Open Access Article
Open Access ArticleTranscription between human-readable synthetic descriptions and machine-executable instructions: an application of the latest pre-training technology
Zheni
Zeng†
 a,
Yi-Chen
Nie†
b,
Ning
Ding†
a,
Qian-Jun
Ding
b,
Wei-Ting
Ye
b,
Cheng
Yang
c,
Maosong
Sun
a,
Weinan
E
d,
Rong
Zhu
*b and
Zhiyuan
Liu
a,
Yi-Chen
Nie†
b,
Ning
Ding†
a,
Qian-Jun
Ding
b,
Wei-Ting
Ye
b,
Cheng
Yang
c,
Maosong
Sun
a,
Weinan
E
d,
Rong
Zhu
*b and
Zhiyuan
Liu
 *a
*a
aDepartment of Computer Science and Technology, Tsinghua University, Beijing, China. E-mail: liuzy@tsinghua.edu.cn
bCollege of Chemistry and Molecular Engineering, Peking University, Beijing, China. E-mail: rongzhu@pku.edu.cn
cSchool of Computer Science, Beijing University of Posts and Telecommunications, Beijing, China
dCenter for Machine Learning Research and School of Mathematical Sciences, Peking University AI for Science Institute, Beijing, China
First published on 24th August 2023
Abstract
AI has been widely applied in scientific scenarios, such as robots performing chemical synthetic actions to free researchers from monotonous experimental procedures. However, there exists a gap between human-readable natural language descriptions and machine-executable instructions, of which the former are typically in numerous chemical articles, and the latter are currently compiled manually by experts. We apply the latest technology of pre-trained models and achieve automatic transcription between descriptions and instructions. We design a concise and comprehensive schema of instructions and construct an open-source human-annotated dataset consisting of 3950 description–instruction pairs, with 9.2 operations in each instruction on average. We further propose knowledgeable pre-trained transcription models enhanced by multi-grained chemical knowledge. The performance of recent popular models and products showing great capability in automatic writing (e.g., ChatGPT) has also been explored. Experiments prove that our system improves the instruction compilation efficiency of researchers by at least 42%, and can generate fluent academic paragraphs of synthetic descriptions when given instructions, showing the great potential of pre-trained models in improving human productivity.
1 Introduction
AI in chemistry is an emerging interdisciplinary field that has achieved impressive results in various scenarios.1–3 Take chemical synthesis as an example; robotic synthesis systems have recently been developed to perform chemical synthetic actions following formatted instructions to free researchers from monotonous experimental procedures.4 Nevertheless, these systems are only capable of executing deterministic instructions manually compiled by human experts, while synthetic experimental procedures in the real world are typically described with natural language in numerous chemical articles. Due to the gap between machine-executable instructions and human-readable descriptions, it is exceedingly labor-intensive to repeatedly record the same procedures in two sets of language systems. This calls for the development of automatic transcription of synthetic procedures between descriptions in the human cognitive space and instructions in the machine operative space.Specifically regarding the benefits of transcription, there exists a large body of chemical literature describing experimental procedures for synthetic reactions. Once we have a description-to-instruction (D2I) transcription system, the vast amount of synthetic knowledge can be documented into the instruction library efficiently to enhance the robotic synthesis platforms. Correspondingly, discrete instruction options can be predicted and prompted easier than natural language descriptions. Once we have an instruction-to-description (I2D) transcription system, it is possible for chemical researchers to program instructions quickly instead of manually writing natural language descriptions in the literature.
However, transcription requires a high level of natural language intelligence and chemical knowledge. For instance, the flexibility and complexity of natural language stump the intuitive solutions with low generalizability, including natural language processing (NLP) modules for specific property extraction5 and manually designed rule mapping.6 Meanwhile, it is also challenging for general AI systems without chemical expertise to recognize the chemical terms, mine and complete implicit conditions, and master the grammar of instructions.
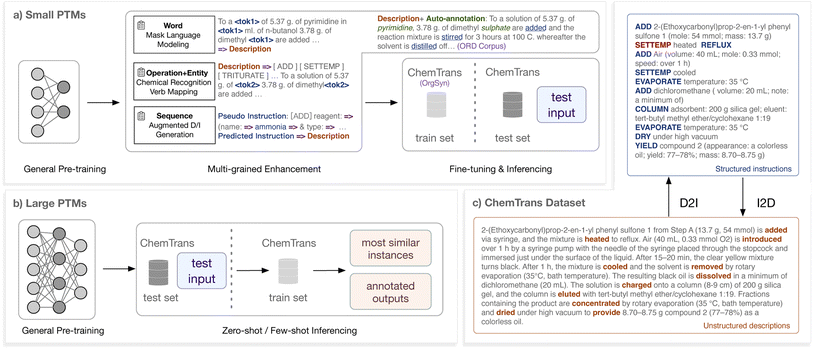
In response to the above challenges, we propose a knowledgeable transcription system equipped with pre-trained models (PTMs). There are three key elements to achieve this system: the task dataset, the PTM, and the knowledge enhancement.
1.1 Task dataset
To activate and evaluate the data-driven deep learning models better, a dataset for synthetic transcription has to be constructed. We first define a concise and complete instruction schema for chemical synthetic actions according to statistical information and expert knowledge, containing 16 types of operations with 18 types of corresponding hierarchical arguments, with which the experimental procedures can be clearly expressed and the target of transcription is perspicuous. Following this schema, we construct an open-source human-annotated dataset, ChemTrans, consisting of 3950 pieces of OrgSyn paragraphs and their formatted synthetic instructions, with 9.2 operations in each instruction on average.1.2 Pre-trained model
To understand and generate flexible and complex natural language, we apply the pre-trained language models as the backbone of our system. PTMs have already been proven to acquire versatile knowledge implicitly and scattered in the unlabeled data,7 and have achieved impressive performances in various tasks, such as the open-domain chatting with ChatGPT. In this paper, we explore two solutions: (1) the fine-tuned models based on T5,8 an encoder–decoder PTM; (2) the directly used large-scale models‡ represented by GPT-3.5. We prove the significance of applying PTMs in the synthetic transcription scenario.1.3 Knowledge enhancement
To enhance the system with rich knowledge as human experts when facing transcription tasks, we specifically designed several training tasks to teach the model multi-grained chemical domain knowledge. (1) Word-level: we conduct masked language modeling by randomly masking words in chemical articles and learning to predict them. (2) Entity-level: we train the model to recognize all chemical entities obtained with SciSpacy tools9 in the given articles. (3) Operation-level: we also enable the model to identify the trigger words or phrases of synthetic actions and accomplish verb-operation mapping constructed by manually crafted rules. (4) Sequence-level: we conduct decoder language modeling on the augmented pseudo instructions specially for the D2I model to learn unified instruction grammar. On the other hand, we apply the description–instruction pairs generated in D2I to the I2D model training to learn the language style of synthetic descriptions.We comprehensively evaluate the performance of our transcription system on the ChemTrans dataset. (1) For both D2I and I2D, our system has achieved an overall satisfying effect. In particular, the system can even achieve next-operation prediction without giving original descriptions. (2) Evaluation metrics, including the BLEU score10 and the newly defined SeqMatch score, unanimously verify the remarkable improvements that the PTMs and multi-grained knowledge enhancement bring. (3) The recently popular large PTMs (e.g., GPT-3.5) are proven to have the basic transcription capability with few or even no instances, while obtaining unsatisfactory performance on chemical details and reliability. This shows that large-scale PTMs have great potential and also challenges in scientific applications. (4) The case study and human annotator test show the practical usage of our model. Users can competently obtain machine-executable results (17+ times faster than human annotators) or human-readable descriptions and improve efficiency even if we require manual verification (42% faster).
We release our code and dataset to encourage more researchers to tap the potential of machine intelligence in chemistry. Besides, we provide the conversion results for over 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 chemical reactions to form a machine-executable instruction library that facilitates automatic chemical synthesis. In the long run, the transcription process is an essential part of the grand picture of fully automatic synthetic chemistry. Meanwhile, implementing this technique is expected to facilitate the standardization and structuralization of the raw data reporting experimental procedures, which is crucial for reproducing and optimizing chemical reactions.
000 chemical reactions to form a machine-executable instruction library that facilitates automatic chemical synthesis. In the long run, the transcription process is an essential part of the grand picture of fully automatic synthetic chemistry. Meanwhile, implementing this technique is expected to facilitate the standardization and structuralization of the raw data reporting experimental procedures, which is crucial for reproducing and optimizing chemical reactions.
2 Results and discussion
2.1 Task formalization and dataset
We define the mutual transcription between descriptions and instructions as mutual sequence-to-sequence tasks. For D2I transcription, the document-level natural language texts that describe multi-step chemical synthesis are fed as input, and synthetic instructions are expected to be formatted and transcribed from the given texts. These instructions can be easily disassembled into machine-executable programs. For I2D transcription, we exchange the input and output of D2I. To formalize the transcription tasks, we first design a schema of synthetic instructions and then construct a dataset facilitating the training process of deep learning systems. We adopt the supplementary information (SI) of Organic Syntheses (OrgSyn)§ as the source corpus to construct the schema and annotate the dataset since the SI paragraphs attached to the primary synthesis articles mention a considerable number of reactants and procedures.Specific to the operation selection, our principle is to choose the frequently appearing meta operations that have clear synthetic meanings (e.g. “move” describes a possible part of an action but has no synthetic meaning) and are indivisible (e.g. “neutralize” can be replaced by other operations such as adding acid/base). We refer to statistical information to guarantee coverage and invite experts to ensure the rigor of the schema. Details are shown in the Methods section.
2.2 Pre-trained models
We conduct experimental comparisons between different model architectures and training settings to verify the effectiveness of PTMs. There are overall two types of PTMs we try: (1) Small PTMs. For models such as T5,8 a text-to-text transformer-based PTM with the encoder–decoder architecture, they can be applied to downstream tasks by transfer learning. In this paper, we further train the small PTMs with knowledge enhancement which is introduced in the next subsection, and then fine-tune the models on the ChemTrans dataset. (2) Large PTMs. For models such as GPT-3.5 (chat completion mode),11 they are proven to be good few-shot learners and can finish downstream tasks given a few instances. Meanwhile, the tuning for large PTMs is time-consuming. In this paper, we directly provide the large PTM instances and input text waiting for transcription.As shown in Table 1, we study various types of models for transcription:
| Model | D2I | I2D | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SM-A | SM-O | BLEU-2 | BLEU-4 | EM | Distinct-4 | ROUGE-4 | BLEU-2 | BLEU-4 | |
| Transformer | 21.57 | 57.91 | 44.88 | 27.19 | 0 | 8.308 | 0.517 | 5.210 | 0.365 |
| Transformer+ | 22.43 | 58.45 | 44.97 | 27.97 | 0 | 18.36 | 1.225 | 8.168 | 0.933 |
| GPT-3.5 | 0.441 | 4.471 | 7.520 | 0.931 | 0 | 67.99 | 5.261 | 10.83 | 2.920 |
| 3-shot | 37.53 | 66.96 | 59.69 | 44.91 | 4.937 | 56.51 | 13.39 | 20.41 | 8.816 |
| 3-shot* | 45.11 | 70.45 | 62.84 | 50.16 | 6.709 | 59.26 | 15.06 | 23.19 | 10.69 |
| GPT-3.5-chat | 2.708 | 35.49 | 14.17 | 2.718 | 0 | 74.62 | 3.016 | 6.423 | 1.982 |
| 3-shot | 25.75 | 57.99 | 49.25 | 31.92 | 0.719 | 70.70 | 8.619 | 15.73 | 5.486 |
| 3-shot* | 34.88 | 62.28 | 55.57 | 40.45 | 3.249 | 69.59 | 10.33 | 17.96 | 6.913 |
| Ours-base | 65.85 | 84.69 | 74.36 | 65.53 | 18.31 | 56.87 | 20.27 | 27.98 | 15.29 |
| w/o seq | 65.40 | 84.36 | 74.33 | 65.39 | 18.99 | 54.80 | 18.55 | 26.36 | 13.81 |
| w/o o+e | 65.31 | 84.14 | 74.23 | 65.37 | 19.92 | 57.09 | 20.62 | 27.87 | 15.27 |
| w/o word | 65.14 | 83.80 | 74.63 | 65.78 | 18.82 | 53.24 | 20.88 | 27.43 | 15.28 |
| w/o multi (T5) | 64.13 | 83.10 | 74.05 | 65.03 | 18.65 | 54.78 | 18.33 | 26.02 | 13.50 |
| Ours-large | 67.12 | 85.41 | 75.89 | 67.33 | 22.36 | 57.17 | 21.82 | 29.54 | 16.55 |
| w/o seq | 66.29 | 84.45 | 75.32 | 66.83 | 19.33 | 55.58 | 20.19 | 27.66 | 15.02 |
| w/o o+e | 66.93 | 85.15 | 75.76 | 67.21 | 21.73 | 54.16 | 22.05 | 28.75 | 16.39 |
| w/o word | 65.82 | 84.17 | 74.60 | 66.11 | 19.37 | 55.63 | 22.47 | 28.97 | 16.51 |
| w/o multi (T5) | 65.50 | 83.85 | 74.36 | 65.83 | 19.24 | 56.20 | 18.92 | 26.27 | 13.96 |
Transformer refers to a simple model without any pre-training tasks, and with the same parameter scale as the previous framework for a similar transcription task.12 The Transformer+ variant has been involved in all grains of knowledgeable enhancement. Both of them are tuned and evaluated on ChemTrans.
D2IPTM and I2DPTM use the small PTM T5 as the backbone model. T5 (the w/o multi variant) is not involved in knowledgeable enhancement tasks. For the other variants, w/o seq is knowledge-enhanced on tasks except for the augmented generation; w/o o+e is enhanced on tasks except the verb mapping and chemical entity recognition; w/o word is only enhanced on the augmented generation. The original versions of D2IPTM and I2DPTM are fully involved in all the knowledgeable enhancement tasks.
GPT-3.5 (text-davinci-003 completion mode) and GPT-3.5-chat (gpt-3.5-turbo chat completion mode) are adopted as representative large PTMs, and all the versions are not specially tuned before being evaluated on ChemTrans. The original version displays the zero-shot performance; 3-shot is given 3 randomly picked training instances, and 3-shot* is given 3 training instances that have the highest similarities with the current testing instance. Notice that for D2I, the zero-shot model has never seen the grammar of instructions, therefore we provide the schema information in the task prefix. Details are shown in the Methods section.
It is noticed that in the table, boldfaced numbers indicate a significant advantage over the T5 results of more than 1 point in the one-sided t-test with p-value < 0.02.
2.3 Knowledge enhancement tasks
Inspired by human researchers transcribing with the assistance of multiple types of knowledge (as we have described in the introduction), we design the following four knowledge enhancement tasks that are associated with different granularities but are conducted simultaneously. Small PTMs are trained on these tasks after their general pre-training but before the task-oriented fine-tuning.2.4 Evaluation metrics
For I2D transcription, we adopt the BLEU score10 and ROUGE score13 for our evaluation, which are both traditional and popular metrics for sequence-to-sequence generation tasks such as machine translation. BLEU focuses on precision while ROUGE cares more about the recall of the generation. We also adopt the Distinct unigram14 to evaluate the diversity of the generated results.For D2I transcription, we propose a new solution which treats the predicted and the labeled operations as a sequence matching task, since it is hard to specify the location of the predicted item by rules and to count the accuracy. This is similar to protein sequence alignment, and one popular solution is dynamic programming, an efficient technique that helps solve problems with overlapping subproblems or optimal substructures. When conducting dynamic programming, the reward rules for matching and mismatching have to be specified. We try two different reward rules, among which SeqMatch-O focuses on the performance of operation prediction, and SeqMatch-A further takes the performance of argument prediction into consideration.
To be specific, SeqMatch-O is to give 1 point for the matched position and 0 for the missed/unmatched/redundant position, which only cares about the quality of operation classification. The sum of the reward scores is divided by the average length of the prediction sequence and ground-truth sequence. For instance, as shown in Fig. 3, the system predicts a 5-operation sequence while the answer is a 6-operation sequence, and the longest matching subsequence is with 4 operations. Therefore, the sum of reward scores is 4 and the average length of sequences is 5.5, and we get SeqMatch − O = 0.727. Further, SeqMatch-A is to change the reward point to the average normalized BLEU score of the corresponding arguments for the current operation. In the same instance as above, we calculate the BLEU score for the text of each argument, and also divide the sum by the average number of predicted arguments and the answer arguments. In this way, we get the reward score for the first position (ADD) as 0.444 to replace the original 1.
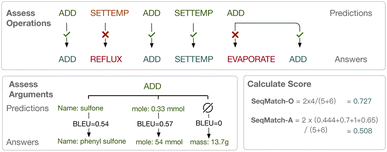 | ||
| Fig. 3 The new evaluation metrics for the task are proposed, assessing operations and arguments separately. | ||
In addition, we also adopt the BLEU score and ExactMatch score for D2I evaluation. BLEU score measures the proportion of n-grams (n words in a row) on matches. However, the n-grams in the generated instructions are not equally important, such as a bracket delimiter and an operation. We mainly consider the operations and the key arguments, therefore we believe that the SeqMatch score is more reasonable. As for ExactMatch, we provide the proportion of the perfectly predicted items. To avoid the influence of unimportant factors such as capitalization, spaces, reagent addition order, etc., we regard the results that have the difflib similarity with answers higher than 0.95 as perfect predictions.
2.5 Results analysis
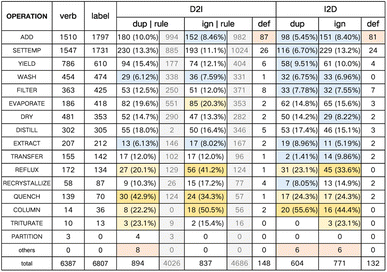
We analyze the experiment results from the following aspects: the overall performance of transcription tasks, the numerical result comparison between different settings, and the impact of the amount of training data. Lastly, we provide human evaluation on a case study.Specifically, Fig. 4 shows the operation-level statistics of D2IPTM-large and I2DPTM-large on the ChemTrans testing set. There are altogether 3 types of error: (1) duplicated: the predicted operation does not exist in the answer, and is skipped in dynamic programming matching; (2) ignored: the operation mentioned in the original paragraph has not been predicted successfully; (3) defective: the matched operation has a low quality of argument prediction, with the BLEU score lower than 0.6. To verify the challenge of ChemTrans and the necessity of adopting deep learning models, we also adopt the rule-based system for pre-training verb-operation mapping to automatically recognize operation sequences for D2I. Since this method cannot process the argument extraction, the defective error is therefore meaningless. The other 2 types of error are shown in the grey cubes of Fig. 4, which are much more than what D2IPTM-large makes.
Intuitively, the small PTM with knowledge enhancement can successfully read and transcribe most of the basic operations with any type of overall error rate lower than 15%. For the 6807 testing set operations in total, there are 894 duplicated, 837 ignored, and 148 defective error operations. The operations WASH and EXTRACT are processed quite well, which may benefit from the explicitness of the related descriptions. In contrast, several operations bring challenges to our system causing both many duplicated and ignored errors, including REFLUX, QUENCH, and COLUMN. The operation REFLUX has similar keywords and expressions to SETTEMP, and therefore is easy to be misjudged as other operations. Other operations with small sample sizes (fewer than 100 in the testing set) still have room for improvement. As for defective argument prediction, the operation ADD uses extracted reagent information as arguments, which are diverse in terms of expressions and may be hard to recognize in some cases.
For I2D transcription, large PTMs show their basic capability of describing synthetic instructions in fluent and professional natural language, while it is difficult to accurately generate the description style we need. Meanwhile, too powerful generation ability leads to over-imagination of large PTMs, and sometimes it deviates from the given instructions. In comparison, small PTMs are fine-tuned to strictly follow instructions during generation. Similarly, models without pre-training are not up to the task and cannot even generate readable paragraphs.
We also provide the operation-level analysis by reversely transcribing the I2D generated results into instructions and comparing with the initial inputs. From Fig. 4 we can see that the performance of the two tasks shows a very similar distribution. I2D makes obviously less duplicated error, of which one possible reason is that the predicted descriptions do not involve extraneous details discarded by the instructions, and the reversely transcribed instructions are naturally concise without much duplication.
(1) Pre-training plays a vital role in the mutual transcription tasks. For D2I, the tuned transformer performs much worse than PTMs, showing unsatisfactory language understanding capability. For I2D, moreover, it even fails to generate reasonable and coherent long paragraphs considering the too-low ROUGE-4 and BLEU-4 scores. In contrast, the PTMs show an overall impressive performance.
(2) The pre-training-fine-tuning paradigm is more suitable for special tasks in specific domains when a small set of data is available. For the large PTMs which are not tuned in a targeted manner, more instances (especially the highly related instances) can help them perform better, while the number of instances is limited by the length of the allowed input. Especially for the I2D task, large PTMs generate readable and reasonable results which are not similar to the ground-truth answers that we expected. Notice that GPT-3.5-chat performs worse than GPT-3.5, and this may also be caused by the overly flexible dialog capability, leading to a reduced ability to follow task instructions. Therefore, the tuned small PTMs are better choices for our task.
(3) Multi-grained knowledge enhancement is proven to be effective. Compared with the strong T5 models involved in general pre-training, the D2IPTM we propose performs better under both base and large settings. The ablation study has shown that the knowledgeable training tasks at each granularity we propose are necessary, contributing to performance improvement evenly for D2I, and the sequence-level augmentation has the most critical impact for I2D.
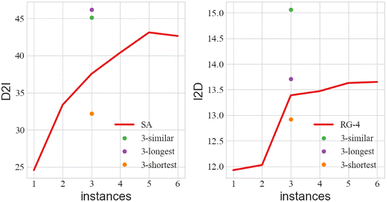
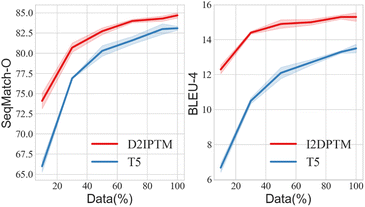
Besides, it is worth mentioning that randomly picked instances are not the best choice, since they may not cover the operations that we currently need. The 3-shot* version that finds the most similar training instances can partly solve this problem. Meanwhile, we find out combinations of three training instances that can cover all of the operations, in which the shortest have altogether 19 operations, and the longest has 68. From Fig. 5 we can see that the longest combination with more complicated instances carries more information, and this helps the large PTM achieve better D2I. Similar instances can further provide hints about language style, and bring improvement, especially for I2D. But overall, there is still a gap between the results of the large PTMs and the small fine-tuned models.
| Model | HMM | T5-large | Ours |
|---|---|---|---|
| Micro-F1 | 30.1 | 43.8 | 47.0 |
| Macro-F1 | 7.9 | 10.9 | 16.3 |
We conduct this experiment still on the ChemTrans dataset. Given the beginning part of the operation sequences (no shorter than 4 operations), we require the models to predict the next synthetic operation. For state transitions between discrete operations, the traditional machine learning algorithm hidden Markov model (HMM15) is a good choice. For the PTM methods, we test T5-large and our D2IPTM-large on this task, directly generating the names of the next operations. As shown in the results, PTMs can comprehend the synthetic process and predict the operations much better than the traditional HMM. With the knowledge enhancement, our model shows an even greater improvement, and can predict the next operation precisely in nearly half of the cases.
Observing the generated results, we can see that our model also successfully learns some general rules, and here we provide several instances for commonly generated combinations:
* ADD sodium/bromine speed: slowly
* QUENCH → EXTRACT → DRY
* ADD → SETTEMP overnight → FILTER → WASH
The above combinations are chemically reasonable. For example, it is an important safety reminder that introducing hazardous substances should be done slowly. It is also a commonly coupled operation to extract with ether or other organic solvents after quenching with aqueous solutions and then drying with anhydrous magnesium sulfate or other dehydrates. This indicates that the latest pre-training technology may have greater potential in assisting synthetic chemistry.
2.6 Manual evaluation
For a more comprehensive and flexible evaluation of our schema and model, we randomly sample several input paragraphs from the testing set and perform a manual inspection of the output results. Detailed examples are shown in Fig. 7.We then try other similar methods for synthetic description–instruction transcription. Reversely generating descriptions has not been explored much, while there exist some D2I systems. RXN12 refers to a text-to-procedure deep learning system based on a smaller transformer, and ChemIDE16 is a pure rule-based procedure extraction system. The two systems have different schemas from ours, but we can still observe their outputs manually. As shown in Fig. 8, the rule-based ChemIDE has missed lots of information about reagents (e.g., the main reactant in the first sentence), and the recall for operation recognition is also quite low (e.g., DRY and YIELD at the end). RXN shows an overall much better performance, while still makes some mistakes, such as recognizing “step A” as a reagent. Our system provides the most complete necessary information and parameters, and the expression of our schema is also very clear. For example, silica gel is used for column chromatography, which is far from simply adding the adsorbent into the system, and this information is only successfully expressed under our schema.
We also compare our automatically generated instruction library with the original workup records in the Open Reaction Database (ORD) which will be introduced in the Methods section. For a paragraph of experimental procedures, only a few instructions “CONCENTRATION – WASH – DRY – FILTRATION – CONCENTRATION” are recorded, and the details are simply provided in the form of original text segments (e.g., “wash with 0.5 M hydrochloric acid that”). In contrast, our system successfully recognizes the details for the related reagents, and provides the complete “ADD – ADD – SETTEMP – EVAPORATE – ADD – WASH – DRY – FILTER – EVAPORATE – COLUMN” pipeline with corresponding arguments (e.g., the name, concentration and mass for the three washing reagents). The instructions in our library have much higher practical value for automatic synthesis than existing records.
2.7 Discussion
Our system makes it possible to automatically make use of the mass articles in databases and instruct machines to carry out the preparation of known compounds. Meanwhile, it can conduct assisted writing in the field of synthetic chemistry to significantly improve the efficiency of researchers. To the best of our knowledge, this is the first fully open-source work (including data, code and models) on automated synthetic transcription, which may become a significant benchmark to inspire the standardized recording of synthetic actions based on our schema, and the development of an automatic synthetic database.Still, there is space for improvement in our system. Currently, the structured generation results need to be further compiled to form the lower computer instructions, like the temperature argument “cold (1–4°)” has to be translated to specific temperature-controlling actions. Besides, the schema set we designed can only express single reaction streamlines. For more complicated situations (e.g. different fractions are multi-step processed separately and then combined), temporarily our benchmark does not have a good solution and thus ignores them. With more powerful large PTMs at hand in the future, the system is supposed to have the ability to decompose complex problems into sub-problems. Considering the impressive cross-modal comprehension capability of the recent large models (e.g., GPT-4 (ref. 17)), heterogeneous information including molecule images and reaction formulae is also expected to be recognized by the model to alleviate the information loss problem mentioned in the case study, and previous research has already explored the heterogeneous bridging problem.18
3 Methods
3.1 Related work
Automatic chemical synthesis dates back half a century,19,20 targeting highly specialized substances. To free human researchers from labor-intensive actions, comprehensive platforms which can handle multiple synthetic paradigms are proposed to assist organic synthesis.21 Current robotic systems can achieve dozens of different reactions requiring multiple synthesis steps.22 Researchers go a step further and expect the systems to have automatic analysis capabilities, such as searching for new reactivity.23 Given a target compound, the systems predict the intermediate,24 search the templates and plan the synthetic actions automatically.25 For better compatibility with information such as human instructions, literature knowledge, and reaction templates, formalization of the synthetic scheme is necessary, thus a chemical programming language is proposed.26 Capturing and coding the synthesis information in articles are also emphasized.6Specific to reading the literature and extracting chemical information, many algorithms rely on the manually designed rules, recognizing chemical attributes sentence by sentence.27 This is proven to be practical and necessary when there is no large dataset for conducting supervised learning,16 but it still requires manual modification. Machine learning methods are further adopted into the processing pipeline,28–30 improving the performance of chemical, operation, and relation recognition to some extent.
However, for the development of machine reading, the breakthrough comes in the era of deep learning. Rule-based methods still play an important role in the interpretation improvement,31 while the deep learning models achieve satisfying performance in most of the NLP scenarios, including the language–action transcription that we are concerned about.12 Especially in recent years, the bigger models with transformer32 blocks and the better pre-training methods show their power and achieve surprising performances in various NLP tasks, including event detection and argument extraction33,34 which has some similarities to our task. For natural language understanding, PTMs including BERT,35 XLNet,36 and RoBERTa37 finish some tasks at a comparable level with human beings. For natural language generation, PTMs such as GPT-2,38 BART,39 and T5 (ref. 8) are proven to be effective while they still have a lot of room for improvement.
In this work, we require the model to decode in a pre-defined instruction space. Machine reading systems equipped with deep learning have been proven to be effective for comprehending cognitive content and generating specific operations including the utilization of search engines.40 To achieve formatted generation, the decoding process of models can be restricted in different ways, such as the pointer mechanism in NER tasks,41 and slot-filling modules in text-to-SQL tasks.42 More often, the models autonomously learn the ability of formatted output through pre-training on large-scale domain data.43 Therefore, we choose the neat and elegant approach to provide text input and formatted output for the deep learning models. The experiment results and cases show that PTMs perfectly decode in the rule we define.
3.2 Corpus
OrgSyn collects the publication of reliable methods for the preparation of organic compounds. The methods are usually described in the SI of articles in the chemical synthesis domain. Procedures in OrgSyn are open to the public and can be easily accessed. Besides, these procedures are peer-reviewed and reliable, thus they can be used as benchmarks in the evaluation of our NLP models and automatic synthesis. Permission for our research and subsequent annotation has already been provided by OrgSyn.Data from the ORD is open-accessed.¶ We download all the data, filter out short segments (string length less than 100) and meaningless characters such as URLs, timestamps, chemical formulae, etc., and perform deduplication operations (delete if the similarity of adjacent paragraphs is greater than 0.8). In this way, we get 160m pieces of text describing synthetic actions in a similar style to OrgSyn text. We use 251![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689 paragraphs with 100.5 words on average for the multi-grained knowledgeable pre-training. The corpus is also applied for constructing a machine-executable instruction library. We generate the synthetic instructions of 50
689 paragraphs with 100.5 words on average for the multi-grained knowledgeable pre-training. The corpus is also applied for constructing a machine-executable instruction library. We generate the synthetic instructions of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pieces of ORD data that ensure grammatical plausibility. The generated instructions can be queried with the chemical reactions or expected products, and are much more detailed than existing pipeline records in the original database. This library can be automatically enlarged easily in the future.
000 pieces of ORD data that ensure grammatical plausibility. The generated instructions can be queried with the chemical reactions or expected products, and are much more detailed than existing pipeline records in the original database. This library can be automatically enlarged easily in the future.
3.3 Schema and dataset
For the definition of the ChemTrans schema actions, we first present the top 100 high-frequency verbs in our corpus in Table 3, which cover over 80% of all the detected 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 verb occurrences. For all these frequently mentioned verbs, we filter out 17.0% of the verb mentions that are unrelated to the synthesis operations. For the other 83.0% of the verb mentions, we further discard 3.3% of them that are chemical instrument actions difficult to achieve automatically (e.g., “connected”), or relatively unnecessary in our scenes (e.g., “evacuate” is discarded since experiments were operated under a nitrogen atmosphere as default in our system, and other experimental setups we might express by adding gas). Eventually, we conclude the 16 operations in our schema from the retained verb mentions, among which 67.5% accurately correspond to an operation, and 12.2% vaguely correspond to one or more possible operations. We also supplement several operations based on the practical experience of experts, including COLUMN, TRITURATE, and PARTITION.
400 verb occurrences. For all these frequently mentioned verbs, we filter out 17.0% of the verb mentions that are unrelated to the synthesis operations. For the other 83.0% of the verb mentions, we further discard 3.3% of them that are chemical instrument actions difficult to achieve automatically (e.g., “connected”), or relatively unnecessary in our scenes (e.g., “evacuate” is discarded since experiments were operated under a nitrogen atmosphere as default in our system, and other experimental setups we might express by adding gas). Eventually, we conclude the 16 operations in our schema from the retained verb mentions, among which 67.5% accurately correspond to an operation, and 12.2% vaguely correspond to one or more possible operations. We also supplement several operations based on the practical experience of experts, including COLUMN, TRITURATE, and PARTITION.
| Type | Frequency | Verb |
|---|---|---|
| Unrelated | 7056 | Reduced, allowed, using/used, room, followed, continued, prepared, remaining/remains, becomes, stand, begins, passed, desired, described, based, required, carried, turns, repeated, taken, adjusted, rise |
| Discarded | 1374 | Connected, attached, immersed, sealed, shaken, evacuated, capped, packed, stored, discarded |
| Accurate | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 054 054 |
Added/adding, stirred/stirring/stir, washed, dried/flame-dried/drying, cooled/cool/cooling, equipped, charged, filtered, resulting, concentrated, extracted, transferred, distilled, fitted, give/gives/giving, placed, containing/contains, dissolved, afford/affords, poured, combined, obtained, evaporated, warm/warmed, rinsed, yielding/yield, introduced, quenched, recrystallized, boiling, eluted, maintain, filled, decanted, dropping, inserted |
| Vague | 5054 | Removed/remove, heated, collected, separated, flushed, purified, diluted, kept, treated, provide, purged, saturated, separates |
Compared with other chemical synthetic action schemas, our schema selects operations with appropriate granularity and therefore achieves a balance between good coverage and concise structure, of which the latter can promise the operability of the downstream automatic synthetic platforms. Take the verb “neutralize” for example. Considering this action is not a frequent operation and can be further decomposed and expressed with more basic operations such as ADD, we select to ignore such verbs to simplify the framework without compromising the coverage. In this way, there is either no need for the platforms to further compile “neutralize” into concrete operations.
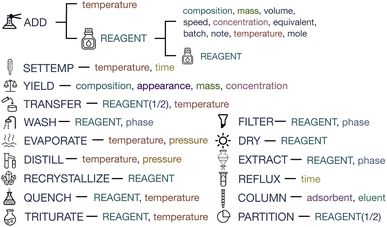
SciSpacy9 Part-of-Speech tagging tool is applied to count the verbs that appear in the text to be annotated. We analyze, combine and filter the verbs and decide on 16 basic actions for chemical synthesis as shown in Fig. 2. Meanwhile, the reagent is defined as a special item that is usually the basis of the reaction systems. Here we explain all the schema items:
REAGENT: solutions, gas, or other substances that join in the reaction should be annotated as REAGENT. A mixture reagent is composed of several pure reagents.
ADD: any operations that introduce effective substances into the reaction system are supposed to be an ADD (e.g. inject, charge, dissolve). The construction of the initial reaction system is also regarded as an ADD operation.
WASH: this operation is usually targeted at a specific phase (e.g. organic phase) of the reagent. Keywords include “wash” and “rinse”.
EXTRACT: similar to WASH, the EXTRACT operation requires phase and reagent arguments.
DRY: this operation is usually finished with the reagent or the heating equipment.
FILTER: this operation leads to the difference in the object of follow-up operations (e.g. the residue of filtration).
COLUMN: this operation is supposed to define the adsorbent and eluent, and keywords include “column” and “chromatography”.
DISTILL/EVAPORATE: distillation usually provides temperature and pressure. Evaporation also focuses on the two arguments, while emphasizing the removal of extra substances, such as concentrating the solutions.
SETTEMP: this operation is accompanied by stirring. The default temperature is the room temperature if not defined.
REFLUX: this operation also sets quite a high temperature, with the hint of keywords like “reflux” and “boil”.
QUENCH/TRITURATE: the quench operation is used to terminate the reaction, with keywords like “quench” and “pour”. The trituration operation is for purification, mashing the solid in the reagent. Both are concerned about the reagent and temperature arguments.
RECRYSTALLIZE/PARTITION: the two operations should be explicitly stated in the text to recrystallize the given reagent or to be partitioned between reagents 1 and 2.
TRANSFER: reagent 1 is transferred into reagent 2, usually under the given temperature.
YIELD: this operation usually appears at the end of synthesis descriptions, providing the product, appearance, purity, and other information.
All the annotators we hire have passed the TEM8, majored in chemistry-related disciplines, or participated in Chemistry Olympiads. As Fig. 1b shows, the arguments and operations are selected by the cursor, tagged with their types, and then linked with the arrows representing hierarchical relations. We sample and check 20% of the labeled results. The sampled item is required to be revised if the operation-level accuracy is lower than 90%, and a small batch of the items is required to be relabeled if lower than 70%.
For the convenience and accuracy of labeling, the annotators are required to label the operations and arguments in an extractive form. The arguments are linked with corresponding reagents or operations. We pre-process the JSON file, filter out those isolated labels and those items that do not meet the schema correspondence, and then transcribe the hierarchical relationship into sequential text. If the labels are too sparse (the generated output is shorter than 0.3 multiplied by the length of input), or the correspondence error appears more than twice, then the paragraph is abandoned.
3.4 Model settings
The transformer model we implement is a 4-layer transformer model with a hidden size of 256, following the setting of the previous framework.12The backbone model T5 (ref. 8) is one of the representative sequence-to-sequence models. For the T5-base model, there are 12 encoder layers and 12 decoder layers with a hidden size of 768, and altogether 220m parameters. For the T5-large model, there are 24 encoder layers and 24 decoder layers with a hidden size of 1024, and altogether 770m parameters. The simple baseline transformer model is a smaller T5, with the encoder and decoder layer number as 4 and hidden size as 256, which is comparable with the model applied in RXN.12
We transcribe various NLP tasks into a unified sequence-to-sequence format and distinguish them with the prefix prompts, which facilitates multi-task training. In our multi-grained knowledgeable pre-training, the combined task (verb mapping and chemical recognition) is conducted on two-thirds of the encoder–decoder pre-training data, and masked language modeling is conducted on the rest. The instruction generation for the decoder training is conducted simultaneously while on separate fake data. From Fig. 1c we can observe that the automatic labeling sometimes makes mistakes, such as missing/redundant operation verbs or chemical recognition (in the color of dark red), while overall providing worth-studying information for comprehension of operations and chemical substances.
For the large-scale PTMs, we set the prefix as follows:
D2I grammar: Use “[ ]”, “&” and “:” to mark operations, split different segments and split the name and value of arguments, such as “[OPERATION] ARGUMENT1 NAME: VALUE1 & ARGUMENT2 NAME: VALUE2 &”. Operations include ADD, WASH, FILTER, DRY, EXTRACT, RECRYSTALLIZE, QUENCH, PARTITION, TRANSFER, YIELD, DISTILL, EVAPORATE, COLUMN, SETTEMP and REFLUX. Arguments include time, temperature, phase, reagent, mass, composition, speed, mole, batch, volume, concentration and note. Notice that the grammar rule is only provided for zero-shot D2I transcription.
Zero-shot D2I: Generate the synthetic instructions according to the given descriptions.
Zero-shot I2D: Generate the synthetic description according to the given instructions.
3-shot and 3-shot*:  + Refer to the following instances. INSTRUCTION: …DESCRIPTION: …
+ Refer to the following instances. INSTRUCTION: …DESCRIPTION: …
Chat wrapper: You are now a synthetic literature writing/instruction generating assistant. +  + Now the INSTRUCTION/DESCRIPTION is given. In any cases, please generate the DESCRIPTION/INSTRUCTION.
+ Now the INSTRUCTION/DESCRIPTION is given. In any cases, please generate the DESCRIPTION/INSTRUCTION.
3.5 Training settings
We implement our method in the PyTorch44 framework, and adopt the HuggingFace transformers.45 For the multi-grained knowledgeable pre-training and fine-tuning of the models, we take the AdamW optimizer46 which is suitable for most of the PTMs in the T5 backbone. Reported scores are the average scores under 5 different random seeds. Following the setting in the original T5, we simply mix different knowledge enhancement data sets together, and treat these tasks equally. As for the proportion of tasks, we equally divided the pre-training data to create different task sets.In the stage of multi-grained knowledgeable pre-training, the corpus scale is comparably large, and the empirical batch size for multi-task training is bigger than fine-tuning. Therefore, we set the batch size as 256, with the default learning rate as 1e − 3. Since other tasks pay attention to the whole model while the instruction generation task only focuses on the decoder, the two parts calculate the loss and do back-propagation separately, and the second part is multiplied by the coefficient 0.1. In the stage of fine-tuning, since the dataset is small, we adjust the batch size to 16, with the common learning rate 1e − 4. These two hyper-parameters have been tried and searched within the vicinity and the best have been chosen.
The multi-grained knowledgeable pre-training stage takes 1000 steps and runs the data with only one epoch. For the fine-tuning period, the models are evaluated every epoch with the validation set, generated by greedy search and compared by the SeqMatch-A score. The maximum number of epochs is 20, and the early stop number is 3. Note that for the transformer baseline, due to lack of pre-training, the learning rate is set as 5e − 4 and the maximum epoch as 50 to ensure the convergence.
The greedy decoding strategy is applied for all the periods except for testing. When evaluating with the testing set or during the interaction, we set the model decoder beam size as 3.
For the practical test, 20 input paragraphs are randomly picked. The three researchers are asked to: (1) read the raw data, manually pick out the operations and arguments, and mark their types (to imitate the manual programming process for automatic chemical synthesis platforms); (2) read both the raw data and the instructions transcribed by our system, and correct the wrong parts (to imitate the manual verification of automatic transcription). Researchers first finish the written-from-scratch setting for paragraphs 1–10, and try the revision setting for paragraphs 11–20. The sequence for the two settings is exchanged then to reduce the effect of proficiency.
4 Conclusions
In this paper, we propose the solutions of transcription between human-readable descriptions and machine-executable instructions, a practical scenario in synthetic chemistry. We provide the task definition, the instruction schema, and a human-annotated dataset, ChemTrans, in the hope to draw more attention from the machine intelligence community to this interdisciplinary field. To apply the latest pre-training technology, we try two approaches, the small PTMs and large PTMs, on the mutual transcription tasks. For the former, we design the multi-grained knowledge enhancement method aiming at improving the capability of recognizing and generating chemical entities and operations/descriptions. Our experimental study has demonstrated the effectiveness of the proposed method. In the future, we will try to further improve the benchmark settings, the model architectures, and training methods. The potential of predicting synthetic actions and designing retrosynthesis pathways will be explored. Also, our system may get access to the actual system for use, providing practical help for researchers.Data availability
All data that support the findings of this study are available and have been deposited in Google Drive (https://drive.google.com/drive/folders/1AT-1uUR5Ev5d8fxX2FFrIZSnQ1-jbiG6?usp=sharing). The code of this study can be obtained from GitHub https://github.com/thunlp/ChemTrans. The zip file of the code can be downloaded via the Google Drive link above.Author contributions
Conceptualization: Weinan E, Rong Zhu, Zhiyuan Liu; data curation: Yi-Chen Nie, Ning Ding, Wei-Ting Ye; investigation: Yi-Chen Nie, Ning Ding, Zheni Zeng; methodology: Zheni Zeng, Ning Ding; software: Zheni Zeng; supervision: Rong Zhu, Zhiyuan Liu, Cheng Yang; validation: Zheni Zeng, Qian-Jun Ding; visualization: Zheni Zeng, Ning Ding; writing—original draft: Zheni Zeng, Ning Ding; writing—review & editing: Qian-Jun Ding, Wei-Ting Ye, Zhiyuan Liu, Rong Zhu, Weinan E, Maosong Sun.Conflicts of interest
There are no conflicts to declare.References
- G. Chen, P. Chen, C.-Y. Hsieh, C.-K. Lee, B. Liao, R. Liao, W. Liu, J. Qiu, Q. Sun, J. Tang, et al., arXiv, 2019, preprint, arXiv:1906.09427 Search PubMed.
- A. F. de Almeida, R. Moreira and T. Rodrigues, Nat. Rev. Chem, 2019, 3, 589–604 CrossRef.
- W. P. Walters and M. Murcko, Nat. Biotechnol., 2020, 38, 143–145 CrossRef CAS PubMed.
- B. Burger, P. M. Maffettone, V. V. Gusev, C. M. Aitchison, Y. Bai, X. Wang, X. Li, B. M. Alston, B. Li and R. Clowes, et al. , Nature, 2020, 583, 237–241 CrossRef CAS PubMed.
- Q. Zhu, F. Zhang, Y. Huang, H. Xiao, L. Zhao, X. Zhang, T. Song, X. Tang, X. Li and G. He, et al. , Natl. Sci. Rev., 2022, 9, nwac190 CrossRef CAS PubMed.
- S. Rohrbach, M. Šiaučiulis, G. Chisholm, P.-A. Pirvan, M. Saleeb, S. H. M. Mehr, E. Trushina, A. I. Leonov, G. Keenan and A. Khan, et al. , Science, 2022, 377, 172–180 CrossRef CAS PubMed.
- X. Han, Z. Zhang, N. Ding, Y. Gu, X. Liu, Y. Huo, J. Qiu, Y. Yao, A. Zhang and L. Zhang, et al. , AI Open, 2021, 2, 225–250 CrossRef.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, et al. , J. Mach. Learn. Res., 2020, 21, 1–67 Search PubMed.
- M. Neumann, D. King, I. Beltagy and W. Ammar, Proceedings of the 18th BioNLP Workshop and Shared Task, 2019, pp. 319–327 Search PubMed.
- K. Papineni, S. Roukos, T. Ward and W.-J. Zhu, Proceedings of the 40th annual meeting of the Association for Computational Linguistics, 2002, pp. 311–318 Search PubMed.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry and A. Askell, et al. , Adv. Neural Inf. Process., 2020, 33, 1877–1901 Search PubMed.
- A. C. Vaucher, F. Zipoli, J. Geluykens, V. H. Nair, P. Schwaller and T. Laino, Nat. Commun., 2020, 11, 1–11 CrossRef PubMed.
- C.-Y. Lin, Text summarization branches out, 2004, pp. 74–81 Search PubMed.
- J. Li, M. Galley, C. Brockett, J. Gao and W. B. Dolan, Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2016, pp. 110–119 Search PubMed.
- S. R. Eddy, Curr. Opin. Struct. Biol., 1996, 6, 361–365 CrossRef CAS PubMed.
- S. H. M. Mehr, M. Craven, A. I. Leonov, G. Keenan and L. Cronin, Science, 2020, 370, 101–108 CrossRef CAS PubMed.
- OpenAI arXiv , 2023, preprint, arXiv:2303.08774, DOI:10.48550/arXiv.2303.08774.
- Z. Zeng, Y. Yao, Z. Liu and M. Sun, Nat. Commun., 2022, 13, 1–11 CAS.
- R. Merrifield, Science, 1965, 150, 178–185 CrossRef CAS PubMed.
- G. Alvarado-Urbina, G. M. Sathe, W.-C. Liu, M. F. Gillen, P. D. Duck, R. Bender and K. K. Ogilvie, Science, 1981, 214, 270–274 CrossRef CAS PubMed.
- S. V. Ley, D. E. Fitzpatrick, R. J. Ingham and R. M. Myers, Angew. Chem., Int. Ed., 2015, 54, 3449–3464 CrossRef CAS PubMed.
- D. Angelone, A. J. Hammer, S. Rohrbach, S. Krambeck, J. M. Granda, J. Wolf, S. Zalesskiy, G. Chisholm and L. Cronin, Nat. Chem., 2021, 13, 63–69 CrossRef CAS PubMed.
- J. M. Granda, L. Donina, V. Dragone, D.-L. Long and L. Cronin, Nature, 2018, 559, 377–381 CrossRef CAS PubMed.
- J. Xu, Y. Zhang, J. Han, H. Qiao, J. Tang, S. Xi, B. Sun, S. Zhai, X. Wang, Y. Wu, et al.,ChemRxiv, 2021, preprint, DOI:10.26434/chemrxiv-2021-1bhnc.
- C. W. Coley, D. A. Thomas III, J. A. Lummiss, J. N. Jaworski, C. P. Breen, V. Schultz, T. Hart, J. S. Fishman, L. Rogers and H. Gao, et al. , Science, 2019, 365, eaax1566 CrossRef CAS PubMed.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson and D. Angelone, et al. , Science, 2019, 363, eaav2211 CrossRef CAS PubMed.
- M. C. Swain and J. M. Cole, J. Chem. Inf. Model., 2016, 56, 1894–1904 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Tomala, S. Matthews, E. Strubell, A. Saunders, A. McCallum and E. Olivetti, Sci. Data, 2017, 4, 1–9 Search PubMed.
- O. Kononova, H. Huo, T. He, Z. Rong, T. Botari, W. Sun, V. Tshitoyan and G. Ceder, Sci. Data, 2019, 6, 203 CrossRef PubMed.
- P. Shetty, A. C. Rajan, C. Kuenneth, S. Gupta, L. P. Panchumarti, L. Holm, C. Zhang and R. Ramprasad, npj Comput. Mater., 2023, 9, 52 CrossRef PubMed.
- M. Saeidi, Interpretation of Natural Language Rules in Conversational Machine Reading, Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, 2018 Search PubMed.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, Ł. Kaiser and I. Polosukhin, Advances in neural information processing systems, 2017, vol. 30 Search PubMed.
- R. Li, W. Zhao, C. Yang and S. Su, Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, 2021, pp. 2625–2635 Search PubMed.
- R. Li, W. Zhao, C. Yang and S. Su, Proceedings of the 45th International ACM SIGIR Conference on Research and Development in Information Retrieval, 2022, pp. 1110–1121 Search PubMed.
- J. D. M.-W. C. Kenton and L. K. Toutanova, Proceedings of NAACL-HLT, 2019, pp. 4171–4186 Search PubMed.
- Z. Yang, Z. Dai, Y. Yang, J. Carbonell, R. R. Salakhutdinov and Q. V. Le, Advances in neural information processing systems, 2019, vol. 32 Search PubMed.
- Y. Liu, M. Ott, N. Goyal, J. Du, M. Joshi, D. Chen, O. Levy, M. Lewis, L. Zettlemoyer and V. Stoyanov, A Robustly Optimized BERT Pretraining Approach, arXiv, 2019, preprint, DOI:10.48550/arXiv.1907.11692.
- A. Radford, J. Wu, R. Child, D. Luan, D. Amodei, I. Sutskever, et al., Language Models are Unsupervised Multitask Learners, OpenAI blog, 2019, 1, 9, https://d4mucfpksywv.cloudfront.net/better-language-models/language-models.pdf Search PubMed.
- M. Lewis, Y. Liu, N. Goyal, M. Ghazvininejad, A. Mohamed, O. Levy, V. Stoyanov and L. Zettlemoyer, Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, 2020, pp. 7871–7880 Search PubMed.
- R. Nakano, J. Hilton, S. Balaji, J. Wu, L. Ouyang, C. Kim, C. Hesse, S. Jain, V. Kosaraju and W. Saunders, WebGPT: Browser-assisted question-answering with human feedback, arXiv, 2021, preprint, DOI:10.48550/arXiv.2112.09332.
- H. Yan, T. Gui, J. Dai, Q. Guo, Z. Zhang and X. Qiu, Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing, 2021, vol. 1, pp. 5808–5822 Search PubMed.
- T. Yu, Z. Li, Z. Zhang, R. Zhang and D. Radev, Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2018, vol. 2, pp. 588–594 Search PubMed.
- Z. Feng, D. Guo, D. Tang, N. Duan, X. Feng, M. Gong, L. Shou, B. Qin, T. Liu, D. Jiang, et al., Findings of the Association for Computational Linguistics: EMNLP 2020, 2020, pp. 1536–1547 Search PubMed.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, et al., Advances in neural information processing systems, 2019, vol. 32 Search PubMed.
- T. Wolf, L. Debut, V. Sanh, J. Chaumond, C. Delangue, A. Moi, P. Cistac, T. Rault, R. Louf, M. Funtowicz, J. Davison, S. Shleifer, P. von Platen, C. Ma, Y. Jernite, J. Plu, C. Xu, T. L. Scao, S. Gugger, M. Drame, Q. Lhoest and A. M. Rush, Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, 2020, pp. 38–45 Search PubMed.
- I. Loshchilov and F. Hutter, International Conference on Learning Representations, 2019 Search PubMed.
Footnotes |
| † These authors contributed equally to this work. |
| ‡ https://platform.openai.com/docs/model-index-for-researchers. |
| § https://www.orgsyn.org/. |
| ¶ https://github.com/open-reaction-database/ord-data. |
| This journal is © The Royal Society of Chemistry 2023 |