Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Force-field-enhanced neural network interactions: from local equivariant embedding to atom-in-molecule properties and long-range effects†
Thomas
Plé
*,
Louis
Lagardère
* and
Jean-Philip
Piquemal
 *
*
Sorbonne Université, LCT, UMR 7616 CNRS, F-75005, Paris, France. E-mail: thomas.ple@sorbonne-université; louis.lagardere@sorbonne-université; jean-philip.piquemal@sorbonne-université
First published on 3rd October 2023
Abstract
We introduce FENNIX (Force-Field-Enhanced Neural Network InteraXions), a hybrid approach between machine-learning and force-fields. We leverage state-of-the-art equivariant neural networks to predict local energy contributions and multiple atom-in-molecule properties that are then used as geometry-dependent parameters for physically-motivated energy terms which account for long-range electrostatics and dispersion. Using high-accuracy ab initio data (small organic molecules/dimers), we trained a first version of the model. Exhibiting accurate gas-phase energy predictions, FENNIX is transferable to the condensed phase. It is able to produce stable Molecular Dynamics simulations, including nuclear quantum effects, for water predicting accurate liquid properties. The extrapolating power of the hybrid physically-driven machine learning FENNIX approach is exemplified by computing: (i) the solvated alanine dipeptide free energy landscape; (ii) the reactive dissociation of small molecules.
1. Introduction
In large-scale simulations, interactions between atoms cannot generally be computed from first principles because of the high numerical cost of quantum methods. Instead, they are generally modeled using force fields (FFs) that postulate a physically-motivated functional form of the potential energy and are parameterized in order to match ab initio energies and/or reproduce experimental data. The most widespread FFs are the so-called classical force fields (such as AMBER1 or CHARMM2) which use a combination of fixed-charge Coulomb potential and Lennard-Jones interactions to model the inter-molecular potential. These models are extremely efficient numerically, allowing the simulation of very large systems over long time scales. Their simple functional form, however, lacks polarization and many-body effects which can be critical to correctly describe some systems (for example solvation in a polar solvent, pi-stacking or complex protein structures3). More advanced force fields – such as AMOEBA,4 TTM,5 CHARMM Drude,6 ARROW7 or SIBFA8,9 – have thus been developed in order to explicitly include these effects. These polarizable force fields (PFFs)10,11 are much more flexible and accurate but are significantly costlier. Nonetheless, advances in high-performance computing (HPC), the increase in GPU (Graphical Processing Units) availability and recent methodological developments (advanced iterative solvers) now allow large-scale PFF simulations.12 Both classical and polarizable FFs however assume a fixed connectivity between atoms (i.e. covalent bonds cannot be broken), making them unsuitable to study chemical reactions. Some reactive force fields – such as ReaxFF13 or Empirical Valence Bond14 – are actively being developed but are generally specialized towards a relatively narrow class of systems.From this brief overview of the domain of force fields, it is clear that a general, many-body reactive model is highly desirable but its design remains an outstanding challenge for the current frameworks. In recent years, considerable attention and resource have been devoted to the development of machine-learning potentials that promise to bridge the accuracy and generality gap between force fields and ab initio methods. These models use flexible functional forms from the domain of machine-learning (such as deep neural networks,15–17 body-ordered expansions18–20 or kernel models21,22) in order to accurately fit ab initio energies, with a numerical cost comparable to standard FFs. A large variety of such models have been developed over the last few years (for example the HD-NNP,15 ANI,16 AIMNet,23,24 DeePMD,25 ACE,18 sGDML,21 Tensormol-0.1,26 Nequip,17etc…) and have been applied to small molecular systems,16 periodic crystals27,28 and more general condensed-phase systems.29,30 Among these, the ANI models occupy a particular place as they aim to provide a generic pre-trained potential for a whole class of organic molecules. In this work, we follow a similar strategy.
In order to respect the inherent symmetries of molecular systems, most architectures are designed to be invariant with respect to rotations, translations and permutation of identical atoms. These models have shown good accuracy on many systems but usually require large amounts of data to be trained on (of the order of a million molecular configurations). More recently, equivariant models (for example Nequip,17 Allegro,31 SpookyNet,32 UNiTE33 or GemNet34) have attracted much attention because of their impressive data efficiency and their ability to generalize more accurately to out-of-distribution configurations.17,35
Most ML models, however, assume a purely local functional form and tend to neglect or implicitly account for long range effects from the training data. The accurate description of long-range interactions is however critical to correctly simulate condensed-phase systems and to describe the structure of large molecular formations (e.g. protein or DNA structure36,37). The framework of message-passing neural networks38,39 in principle allows to describe long-range effects by iteratively exchanging information with neighbouring atoms. This approach however have been shown to pose difficulties when applied to large systems as this iterative process is not well suited for parallel architectures because of the associated communications that are required. Furthermore, very long-range effects such as electrostatic interactions would require a large number of iterations (and the molecular graph to be connected) to be captured by a message-passing model, thus imposing a high computational cost.31 Another paradigm to account for long-range effects is the use of global descriptors (for example the Coulomb matrix40) that couple all degrees of freedom without imposing any locality prior. These descriptors are usually used in conjunction with kernel-based models21,41 and where shown to be accurate and data-efficient for small to medium-sized molecules and crystals. Although some recent progress have been made to apply these models to larger systems,42 they cannot tackle systems larger than a few hundreds of atoms due to the 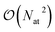 scaling of the global descriptor size. Some other pure ML multi-scale models are currently being developed (for example the LODE descriptor43,44) but this area is still in its infancy. On the other hand, quantum perturbation theory (for example Symmetry Adapted Perturbation Theory, SAPT) gives solid grounds to the description of long-range effects in terms of classical electrostatics45 which can be well captured in the FF framework (via multipolar Coulomb interactions and dispersion effects for example46). It thus seems advantageous to combine an ML model – which excel at predicting short-range properties – with long-range FF interactions, in order to obtain the best of both approaches. A few models applying this idea have recently been developed (for example HDNNP-Gen4,47 PhysNet,48 SpookyNet,32 ANIPBE0-MLXDM,49 q-AQUA-pol50 and others19,29,51,52) and have shown good results across multiple systems. Hybrid FF/ML models thus provide a promising route to more physics-aware ML potentials.
scaling of the global descriptor size. Some other pure ML multi-scale models are currently being developed (for example the LODE descriptor43,44) but this area is still in its infancy. On the other hand, quantum perturbation theory (for example Symmetry Adapted Perturbation Theory, SAPT) gives solid grounds to the description of long-range effects in terms of classical electrostatics45 which can be well captured in the FF framework (via multipolar Coulomb interactions and dispersion effects for example46). It thus seems advantageous to combine an ML model – which excel at predicting short-range properties – with long-range FF interactions, in order to obtain the best of both approaches. A few models applying this idea have recently been developed (for example HDNNP-Gen4,47 PhysNet,48 SpookyNet,32 ANIPBE0-MLXDM,49 q-AQUA-pol50 and others19,29,51,52) and have shown good results across multiple systems. Hybrid FF/ML models thus provide a promising route to more physics-aware ML potentials.
In this paper, we propose a general framework for building force-field-enhanced ML models. We leverage the latest advances in local equivariant neural networks in order to accurately predict short-range energy contributions as well as multiple atom-in-molecule properties that are then used to dynamically parameterize QM-inspired FF energy terms that account for long-range interactions. We show that this architecture allows for highly transferable models that are able to accurately generalize on large molecular systems, as well as in the condensed phase after being trained on small monomers and dimers only. This paper is organized as follows. Section 2 describes the model architecture, from the Allegro31 equivariant embedding to the output and physics modules. It also describes the particular FF terms that we used for the pretrained model, named FENNIX-OP1, that we provide with this work. Section III focuses on the FENNIX-OP1 model and provides details on its construction, its target properties, the datasets used for training and the different training stages that were required. In Section 4, we validate the model via several applications. First, we show that the model predicts accurate dissociation energy curves of some simple molecules. We then compute structural properties of liquid water in molecular dynamics simulations including nuclear quantum effects (NQEs) via the recently developed adaptive quantum thermal bath (adQTB).53,54 Indeed, since the model is trained purely on ab initio data, the explicit inclusion of NQEs is critical for the correct calculation of thermodynamical properties, as was shown in numerous previous studies.55,56 For this purpose, the adQTB was shown to provide robust approximations of NQEs while being numerically affordable (similar to a classical MD) and thus constitutes a very efficient tool for quickly testing ML models. We then show that the model produces stable dynamics of alanine dipeptide in solution and provides a qualitatively correct description of the torsional free energy profile (computed using an enhanced sampling method57); as well as stable dynamics of the 1FSV protein in gas phase. Finally, Section 5 provides some conclusions and outlooks for future extensions of the model.
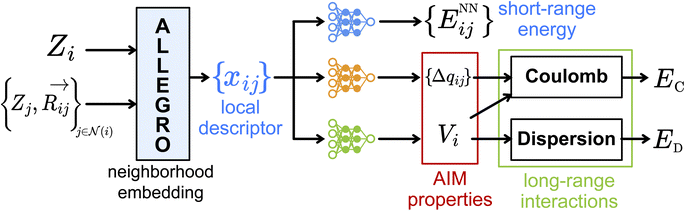
2. Model architecture
The FENNIX (Force-field-Enhanced Neural Network Interactions) model is based on a local multi-output equivariant model that processes atomic neighborhoods and predicts multiple atomic or pairwise properties. As an example for this work, we will present a model that outputs local pairwise energy contributions, charges and atomic volumes. The output is subsequently enriched by a “physical” module that computes force field terms such as electrostatic and dispersion energy terms. The core of our model is a slightly modified version of the Allegro local equivariant model presented in ref. 31. We use the Allegro model as a general embedding of atomic pairs which is then fed into independent neural networks that predict the target properties. The choice of the Allegro model is mostly motivated by two of its properties: (i) it is strictly local and thus allows for favourable scaling with system size and efficient parallelization; (ii) it is equivariant and thus allows the prediction of tensorial properties such as atomic multipoles (which are not used in this work but will be the focus of future improvements of the model). A compact flow diagram of the model is shown in Fig. 1.In this section, we will briefly describe the Allegro architecture and our modifications to the model. The theoretical analysis of this architecture was thoroughly done in the original paper31 so that we will only review here the main points necessary to the understanding of the model and our improvements to the architecture. We will then present the output module that processes the Allegro embedding. Finally, we will describe the physical module and the particular functional form for the FENNIX-OP1 potential energy surface that we used in this work.
2.1 Equivariant embedding using the Allegro architecture
 defined by all the atoms located at a distance shorter than a cutoff radius rc from atom i:
defined by all the atoms located at a distance shorter than a cutoff radius rc from atom i: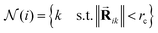 | (1) |
![[R with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/b_char_0052_20d1.gif) ik =
ik = ![[R with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/b_char_0052_20d1.gif) k −
k − ![[R with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/b_char_0052_20d1.gif) i the vector going from the position of atom i to atom k. The first part of the descriptor xij is built so that it is invariant under the action of certain geometric symmetries (i.e. global rotations, translations and inversions of the system). On the other hand, the second descriptor Vijnlp is composed of features that are equivariant with respect to these symmetries. These features take the form of tensors that are labeled with a channel index n ∈ 1, …, Nchannels, a rotational index l ∈ 0, 1, …, lmax and a parity index p ∈ −1, 1. The rotational index indicates how the tensor transforms under rotation operations: l = 0 corresponds to scalar/invariant quantities, l = 1 corresponds to vector-like objects and we refer to l ≥ 2 objects as higher-order tensors. The parity index, on the other hand, indicates how the tensor's sign changes under inversion of the coordinate system. The channel index simply allows the model to process multiple features of same l and p indices. In our implementation of the Allegro model, these tensorial objects are handled by the
i the vector going from the position of atom i to atom k. The first part of the descriptor xij is built so that it is invariant under the action of certain geometric symmetries (i.e. global rotations, translations and inversions of the system). On the other hand, the second descriptor Vijnlp is composed of features that are equivariant with respect to these symmetries. These features take the form of tensors that are labeled with a channel index n ∈ 1, …, Nchannels, a rotational index l ∈ 0, 1, …, lmax and a parity index p ∈ −1, 1. The rotational index indicates how the tensor transforms under rotation operations: l = 0 corresponds to scalar/invariant quantities, l = 1 corresponds to vector-like objects and we refer to l ≥ 2 objects as higher-order tensors. The parity index, on the other hand, indicates how the tensor's sign changes under inversion of the coordinate system. The channel index simply allows the model to process multiple features of same l and p indices. In our implementation of the Allegro model, these tensorial objects are handled by the 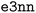 python package58 which provides high-level classes that represent them and easy-to-use functions to manipulate and combine them while preserving symmetries and global equivariance.
python package58 which provides high-level classes that represent them and easy-to-use functions to manipulate and combine them while preserving symmetries and global equivariance.
In the following paragraphs, we describe how initial two-body features are computed, how features from neighbors are combined through Nlayers layers of interactions to enrich them with many-body information and how they are filtered at each layer to control the size of the embedding.
2.1.1.1 Initial two-body features. The Allegro model starts by decomposing each interatomic vector
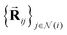 into fingerprints that are more suitably processed by the network. The interatomic distance Rij is projected onto a radial basis B(Rij) = [B1(Rij), …, BNbasis(Rij)] (we use the Bessel basis function with a polynomial envelope59 that we normalize as in the original paper) and we compute the two-body scalar embedding as:
into fingerprints that are more suitably processed by the network. The interatomic distance Rij is projected onto a radial basis B(Rij) = [B1(Rij), …, BNbasis(Rij)] (we use the Bessel basis function with a polynomial envelope59 that we normalize as in the original paper) and we compute the two-body scalar embedding as: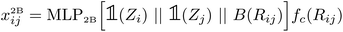 | (2) |
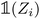 (resp.
(resp. 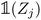 ) is a vector representing the chemical species of the source atom i (resp. destination atom j). In the original paper
) is a vector representing the chemical species of the source atom i (resp. destination atom j). In the original paper 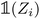 is a direct one-hot encoding of the atomic number Zi, meaning that one has to fix in advance the number of species the model will be able to process (the one-hot encoding then defines a simple orthonormal basis which has the same dimensions as the chosen number of species). In Section 2.1.2, we propose to modify this one-hot encoding by a positional encoding of coordinates in the periodic table which allows for more flexibility in the treatment of atomic species.
is a direct one-hot encoding of the atomic number Zi, meaning that one has to fix in advance the number of species the model will be able to process (the one-hot encoding then defines a simple orthonormal basis which has the same dimensions as the chosen number of species). In Section 2.1.2, we propose to modify this one-hot encoding by a positional encoding of coordinates in the periodic table which allows for more flexibility in the treatment of atomic species.
We obtain the two-body equivariant features by projecting the unit vector ![[R with combining circumflex]](https://www.rsc.org/images/entities/i_char_0052_0302.gif) ij =
ij = ![[R with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/b_char_0052_20d1.gif) ij/Rij onto a basis of real spherical harmonics Yijlp. We then mix them with radial information with a linear embedding on Nchannels channels:
ij/Rij onto a basis of real spherical harmonics Yijlp. We then mix them with radial information with a linear embedding on Nchannels channels:
| Vijnlp,2B = [MLPembed2B(xij2B)]nlpYijlp | (3) |
2.1.1.2 Interaction with the local environment. The two-body embedding (xij2B,Vijnlp,2B) is then processed through multiple “interaction” layers that allow to combine information with other atoms in the vicinity of atom i. Each interaction layer starts by building a global equivariant neighborhood embedding for atom i from the current scalar embeddings xik and the spherical harmonics projections Yiklp:
 | (4) |
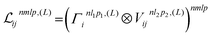 | (5) |
2.1.1.3 Feature filtering and channel mixing. Finally, the latent space is filtered to obtain the new pairwise embedding. The scalar embedding is combined with the scalar part of the latent space (with every channels and all multiplicities concatenated) to obtain:
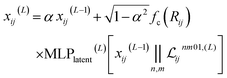 | (6) |
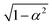 ) is chosen to enforce normalization (see ESI of ref. 31). In our implementation, the value of α can be set as a hyperparameter (for example to the value
) is chosen to enforce normalization (see ESI of ref. 31). In our implementation, the value of α can be set as a hyperparameter (for example to the value 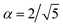 proposed in the original Allegro paper) or can be optimized independently for each layer during the training procedure.
proposed in the original Allegro paper) or can be optimized independently for each layer during the training procedure.
The new equivariant features are obtained by linearly combining the elements of the latent space with same indices (l, p) from all channels and multiplicities:
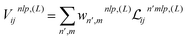 | (7) |
The output features of the last layer (xij(Nlayers),Vijnlp,(Nlayers)) compose the many-body embedding of our model which is passed to the output module to predict the different atomic or pairwise properties that the model is trained on.
In this work, we propose to use a positional encoding that encodes coordinates in the periodic table using sine and cosine functions of different frequencies. It is inspired from ref. 61 where it is used to encode the position of words in sentences in the context of natural language processing. The column index c is encoded as a vector ec of dimension dcol as:
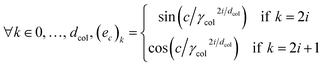 | (8) |
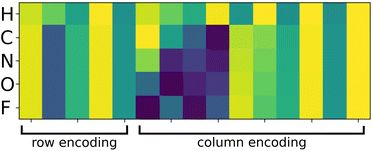
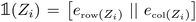 . Fig. 2 shows a heatmap of the positional encoding of the species H,C,N,O and F (from top to bottom). We see that the first five columns are the same for all the heavy atoms as they represent the row encoding (the second row of the periodic table in this case) while they are different from the first line corresponding to the Hydrogen. We also see that the last ten columns are different for all the species shown here as they are all on different columns of the periodic table. The motivation behind using this positional encoding is that we hypothesized that having similar encodings for species sharing a row or a column might help with generalization and allow to transfer learned knowledge from a species to another, thus requiring less training data. Interestingly, the positional embedding defines a smooth function of the coordinates in the periodic table (of which integer coordinates are actual chemical elements) and allows to “interpolate” between chemical species. Furthermore, as stated in ref. 61 the encoding for index c + k can be represented as a linear function of the encoding for index c, which might further help with inferring similarities. Additional features such as the ionization state could be encoded in the same manner (though we restrict ourselves to neutral atoms in this work, thus only requiring the knowledge of chemical species).
. Fig. 2 shows a heatmap of the positional encoding of the species H,C,N,O and F (from top to bottom). We see that the first five columns are the same for all the heavy atoms as they represent the row encoding (the second row of the periodic table in this case) while they are different from the first line corresponding to the Hydrogen. We also see that the last ten columns are different for all the species shown here as they are all on different columns of the periodic table. The motivation behind using this positional encoding is that we hypothesized that having similar encodings for species sharing a row or a column might help with generalization and allow to transfer learned knowledge from a species to another, thus requiring less training data. Interestingly, the positional embedding defines a smooth function of the coordinates in the periodic table (of which integer coordinates are actual chemical elements) and allows to “interpolate” between chemical species. Furthermore, as stated in ref. 61 the encoding for index c + k can be represented as a linear function of the encoding for index c, which might further help with inferring similarities. Additional features such as the ionization state could be encoded in the same manner (though we restrict ourselves to neutral atoms in this work, thus only requiring the knowledge of chemical species).
2.2 Output module
After computing the embedding from the Allegro model, we use it as input for independent MLPs for each target property. In the following, we will simply denote (xij,Vijnlp) the output from the last Allegro layer (thus dropping the (L) layer index). The current implementation also allows some modularity in the composition of inputs and on the operations done on the outputs. For example, the input can exploit either the scalar embedding to obtain invariant properties via a standard MLP as| oij = MLPout[xij], | (9) |
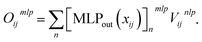 | (10) |
For atom-wise properties, the pairwise outputs are simply summed up on the central atom. For properties that should sum up to zero (for example partial atomic charges), the outputs oij and oji can be antisymmetrized (which for partial charges is equivalent to charge exchange between neighbouring atom pairs). Furthermore, in order to impose constraints on invariant outputs, a final activation function can optionally be applied. This is for example useful when the output targets a positive quantity (for instance an atomic volume) for which we can apply a softplus function, a probability for which we may apply a sigmoid function or a discrete probability distribution (in the case of multidimensional oij) for which a softmax function can be used.
Further modifications can optionally be applied. For instance, the input can be filtered according to an additional shorter-range cutoff for example one distinguishing between bonded and non-bonded pairs. Finally, the two-body embedding xij2B can be used in place of or concatenated to (as it is done in the FENNIX-OP1 model) the final embedding to use as input. This allows the output MLP to easily access simple pairwise information and should let the Allegro embedding specialize in finer many-body correlations. This compositional approach allows for a great flexibility in the model's output, which is especially useful when experimenting with a ML parametrization of physical models.
The output module for the FENNIX-OP1 model is composed of three targets: a local energy contribution 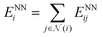 (which is a simple scalar output), an atomic partial charge
(which is a simple scalar output), an atomic partial charge 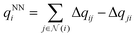 (through antisymmetrized charge exchange) and an atomic volume vNNi (constrained to be positive).
(through antisymmetrized charge exchange) and an atomic volume vNNi (constrained to be positive).
2.3 Physics module and energy functional form
Finally, the physics module uses the results from the output module and feed them into physically-motivated models to enrich the output.In the case of FENNIX-OP1, the force field module is composed of an electrostatic energy term ECij and a pairwise dispersion term EDij. The functional form of FENNIX-OP1 is then given by:
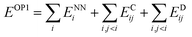 | (11) |
The first term ENNi is the neural network contribution that accounts for short-range interactions that we introduced in Section 2.2. We model the electrostatic interaction ECijvia a Coulomb potential with fluctuating charges and the Piquemal charge penetration model:62
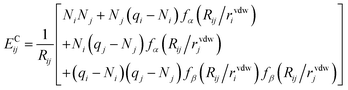 | (12) |
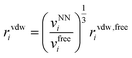 | (13) |
Finally, the dispersion interaction EDij is computed using the pairwise Tkatchenko–Scheffler model:63
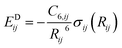 | (14) |
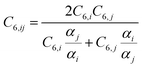 | (15) |
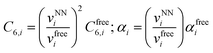 | (16) |
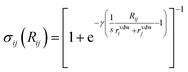 | (17) |
We furthermore mention that the physics module is not limited in principle to compute energy terms. The aim of this module is to be a general physical interface between the ML embedding and the final target properties. For example, we use it in the FENNIX-OP1 model to correct for ML-predicted charge exchanges that would deplete the valence shell of an atom. The implementation also provides charge exchange via a simple bond-capacity model64 that leverages ML-predicted atom-in-molecule electronegativities and which capabilities will be explored in future iterations of the FENNIX model. Each physical model is implemented as a simple Pytorch Module that takes as input a dictionary which contains previously computed properties. It then adds its contribution and outputs the enriched dictionary. These physical submodules can then easily be chained, and it facilitates the implementation of additional models.
3. Datasets and training procedure
In this section, we start by providing some details on the construction of FENNIX-OP1 model. We then review the datasets that we used for training the model and its target properties. Finally, we will focus on the non-trivial task of training a multi-output FENNIX model.3.1 FENNIX-OP1 model architecture
In the FENNIX-OP1, chemical species are encoded using the positional encoding defined in Section 2.1.2, with 5 dimensions for the row encoding and 10 dimensions for the column encoding. We use Nbasis = 10 Bessel basis functions for the radial embedding, with a cutoff distance rc = 5.2 Å and a smoothing parameter p = 3 for the polynomial envelope. We used 256 features for the scalar embedding and 10 channels with a maximum rotational index lmax = 2 for all equivariant features. The two-body scalar MLP (MLP2B) is composed of two hidden layers with 64 and 128 neurons respectively with SiLU activation function. The two-body embedding MLP (MLPembed2B) for the initial equivariant features is a simple linear projection of the two-body embedding with no activation function. The embedding is constructed using 3 Allegro layers. In each layer, the embedding MLP (MLPembed(L)) is a simple linear projection with no activation function and the latent MLP (MLPlatent(L)) contains two hidden layers both with 256 neurons and SiLU activation function. The mixing coefficient α is initially set to the original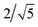 and is optimized independently for each layer during training.
and is optimized independently for each layer during training.
The output module is composed of three independent identical MLPs for the short-range energy contribution, the charge exchange and the atomic volume. The input of these MLPs is the concatenation of the two-body scalar embedding and the final scalar embedding. They have 5 hidden layers with 256, 128, 64, 32 and 16 neurons. In total, the model has approximately 1.2 million parameters.
The output module also has a “constant” submodule that simply provides the atomic reference energies. Importantly, we used the CCSD(T) isolated atom energies as a reference instead of the average atomic energy over the dataset that is typically advised when training a ML model. Indeed, this reference energy has a physical meaning – which is the energy of an atom when it has no neighbors to interact with – and is not solely a convenience parameter for facilitating the training. In particular, this choice has important consequences for the description of molecular dissociation,65 as will become clear in Section 4.1.
Finally, the physics module is composed of four submodules: a charge correction module, a charge scaling module, the Coulomb interaction module and the dispersion module. These last two modules implement the physical interactions using the corresponding equations described in Section 2.3. The charge scaling module simply scales all charges by a constant factor in order to compensate for the lack of higher-order multipoles in the permanent electrostatics. The charge correction module antisymmetrizes the charge exchanges and ensures that atoms do not unphysically deplete their valence shell. Indeed, if we assume that only valence electrons can be transferred, an atom cannot have a partial charge larger than +Ni (which is particularly important for the charge penetration model that we use). This constraint is not ensured by the neural network model so we need to enforce it a posteriori. The constraint is achieved by transferring back some electrons from neighbouring atoms that drained too much charge. First the unphysical “hole” in the valence shell is computed using a smooth function (to ensure smooth gradients of the final coulomb energy) on the basis that an atom cannot lose more than 95 percent of its valence electrons. Charges that sum up to the valence hole are then transferred from neighbouring atoms that took charges, proportional to the quantity drained. This procedure should enforce the constraint in a single iteration for most non-pathological cases. Reassuringly however, the charge correction is almost never needed after training (i.e. the valence hole is almost always zero), even in condensed-phase MD simulations for which the model was not explicitly trained.
3.2 Datasets and target properties
For the FENNIX-OP1 model, we chose to reproduce high-level coupled-cluster energies and we thus selected three of the few freely-available and generalist datasets that provide such data: the ANI-1ccx16 dataset, the DES370K66 dataset and the q-AQUA dimers dataset.20The ANI-1ccx dataset provides approximately 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 CCSD(T)/CBS total energies of various neutral monomers and dimers (composed of the elements H,C,N,O) in equilibrium and out-of-equilibrium configurations. It also provides atomic forces at DFT level that were crucial to speed-up the beginning of the training procedure. Importantly, it provides partial charges and atomic volumes obtained from a Minimal Basis Iterative Stockholder67 (MBIS) partitioning of the DFT electronic density that were used to parameterize the force field terms.
000 CCSD(T)/CBS total energies of various neutral monomers and dimers (composed of the elements H,C,N,O) in equilibrium and out-of-equilibrium configurations. It also provides atomic forces at DFT level that were crucial to speed-up the beginning of the training procedure. Importantly, it provides partial charges and atomic volumes obtained from a Minimal Basis Iterative Stockholder67 (MBIS) partitioning of the DFT electronic density that were used to parameterize the force field terms.
The DES370K dataset is composed of approximately 370![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 CCSD(T)/CBS interaction energies of diverse dimers in various configurations. It comprises both neutral and charged molecules. The latter were discarded for the training of the FENNIX-OP1 model as out-of-scope for this study. It also provides SAPT decompositions of the interaction energies that we used to regularize the Coulomb interactions.
000 CCSD(T)/CBS interaction energies of diverse dimers in various configurations. It comprises both neutral and charged molecules. The latter were discarded for the training of the FENNIX-OP1 model as out-of-scope for this study. It also provides SAPT decompositions of the interaction energies that we used to regularize the Coulomb interactions.
The q-AQUA dimers dataset is composed of more than 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 water dimer interaction energies at CCSD(T)/CBS level. We used this dataset in the end of the training in order to fine-tune the model for handling water.
000 water dimer interaction energies at CCSD(T)/CBS level. We used this dataset in the end of the training in order to fine-tune the model for handling water.
The FENNIX-OP1 model has then three target properties that use the available data: the CCSD(T) total energy of the system as modelled by VOP1 described in Section 2.3, the MBIS partial charges qNNi and the ratio of MBIS volumes to free atom volumes vNNi/vfreei.
3.3 Training procedure
The training of a multi-output force-field-enhanced neural network revealed to be a non-trivial task. Indeed, the inter-dependencies between target properties (for example partial charges and the electrostatic contribution to the total energy) seemed to pose difficulties for the standard optimization methods, which implied that a brute-force optimization of the whole model would not give satisfactory results. To overcome this difficulty, we resorted to a training procedure in multiple stages that is described in the following of this section.Furthermore, in order to obtain a final model that was stable when performing molecular dynamics, we found that strong regularization was needed, both in the form of standard weight decay and also physically-motivated loss contributions, as detailed later. Throughout, we used the AdamW optimizer68 implemented in Pytorch, as its algorithm for weight decay provides one of the best compromise between training speed and accuracy. We used a fairly strong weight decay parameter of 0.5 for all the parameters in the output module and no weight decay for the Allegro parameters. In all stages, we used the same random 10% of the dataset as a validation set and optimized the model using mini-batches of 256 configurations from the ANI-1ccx and 64 configurations from DES370K and q-AQUA when they are used.
The training procedure for the FENNIX-OP1 model required four stages. In the first stage, the model was trained to reproduce DFT total energies and forces using the short-range contribution VNN only. We also trained at the same time the charges and atomic volumes to reproduce the MBIS targets. At this stage, only the ANI-1ccx dataset was used. The loss function for this stage is given by:
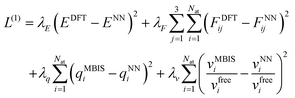 | (18) |
In the second stage, we freeze the embedding parameters and output MLPs for charge and volumes and activate the Coulomb and dispersion energy terms. We then retrain the short-range energy MLP so that the full VOP1 of eqn (11) reproduces DFT energies and forces. The loss function for this stage is:
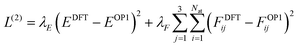 | (19) |
In the third stage, the embedding and charge and volumes MLPs are frozen again and the energy MLP is finally retrained to reproduce CCSD(T) energies from both ANI-1ccx total energies and DES370K interaction energies. We used the same type of mean-square loss functions with λE = 0.1 and λDES = 5. Again, we train the energy MLP until the error is close to the previous stage, unfreeze the whole model and optimize again all the parameters. For this stage, we also optimize the parameters from the physical module in order to reach the lowest error possible. We stop the training when the learning rate reaches 10−5.
In the last training stage, we refine the model for water by including the q-AQUA water dimers interaction energies in the loss function. We also generated batches of randomly deformed water monomers (with very large deformations) and trained the model to reproduce forces from the highly accurate Partridge–Schwenke potential energy surface (that was itself fitted on high-accuracy coupled-cluster data). This was particularly helpful to reproduce the molecule's bending energy surface away from equilibrium where fewer data are available in the ANI-1ccx dataset.
At the end of the training procedure, the model reached a root mean square error (RMSE) of less than 4 kcal mol−1 for CCSD(T) total energies on both validation and training sets of the ANI-1ccx dataset. While it was possible to reach much lower errors with less regularized models (less than 1 kcal mol−1), we found that they were unstable when performing MD and concluded that they were overfitting the data. We thus favoured a more strongly regularized, perhaps slightly underfitted model that allowed for better generalization in the condensed phase. The model also reached a RMSE of about 0.35 kcal mol−1 on both DES370K and q-AQUA datasets. Finally, it reached a RMSE of about 0.017e for charges and about 0.017 for volume ratios. As for total energies, less regularized models allowed for much lower errors (less than 0.008e for charges for example) but with visible overfitting, leading to irregular coulomb interactions and unstable dynamics.
As a validation for the water interactions, we used the standard energy benchmarks when training force fields for water. The model gives a RMSE of 0.13 kcal mol−1 on the Smith dimer set69 which are representative of the most stable water dimer configurations. This low error is not surprising as the Smith dimers are very close to configurations present in the q-AQUA dataset on which the model was trained. For a more challenging test, we used a set of typical water clusters up to hexamers. For these, the model achieved a RMSE lower than 2 kcal mol−1, which is comparable to our recent Q-AMOEBA model56 which was specifically trained to reproduce these energies. In the next section, we investigate more thoroughly the validity, transferability and robustness of the model up to the unforgiving test of condensed-phase molecular dynamics including nuclear quantum effects.
4. Model validation
In this section, we validate the FENNIX-OP1 model using a few examples of applications. First, we compute bond dissociation energy profiles of typical small molecules and show that the model is able to consistently break covalent bonds. Then, we show that the model is able to produce stable and accurate MD simulations of condensed-phase water including nuclear quantum effects. For this study, we included nuclear quantum effects using the adaptive quantum thermal bath (adQTB) method introduced in ref. 53. We showed in previous studies54 that the adQTB provides an efficient and accurate alternative to path integrals – the gold standard method for including NQEs in MD simulations – at a cost similar to classical MD. For these two examples, we compare our results to the ANI models16 that have a comparable scope as FENNIX-OP1 in terms of chemical diversity and were trained on similar datasets (note that we used the ANI models as provided in the torchANI package and did not re-train them). Finally, we show that FENNIX-OP1 is able to produce stable dynamics of organic molecules solvated in water and provides a good qualitative description of the torsional free energy landscape of the alanine dipeptide in solution.All calculations for this work were performed on an Nvidia A100 GPU using our custom TorchNFF package that is built on top of Pytorch70 and provides the implementation for FENNIX as well as a simple MD algorithm for NVT simulations in periodic boundary conditions (with Ewald summation for electrostatic interactions) and an efficient Pytorch implementation of the adQTB. TorchNFF is in an early development version and is thus not yet optimized. For example, a FENNIX-OP1 simulations of a box of 216 molecules of water in periodic boundary conditions can currently reach about 0.8 ns day−1 of adQTB simulation (with a timestep of 0.5 fs) on a single A100 GPU.
4.1 Bond dissociation energy profiles
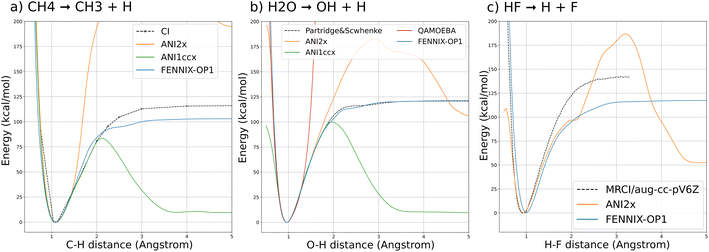
We first validate the training and the choice of reference energy on potential energy curves for the dissociation of covalent bonds in small molecules. This kind of bond breaking is a fundamental step in the process of many chemical reactions, for example in enzyme-catalyzed reactions,71 and is key to the description of mass spectroscopy experiments.72,73 They are however difficult to accurately model: force fields usually forbid them by design (by assigning a fixed connectivity) and ab initio approaches require the use of expensive multi-reference calculations to correctly describe the dissociation process. Their practical description thus usually requires the use of specifically designed force fields (such as ReaxFF) or low-dimensional potential energy surfaces.Fig. 3 shows the potential energy curves for (a) the dissociation of a hydrogen atom in methane; (b) the asymmetric dissociation of the water molecule; (c) the dissociation of the H–F molecule computed with FENNIX-OP1 and compared with reference quantum calculations from the literature (multi-reference CI/6-31G** from ref. 74 for CH4, Partridge-Scwhenke PES75 that was fitted on CCSD(T) data and multi-reference CI/aug-cc-pV6Z from ref. 76 for the H–F molecule) and the ANI models. We see that FENNIX-OP1 consistently captures a smooth dissociation curve thanks to the physically-motivated choice of reference energy. On the other hand, the ANI models, for which the reference energy was set according to average values over the dataset, fail to reproduce the dissociation curves for large distances. FENNIX-OP1 agrees particularly well with the reference for water as the Partridge–Schwenke PES was included in the training set. For the other two molecules, it tends to underestimate the dissociation barrier. Interestingly, while the training data did not contain covalent interaction energies for F, the model is still able to reasonably generalize its prediction. In Appendix, we describe how the model learned a generic representation of dissociation energy profiles, independently of chemical species. The knowledge of the chemical species involved in the bond (via the chemical encoding) then provides the details of the energy curve such as the equilibrium bond length and the height of the dissociation barrier, with some transferability across the periodic table thanks to the positional encoding introduced in Section 2.1.2.
4.2 Structural and spectroscopic properties of liquid water
In order to test the robustness of the model, we performed molecular dynamics simulations of liquid water. Since the model was fitted purely on ab initio data, it was critical to explicitly include nuclear quantum effects in order to accurately compute thermodynamical properties.55 We used the adaptive quantum thermal bath method to include NQEs,53 as it was previously shown to be a robust alternative to the more costly path integrals MD.54 All simulations were performed for a box of 216 molecules in the NVT ensemble at 300K and experimental density, with the BAOAB integrator78 and a timestep of 0.5 fs. Coulomb interactions in periodic boundary conditions were handled using the Ewald summation method,79 that we directly implemented using PyTorch operations so that we can leverage its automatic differentiation capabilities to obtain atomic forces. We equilibrated the system for 100 ps, which was sufficient to thermalize the system and converge the adQTB parameters. We then computed the radial distribution functions (RDFs) from 1 ns of MD simulation. Fig. 4 shows the partial RDFs of water simulated using adQTB and classical MD with the FENNIX-OP1 model compared to experimental results from ref. 77. As a baseline, we also compare to ANI-2x results with adQTB MD. We see that FENNIX-OP1 is able to accurately capture the subtle structure of liquid water and agrees well with experimental results when nuclear quantum effects are included. This is quite remarkable as the model was not explicitly trained on condensed phase reference data. On the other hand, ANI-2x predicts a largely over-structured liquid, which was shown to be mainly due to the insufficient quality of the DFT reference used for training the model.80 The ANI-1ccx model, that was trained with CCSD(T) reference data, produced very accurate radial distribution functions at the beginning of the simulations, thus validating the necessity of accurate CCSD(T) reference data to be able to describe liquid water. It was however not stable for simulation times longer than a few tens of picoseconds (even in classical MD with a smaller 0.1 fs timestep). As pointed out in ref. 80, this can be affected to the lack of long-range effects that are important to maintain the structure of the liquid. Furthermore, we show in ESI,† that neglecting the long-range tail of the dispersion interactions in FENNIX-OP1 leads to visible distortions of the radial distribution functions. The sensitivity of the structural results even to these relatively small interactions (compared to the larger Coulomb interactions) reinforces the necessity to accurately capture long-range interactions with physics-informed models.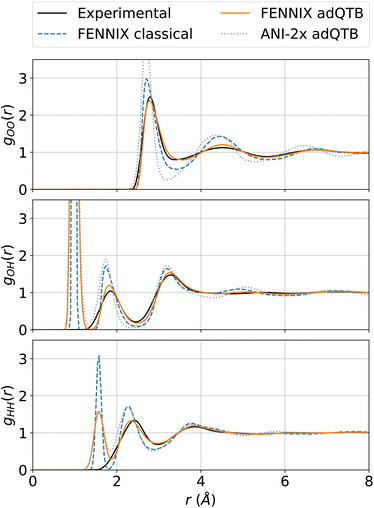 | ||
| Fig. 4 Partial radial distribution functions of liquid water at 300 K simulated using classical MD and adQTB MD with the FENNIX-OP1 model, compared to experimental results from ref. 77 and ANI-2x (adQTB) results. | ||
We then explored the impact of deuteration on the structural properties. Such thermodynamical isotope effects arise purely from NQEs (since the classical Boltzmann distribution does not depend on the mass of the atoms) and can be used to experimentally probe the impact of quantum effects on chemical processes. In liquid water, deuteration tends to strengthen the liquid's structure, resulting in slightly sharper peaks in the O–O radial distribution function.77,81,82 Reproducing this effect is a stringent test for the interaction model as it requires to accurately capture the subtle balance of competing quantum effects that affect the hydrogen bonds,82,83 which is challenging even for accurate DFT functionals.84 We showed in earlier works that, provided an accurate potential energy surface, the adQTB is able to reliably capture these isotope effects.54,56Fig. 5 shows the O–O radial distribution function of light and heavy water at 300 K computed using FENNIX-OP1 and compared to experimental results from ref. 81. The model captures the strengthening of the structure with deuteration but seems to amplify the effect, which could indirectly indicate slightly too weak hydrogen bonds.56,82,83
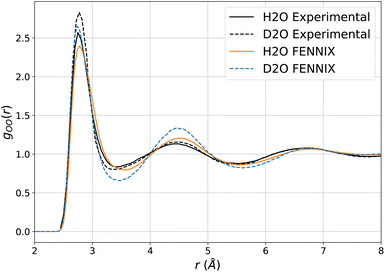 | ||
| Fig. 5 O–O radial distribution computed using adQTB and the FENNIX-OP1 model for light and heavy water, compared to experimental results from ref. 81. | ||
Additionally, FENNIX-OP1 predicts an enthalpy of vaporization for light water around 13 kcal mol−1, which is slightly overestimated compared to the experimental result of 10.51 kcal mol−1.85 This paradoxically indicates a tendency of the model to form too strong hydrogen bonds in the condensed phase (further investigations will be required). We expect that training the model using interaction energies from larger molecular clusters would improve the results overall. Indeed, it was recently shown with the q-AQUA(-pol)20,50 and MB-pol(2023)19 water models that including interaction energies of water clusters containing at least up to four molecules in the training set was necessary to obtain state-of-the-art results on both gas-phase and condensed-phase properties.
Contrary to ML models that only predict interaction energies, FENNIX provides environment-dependent atomic charges, enabling us to estimate infrared spectra through the computation of time correlation functions of the (nonlinear) total dipole moment from the adQTB dynamics. The results, shown in ESI,† are in qualitative agreement with experimental data for the main features, albeit a 150 cm−1 red-shift of the bending peak (similar to the q-SPC/Fw model86) and a broadening of low-frequency features. Comparison to spectra computed using classical MD shows that the model captures the typical red-shift of the stretching peak due to nuclear quantum effects.87–89 Still, better accuracy on liquid water's IR spectrum are obtained by models specifically targeting water, for example in ref. 90, that include a refined many-body dipole moment surface. Furthermore, the FENNIX-OP1 spectra lack typical features produced by the slow dynamics of induced dipole.56,87,91 Even though FENNIX-OP1 is able to capture local polarization effects via fluctuating charges, this subtle many-body effect cannot be reproduced without an explicit treatment of long-range polarization, as is done for example in (Q-)AMOEBA,56,92 SIBFA9 or ARROW.7 Future iterations of FENNIX will then focus on refining the physical model for including such interactions and better reproducing the system's dipole moment and its evolution in a framework including multipolar electrostatics.
4.3 Enhanced sampling of the torsional free energy profile of solvated alanine dipetide
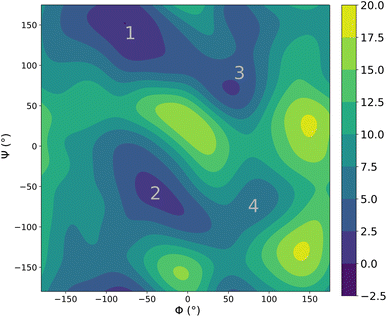
Lastly, we performed molecular dynamics simulations (using enhanced sampling) of alanine dipetide explicitly solvated in a cubic box of water of length 30 Å (with periodic boundary conditions). The alanine dipeptide is a fundamental building block for larger biomolecules and its accurate description is thus critical. This system is also a typical benchmark for both interaction models and enhanced sampling methods and was recently shown to be extremely challenging for ML potentials.35 It thus constitutes an interesting test case for our potential.Fig. 6 shows the joint free energy profile, obtained after 3 ns of classical well-tempered metadynamics93 simulation (performed using the PLUMED library94) with the FENNIX-OP1 model, for the two dihedral angles defining the torsional degrees of freedom of the molecule. The model correctly assigns most of the probability to the two expected energy basins denoted 1 and 2 on Fig. 6. It was also able to explore the less probable states denoted 3 and 4 in the figure. The model however seems to underestimate the barrier at Φ = 0, thus allowing too many transitions between states 1 and 2 to 3 and 4. We note that this simulation was performed using classical MD, as the adQTB is not directly compatible with enhanced sampling methods. It would be interesting to explore the impact of nuclear quantum effects on this energy profile. Indeed, as we showed above, nuclear quantum effects drastically modify the structure of water. Since the torsional free energy profile is strongly affected by the solvent (see the difference between solvated and gas phase alanine dipeptide for example in ref. 95), we can expect important changes when going from classical to quantum dynamics. This calculation would require long and costly path-integrals simulations or the development of adequate enhanced sampling techniques for the adQTB, that we leave for future works.
4.4 Perspective: gas-phase simulation of a protein
As a perspective, we pushed the model towards larger molecular structures that are not included in the training data. We performed molecular dynamics, including nuclear quantum effects, of the 26-residue 1FSV protein96 in gas phase. It contains around 500 atoms with a few charged groups. Since the model is not designed to handle ionic species, we first performed a simulation where we neutralized each charged group (by addition or subtraction of a proton). We ran an adQTB MD simulation for 500 ps with a timestep of 0.5 fs. Starting from the PDB structure, the dynamics was stable and the protein folded to a more compact form typical of the gas phase. The RMSD of the backbone with respect to the PDB structure stabilized after around 150 ps of simulation to a value of ∼3.2 Å, as shown in Fig. 7. This fast structural rearrangement is mostly driven by the long-range interactions present in the model whose magnitude is greater in gas-phase compared to condensed-phase due to a lack of screening by the solvent. These preliminary results illustrate the robustness of the force-field-enhanced ML approach, able to generalize to much larger and complex systems than that included in the training set.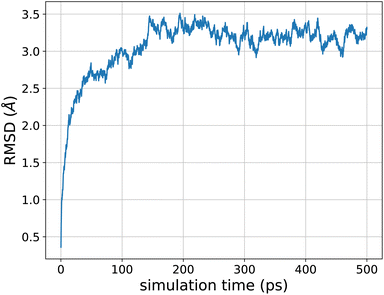 | ||
| Fig. 7 Root-mean-square deviation of the 1FSV protein with respect to the PDB structure during a 500 ps adQTB MD simulation in gas phase with the FENNIX-OP1 model. | ||
Even though the model was not trained on charged species, we ran a short MD simulation of the protein in solution by explicitly distributing the ionic charges among the atoms of the charged groups. After a few picoseconds, however, the simulation displayed instabilities due to unphysical proton transfers around the charged groups and too large Coulomb interactions that collapsed the molecule. In order to produce quantitative results and stable dynamics in this context, next iterations of the FENNIX model will need to explicitly handle charged systems, which will require a more thorough dataset. The recently introduced SPICE dataset97 could be a good complement to the ones already used in this work as it provides reference data for many charged molecules and focuses on biological systems (however at the lower quality DFT level). Furthermore, it will require finer description of molecular interactions through the inclusion of more advanced energy terms (such as multipolar electrostatics and polarization).
5. Conclusion and outlooks
We introduced FENNIX, a general class of models representing molecular interactions using an hybrid ML/FF approach. It builds upon the latest equivariant architectures to construct an embedding of the local chemical environment which is fed into a multi-output module predicting atom-in-molecule properties. The latter allows for a flexible design of potential energy terms. We apply this strategy to design a specific model: FENNIX-OP1, capturing both short-range interactions, with a dedicated ML energy term, and long-range ones using predicted atomic volumes and charges that parametrize both a charge penetration corrected electrostatic energy and a Tkatchenko–Scheffler like dispersion one. FENNIX-OP1 is first trained on both the ANI-1ccx and the DES370K datasets which contain monomers, dimers and a few multimeric structures focusing on small neutral organic molecules. It is then refined for water using the q-AQUA dimers dataset and the highly accurate Partridge–Schwenke potential energy surface for a final RMSE of 0.35 kcal mol−1 on interaction energies. We then showed that the model is stable during molecular dynamics (including NQEs via adQTB) and able to generalize to condensed phase (despite the lack of reference data), capturing structural properties of bulk water and solvated organic molecules. More precisely, it qualitatively reproduces the torsional free energy profile of the solvated alanine dipeptide using enhanced sampling techniques and yields stable trajectories of the 1FSV protein in gas phase. Interestingly, we showed that the model learned some generic concepts (such as dissociation profiles) independently of the chemical species, which are then specifically refined for each element – with some transferability across the periodic table – thanks to the continuous encoding of their positions in the periodic table. The good accuracy of the model for bond dissociations in simple molecules is a promising first step toward the general description of more complex chemical reactions.All in all, this work shows the extrapolating power of hybrid physically-driven machine learning approaches and paves the way to even more refined models using enriched data sets (in particular for charged systems) and additional energy terms in the spirit of advanced polarizable force-fields such as SIBFA that include multipolar electrostatics and many-body polarization and dispersion. These will enable the study of reactive complex systems such as proton transfers between DNA pairs or enzyme catalyzed reactions where such an accurate description of molecular interactions is mandatory. An important question going forward is to what extent general and reactive models can compete with specialized potentials. Indeed, specialized models19,20,50 can achieve very good accuracy on multiple properties of the system they target, that general ones (such as FENNIX-OP1) currently do not match. On the other hand, the flexibility of general models enables the simulation of more complex systems involving a large variety of constituents, as is often the case in biochemistry. A promising strategy that could provide a solid middle-ground is the fine-tuning approach where a general model is slightly re-trained with limited data to better represent a specific target system. Further work will focus on the development and use of more general datasets (such as the SPICE dataset97), the extension of the FENNIX approach and its inclusion in the optimized Deep-HP platform98,99 present within the Tinker-HP package.12,100
Appendix: dissociations across chemical space
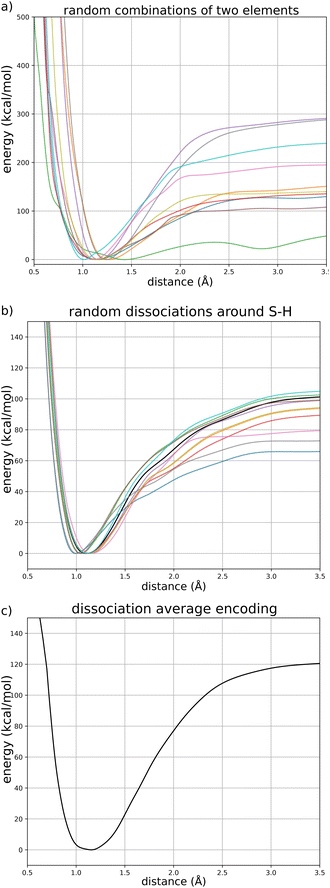
The goal of this section is to illustrate that FENNIX-OP1 learned a generic representation of dissociation energy profiles (independently of the chemical species involved) and that the positional encoding of chemical species provides the specific properties of each bond (for example the equilibrium distance and bond dissociation energy). To this end, we present three numerical experiments shown in Fig. 8: (a) dissociation curves for random couples of chemical element; (b) S–H dissociation with the periodic table coordinates of S perturbed by Gaussian noise; (c) generic dissociation curve for the average encoding.Fig. 8a shows dissociation curves for ten random couples of chemical elements (from H to Xe). Even if most of these elements were not included in the training data, FENNIX-OP1 predicts meaningful energy profiles with a clear minimum and short-range repulsion, which shows that the general shape of the dissociation curve is not tied to the chemical encoding.
We then exploited the continuity of the chemical encoding with respect to coordinates in the periodic table in order to show its impact on a specific dissociation curve. Fig. 8b show the dissociation profile of S–H (in black) as well as other profiles obtained by shifting the coordinates of S in the periodic table by a two-dimensional Gaussian random vector (with mean 0 and standard deviation 1). We see that moving this way in the periodic table produces similarly-shaped energy profiles but with different bond parameters (bond length, dissociation energy, frequency at equilibrium…) which are dictated by the specific value of the encoding.
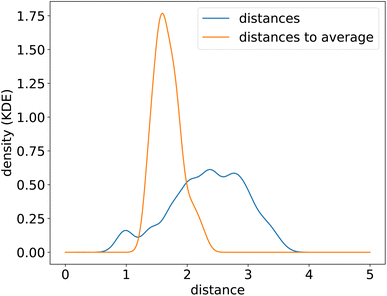
To uncover the generic energy profile learned by the model, we show in Fig. 8c the homonuclear dissociation energy profile of a fictitious element which encoding is the average encoding of elements from H to Xe. In the encoding space, this average element is roughly at equal distance from all the other elements, and closer in average to all elements than true elements are to each other, as can be seen from the distributions of distances in the encoding space shown in Fig. 9. The dissociation curve shown in Fig. 8c can thus be thought of as the prototypical energy profile learned by the model, independently of the chemical species.
Author contributions
T. P. performed simulations and contributed new code; T. P., L. L., J. P. P. contributed new methodology; T. P., L. L., J. P. P. contributed analytical tool; T. P., L. L., J. P. P. analyzed data. T. P., L. L., J. P. P. wrote the paper; J. P. P. designed the research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was made possible thanks to funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No. 810367), project EMC2. Computations have been performed at GENCI (IDRIS, Orsay, France and TGCC, Bruyères le Chatel) on grant no A0130712052.References
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, Development and testing of a general amber force field, J. Comput. Chem., 2004, 25, 1157 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes and I. Vorobyov, et al., Charmm general force field: a force field for drug-like molecules compatible with the charmm all-atom additive biological force fields, J. Comput. Chem., 2010, 31, 671 CAS.
- J. Melcr and J.-P. Piquemal, Accurate biomolecular simulations account for electronic polarization, Front. Mol. Biosci., 2019, 6, 143 CrossRef CAS PubMed.
- J. W. Ponder, C. Wu, P. Ren, V. S. Pande, J. D. Chodera, M. J. Schnieders, I. Haque, D. L. Mobley, D. S. Lambrecht and R. A. DiStasio Jr, et al., Current status of the amoeba polarizable force field, J. Phys. Chem. B, 2010, 114, 2549 CrossRef CAS PubMed.
- G. S. Fanourgakis and S. S. Xantheas, Development of transferable interaction potentials for water. v. extension of the flexible, polarizable, thole-type model potential (TTM3-F, v. 3.0) to describe the vibrational spectra of water clusters and liquid water, J. Chem. Phys., 2008, 128, 074506 CrossRef PubMed.
- J. A. Lemkul, J. Huang, B. Roux and A. D. MacKerell Jr, An empirical polarizable force field based on the classical drude oscillator model: development history and recent applications, Chem. Rev., 2016, 116, 4983 CrossRef CAS PubMed.
- L. Pereyaslavets, G. Kamath, O. Butin, A. Illarionov, M. Olevanov, I. Kurnikov, S. Sakipov, I. Leontyev, E. Voronina and T. Gannon, et al., Accurate determination of solvation free energies of neutral organic compounds from first principles, Nat. Commun., 2022, 13, 1 Search PubMed.
- N. Gresh, G. A. Cisneros, T. A. Darden and J.-P. Piquemal, Anisotropic, polarizable molecular mechanics studies of inter-and intramolecular interactions and ligand- macromolecule complexes. a bottom-up strategy, J. Chem. Theor. Comput., 2007, 3, 1960 CrossRef CAS PubMed.
- S. Naseem-Khan, L. Lagardère, C. Narth, G. A. Cisneros, P. Ren, N. Gresh and J.-P. Piquemal, Development of the quantum inspired sibfa many-body polarizable force field: I. enabling condensed phase molecular dynamics simulations, J. Chem. Theory Comput., 2022, 18(6), 3607–3621 CrossRef CAS PubMed.
- Y. Shi, P. Ren, M. Schnieders and J.-P. Piquemal, Polarizable force fields for biomolecular modeling, in Reviews in Computational Chemistry, John Wiley and Sons, Ltd, 2015, ch. 2, vol. 28, pp. 51–86, DOI:10.1002/9781118889886.ch2.
- Z. Jing, C. Liu, S. Y. Cheng, R. Qi, B. D. Walker, J.-P. Piquemal and P. Ren, Polarizable force fields for biomolecular simulations: recent advances and applications, Annu. Rev. Biophys., 2019, 48, 371 CrossRef CAS PubMed.
- O. Adjoua, L. Lagardère, L.-H. Jolly, A. Durocher, T. Very, I. Dupays, Z. Wang, T. J. Inizan, F. Célerse, P. Ren, J. W. Ponder and J.-P. Piquemal, Tinker-HP: accelerating molecular dynamics simulations of large complex systems with advanced point dipole polarizable force fields using gpus and multi-gpu systems, J. Chem. Theor. Comput., 2021, 17, 2034 CrossRef CAS PubMed.
- A. C. Van Duin, S. Dasgupta, F. Lorant and W. A. Goddard, ReaxFF: a reactive force field for hydrocarbons, Annu. Rev. Biophys., 2001, 105, 9396 CAS.
- A. Warshel and R. M. Weiss, An empirical valence bond approach for comparing reactions in solutions and in enzymes, J. Am. Chem. Soc., 1980, 102, 6218 CrossRef CAS.
- K. Shakouri, J. Behler, J. Meyer and G.-J. Kroes, Accurate neural network description of surface phonons in reactive gas–surface dynamics: N2+ Ru (0001), J. Phys. Chem. Lett., 2017, 8, 2131 CrossRef CAS PubMed.
- J. S. Smith, O. Isayev and A. E. Roitberg, Ani-1: an extensible neural network potential with dft accuracy at force field computational cost, Chem. Sci., 2017, 8, 3192 RSC.
- S. Batzner, A. Musaelian, L. Sun, M. Geiger, J. P. Mailoa, M. Kornbluth, N. Molinari, T. E. Smidt and B. Kozinsky, E (3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials, Nat. Commun., 2022, 13, 1 Search PubMed.
- R. Drautz, Atomic cluster expansion for accurate and transferable interatomic potentials, Phys. Rev. B: Condens. Matter Mater. Phys., 2019, 99, 014104 CrossRef CAS.
- X. Zhu, M. Riera, E. F. Bull-Vulpe and F. Paesani, MB-pol (2023): sub-chemical accuracy for water simulations from the gas to the liquid phase, J. Chem. Theory Comput., 2023, 3551–3566 CrossRef CAS PubMed.
- Q. Yu, C. Qu, P. L. Houston, R. Conte, A. Nandi and J. M. Bowman, q-aqua: a many-body ccsd (t) water potential, including four-body interactions, demonstrates the quantum nature of water from clusters to the liquid phase, J. Phys. Chem. Lett., 2022, 13, 5068 CrossRef CAS PubMed.
- S. Chmiela, H. E. Sauceda, K.-R. Müller and A. Tkatchenko, Towards exact molecular dynamics simulations with machine-learned force fields, Nat. Commun., 2018, 9, 1 CrossRef CAS PubMed.
- F. Bigi, S. N. Pozdnyakov, and M. Ceriotti, Wigner kernels: body-ordered equivariant machine learning without a basis, arXiv, 2023, preprint, arXiv:2303.04124, DOI:10.48550/arXiv.2303.04124.
- R. Zubatyuk, J. S. Smith, J. Leszczynski and O. Isayev, Accurate and transferable multitask prediction of chemical properties with an atoms-in-molecules neural network, Sci. Adv., 2019, 5, eaav6490 CrossRef CAS PubMed.
- R. Zubatyuk, J. S. Smith, B. T. Nebgen, S. Tretiak and O. Isayev, Teaching a neural network to attach and detach electrons from molecules, Nat. Commun., 2021, 12, 1 CrossRef PubMed.
- L. Zhang, J. Han, H. Wang, R. Car and E. Weinan, Deep potential molecular dynamics: a scalable model with the accuracy of quantum mechanics, Phys. Rev. Lett., 2018, 120, 143001 CrossRef CAS PubMed.
- K. Yao, J. E. Herr, D. Toth, R. Mckintyre and J. Parkhill, The tensormol-0.1 model chemistry: a neural network augmented with long-range physics, Chem. Sci., 2018, 9, 2261 RSC.
- F. Faber, A. Lindmaa, O. A. von Lilienfeld and R. Armiento, Crystal structure representations for machine learning models of formation energies, Int. J. Quantum Chem., 2015, 115, 1094 CrossRef CAS.
- Y. Lysogorskiy, C. v. d. Oord, A. Bochkarev, S. Menon, M. Rinaldi, T. Hammerschmidt, M. Mrovec, A. Thompson, G. Csányi and C. Ortner, et al., Performant implementation of the atomic cluster expansion (pace) and application to copper and silicon, npj Comput. Mater., 2021, 7, 1 CrossRef.
- L. Zhang, H. Wang, M. C. Muniz, A. Z. Panagiotopoulos, R. Car and W. E, A deep potential model with long-range electrostatic interactions, J. Chem. Phys., 2022, 156, 124107 CrossRef CAS PubMed.
- B. Cheng, E. A. Engel, J. Behler, C. Dellago and M. Ceriotti, Ab initio thermodynamics of liquid and solid water, Proc. Natl. Acad. Sci. U.S.A., 2019, 116, 1110 CrossRef CAS PubMed.
- A. Musaelian, S. Batzner, A. Johansson, L. Sun, C. J. Owen, M. Kornbluth and B. Kozinsky, Learning local equivariant representations for large-scale atomistic dynamics, Nat. Commun., 2023, 14, 579 CrossRef CAS PubMed.
- O. T. Unke, S. Chmiela, M. Gastegger, K. T. Schütt, H. E. Sauceda and K.-R. Müller, Spookynet: learning force fields with electronic degrees of freedom and nonlocal effects, Nat. Commun., 2021, 12, 1 CrossRef PubMed.
- Z. Qiao, A. S. Christensen, M. Welborn, F. R. Manby, A. Anandkumar and Miller, Informing geometric deep learning with electronic interactions to accelerate quantum chemistry, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(31), e2205221119 CrossRef CAS PubMed.
- J. Gasteiger, F. Becker and S. Günnemann, Gemnet: universal directional graph neural networks for molecules, Adv. Neural Inf. Process. Syst., 2021, 34, 6790 Search PubMed.
- X. Fu, Z. Wu, W. Wang, T. Xie, S. Keten, R. Gomez-Bombarelli and T. Jaakkola, Forces are not enough: benchmark and critical evaluation for machine learning force fields with molecular simulations, arXiv, 2022, preprint, arXiv:2210.07237, DOI:10.48550/arXiv.2210.07237.
- M. M. Gromiha and S. Selvaraj, Importance of long-range interactions in protein folding, Biophys. Chem., 1999, 77, 49 CrossRef CAS PubMed.
- D. M. York, T. A. Darden and L. G. Pedersen, The effect of long-range electrostatic interactions in simulations of macromolecular crystals: a comparison of the ewald and truncated list methods, J. Chem. Phys., 1993, 99, 8345 CrossRef CAS.
- J. Gilmer, S. S. Schoenholz, P. F. Riley, O. Vinyals and G. E. Dahl, Neural message passing for quantum chemistry, in International conference on machine learning, PMLR, 2017, pp. 1263–1272 Search PubMed.
- M. Haghighatlari, J. Li, X. Guan, O. Zhang, A. Das, C. J. Stein, F. Heidar-Zadeh, M. Liu, M. Head-Gordon and L. Bertels, et al., Newtonnet: a newtonian message passing network for deep learning of interatomic potentials and forces, Digital Discovery, 2022, 1, 333 RSC.
- M. Rupp, A. Tkatchenko, K.-R. Müller and O. A. Von Lilienfeld, Fast and accurate modeling of molecular atomization energies with machine learning, Phys. Rev. Lett., 2012, 108, 058301 CrossRef PubMed.
- P. O. Dral, A. Owens, S. N. Yurchenko and W. Thiel, Structure-based sampling and self-correcting machine learning for accurate calculations of potential energy surfaces and vibrational levels, J. Chem. Phys., 2017, 146, 244108 CrossRef PubMed.
- S. Chmiela, V. Vassilev-Galindo, O. T. Unke, A. Kabylda, H. E. Sauceda, A. Tkatchenko and K.-R. Müller, Accurate global machine learning force fields for molecules with hundreds of atoms, Sci. Adv., 2023, 9, eadf0873 CrossRef PubMed.
- A. Grisafi and M. Ceriotti, Incorporating long-range physics in atomic-scale machine learning, J. Chem. Phys., 2019, 151, 204105 CrossRef PubMed.
- A. Grisafi, J. Nigam and M. Ceriotti, Multi-scale approach for the prediction of atomic scale properties, Chem. Sci., 2021, 12, 2078 RSC.
- K. Szalewicz, Symmetry-adapted perturbation theory of intermolecular forces, Wiley Interdiscip. Rev. Comput. Mol. Sci., 2012, 2, 254 CrossRef CAS.
- S. Naseem-Khan, N. Gresh, A. J. Misquitta and J.-P. Piquemal, Assessment of SAPT and supermolecular eda approaches for the development of separable and polarizable force fields, J. Chem. Theory Comput., 2021, 17, 2759 CrossRef CAS PubMed.
- T. W. Ko, J. A. Finkler, S. Goedecker and J. Behler, A fourth-generation high-dimensional neural network potential with accurate electrostatics including non-local charge transfer, Nat. Commun., 2021, 12, 1 CrossRef PubMed.
- O. T. Unke and M. Meuwly, Physnet: a neural network for predicting energies, forces, dipole moments, and partial charges, J. Chem. Theory Comput., 2019, 15, 3678 CrossRef CAS PubMed.
- N. T. P. Tu, N. Rezajooei, E. R. Johnson and C. Rowley, A neural network potential with rigorous treatment of long-range dispersion, Digital Discovery, 2023,(3), 718–727 RSC.
- C. Qu, Q. Yu, P. L. Houston, R. Conte, A. Nandi and J. M. Bowman, Interfacing q-aqua with a polarizable force field: the best of both worlds, J. Chem. Theory Comput., 2023, 19(12), 3446–3459 CrossRef CAS PubMed.
- L. Yang, J. Li, F. Chen and K. Yu, A transferrable range-separated force field for water: combining the power of both physically-motivated models and machine learning techniques, J. Chem. Phys., 2022, 157, 214108 CrossRef CAS PubMed.
- J. M. Bowman, C. Qu, R. Conte, A. Nandi, P. L. Houston and Q. Yu, δ-machine learned potential energy surfaces and force fields, J. Chem. Theory Comput., 2022, 19, 1 CrossRef PubMed.
- E. Mangaud, S. Huppert, T. Plé, P. Depondt, S. Bonella and F. Finocchi, The fluctuation–dissipation theorem as a diagnosis and cure for zero-point energy leakage in quantum thermal bath simulations, J. Chem. Theory Comput., 2019, 15, 2863 CrossRef CAS PubMed.
- N. Mauger, T. Plé, L. Lagardère, S. Bonella, É. Mangaud, J.-P. Piquemal and S. Huppert, Nuclear quantum effects in liquid water at near classical computational cost using the adaptive quantum thermal bath, J. Phys. Chem. Lett., 2021, 12(34), 8285–8291 CrossRef CAS PubMed.
- L. Pereyaslavets, I. Kurnikov, G. Kamath, O. Butin, A. Illarionov, I. Leontyev, M. Olevanov, M. Levitt, R. D. Kornberg and B. Fain, On the importance of accounting for nuclear quantum effects in ab initio calibrated force fields in biological simulations, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 8878 CrossRef CAS PubMed.
- N. Mauger, T. Plé, L. Lagardère, S. Huppert and J.-P. Piquemal, Improving condensed-phase water dynamics with explicit nuclear quantum effects: the polarizable Q-AMOEBA force field, J. Phys. Chem. B, 2022, 126, 8813 CrossRef CAS PubMed.
- A. Barducci, M. Bonomi and M. Parrinello, Metadynamics, Wiley Interdiscip. Rev.-Comput. Mol. Sci., 2011, 1, 826 CrossRef CAS.
- M. Geiger and T. Smidt, e3nn: euclidean neural networks, arXiv, 2022, preprint, arXiv:2207.09453, DOI:10.48550/arXiv.2003.03123.
- J. Gasteiger, J. Groß, and S. Günnemann, Directional message passing for molecular graphs, arXiv, preprint, arXiv:2003.03123, 2020 Search PubMed.
- S. Takamoto, S. Izumi and J. Li, Teanet: universal neural network interatomic potential inspired by iterative electronic relaxations, Comput. Mater. Sci., 2022, 207, 111280 CrossRef CAS.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, Ł. Kaiser and I. Polosukhin, Attention is all you need, Adv. Neural Inf. Process. Sys., 2017, 30, 5998–6008 Search PubMed.
- J.-P. Piquemal, N. Gresh and C. Giessner-Prettre, Improved formulas for the calculation of the electrostatic contribution to the intermolecular interaction energy from multipolar expansion of the electronic distribution, J. Phys. Chem. A, 2003, 107, 10353 CrossRef CAS PubMed.
- A. Tkatchenko and M. Scheffler, Accurate molecular van der waals interactions from ground-state electron density and free-atom reference data, Phys. Rev. Lett., 2009, 102, 073005 CrossRef PubMed.
- P. P. Poier and F. Jensen, Describing molecular polarizability by a bond capacity model, J. Chem. Theor. Comput., 2019, 15, 3093 CrossRef CAS PubMed.
- I. Batatia, S. Batzner, D. P. Kovács, A. Musaelian, G. N. Simm, R. Drautz, C. Ortner, B. Kozinsky and G. Csányi, The design space of e (3)-equivariant atom-centered interatomic potentials, arXiv, 2022, preprint, arXiv:2205.06643, DOI:10.48550/arXiv.2205.06643.
- A. G. Donchev, A. G. Taube, E. Decolvenaere, C. Hargus, R. T. McGibbon, K.-H. Law, B. A. Gregersen, J.-L. Li, K. Palmo and K. Siva, et al., Quantum chemical benchmark databases of gold-standard dimer interaction energies, Sci. Data, 2021, 8, 1 Search PubMed.
- T. Verstraelen, S. Vandenbrande, F. Heidar-Zadeh, L. Vanduyfhuys, V. Van Speybroeck, M. Waroquier and P. W. Ayers, Minimal basis iterative stockholder: atoms in molecules for force-field development, J. Chem. Theory Comput., 2016, 12, 3894 CrossRef CAS PubMed.
- I. Loshchilov and F. Hutter, Decoupled weight decay regularization, arXiv, 2017, preprint, arXiv:1711.05101, DOI:10.48550/arXiv.1711.05101.
- B. J. Smith, D. J. Swanton, J. A. Pople, H. F. Schaefer III and L. Radom, Transition structures for the interchange of hydrogen atoms within the water dimer, J. Chem. Phys., 1990, 92, 1240 CrossRef CAS.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Kopf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai, and S. Chintala, Pytorch: an imperative style, high-performance deep learning library, in Advances in Neural Information Processing Systems 32, ed. by H. Wallach, H. Larochelle, A. Beygelzimer, F. d' Alché-Buc, E. Fox and R. Garnett, Curran Associates, Inc., 2019, pp. 8024–8035 Search PubMed.
- A. Warshel, P. K. Sharma, M. Kato, Y. Xiang, H. Liu and M. H. Olsson, Electrostatic basis for enzyme catalysis, Chem. Rev., 2006, 106, 3210 CrossRef CAS PubMed.
- T. Baer and W. L. Hase, Unimolecular reaction dynamics: theory and experiments, Oxford University Press on Demand, 1996, vol. 31 Search PubMed.
- E. De Hoffmann and V. Stroobant, Mass spectrometry: principles and applications, John Wiley & Sons, 2007 Search PubMed.
- D. M. Hirst, An ab initio potential energy surface for the reaction ch4 → ch3+ h, Chem. Phys. Lett., 1985, 122, 225 CrossRef CAS.
- H. Partridge and D. W. Schwenke, The determination of an accurate isotope dependent potential energy surface for water from extensive ab initio calculations and experimental data, J. Chem. Phys., 1997, 106, 4618 CrossRef CAS.
- H. Duohui, W. Fanhou, Y. Junsheng, C. Qilong and W. Mingjie, Mrci study on potential energy curves and spectroscopic properties of hf molecule, Spectrochim. Acta, Part A, 2014, 128, 163 CrossRef PubMed.
- A. K. Soper, The radial distribution functions of water as derived from radiation total scattering experiments: is there anything we can say for sure?, Int. Sch. Res. Notices, 2013, 2013, 279463 Search PubMed.
- B. Leimkuhler and C. Matthews, Rational construction of stochastic numerical methods for molecular sampling, Appl. Math. Res. eXpress, 2012 DOI:10.1093/amrx/abs010.
- P. P. Ewald, Ewald summation, Ann. Phys., 1921, 369, 1 CrossRef.
- D. Rosenberger, J. S. Smith and A. E. Garcia, Modeling of peptides with classical and novel machine learning force fields: a comparison, J. Phys. Chem. B, 2021, 125, 3598 CrossRef CAS PubMed.
- A. Soper and C. Benmore, Quantum differences between heavy and light water, Phys. Rev. Lett., 2008, 101, 065502 CrossRef CAS PubMed.
- M. Ceriotti, W. Fang, P. G. Kusalik, R. H. McKenzie, A. Michaelides, M. A. Morales and T. E. Markland, Nuclear quantum effects in water and aqueous systems: experiment, theory, and current challenges, Chem. Rev., 2016, 116, 7529 CrossRef CAS PubMed.
- X.-Z. Li, B. Walker and A. Michaelides, Quantum nature of the hydrogen bond, Proc. Natl. Acad. Sci. U.S.A., 2011, 108, 6369 CrossRef CAS.
- C. Li, F. Paesani and G. A. Voth, Static and dynamic correlations in water: comparison of classical ab initio molecular dynamics at elevated temperature with path integral simulations at ambient temperature, J. Chem. Theory Comput., 2022, 18, 2124 CrossRef CAS PubMed.
- W. Wagner and A. Pruss, International equations for the saturation properties of ordinary water substance. Revised according to the international temperature scale of 1990. addendum to j. phys. chem. ref. data 16, 893 (1987), J. Phys. Chem. Ref. Data, 1993, 22, 783 CrossRef CAS.
- F. Paesani, W. Zhang, D. A. Case, T. E. Cheatham III and G. A. Voth, An accurate and simple quantum model for liquid water, J. Chem. Phys., 2006, 125, 184507 CrossRef PubMed.
- S. Habershon, T. E. Markland and D. E. Manolopoulos, Competing quantum effects in the dynamics of a flexible water model, J. Chem. Phys., 2009, 131, 024501 CrossRef PubMed.
- R. L. Benson, G. Trenins and S. C. Althorpe, Which quantum statistics–classical dynamics method is best for water?, Faraday Discuss., 2020, 221, 350 RSC.
- T. Plé, S. Huppert, F. Finocchi, P. Depondt and S. Bonella, Anharmonic spectral features via trajectory-based quantum dynamics: a perturbative analysis of the interplay between dynamics and sampling, J. Chem. Phys., 2021, 155, 104108 CrossRef PubMed.
- H. Liu, Y. Wang and J. M. Bowman, Quantum calculations of the ir spectrum of liquid water using ab initio and model potential and dipole moment surfaces and comparison with experiment, J. Chem. Phys., 2015, 142(19), 194502 CrossRef PubMed.
- R. Impey, P. Madden and I. McDonald, Spectroscopic and transport properties of water: model calculations and the interpretation of experimental results, Mol. Phys., 1982, 46, 513 CrossRef CAS.
- P. Ren and J. W. Ponder, Polarizable atomic multipole water model for molecular mechanics simulation, J. Phys. Chem. B, 2003, 107, 5933 CrossRef CAS.
- A. Barducci, G. Bussi and M. Parrinello, Well-tempered metadynamics: a smoothly converging and tunable free-energy method, Phys. Rev. Lett., 2008, 100, 020603 CrossRef PubMed.
- the PLUMED consortium, Promoting transparency and reproducibility in enhanced molecular simulations, Nat. Methods, 2019, 16, 670 CrossRef PubMed.
- P. E. Smith, The alanine dipeptide free energy surface in solution, J. Chem. Phys., 1999, 111, 5568 CrossRef CAS.
- B. I. Dahiyat and S. L. Mayo, De novo protein design: fully automated sequence selection, Science, 1997, 278, 82 CrossRef CAS PubMed.
- P. Eastman, P. K. Behara, D. L. Dotson, R. Galvelis, J. E. Herr, J. T. Horton, Y. Mao, J. D. Chodera, B. P. Pritchard and Y. Wang, et al., Spice, a dataset of drug-like molecules and peptides for training machine learning potentials, Sci. Data, 2023, 10, 11 CrossRef CAS PubMed.
- T. Jaffrelot Inizan, T. Plé, O. Adjoua, P. Ren, H. Gökcan, O. Isayev, L. Lagardère and J.-P. Piquemal, Scalable hybrid deep neural networks/polarizable potentials biomolecular simulations including long-range effects, Chem. Sci., 2023, 14, 5438–5452 RSC.
- T. Plé, N. Mauger, O. Adjoua, T. Jaffrelot-Inizan, L. Lagardère, S. Huppert and J.-P. Piquemal, Routine molecular dynamics simulations including nuclear quantum effects: from force fields to machine learning potentials, J. Chem. Theory Comput., 2023, 19(5), 1432–1445 CrossRef PubMed.
- L. Lagardère, L.-H. Jolly, F. Lipparini, F. Aviat, B. Stamm, Z. F. Jing, M. Harger, H. Torabifard, G. A. Cisneros, M. J. Schnieders, N. Gresh, Y. Maday, P. Y. Ren, J. W. Ponder and J.-P. Piquemal, Tinker-HP: a massively parallel molecular dynamics package for multiscale simulations of large complex systems with advanced point dipole polarizable force fields, Chem. Sci., 2018, 9, 956 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Contains the results for infrared spectra of liquid water computed using the FENNIX-OP1 model. See DOI: https://doi.org/10.1039/d3sc02581k |
| This journal is © The Royal Society of Chemistry 2023 |