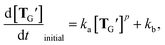Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Minimal RNA self-reproduction discovered from a random pool of oligomers†
Ryo
Mizuuchi
 *ab and
Norikazu
Ichihashi
*ab and
Norikazu
Ichihashi
 cde
cde
aDepartment of Electrical Engineering and Bioscience, Faculty of Science and Engineering, Waseda University, Shinjuku, Tokyo 162-8480, Japan. E-mail: mizuuchi@waseda.jp
bJST, FOREST, Kawaguchi, Saitama 332-0012, Japan
cKomaba Institute for Science, The University of Tokyo, Meguro, Tokyo 153-8902, Japan
dDepartment of Life Science, Graduate School of Arts and Science, The University of Tokyo, Meguro, Tokyo 153-8902, Japan
eUniversal Biology Institute, The University of Tokyo, Meguro, Tokyo 153-8902, Japan
First published on 20th June 2023
Abstract
The emergence of RNA self-reproduction from prebiotic components would have been crucial in developing a genetic system during the origins of life. However, all known self-reproducing RNA molecules are complex ribozymes, and how they could have arisen from abiotic materials remains unclear. Therefore, it has been proposed that the first self-reproducing RNA may have been short oligomers that assemble their components as templates. Here, we sought such minimal RNA self-reproduction in prebiotically accessible short random RNA pools that undergo spontaneous ligation and recombination. By examining enriched RNA families with common motifs, we identified a 20-nucleotide (nt) RNA variant that self-reproduces via template-directed ligation of two 10 nt oligonucleotides. The RNA oligomer contains a 2′–5′ phosphodiester bond, which typically forms during prebiotically plausible RNA synthesis. This non-canonical linkage helps prevent the formation of inactive complexes between self-complementary oligomers while decreasing the ligation efficiency. The system appears to possess an autocatalytic property consistent with exponential self-reproduction despite the limitation of forming a ternary complex of the template and two substrates, similar to the behavior of a much larger ligase ribozyme. Such a minimal, ribozyme-independent RNA self-reproduction may represent the first step in the emergence of an RNA-based genetic system from primordial components. Simultaneously, our examination of random RNA pools highlights the likelihood that complex species interactions were necessary to initiate RNA reproduction.
Introduction
The first genetic system before the emergence of life may have been based on RNA, because RNA can simultaneously carry genetic information and catalyze chemical reactions.1,2 This “RNA World” hypothesis is supported by the observation that all genetically encoded proteins are synthesized by RNA in the ribosome.3 A crucial aim in the quest for an RNA-based genetic system is to find self-reproducing RNA molecules.4–6 A potential mechanism for RNA reproduction is template-directed polymerization of nucleotides, i.e., replication, as observed in extant life. However, despite significant progress in improving non-enzymatic or ribozyme-catalyzed RNA polymerization,7–10 the self-replication of these systems remains challenging. Previous studies have therefore explored alternative, simpler mechanisms for RNA self-reproduction through the assembly of oligonucleotides.6,11,12 In view of recent clarification, we use the term “reproduction” to denote RNA copying in general by distinguishing canonical “replication” that follows template-directed polymerization chemistry.6,12RNA reproduction has been demonstrated for ligase and recombinase ribozymes.13–16 For example, a ligase ribozyme derived from the R3C ligase ribozyme17 catalyzes the joining of two RNA substrates as a template to form a sequence identical to itself.13 This ribozyme is the simplest self-reproducing RNA known to date in terms of its length (61 nt) and the number of components (two fragments: 13 and 48 nt). However, the ribozyme is still relatively large and was rationally designed, and it remains unclear how such a ribozyme and its components could have been prevalent in prebiotically accessible RNA mixtures, which were likely dominated by shorter (up to ∼20 nt) and random oligonucleotides.18,19 The ligase ribozyme also requires 5′-triphosphate activation, which necessitates an additional set of complex reactions.20 Consequently, it has been proposed that the template-based self-reproduction of short RNA molecules independent of complex ribozymes may have emerged first in the RNA World.21,22
Self-reproduction of short nucleic acids has been studied mainly using DNA. Previous studies demonstrated the autocatalytic reproduction of chemically modified DNA oligonucleotides through template-directed ligation, although the reproduction was severely hindered by two tightly bound templates (or a template and its identical product after ligation).23–25 A recent study employed temperature cycling to overcome such template inhibition in the reproduction of chemically activated DNA.26 Despite these efforts with DNA, the self-reproduction of short RNA oligomers is currently missing. Moreover, temperature cycling would be incompatible with RNA because, unlike DNA, RNA easily degrades at high temperature, which is accelerated by divalent metal ions27,28 that commonly enhance ribozyme catalysis28,29 as well as template-directed RNA synthesis.30,31
Template-directed ligation of short RNA is achieved in the laboratory using terminal activation such as with 2′,3′-cyclic phosphate (>p),30 which readily forms in prebiotically plausible environments,32,33 while recombination occurs directly—or in combination with spontaneous >p formation through hydrolysis of RNA, termed α/α′ mechanisms.31,34 Notable are recent studies that demonstrated that pools of short random RNA can undergo diverse intermolecular ligation and recombination, presumably in a templated manner.31,35 In these populations, RNA products that form efficiently or that self-amplify are expected to be enriched. Thus, a close examination of the enriched products may lead to the discovery of efficient RNA reproduction via template-directed ligation or recombination. The identification of such reproducing RNA would also provide insights into the likelihood of the emergence of self-reproduction out of random chemistry.
In this study, we first examined spontaneous ligation and recombination reactions in pools of short random RNAs and found that they can be detected more quickly than previously demonstrated. We observed the enrichment of RNA families with common motifs in multiple RNA pools. Subsequent analyses of the most enriched products and their variants led us to find a short (20 nt) RNA oligomer that can self-reproduce via template-directed ligation of two 10 nt substrates. The RNA contains a 2′–5′ phosphodiester bond, a linkage usually generated during non-enzymatic RNA synthesis.36–38 Partly due to the non-canonical linkage, the RNA circumvented its dimerization and displayed a potential for exponential reproduction in an isothermal environment, although the restricted formation of an active complex with the substrates limited the amplification. Its autocatalytic properties and structures are somewhat similar to those of the previously developed self-reproducing ligase ribozyme.13 These results demonstrate the first example of minimal RNA self-reproduction independent of a ribozyme and also help understand the dynamics of primordial, random RNA pools.
Results and discussion
Incubation of short random RNA pools
We investigated reactions in fully random 20 nt RNA (N20), which was previously shown to undergo both ligation and recombination if pre-activated with >p.35 We prepared N20 and N20>p pools containing ∼3 × 1014 molecules to cover all possible ∼1012 sequences of 20 nt with redundancy (∼300 copies). Previous studies detected ligation and recombination in 16–20 nt random RNA pools (5–100 μM) only after incubation for months or longer times in ice.31,35 However, we found that, in the presence of high concentration (100 mM) of MgCl2, which promotes >p-mediated template-directed ligation and recombination,30,31 both N20 and N20>p pools (50 μM) generated detectable >20 nt products after just a 2 day incubation, as visualized by denaturing polyacrylamide gel electrophoresis (PAGE) (Fig. 1A). Note that degraded fragments in the initial pools may also have contributed to the reactions.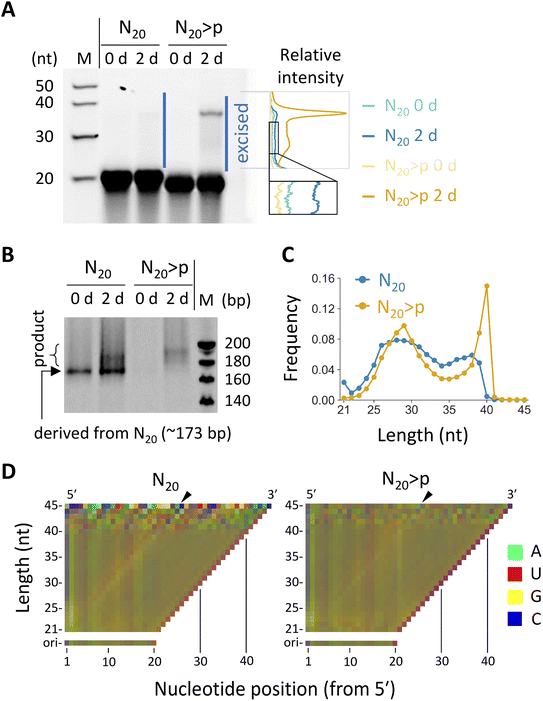 | ||
| Fig. 1 Reactivity of N20 and N20>p RNA pools. (A) Incubation of 50 μM N20 or N20>p in 100 mM MgCl2 at 22 °C and pH 8.0 for 2 days, analyzed by 20% denaturing PAGE. RNA products (ca. 21–45 nt) were excised and subjected to RT-PCR. Note that RNA with >p migrates slightly faster. Relative band intensities of the indicated region are shown on the right. The dependence of band intensity on RNA length was not calibrated. (B) RT-PCR products analyzed by 15% native PAGE. Different PCR cycles were applied to the N20 and N20>p samples. (C) Length distribution of 21–45 nt products detected multiple times in the HTS analyses. (D) Nucleotide compositions of the products with each length for N20 (left) and N20>p (right) pools. The frequency of each nucleotide was represented by a linear combination of RGB values as in the previous study.35 The compositions of the original 20 nt pools were displayed for comparison. Arrowheads indicate putative ligation junctions. | ||
To examine sequences enriched in the random RNA pools, we excised the elongated products (ca. 21–45 nt) in both N20 and N20>p pools from a denaturing polyacrylamide gel and subjected them to RT-PCR and high-throughput sequencing (HTS). The RT-PCR was performed using the SMARTer technology, via poly-A tailing and following template switching during reverse transcription. We detected PCR products for both N20 and N20>p pools only if they were pre-incubated for two days, confirming recombination and ligation during the incubation (Fig. 1B). From the HTS data, we analyzed 374![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357 and 412
357 and 412![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 461 reads of 21–45 nt products that were detected at least twice for the N20 and N20>p pools, respectively. The majority of the products derived from the N20 pool were 24–39 nt (95%) with a sharp drop-off above 39 nt (Fig. 1C), indicating that they were generated primarily by recombination, because a single recombination of two 20 nt RNAs could lead to a 21–39 nt product. On the other hand, the products in the N20>p pool were predominantly 24–40 nt (98%) with a sharp peak at 40 nt (Fig. 1C), suggesting that both recombination and ligation operated in the pool. It should be noted that recombination could occur either directly or indirectly via ligation on >p of a hydrolyzed RNA.31 The nucleotide compositions in the <40 nt products of the random RNA pools displayed a slight enrichment in G at the both sides of a putative ligation junction between a cleaved RNA>p (<20 nt) and a 20-mer (Fig. 1D and S1,† in the direction indicated by black arrowheads), which was more evident in the N20 pool than in the N20>p pool. The results contrast with the previous studies that incubated random RNAs in ice and without MgCl2, where cytosine and/or uracil were particularly enriched as putative phosphate donors.31,35 The predicted secondary structures of the products tended to be more stable than those of random sequences of the same sizes and nucleotide compositions (Fig. S2†), consistent with a previous study.35
461 reads of 21–45 nt products that were detected at least twice for the N20 and N20>p pools, respectively. The majority of the products derived from the N20 pool were 24–39 nt (95%) with a sharp drop-off above 39 nt (Fig. 1C), indicating that they were generated primarily by recombination, because a single recombination of two 20 nt RNAs could lead to a 21–39 nt product. On the other hand, the products in the N20>p pool were predominantly 24–40 nt (98%) with a sharp peak at 40 nt (Fig. 1C), suggesting that both recombination and ligation operated in the pool. It should be noted that recombination could occur either directly or indirectly via ligation on >p of a hydrolyzed RNA.31 The nucleotide compositions in the <40 nt products of the random RNA pools displayed a slight enrichment in G at the both sides of a putative ligation junction between a cleaved RNA>p (<20 nt) and a 20-mer (Fig. 1D and S1,† in the direction indicated by black arrowheads), which was more evident in the N20 pool than in the N20>p pool. The results contrast with the previous studies that incubated random RNAs in ice and without MgCl2, where cytosine and/or uracil were particularly enriched as putative phosphate donors.31,35 The predicted secondary structures of the products tended to be more stable than those of random sequences of the same sizes and nucleotide compositions (Fig. S2†), consistent with a previous study.35
Identification of enriched RNA families
If the RNA products were synthesized by previously identified ligation or recombination mechanisms,30,31,34 20 nt sequences in the original pools should remain intact at the 5′ or 3′ end of the products, consistent with the enrichment of specific nucleotides at the putative junctions (Fig. 1D and S1†). Thus, we grouped the most abundant 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 products from each pool of N20 and N20>p into families based on sequence similarity around the 5′ or 3′ terminus. Products differ from the most abundant sequence of each family by seven or fewer edits for the 21 nucleotides at each end. When grouping N20-derived products by their 3′ ends, we observed a highly enriched family, named N20-f1, that comprised ∼1.5% of all analyzed products. This family was 2.4-fold more abundant than the second most enriched family (Fig. 2A). The N20-f1 family consists of 93 sequences that were well aligned at the 3′ end (Fig. 2B). More than 80% of them contained common nucleotides at positions 1, 2, 4, 13–15, 17, and 19–25 from the 3′ end (indicated by the black lines), while nucleotides at other positions were relatively random. Likewise, when grouping the N20>p-derived products by their 3′ ends, we found an enriched family with a similar set of sequences, N20>p-f1 (Fig. 2B). Although N20>p-f1 was the most abundant in the pool, the frequency was comparable to other low-rank families and comprised ∼0.6% of the analyzed products (Fig. 2A). 17 sequences were commonly found in both N20-f1 (18%) and N20>p-f1 (43%). The enrichment of specific families was less clear when grouped by the 5′ end (Fig. 2A). Other high-ranked families are described in Fig. S4;† some of them have similar nucleotide compositions to N20-f1 and N20>p-f1. We also note that in the same analyses using the synthetic sequences (Fig. S2†), unsurprisingly, the most enriched families represented only ∼0.2% for each set, and their components did not align at all.
000 products from each pool of N20 and N20>p into families based on sequence similarity around the 5′ or 3′ terminus. Products differ from the most abundant sequence of each family by seven or fewer edits for the 21 nucleotides at each end. When grouping N20-derived products by their 3′ ends, we observed a highly enriched family, named N20-f1, that comprised ∼1.5% of all analyzed products. This family was 2.4-fold more abundant than the second most enriched family (Fig. 2A). The N20-f1 family consists of 93 sequences that were well aligned at the 3′ end (Fig. 2B). More than 80% of them contained common nucleotides at positions 1, 2, 4, 13–15, 17, and 19–25 from the 3′ end (indicated by the black lines), while nucleotides at other positions were relatively random. Likewise, when grouping the N20>p-derived products by their 3′ ends, we found an enriched family with a similar set of sequences, N20>p-f1 (Fig. 2B). Although N20>p-f1 was the most abundant in the pool, the frequency was comparable to other low-rank families and comprised ∼0.6% of the analyzed products (Fig. 2A). 17 sequences were commonly found in both N20-f1 (18%) and N20>p-f1 (43%). The enrichment of specific families was less clear when grouped by the 5′ end (Fig. 2A). Other high-ranked families are described in Fig. S4;† some of them have similar nucleotide compositions to N20-f1 and N20>p-f1. We also note that in the same analyses using the synthetic sequences (Fig. S2†), unsurprisingly, the most enriched families represented only ∼0.2% for each set, and their components did not align at all.
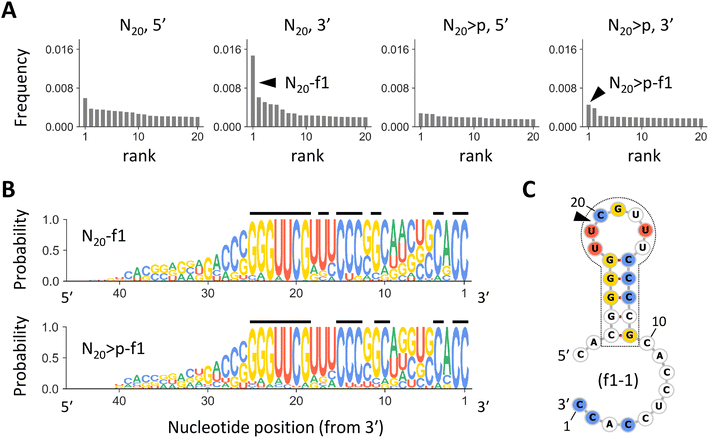 | ||
| Fig. 2 The most enriched RNA families. (A) Frequencies of the most enriched 20 RNA families in analyzed N20 or N20>p-derived products, sorted in descending order. Each panel represents families constructed based on sequence similarity around 5′ or 3′ terminus for N20 or N20>p. The arrowheads indicate N20-f1 and N20>p-f1. (B) Nucleotide compositions in sequences of N20-f1 (top) and N20>p-f1 (bottom). The sequence logos show the probability of each nucleotide at each position, calculated by ignoring the redundancy of each sequence (Fig. S3†). The black lines above indicate sites where a specific nucleotide is detected with a probability of >0.8. (C) A predicted secondary structure of f1-1, the most abundant sequence in N20-f1. Nucleotides detected with a probability of >0.8 are colored according to panel B. The commonly observed stem-loop structure is enclosed in the dotted line. The arrowhead indicates the putative recombination junction. | ||
RNA sequences in N20-f1 and N20>p-f1 displayed a common stem-loop structure at positions 11–27 nucleotides from the 3′ end, with five consecutive base pairs and a seven-base loop (Fig. 2C). The stem-loop region contained the majority of the commonly observed nucleotides, as represented in the most dominant sequence in N20-f1, named f1-1. Secondary structural prediction showed the same stem-loop structure at the same positions in 68% and 52% of ≥27 nt sequences in N20-f1 and N20>p-f1, respectively. In addition, only 7% of the RNAs in either family could form more than five base pairs in the stem region, underscoring the dominance of the specific stem-loop structure.
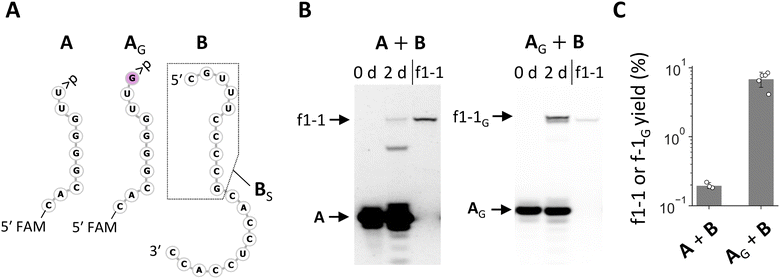
The enrichment of RNA families with shared nucleotides and structures in the random RNA pools encouraged us to investigate how these sequences could have been synthesized. As they were observed in both N20 and N20>p pools, they should form via recombination. The conserved 3′ region in the RNA of varying lengths, in conjunction with the current understanding of recombination mechanisms, suggests a two-step α/α′ recombination, wherein hydrolysis forms >p at the 3′ end of one RNA, followed by ligation of the 5′-OH of another RNA to the >p.31 If the ligating RNA is 20 nt long, as in the original pools, the probable recombination junction was between the oft-observed C and U at positions 20 and 21 from the 3′ end. We first tested whether f1-1 (29 nt) can form through this mechanism by splitting f1-1 into the first 9 nt attached with >p (i.e., fragment A) and the remaining 20 nt (i.e., fragment B) (Fig. 3A) so they could undergo ligation, the second step of α/α′ recombination. In a 2 day incubation of A and B, we detected f1-1 with ∼0.2% yield (Fig. 3B and C). It is important to note that this reaction may not strictly reflect what happened in the original random RNA pools because other RNAs could have been involved.
We also tested recombination directly by attaching 11 nt random nucleotides to A (AN11). Incubation of AN11 with B did generate a distinguishable product whose length is similar to—but slightly longer than—f1-1 (Fig. S5A and S5B†). Sequence analysis of the product revealed that it was predominantly f1-1 with a G inserted between positions 20 and 21, named f1-1G (Fig. S5C†). We confirmed that the addition of a G at the 3′ end of A (AG) (Fig. 3A) significantly enhanced its ligation with B (Fig. 3B and C). We also examined the effect of other nucleotides A, U, or C at the same position (AA, AU, or AC) for ligation with B. The fragment AA exhibited improved ligation but less efficiently so than AG, whereas AU and AC did not show enhanced ligation (Fig. S6†). These variant RNAs were not detected in the products derived from the N20 and N20>p pools, despite only a single nucleotide difference from f1-1 and high capacity for synthesis, highlighting the difficulty of understanding reactions in random RNA mixtures based on an examination of only a small number of isolated RNAs.
Discovery of a minimal self-reproducing RNA
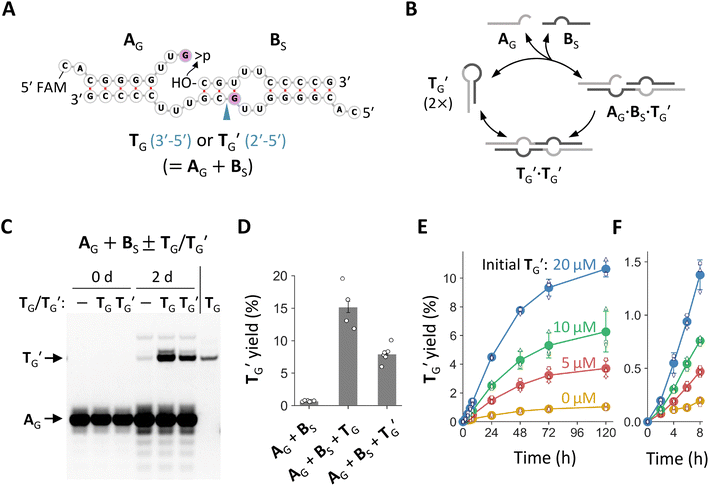
We noticed that the common stem-loop structure in N20-f1 and N20>p-f1 (Fig. 2C) and their variants with the G insertion could catalyze the ligation between the 5′ and 3′ regions of themselves as a template, i.e., self-reproduction (Fig. S7A,†4A, and B). In particular, nucleotide pairings around the ligation junctions upon ternary complex formation could enhance the ligation by positioning the termini of the two RNA substrates more proximally. We tested this hypothesis using the stem-loop regions of f1-1 and its variant with G at the ligation site, named T and TG, respectively (Fig. S7A† and 4A). We incubated 20 μM each of the 5′ regions with >p (A or AG) and the 3′ region (BS, the first 10 nt of B) for 2 days in the absence or presence of 20 μM T or TG. Whereas T improved ligation between A and BS only slightly (∼1.4 fold) (Fig. S7B and S7C†), TG enhanced ligation between AG and BS far more noticeably (∼21-fold) (Fig. 4C and D), demonstrating possible self-reproduction. We also tested the same reaction using AA, AU, and AC and corresponding templates (TA, TU, and TC, respectively) instead of AG and TG (Fig. S8†). Although TA and TU catalyzed ligations between AA or AU and BS, their spontaneous ligations relative to the template-directed reactions were more productive than that of AG and BS. The fragment TC did not affect the ligation between AC and BS.Ligation between >p of AG and BS could generate two possible phosphodiester bonds, either 3′–5′ or 2′–5′ linkages (Fig. 4A). Using ribonuclease (RNase) T1, which selectively cleaves G3′-p-5′N linkages of unpaired nucleotides, we determined that the ligation catalyzed by TG primarily formed a 2′–5′ linkage (Fig. S9†). Next, we prepared TG containing a 2′–5′ linkage at the ligation junction and named it TG′. We confirmed that TG′ catalyzed the same ligation reaction to generate more of itself (Fig. 4C and S9†), demonstrating true self-reproduction (Fig. 4B), although the extent of catalysis was approximately half than that of TG (Fig. 4D). Whereas previous studies found that RNA containing a fraction of 2′–5′ linkages can assist non-enzymatic RNA polymerization39 and retain functions as aptamers or ribozymes,40 our study further showed that such RNA can also self-reproduce. A time course experiment revealed the gradual appearance of TG′, with the reaction slowing after a 2 day (48 h) incubation (Fig. 4E, F and S10†). The yield of TG′ was positively increased with the concentration of initial TG′, demonstrating its autocatalytic ability. The ligation between >p of AG and BS was confirmed by control reactions performed in the absence of >p or BS, which showed negligible TG′ reproduction (Fig. S11†). We also found that the self-reproduction of TG′ was substantially enhanced at high concentration of Mg2+ (100 mM MgCl2) and temperatures around 22 °C (Fig. S12†), the condition used for incubating the original random RNA pools (Fig. 1A).
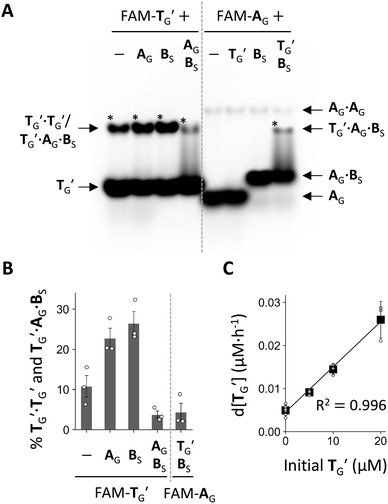
Next, we examined the formation of higher-order complexes among AG, BS, and TG′ by native PAGE after co-incubating one, two, or three of these RNAs containing fluorescently labeled TG′ (FAM-TG) or AG (FAM-AG) for 6 h (Fig. 5A). In this experiment, AG contained a monophosphate (-p) instead of >p at the 3′ end to preclude ligation to BS (Fig. S11†). When incubating only TG′, we found that the majority of TG′ existed as a TG′ monomer, with only a fraction (∼11%) forming a TG′·TG′ dimer (Fig. 5B). A TG′·TG′ dimer is presumably a simple self-complementary template dimer (Fig. S14†), but two TG′ molecules may also interact by forming a kissing loop. The prevention of the formation of a TG′·TG′ dimer was partly due to the 2′–5′ linkage, which significantly reduced the dimerization of TG′ (Fig. S13†), consistent with previous studies showing the diminished thermal stability of RNA duplexes in the presence of 2′–5′ linkages.40,41 The amount of TG′·TG′ increased to 23–27% in the presence of either AG or BS. However, in the presence of both AG and BS, the total amount of the TG′·TG′ dimer and a TG′·AG·BS ternary complex decreased to ∼3.8%. When incubating the three RNA molecules with FAM-AG, we detected the formation of a comparable amount of the TG′·AG·BS complex. In addition, we found that the majority (∼80%) of AG was bound to BS, and thus most of the substrates were not freely available, which could explain the low percentage of the TG′·AG·BS complex formation and the limited self-reproduction of TG (Fig. 4E).
The high availability of TG′ as a monomer implies its potential to undergo non-linear amplification by circumventing the strong association of two self-complementary TG′ molecules that form after ligation of AG and BS (Fig. 4B). A common way of examining such a possibility for a template (or an autocatalyst) is to fit the initial rate of its own production to the model of self-reproduction:13,14,23,24,42
The RNA molecule TG′ shares many similarities with the previously engineered 61 nt self-reproducing ligase ribozyme,13 although they catalyzed different ligation chemistries (Fig. S14†). The ribozyme catalyzes the attack of the 3′-OH of an RNA substrate on a 5′ triphosphate of another substrate in a template-directed manner and generates a ligated product identical to the ribozyme. Its self-reproduction was limited because of the strong association of the two substrates, as is also observed in TG′ (Fig. 5A). Nevertheless, both systems exhibited high apparent autocatalytic reaction order (∼1) in an isothermal environment as a consequence of the weak self-binding of the templates, compared to other nucleotide-based template-directed self-reproduction systems that showed an order of ∼0.5.23–25 This could be partly attributed to the intramolecular structural formation of a template, G:U wobble pairs that can facilitate template-directed ligation while supporting dissociation of a duplex,43 and multiple thermodynamically unfavorable bulges in a dimer,44 all of which are commonly observed in both TG′ and the ligase ribozyme (Fig. S14†).
The limited self-reproduction of TG′ resulted from multiple factors. First, the 2′–5′ linkage, while reducing the dimerization of TG′, decreased the ligation efficiency (Fig. 4D). Second, TG′ did not efficiently form an active complex with the substrates AG and BS because most of the two substrates bound to each other and were not freely available (Fig. 5A). These limitations may be overcome if strong chemical activation is adopted instead of >p or in environments that periodically experience low pH, high temperatures, or low MgCl2 concentrations, which destabilize RNA–RNA interactions (e.g., the association of substrates).45–47 Alternatively, as demonstrated for a self-reproducing ligase ribozyme,48 directed evolution with TG′ as the parent RNA may also identify highly efficient reproduction of oligonucleotides in a constant environment. It was shown that only a slight difference, including two critical mutations, was sufficient to convert the original ligase ribozyme13 (Fig. S14†) into a continuously self-reproducible RNA.49 Thus, it is conceivable that there may be a short RNA oligonucleotide capable of unlimited self-reproduction, in a sequence space accessible from TG′ by natural selection.
Conclusions
We demonstrated a form of minimal RNA self-reproduction driven by prebiotically plausible chemistry, providing a potential missing link between abiotic oligomers and the eventual emergence of a genetic system. The 20 nt RNA, TG′, accelerated >p-dependent ligation between two 10 nt substrates, AG and BS, as a template for generating identical TG′ molecules (Fig. 4C and S9†). Such self-reproduction of RNA could have occurred in the RNA World because RNA of these lengths can be generated non-enzymatically,18,19 and >p can also be readily formed by spontaneous RNA hydrolysis or with prebiotically plausible reagents.32,33 Although >p is eventually hydrolyzed to monophosphates, in situ reactivation back to >p33 could extend the self-reproduction of TG′, which is currently limited (Fig. 4E). The self-reproduction was also supported by a 2′–5′ phosphodiester bond, which is thought to have been prevalent in primordial RNA pools as generated in typical non-enzymatic RNA synthesis.36–38 Short RNA molecules capable of self-reproduction by template-directed ligation, as shown in the present study, has been proposed as the earliest stage toward the evolution of complex replication ribozymes.21,22 Our results complement this view and help delineate the development of RNA-based genetic systems during the origins of life.Our results also give insights into the dynamics of short random RNA mixtures. From completely random pool of 20-mers, we identified a discrete class of related, enriched sequences of which f1-1 appeared to be a canonical representative. The fragment TG′ is a truncated version of f1-1G, a single-mutation variant of f1-1. Both f1-1G and f1-1 were accessible products in both N20 and N20>p pools explored in the present study. However, while f1-1 was highly enriched in both random RNA pools along with many related sequences (e.g., N20-f1 and N20>p-f1), f1-1G was undetected even at a low frequency. On the other hand, biochemical analyses revealed the superiority of f1-1G to f1-1 for its formation through simple ligation of two substrate fragments (Fig. 3B and C). This discrepancy may imply the involvement of other RNA species for the synthesis of f1-1 in the random RNA pools. In the chaos of primordial soup, it is without question that a complex ecology of chemical reactions must have given rise to enriched species sets.50,51 A previous study also reported the inefficient synthesis of some products isolated from random RNA pools.35 Altogether, our results highlight the difficulty of inferring dominant reactions in random RNA mixtures from the analyses of isolated sequences. Nevertheless, the information obtained from examining the random RNA products was valuable in the discovery of the minimal self-reproducing RNA, which exhibited its highest activity in the original environment where the random RNA pools were exposed (Fig. S12†). Future experiments exploring the synthesis of f1-1, f1-1G, or TG′ in combination with random RNA mixtures would give more insights into the likelihood of the emergence of self-reproduction in a primordial RNA soup.
Data availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.Author contributions
R. M. and N. I. designed the project. R. M. performed experiments, analyzed data, and wrote the paper with comments from N. I.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Niles Lehman for helpful discussion and comments on the manuscript. We also thank Dieter Braun and Bryce Clifton for useful discussion. This research was supported by JSPS KAKENHI (21H05867 to R. M.), JST PRESTO (JPMJPR19KA to R. M.), and JST FOREST (JPMJFR2252 to R. M.).Notes and references
- W. Gilbert, Nature, 1986, 319, 618 CrossRef.
- G. F. Joyce, Nature, 2002, 418, 214–221 CrossRef CAS PubMed.
- P. B. Moore and T. A. Steitz, Cold Spring Harbor Perspect. Biol., 2011, 3, a003780 Search PubMed.
- P. G. Higgs and N. Lehman, Nat. Rev. Genet., 2015, 16, 7–17 CrossRef CAS PubMed.
- G. F. Joyce and J. W. Szostak, Cold Spring Harbor Perspect. Biol., 2018, 10, a034801 CrossRef PubMed.
- P. Pavlinova, C. N. Lambert, C. Malaterre and P. Nghe, FEBS Lett., 2022, 597, 344–379 CrossRef PubMed.
- L. Zhou, S. C. Kim, K. H. Ho, D. K. O. Flaherty, C. Giurgiu, T. H. Wright and J. W. Szostak, Elife, 2019, 8, e51888 CrossRef CAS PubMed.
- J. Attwater, A. Wochner and P. Holliger, Nat. Chem., 2013, 5, 1011–1018 CrossRef CAS PubMed.
- D. P. Horning and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 9786–9791 CrossRef CAS PubMed.
- R. Cojocaru and P. J. Unrau, Science, 2021, 371, 1225–1232 CrossRef CAS PubMed.
- P. Adamski, M. Eleveld, A. Sood, Á. Kun, A. Szilágyi, T. Czárán, E. Szathmáry and S. Otto, Nat. Rev. Chem., 2020, 4, 386–403 CrossRef PubMed.
- S. Ameta, Y. J. Matsubara, N. Chakraborty, S. Krishna and S. Thutupalli, Life, 2021, 11, 308 CrossRef CAS PubMed.
- N. Paul and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12733–12740 CrossRef CAS PubMed.
- E. J. Hayden, G. Von Kiedrowski and N. Lehman, Angew. Chem., Int. Ed., 2008, 47, 8424–8428 CrossRef PubMed.
- T. A. Lincoln and G. F. Joyce, Science, 2009, 323, 1229–1232 CrossRef CAS PubMed.
- N. Vaidya, M. L. Manapat, I. A. Chen, R. Xulvi-Brunet, E. J. Hayden and N. Lehman, Nature, 2012, 491, 72–77 CrossRef CAS PubMed.
- J. Rogers and G. F. Joyce, RNA, 2001, 7, 395–404 CrossRef CAS PubMed.
- P. A. Monnard, A. Kanavarioti and D. W. Deamer, J. Am. Chem. Soc., 2003, 125, 13734–13740 CrossRef CAS PubMed.
- N. Prywes, J. C. Blain, F. Del Frate and J. W. Szostak, Elife, 2016, 5, e17756 CrossRef PubMed.
- H. Lin, E. I. Jiménez, J. T. Arriola, U. F. Müller and R. Krishnamurthy, Angew. Chem., Int. Ed., 2022, 134, e202113625 Search PubMed.
- K. D. James and A. D. Ellington, Origins Life Evol. Biospheres, 1999, 29, 375–390 CrossRef CAS PubMed.
- M. Levy and A. D. Ellington, Nat. Struct. Biol., 2001, 8, 580–582 CrossRef CAS PubMed.
- G. Von Kiedrowski, Angew. Chem., Int. Ed., 1986, 25, 932–935 CrossRef.
- W. S. Zielinski and L. E. Orgel, Nature, 1987, 327, 346–347 CrossRef CAS PubMed.
- G. Von Kiedrowski, B. Wlotzka, J. Helbing, M. Matzen and S. Jordan, Angew. Chem., Int. Ed., 1991, 30, 423–426 CrossRef.
- E. Edeleva, A. Salditt, J. Stamp, P. Schwintek, J. Boekhoven and D. Braun, Chem. Sci., 2019, 10, 5807–5814 RSC.
- Y. Li and R. R. Breaker, J. Am. Chem. Soc., 1999, 121, 5364–5372 CrossRef CAS.
- K. Le Vay, E. Salibi, E. Y. Song and H. Mutschler, Chem. – Asian J., 2020, 15, 214–230 CrossRef CAS PubMed.
- R. Hanna and J. A. Doudna, Curr. Opin. Chem. Biol., 2000, 4, 166–170 CrossRef CAS PubMed.
- A. V. Lutay, E. L. Chernolovskaya, M. A. Zenkova and V. V. Vlassov, Biogeosciences, 2006, 3, 243–249 CrossRef CAS.
- B. A. Smail, B. E. Clifton, R. Mizuuchi and N. Lehman, RNA, 2019, 25, 453–464 CrossRef CAS PubMed.
- C. Gibard, S. Bhowmik, M. Karki, E. K. Kim and R. Krishnamurthy, Nat. Chem., 2018, 10, 212–217 CrossRef CAS PubMed.
- E. Y. Song, E. I. Jiménez, H. Lin, K. Le Vay, R. Krishnamurthy and H. Mutschler, Angew. Chem., Int. Ed., 2021, 60, 2952–2957 CrossRef CAS PubMed.
- A. V. Lutay, M. A. Zenkova and V. V. Vlassov, Chem. Biodiversity, 2007, 4, 762–767 CrossRef CAS PubMed.
- H. Mutschler, A. I. Taylor, A. Lightowlers, G. Houlihan, M. Abramov, P. Herdewijn and P. Holliger, Elife, 2018, 7, e43022 CrossRef PubMed.
- L. E. Orgel, J. Theor. Biol., 1986, 123, 127–149 CrossRef CAS PubMed.
- J. P. Ferris and G. Ertem, Science, 1992, 257, 1387–1389 CrossRef CAS PubMed.
- J. W. Szostak, J. Syst. Chem., 2012, 3, 2 CrossRef CAS.
- T. P. Prakash, C. Roberts and C. Switzer, Angew. Chem., Int. Ed., 1997, 36, 1522–1523 CrossRef CAS.
- A. E. Engelhart, M. W. Powner and J. W. Szostak, Nat. Chem., 2013, 5, 390–394 CrossRef CAS PubMed.
- J. Sheng, L. Li, A. E. Engelhart, J. Gan, J. Wang and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3050–3055 CrossRef CAS PubMed.
- G. von Kiedrowski, in Bioorganic chemistry frontiers, ed. H. Dugas and F. P. Schmidtchen, Springer, Berlin, Heidelberg, 1993, pp. 113–146 Search PubMed.
- L. Zhou, D. K. O'Flaherty and J. W. Szostak, J. Am. Chem. Soc., 2020, 142, 15961–15965 CrossRef CAS PubMed.
- D. H. Mathews, J. Sabina, M. Zuker and D. H. Turner, J. Mol. Biol., 1999, 288, 911–940 CrossRef CAS PubMed.
- A. Mariani, C. Bonfio, C. M. Johnson and J. D. Sutherland, Biochemistry, 2018, 57, 6382–6386 CrossRef CAS PubMed.
- A. Lozoya-Colinas, B. E. Clifton, M. A. Grover and N. V. Hud, ChemBioChem, 2022, 23, e202100495 CrossRef CAS PubMed.
- A. Salditt, L. Karr, E. Salibi, K. Le Vay, D. Braun and H. Mutschler, Nat. Commun., 2023, 14, 1495 CrossRef CAS PubMed.
- M. P. Robertson and G. F. Joyce, Chem. Biol., 2014, 21, 238–245 CrossRef CAS PubMed.
- C. Olea, D. P. Horning and G. F. Joyce, J. Am. Chem. Soc., 2012, 134, 8050–8053 CrossRef CAS PubMed.
- S. A. Kauffman, The origins of order, Oxford University Press, 1993 Search PubMed.
- R. Mizuuchi and N. Lehman, Life, 2019, 9, 20 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01940c |
| This journal is © The Royal Society of Chemistry 2023 |