.37 Therefore, the large difference in viscosity between cell cytosol and organelles can facilitate subcellular bioimaging that results in high brightness, less photodamage, and a large signal-to-noise ratio.Fluorescent probes designed with rotatable single bonds have recently been developed to respond to the viscosity of organelles.38–40 Because PCV-1 is composed of a donor–π–acceptor structure, there are rotatable single bonds (highlighted in purple color, Fig. 3a) present around the vinylene group that connect the electron-donating and electron-accepting moieties. To investigate PCV-1's optical properties in response to viscosity, we collected PCV-1's absorption and emission spectra in water upon the addition of glycerol. Because PCV-1's molecular rotation is likely restricted in solutions of high viscosity, PCV-1 forms a more planar configuration with better conjugation. As a result, PCV-1's absorption peak wavelength showed a 32 nm redshift in glycerol solution compared with that in water (Fig. 3b). Besides, the molar extinction coefficient of PCV-1 in high-viscosity solution is 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 010 M−1 cm−1 (Fig. S8†). Next, we measured PCV-1's emission spectra in solutions with viscosities ranging from 0.9 to 945 cP. As shown in Fig. 3c, PCV-1 initially exhibited relatively weak emission intensity in solutions of low viscosity. However, as the viscosity increased, the molecular rotation was restricted and the excited state energy was primarily released through radiative decay.30 Therefore, PCV-1 presented a significantly enhanced (i.e., 187-fold) emission intensity in glycerol and increased quantum yield (Φ = 1%) in high-viscosity solution (η = 49 cP). By fitting via the Förster–Hoffmann equation, as shown in the inset of Fig. 3c,39,41 PCV-1's emission intensity demonstrated an excellent linear relationship (R2 = 0.98) with viscosity. Meanwhile, its excited-state lifetime was independent of concentration and absorption.42 As shown in Fig. 3d, PCV-1's excited-state lifetime in glycerol was much longer than that in water, which further confirms the different molecular rotation and π-conjugation of PCV-1 in solutions of high and low viscosity.
010 M−1 cm−1 (Fig. S8†). Next, we measured PCV-1's emission spectra in solutions with viscosities ranging from 0.9 to 945 cP. As shown in Fig. 3c, PCV-1 initially exhibited relatively weak emission intensity in solutions of low viscosity. However, as the viscosity increased, the molecular rotation was restricted and the excited state energy was primarily released through radiative decay.30 Therefore, PCV-1 presented a significantly enhanced (i.e., 187-fold) emission intensity in glycerol and increased quantum yield (Φ = 1%) in high-viscosity solution (η = 49 cP). By fitting via the Förster–Hoffmann equation, as shown in the inset of Fig. 3c,39,41 PCV-1's emission intensity demonstrated an excellent linear relationship (R2 = 0.98) with viscosity. Meanwhile, its excited-state lifetime was independent of concentration and absorption.42 As shown in Fig. 3d, PCV-1's excited-state lifetime in glycerol was much longer than that in water, which further confirms the different molecular rotation and π-conjugation of PCV-1 in solutions of high and low viscosity.
To examine whether PCV-1's strong emission could also be attributed to the polarity of solvents, PCV-1's emission spectra were collected in different solvents. As illustrated in Fig. S9a,† PCV-1 showed significantly high emission intensity only in glycerol. When it was dissolved in solvents of different polarities with low viscosity, the observed emission intensity was nearly negligible compared with that in glycerol (Fig. S9b†). Such an infinitesimal polarity effect makes PCV-1 a highly viscosity-sensitive probe in complex biological microenvironments. Moreover, PCV-1's emission intensity remained unchanged after 300 s of irradiation in both low- and high-viscosity solutions (Fig. S10†). This level of photostability is comparable to that of the commercial coumarin 153 dye, confirming that PCV-1 possesses exceptional photostability. Taken together, our photophysical results strongly suggest that PCV-1 is a promising viscosity-responsive probe that is well-suited for imaging highly viscous organelles, such as mitochondria.
Specific localization of PCV-1 in cells
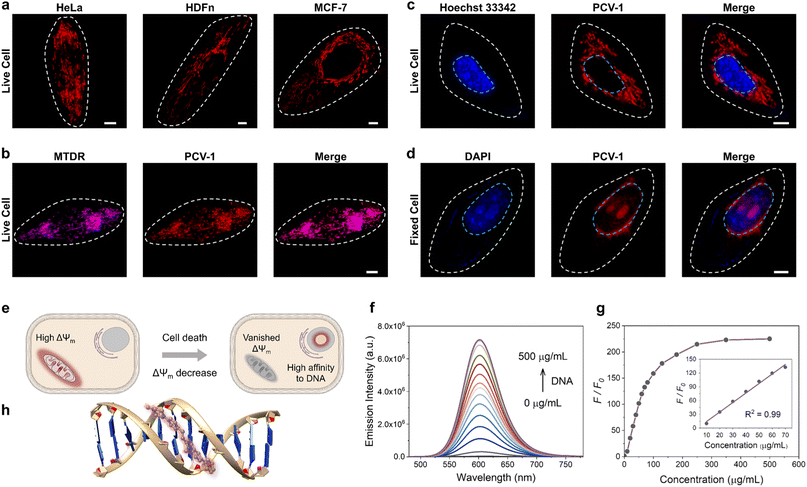
Encouraged by PCV-1's outstanding photophysical properties, we next explored its application in bioimaging. First, PCV-1's toxicity was evaluated using the colorimetric cell viability assay with a Cell Counting Kit-8 (CCK-8). The CCK-8 assay was performed with the highly water-soluble tetrazolium salt WST-8, which can be reduced by dehydrogenase in healthy cells and generate yellow WST-8 formazan. Per those results, the higher activity of dehydrogenase implies greater cell viability. The CCK-8 assay results indicated that PCV-1 had no effect on cell viability at various concentrations after 24 h treatments (Fig. S11†). Next, we performed the fluorescence imaging of PCV-1 via SIM. As depicted in Fig. 4a, the SIM images of PCV-1-stained HeLa cells showed filamentous fluorescence that might indicate mitochondria. The same filamentous fluorescence images were also observed in other cell lines, including fibroblast cells (i.e., HDFn) and breast cancer cells (i.e., MCF-7). Given those observations, co-localization experiments of PCV-1 were conducted with the commercial mitochondria dye MitoTracker Deep Red (MTDR, Fig. 4b), and the lysosome dye LysoTracker Deep Red (LTDR, Fig. S12†). The fluorescence images of PCV-1 showed a higher overlap with MTDR (Pearson's correlation coefficient (PCC) = 0.91, Fig. S13a†) than LTDR (PCC = 0.34, Fig. S13b†), which indicates that PCV-1 can be used as a mitochondrion-targeting probe. To further gauge PCV-1's ability to localize in mitochondria, we investigated its lipophilicity by calculating the n-octanol–water partition coefficient value (i.e., log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). As shown in Fig. S14,† the log
P). As shown in Fig. S14,† the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of PCV-1 was 0.7, thereby indicating that PCV-1 has high lipophilicity. According to Horobin et al.'s theory, a cation with a log
P value of PCV-1 was 0.7, thereby indicating that PCV-1 has high lipophilicity. According to Horobin et al.'s theory, a cation with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value from 0 to 5 has a high probability of mitochondrial localization.43–45 Thus, the co-localization images together with the log
P value from 0 to 5 has a high probability of mitochondrial localization.43–45 Thus, the co-localization images together with the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value demonstrate the mitochondrial localization of PCV-1 in live cells.
P value demonstrate the mitochondrial localization of PCV-1 in live cells.
Afterwards, when we investigated the fluorescence imaging of PCV-1 after drug treatment, we found that PCV-1's fluorescence morphology in dead cells completely differed from that in live cells. As shown in Fig. S15,† no filamentous fluorescence was presented in the cytoplasm region of the dead cells. Instead, PCV-1 was found to gather in the center of cells, where it exhibited elliptical morphology and high fluorescence intensity. Based on these results, we speculated that the new fluorescent region was located in the nucleus and PCV-1 escaped from mitochondria to the nucleus during cell death. To test this hypothesis, we subsequently co-stained it with the nuclei dyes of live cells (i.e., Hoechst 33342) and dead cells (i.e., DAPI). As shown in Fig. 4c, no evident overlap between the blue channel of Hoechst 33342 and the red channel of PCV-1 was observed, which confirmed that PCV-1 was only localized in mitochondria in live cells. In sharp contrast, PCV-1 could also cross the nuclear envelope and displayed strong red fluorescence in fixed cells, which highly overlapped with the blue fluorescence from DAPI, suggesting that PCV-1 can selectively stain the nuclei in dead cells (Fig. 4d). Moreover, the strong red emission of PCV-1 was greatly concentrated in the central nuclear region with spherical morphologies, which indicated that PCV-1 was primarily distributed in the nucleolus.46 To gain further insight into the PCV-1's migration from mitochondria to nucleolus, we acquired in situ real-time fluorescence imaging of PCV-1 in HeLa cells using high-resolution confocal microscopy. As shown in Fig. S16,† upon the addition of NaN3, the escape of PCV-1 from mitochondria to nucleolus was distinctly observed at single-cell level in both fluorescence imaging and bright field imaging. To sum up, we suspected that the different distributions of PCV-1 in live and dead cells were impacted by the mitochondrial membrane potential. As depicted in Fig. 4e, PCV-1 was abundantly enriched in mitochondria due to the cationic quinolinium group in PCV-1 and the highly negative mitochondrial membrane potential in healthy cells. However, in dead cells no longer with mitochondrial membrane potential, PCV-1 targeted the nucleolus via electrostatic attraction and possible intercalation.
To understand the mechanism by which PCV-1 targets the nucleolus in dead cells, we performed a DNA titration experiment using calf thymus DNA (ctDNA). As shown in Fig. S17,† as the concentration of ctDNA in the PCV-1 buffer solution increased, the absorption spectra of PCV-1 showed a clear redshift indicating that PCV-1's π-conjugation has been expanded. Meanwhile, the emission intensity of PCV-1 gradually enhanced with DNA binding, as illustrated in Fig. 4f. When the concentration of ctDNA reached 500 μg mL−1, a 225-fold enhancement of PCV-1's emission intensity was observed (Fig. 4g). In addition, PCV-1's enhanced emission intensity demonstrated an exceptional linear relationship (R2 = 0.99), with the concentration of ctDNA in the range of 10–70 μg mL−1. Based on the properties of PCV-1 in response to viscosity described before, we thus surmise that the light-on behavior of PCV-1 in the presence of DNA can also be due to the restriction of molecular rotation. Therefore, we conducted molecular docking studies between DNA and PCV-1 using the AutoDock Vina software.47 The results of the docking simulation suggested that PCV-1 was strongly bound to the minor grooves of DNA (Fig. 4h). The calculated binding energy was −10.5 kcal mol−1, which is comparable to that of the commercial Hoechst 33342 (−12.2 kcal mol−1) and better than that of a previously reported nuclei probe.12 Overall, the aforementioned imaging, emission titration, and molecular docking results collectively demonstrated that PCV-1 can differentiate live and dead cells via the distinct fluorescence morphologies and intensities of different organelles.
PCV-1's excellence in super-resolution imaging
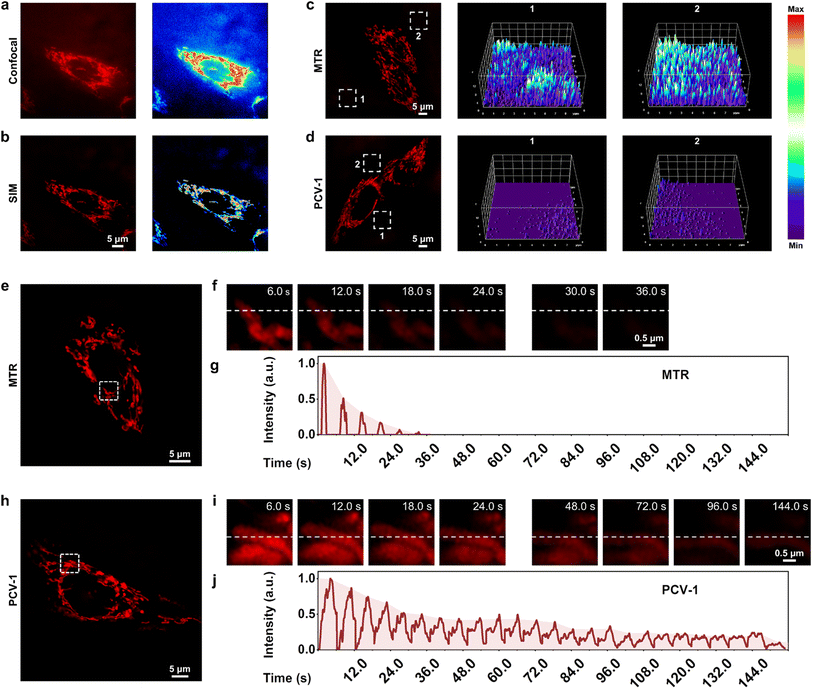
To acquire better fluorescence images at the single-cell level as a means to quantify cell viability, we explored PCV-1's imaging properties in super-resolution imaging. We first compared the fluorescence images of PCV-1 in HeLa cells captured by conventional confocal microscopy and SIM. Confocal imaging exhibited the high spatial overlap of fluorescence due to its low resolution (Fig. 5a), while SIM imaging afforded superior imaging quality, with clearly filamentous mitochondria fluorescence and less background fluorescence (Fig. 5b). Benefiting from the state-of-the-art SIM technology, PCV-1 can visualize previously invisible subcellular details in biological systems.As indicated, PCV-1 showed a large Stokes shift and viscosity-enhanced fluorescence intensity (Fig. 2b and 3c), which can effectively overcome the background noise created by excitation light and increase the signal-to-noise ratio because of high viscosity in mitochondria. Therefore, we chose the commercial mitochondria dye MitoTracker Red (MTR) for comparison to confirm these advantages. The SIM images of HeLa cells labeled with MTR and PCV-1 were taken in the same condition. We randomly picked two regions of background in MTR- and PCV-1-stained SIM images and plotted their corresponding 3D fluorescence intensity distribution maps, as illustrated in Fig. 5c and d. The results elucidated that the background fluorescence of PCV-1-stained SIM images was significantly lower than that of MTR-stained SIM images. Beyond that, PCV-1 showed a higher average fluorescence intensity in mitochondria than using MTR (Fig. S18†). In view of these results, PCV-1 can be expected to effectively improve the signal-to-noise ratio of fluorescence imaging and thus facilitate the study of hyperfine structures of cells.
Resistance to photobleaching is a vital ability of probes that determines whether it can function for long-time fluorescence tracking.24,25 According to the chemical structure and previous fluorescence stability tests, PCV-1 seems to be endowed with a high resistance to photobleaching. To confirm its stability for imaging applications, we performed a photobleaching comparison experiment between MTR and PCV-1 in HeLa cells (Fig. 5e and h). Under 100% laser power mode with the same settings, the fluorescence of MTR became rapidly bleached (Fig. 5f), while PCV-1 exhibited high resistance to photobleaching (Fig. 5i). Specifically, as shown in Fig. 5g and j, the photobleaching lifetime of PCV-1 was more than 144 s, or 4 times longer than that of MTR (i.e., 36 s). Furthermore, when the live HeLa cells stained with MTR and PCV-1 were incubated for 24 h for observation (Fig. S19†), both types of cells exhibited fluorescence in mitochondria at the beginning. On the following day, however, the fluorescence of mitochondria stained with MTR completely disappeared, whereas PCV-1 still showed strong filamentous fluorescence in mitochondria. Thus, we conclude that PCV-1 has superior photostability in live cells.
With all of the outstanding imaging properties described above, including exceptional super-resolution imaging, high signal-to-noise ratio, and great resistance to photobleaching, PCV-1 is expected to be an excellent biological tool for visualizing interactions between organelles at the single-cell level.
Quantifying cell viability using the ORP analysis with PCV-1
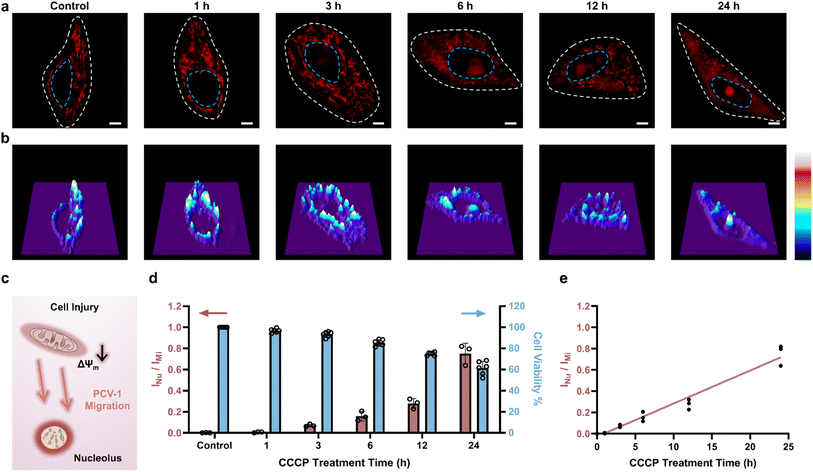
According to the above results, PCV-1 is located in the mitochondria of live cells but escapes to the nucleolus when cells die. Thus, we speculated that there might be a state in which PCV-1 targets both mitochondria and the nucleolus during cell injury. To track the fluorescence morphologies of PCV-1 in the process of cell injury, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a common inducer of mitophagy (i.e., one type of autophagy), was used to damage mitochondria and reduce cell viability. Previous studies have shown that the mitochondrial membrane potential changes rapidly during the CCCP treatment, which results in fluctuations in the fluorescence of a probe.29,48 Thus, to accurately observe the fluorescence imaging, we incubated CCCP-treated HeLa cells to allow the cells to acclimate to the new microenvironment and thereby make the drug work more efficiently.49–51 Next, CCCP-treated HeLa cells stained by PCV-1 were imaged on SIM. As shown in Fig. 6a, upon the addition of CCCP, the filamentous fluorescence of mitochondria broke and became spherical fluorescence in different sizes, thereby indicating that mitophagy had occurred.49,50 As the time of CCCP treatment was lengthened, more damaged mitochondria were observed, meaning that the degree of cell viability was decreased. As expected, PCV-1 was located in the nucleolus in dead cells due to the disappearance of mitochondrial membrane potential. PCV-1 escaped from damaged mitochondria into the nucleolus during cell injury and showed bright red fluorescence in both organelles, also as expected. In particular, SIM images after 24 h treatment of CCCP exhibited remarkably high fluorescence intensity in the nucleolus but less spherical fluorescence in the cytoplasm, which further confirms the exceptionally low cell viability under that condition. Moreover, the corresponding distribution maps of 3D fluorescence intensity under the CCCP treatment at different times (Fig. 6b) allowed the dynamic observation of PCV-1's mitochondria-to-nucleolus migration. The significantly high fluorescence intensities of mitochondria in the control sample gradually diminished during the process of cell injury, while a new high-intensity fluorescent peak emerged in the center of the nuclei. Furthermore, similar migration of PCV-1 from mitochondria to nucleolus has been observed when employing other mitophagy inducers, such as oligomycin A52,53 and erastin,54–56 as shown in Fig. S20.† Taking all those imaging data together, we anticipate that PCV-1's migration ability can be used as a marker of cell viability, as depicted in Fig. 6c.As next-generation methods of quantitative cell biology, the digitization of single cells and subcellular organelles has attracted considerable attention in recent years. Nevertheless, systems of quantitative analysis for measuring cell viability have not been developed. Herein, we propose a new method of image analysis, named organelle ratiometric probing (ORP), which is defined by calculating fluorescence intensity ratios between various morphologies at the subcellular level, including in mitochondria, lysosomes, and endoplasmic reticulum. Having observed the migration of PCV-1 from mitochondria to the nucleolus, we then used the ORP for quantifying cell viability. Among the ORP's merits when using PCV-1, long spatial distances between mitochondria and the nucleolus without direct contact allow accurately quantifying the fluorescence intensities of organelles at the single-cell level using super-resolution imaging. Another merit is that single-channel imaging can significantly reduce instrumental errors and permit reasonable comparisons of the fluorescence intensity of organelles in situ. Last, the PCV's high resistance to photobleaching enables using ORP to observe fluorescence imaging both at multiple times and in the long term without sacrificing any sensitivity.
To digitize SIM images of CCCP-treated HeLa cells, PCV-1's nucleolus-to-mitochondria fluorescence intensity ratios (i.e., ORP ratio) were calculated and displayed in Fig. 6d and S21.† Moreover, to correlate those fluorescence ratios with cell viability, the cell viability of CCCP-treated HeLa cells was also investigated using the conventional CCK-8 assay, as shown in the right blue columns of Fig. 6d. PCV-1 initially exhibited ORP ratios close to 0 owing to the healthy cells with high mitochondrial membrane potential. As treatment with CCCP continued, cell viability gradually decreased, which resulted in an increase in the nucleolus-to-mitochondria fluorescence intensity ratio. Furthermore, as shown in the results of HeLa cells treated with CCCP for 12 h, a fluorescence intensity ratio of 0.3 represents a cell viability of 80%. According to the International Organization for Standardization, a percentage of cell viability less than 80% is considered to indicate cytotoxicity.57 With that knowledge, we chose 0.3 as the cutoff point for our ORP analysis with PCV-1 to assess whether adding a drug would cause cytotoxicity. In addition, when plotting the fluorescence intensity ratios of nucleolus-to-mitochondria against the duration of CCCP treatment (Fig. 6e), the ORP analysis with PCV-1 demonstrated an exceptional linear relationship (R2 = 0.96), providing additional evidence that it is an excellent ratiometric tool for evaluating cell viability. Next, the HeLa cells stained by PCV-1 but without the treatment of CCCP were studied as well. As shown in Fig. S22a,† no migration from mitochondria to the nucleolus was observed, and PCV-1 exhibited stable, strong filamentous fluorescence after 24 h of incubation. Moreover, as shown in Fig. S11,† PCV-1 did not affect cell viability after 24 h of incubation. Therefore, per the ORP analysis, the PCV-1 fluorescence ratios for the SIM images were consistently less than 0.3, which further demonstrates that PCV-1 is non-cytotoxic (Fig. S22b and S23†).
Quantifying cell viability under different concentrations of CCCP treatment
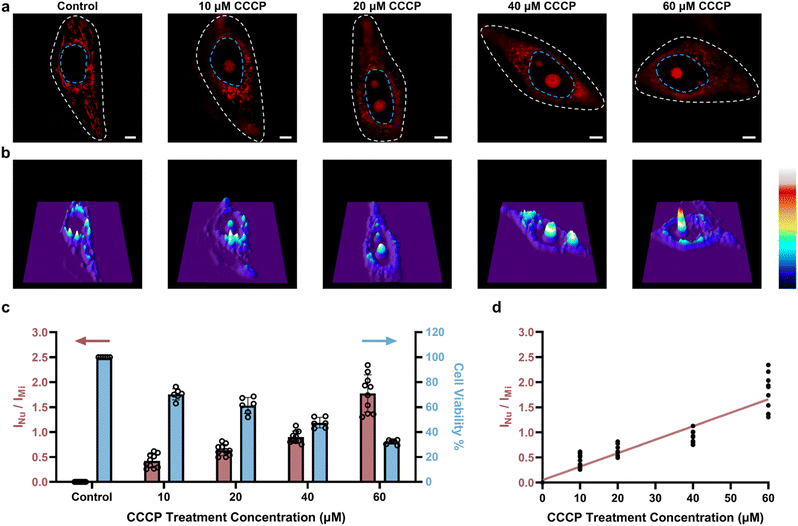
Next, to confirm that PCV-1 is indeed an excellent tool for dynamically visualizing cell viability, we also investigated CCCP concentration-dependent autophagy. As shown in Fig. 7a, as the concentration of CCCP increased, the number of damaged mitochondria increased as well, and more PCV-1 accumulated in the nucleolus, thereby suggesting that autophagy occurred more intensely under pathological conditions.49,50 However, if the concentration of CCCP was too high, then the strong cytotoxicity would destroy the cells, such that PCV-1 displayed only a significantly high fluorescence peak in the nucleolus (Fig. 7b). These results imply that PCV-1 can vividly display the states of cells at different stages in the process of cell injury. Interpreting those SIM images using the ORP analysis with PCV-1 (Fig. S24†) revealed nucleolus-to-mitochondria intensity ratios indicating a growing trend with the increase of the CCCP concentration, as illustrated in Fig. 7c. For the range of each fluorescence ratio, there was a corresponding range of cell viability to match. Thus, when cell viability was less than 80% in the experiment with the treatment of 10 μM CCCP, PCV-1's fluorescence ratio exceeded 0.3, which further confirmed that our assumed cutoff point of 0.3 was appropriate. Additionally, the fluorescence intensity ratios measured by the ORP analysis with PCV-1 also exhibited a decent linear correlation as the concentration of CCCP treatment increased (Fig. 7d). In sum, in super-resolution imaging, PCV-1 together with the ORP analysis allowed visualizing cell injury and quantifying cell viability at the single-cell level.Visualizing cell viability in various routes toward cell death using the ORP analysis
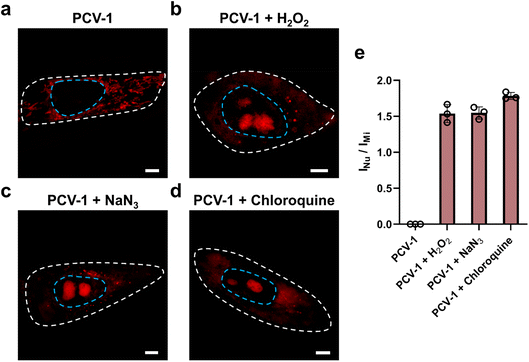
To explore the applications of the ORP analysis in different routes toward cell death, we investigated the SIM images of PCV-1-stained HeLa cells after various drug treatments. One route was hydrogen peroxide (H2O2), a reactive oxygen species that can efficiently inhibit metabolism and cause cell damage by inducing either apoptosis or necrosis depending on the dose.58,59 Another route was sodium azide (NaN3), a highly toxic salt that acts as a potent inhibitor of mitochondrial respiration by hindering the function of cytochrome oxidase and induces cell death.60–62 The last one was chloroquine, an inhibitor of the autophagy–lysosome pathway that has been tested extensively in preclinical cancer models and used as an antimalarial drug.63,64 In all, H2O2, NaN3, and chloroquine were applied to treat HeLa cells to induce cell death.As shown in Fig. 8a, the control group without drug treatment presented clearly filamentous fluorescence of PCV-1 in mitochondria. As the high concentration of H2O2 treatment induced cell necrosis, PCV-1 quickly lit up the nucleolus, whereas the high fluorescence in the cytoplasm reduced rapidly due to the low cell viability under this condition (Fig. 8b). Meanwhile, the NaN3-treated HeLa cells demonstrated spherical fluorescence in both the cytoplasm and nucleolus, thereby suggesting that cell death via apoptosis can be visualized by PCV-1 (Fig. 8c). PCV-1 also allows monitoring the low viability of chloroquine-treated cells, as reflected in intense fluorescence in the center of nuclei (Fig. 8d). To probe cell viability after drug treatments in greater depth, we additionally checked those SIM images in the ORP analysis. All HeLa cells treated with drugs exhibited fluorescence intensity ratios of the nucleolus to the mitochondria exceeding 1.5, as shown in Fig. 8e. With such high fluorescence intensity ratios, we can confidently propose that H2O2, NaN3, and chloroquine are highly cytotoxic, a condition that extremely reduces cell viability.
Last, we explored SIM images of PCV-1 in other cell lines during CCCP-induced mitophagy. As shown in Fig. S25,† PCV-1 migrated from mitochondria to the nucleolus in both HDFn and MCF-7 cells, thereby indicating that it can be adapted to other cell lines to visualize cell viability. In that light, PCV-1 can serve as a versatile tool for rapidly quantifying cell viability in different cells and various pathways toward cell death, including apoptosis, autophagy, and necrosis.
Conclusions
In this study, we rationally designed and synthesized a fluorescent probe, PCV-1, to visualize cell viability under SIM. Because of PCV-1's outstanding photophysical properties and excellent super-resolution imaging performance, its migration from mitochondria to the nucleolus could be dynamically visualized at the single-cell level during cell injury. By extension, harnessing PCV-1's excellent photostability and signal-to-noise ratio and by comparing the fluorescence intensity of the two organelles, mitochondria and nucleoli, we developed a powerful analytical assay, ORP, that we applied to quantitatively analyze and efficiently assess the viability of individual cells, thereby enabling deeper insights into the potential mechanisms of cell death. In our ORP analysis with PCV-1, we identified 0.3 as the cutoff point for assessing whether adding the drug will cause apparent cytotoxicity, which greatly expands the probe's applicability. In the future, combined with machine learning and big data analysis, our proposed ORP analysis with PCV-1 stands to significantly improve the diagnosis of disease, facilitate the development of drugs, and reduce medical costs.Data availability
All experimental details, materials, characterization, synthesis, and methods are included in ESI.†Author contributions
Rui Chen: conceptualization, investigation, methodology, data curation, writing – original draft. Kangqiang Qiu: methodology, conceptualization. Guanqun Han: software. Bidyut Kumar Kundu: software. Guodong Ding: methodology. Yujie Sun: writing – review & editing, supervision, funding acquisition. Jiajie Diao: writing – review & editing, supervision, funding acquisition.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
Yujie Sun acknowledges the support of the National Science Foundation (CHE1955358) and University of Cincinnati. Jiajie Diao is supported by the National Institute of Health (R35GM128837). Research reported in this publication was supported by the Office of the Director, National Institutes of Health under Award Number S10OD030402. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.Notes and references
- R. S. Hotchkiss, A. Strasser, J. E. McDunn and P. E. Swanson, N. Engl. J. Med., 2009, 361, 1570–1583 CrossRef CAS PubMed.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495–516 CrossRef CAS PubMed.
- G. I. Evan and K. H. Vousden, Nature, 2001, 411, 342–348 CrossRef CAS PubMed.
- O. Kepp, L. Galluzzi, M. Lipinski, J. Yuan and G. Kroemer, Nat. Rev. Drug Discovery, 2011, 10, 221–237 CrossRef CAS PubMed.
- M. Tian, Y. Ma and W. Lin, Acc. Chem. Res., 2019, 52, 2147–2157 CrossRef CAS PubMed.
- G. Banfalvi, Apoptosis, 2017, 22, 306–323 CrossRef CAS PubMed.
- J. Muppidi, M. Porter and R. M. Siegel, Curr. Protoc. Immunol., 2004, 59, 3.17.11–13.17.36 Search PubMed.
- M. V. Berridge, P. M. Herst and A. S. Tan, in Biotechnol. Annu. Rev., Elsevier, 2005, vol. 11, pp. 127–152 Search PubMed.
- P. Larsson, H. Engqvist, J. Biermann, E. Werner Rönnerman, E. Forssell-Aronsson, A. Kovács, P. Karlsson, K. Helou and T. Z. Parris, Sci. Rep., 2020, 10, 5798 CrossRef CAS PubMed.
- L. D. Zorova, V. A. Popkov, E. Y. Plotnikov, D. N. Silachev, I. B. Pevzner, S. S. Jankauskas, V. A. Babenko, S. D. Zorov, A. V. Balakireva, M. Juhaszova, S. J. Sollott and D. B. Zorov, Anal. Biochem., 2018, 552, 50–59 CrossRef CAS PubMed.
- D. R. Green and J. C. Reed, Science, 1998, 281, 1309–1312 CrossRef CAS PubMed.
- M. Tian, J. Sun, B. Dong and W. Lin, Angew. Chem., Int. Ed., 2018, 57, 16506–16510 CrossRef CAS PubMed.
- X. Li, M. Tian, G. Zhang, R. Zhang, R. Feng, L. Guo, X. Yu, N. Zhao and X. He, Anal. Chem., 2017, 89, 3335–3344 CrossRef CAS PubMed.
- H. Fang, Y. Chen, S. Geng, S. Yao, Z. Guo and W. He, Anal. Chem., 2022, 94, 17904–17912 CrossRef CAS PubMed.
- L. Wang, R. Chen, G. Han, X. Liu, T. Huang, J. Diao and Y. Sun, Exploration, 2022, 2, 20210215 CrossRef PubMed.
- K. Qiu, W. Zou, H. Fang, M. Hao, K. Mehta, Z. Tian, J. L. Guan, K. Zhang, T. Huang and J. Diao, Nat. Commun., 2022, 13, 4303 CrossRef CAS PubMed.
- H. Fang, S. Geng, M. Hao, Q. Chen, M. Liu, C. Liu, Z. Tian, C. Wang, T. Takebe, J.-L. Guan, Y. Chen, Z. Guo, W. He and J. Diao, Nat. Commun., 2021, 12, 109 CrossRef CAS PubMed.
- Q. Chen, H. Fang, X. Shao, Z. Tian, S. Geng, Y. Zhang, H. Fan, P. Xiang, J. Zhang, X. Tian, K. Zhang, W. He, Z. Guo and J. Diao, Nat. Commun., 2020, 11, 6290 CrossRef CAS PubMed.
- R. Chen, L. Wang, G. Ding, G. Han, K. Qiu, Y. Sun and J. Diao, ACS Sens., 2023, 8, 2068–2078 CrossRef CAS PubMed.
- C. Zhang, Z. Tian, R. Chen, F. Rowan, K. Qiu, Y. Sun, J.-L. Guan and J. Diao, Adv. Drug Delivery Rev., 2023, 199, 114978 CrossRef CAS PubMed.
- C. P. Satori, M. M. Henderson, E. A. Krautkramer, V. Kostal, M. M. Distefano and E. A. Arriaga, Chem. Rev., 2013, 113, 2733–2811 CrossRef CAS PubMed.
- K.-N. Wang, X. Shao, Z. Tian, L.-Y. Liu, C. Zhang, C.-P. Tan, J. Zhang, P. Ling, F. Liu, Q. Chen, J. Diao and Z.-W. Mao, Adv. Sci., 2021, 8, 2004566 CrossRef CAS PubMed.
- H. Fang, S. Yao, Q. Chen, C. Liu, Y. Cai, S. Geng, Y. Bai, Z. Tian, A. L. Zacharias, T. Takebe, Y. Chen, Z. Guo, W. He and J. Diao, ACS Nano, 2019, 13, 14426–14436 CrossRef CAS PubMed.
- K. Qiu, A. Yadav, Z. Tian, Z. Guo, D. Shi, C. K. Nandi and J. Diao, ACS Mater. Lett., 2022, 4, 1565–1573 CrossRef CAS.
- K. Qiu, R. Seino, G. Han, M. Ishiyama, Y. Ueno, Z. Tian, Y. Sun and J. Diao, Adv. Healthcare Mater., 2022, 11, 2102185 CrossRef CAS PubMed.
- R. Yang, X. He, G. Niu, F. Meng, Q. Lu, Z. Liu and X. Yu, ACS Sens., 2021, 6, 1552–1559 CrossRef CAS PubMed.
- M. Tian, J. Zhan and W. Lin, Coord. Chem. Rev., 2022, 451, 214266 CrossRef CAS.
- K. N. Wang, L. Y. Liu, D. Mao, S. Xu, C. P. Tan, Q. Cao, Z. W. Mao and B. Liu, Angew Chem. Int. Ed. Engl., 2021, 60, 15095–15100 CrossRef CAS PubMed.
- M. Tian, J. Sun, B. Dong and W. Lin, Sens. Actuators, B, 2019, 292, 16–23 CrossRef CAS.
- Z. Zou, Q. Yan, S. Ai, P. Qi, H. Yang, Y. Zhang, Z. Qing, L. Zhang, F. Feng and R. Yang, Anal. Chem., 2019, 91, 8574–8581 CrossRef CAS PubMed.
- B. Chen, C. Li, J. Zhang, J. Kan, T. Jiang, J. Zhou and H. Ma, Chem. Commun., 2019, 55, 7410–7413 RSC.
- C. S. Abeywickrama, Chem. Commun., 2022, 58, 9855–9869 RSC.
- A. M. Mastro, M. A. Babich, W. D. Taylor and A. D. Keith, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 3414–3418 CrossRef CAS PubMed.
- J. Dijksterhuis, J. Nijsse, F. A. Hoekstra and E. A. Golovina, Eukaryotic Cell, 2007, 6, 157–170 CrossRef CAS PubMed.
- K. Luby-Phelps, D. L. Taylor and F. Lanni, J. Cell Biol., 1986, 102, 2015–2022 CrossRef CAS PubMed.
- L. Wang, Y. Xiao, W. Tian and L. Deng, J. Am. Chem. Soc., 2013, 135, 2903–2906 CrossRef CAS PubMed.
- Z. Yang, Y. He, J. H. Lee, N. Park, M. Suh, W. S. Chae, J. Cao, X. Peng, H. Jung, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2013, 135, 9181–9185 CrossRef CAS PubMed.
- S.-J. Li, Y.-F. Li, H.-W. Liu, D.-Y. Zhou, W.-L. Jiang, J. Ou-Yang and C.-Y. Li, Anal. Chem., 2018, 90, 9418–9425 CrossRef CAS PubMed.
- H. Zhu, J. Fan, M. Li, J. Cao, J. Wang and X. Peng, Chem. – Eur. J., 2014, 20, 4691–4696 CrossRef CAS PubMed.
- R. Chen, K. Qiu, D. C. Y. Leong, B. K. Kundu, C. Zhang, P. Srivastava, K. E. White, G. Li, G. Han, Z. Guo, C. G. Elles, J. Diao and Y. Sun, Biosens. Bioelectron., 2023, 239, 115604 CrossRef CAS PubMed.
- Y. Song, H. Zhang, X. Wang, X. Geng, Y. Sun, J. Liu and Z. Li, Anal. Chem., 2021, 93, 1786–1791 CrossRef CAS PubMed.
- P. I. H. Bastiaens and A. Squire, Trends Cell Biol., 1999, 9, 48–52 CrossRef CAS PubMed.
- R. W. Horobin and F. Rashid-Doubell, Biotech. Histochem., 2013, 88, 461–476 CrossRef CAS PubMed.
- R. W. Horobin, F. Rashid-Doubell, J. D. Pediani and G. Milligan, Biotech. Histochem., 2013, 88, 440–460 CrossRef CAS PubMed.
- K. Qiu, Y. Liu, H. Huang, C. Liu, H. Zhu, Y. Chen, L. Ji and H. Chao, Dalton Trans., 2016, 45, 16144–16147 RSC.
- T. Mukherjee, V. Soppina, R. Ludovic, Y. Mély, A. S. Klymchenko, M. Collot and S. Kanvah, Org. Biomol. Chem., 2021, 19, 3389–3395 RSC.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- M.-Y. Li, Y.-H. Liu, K. Li, H. Zhang, L. Shi, X.-Y. Liu, K.-K. Yu and X.-Q. Yu, Anal. Methods, 2019, 11, 5750–5754 RSC.
- Q. Chen, X. Shao, Z. Tian, Y. Chen, P. Mondal, F. Liu, F. Wang, P. Ling, W. He, K. Zhang, Z. Guo and J. Diao, Nano Res., 2019, 12, 1009–1015 CrossRef CAS.
- X. Shao, Q. Chen, L. Hu, Z. Tian, L. Liu, F. Liu, F. Wang, P. Ling, Z.-W. Mao and J. Diao, Nano Res., 2020, 13, 2149–2155 CrossRef CAS.
- K. Qiu, Y. Du, J. Liu, J.-L. Guan, H. Chao and J. Diao, Theranostics, 2020, 10, 6072–6081 CrossRef CAS PubMed.
- X. Cen, Y. Chen, X. Xu, R. Wu, F. He, Q. Zhao, Q. Sun, C. Yi, J. Wu, A. Najafov and H. Xia, Nat. Commun., 2020, 11, 5731 CrossRef CAS PubMed.
- D. Narendra, A. Tanaka, D.-F. Suen and R. J. Youle, J. Cell Biol., 2008, 183, 795–803 CrossRef CAS PubMed.
- S. Wen, X. Hu, Y. Shi, J. Han and S. Han, Anal. Chem., 2021, 93, 16887–16898 CrossRef CAS PubMed.
- A. M. Battaglia, R. Chirillo, I. Aversa, A. Sacco, F. Costanzo and F. Biamonte, Cells, 2020, 9, 1505 CrossRef CAS PubMed.
- Y. Xie, W. Hou, X. Song, Y. Yu, J. Huang, X. Sun, R. Kang and D. Tang, Cell Death Differ., 2016, 23, 369–379 CrossRef CAS PubMed.
- J. López-García, M. Lehocký, P. Humpolíček and P. Sáha, J. Funct. Biomater., 2014, 5, 43–57 CrossRef PubMed.
- Y. Saito, K. Nishio, Y. Ogawa, J. Kimata, T. Kinumi, Y. Yoshida, N. Noguchi and E. Niki, Free Radical Res., 2006, 40, 619–630 CrossRef CAS PubMed.
- M. Tochigi, T. Inoue, M. Suzuki-Karasaki, T. Ochiai, C. Ra and Y. Suzuki-Karasaki, Int. J. Oncol., 2013, 42, 863–872 CrossRef CAS PubMed.
- K. Inomata and H. Tanaka, J. Pharmacol. Sci., 2003, 93, 163–170 CrossRef CAS PubMed.
- S. J. Chen, M. E. Bradley and T. C. Lee, Mol. Cell. Biochem., 1998, 178, 141–149 CrossRef CAS PubMed.
- M. W. Bowler, M. G. Montgomery, A. G. W. Leslie and J. E. Walker, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 8646–8649 CrossRef CAS PubMed.
- H. Fang, A. Liu, U. Dahmen and O. Dirsch, Cell Death Dis., 2013, 4, e694 CrossRef CAS PubMed.
- L. E. Gallagher, O. A. Radhi, M. O. Abdullah, A. G. McCluskey, M. Boyd and E. Y. W. Chan, Cell Death Dis., 2017, 8, e3014 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01537h |
| This journal is © The Royal Society of Chemistry 2023 |