Open Access Article
Open Access ArticleImpact of library input on the hit discovery rate in DNA-encoded chemical library selections†
Sara
Puglioli
 a,
Sebastian
Oehler
a,
Luca
Prati
a,
Sebastian
Oehler
a,
Luca
Prati
 a,
Jörg
Scheuermann
a,
Jörg
Scheuermann
 b,
Gabriele
Bassi
a,
Samuele
Cazzamalli
a,
Dario
Neri
*ab and
Nicholas
Favalli
b,
Gabriele
Bassi
a,
Samuele
Cazzamalli
a,
Dario
Neri
*ab and
Nicholas
Favalli
 *a
*a
aPhilochem AG, R&D Department, 8112 Otelfingen, Switzerland. E-mail: nicholas.favalli@philochem.ch; dario.neri@pharma.ethz.ch
bDepartment of Chemistry and Applied Biosciences, Swiss Federal Institute of Technology (ETH Zürich), Zürich, Switzerland
First published on 17th October 2023
Abstract
DNA-encoded chemical libraries (DELs) are powerful drug discovery tools, enabling the parallel screening of millions of DNA-barcoded compounds. We investigated how the DEL input affects the hit discovery rate in DEL screenings. Evaluation of selection fingerprints revealed that the use of approximately 105 copies of each library member is required for the confident identification of nanomolar hits, using generally applicable methodologies.
Introduction
The identification of small organic molecules, which specifically interact with a target protein of interest, represents an important problem in chemical research and a key challenge for drug discovery.1–3 Over the last few decades, DNA-encoded chemical libraries (DELs) have emerged as powerful and cost-effective tools for the discovery of ligands to pharmaceutically relevant proteins.4–8 DELs are large collections of small molecules that are chemically synthesized and covalently linked to cognate DNA sequences, serving as distinctive molecular barcodes. This encoding procedure enables the identification and relative quantification of individual compounds within the library, by means of DNA amplification with the Polymerase Chain Reaction (PCR) and high-throughput DNA sequencing.9–11The applicability of DEL technology as a powerful drug discovery tool has been demonstrated by recent examples of lead candidates which progressed into clinical trials for different pharmaceutical indications.12 Among the most advanced drug prototypes discovered thanks to the DEL technology and that are currently studied in phase I or II clinical trials, it is worth mentioning ligands of Receptor Interacting Protein Kinase 1 (RIPK1; discovered by GSK),13 of soluble Epoxide Hydrolase (sEH; discovered by GSK),14 and of autotaxin (ENPP2; discovered by X-Chem).15
Moreover, a variety of preclinical lead compounds isolated from DELs are being developed, such as PAR2 (AstraZeneca/X-Chem),6 Wip1 (GSK),16 BCATm (GSK),17 and DDR1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 (Roche) binders, and OncoFAP-11 (Philochem).7
18 (Roche) binders, and OncoFAP-11 (Philochem).7
The success rate of a DEL-based drug discovery screening campaign is influenced by multiple factors, including the library chemical purity, encoding fidelity and quality of the protein targets.19–21 Furthermore, affinity capture protocols should be optimized to guarantee reliable selection outcomes. The number of copies per library member used during selections (input) directly impacts on the success rate of DEL screening campaigns.22–24 The definition of a minimum number of input copies for each library member (threshold) represents an important experimental parameter, especially when libraries of very large dimensions (e.g., those containing billions of compounds) are used. It is likely that the threshold for efficient selections may be library-dependent, but only a few studies have addressed this aspect of DEL technology.22–24
We had previously reported that an input threshold of 105 copies per library member was required for the efficient identification of Carbonic Anhydrase IX-binding fragments (sulfonamide derivatives) in one specific DEL.22 Here we present a methodology to define input thresholds on two different well-characterized DELs: NF-DEL (iodo-phenylalanine based library) and SO-DEL (4-aminopyrrolidine-2-carboxylic acid library).25,26 Both yielded novel nanomolar hits for Carbonic Anhydrase IX (CAIX), Human Serum Albumin (HSA) and Non-Structural Protein-14 (NSP14).25,26 NF-DEL and SO-DEL were screened against the targets at different inputs, ranging from 10 million copies to 100 copies per library member per selection.25,27 A threshold of approximately 105 copies per library member was required in order to successfully identify binding fragments (lines in the fingerprint) and unique building block combinations (singletons in the selection fingerprint) against all screened proteins. This finding has an impact on the experimental design of DEL selection campaigns and may influence the screening procedures for very large encoded-compound collections.
Results and discussion
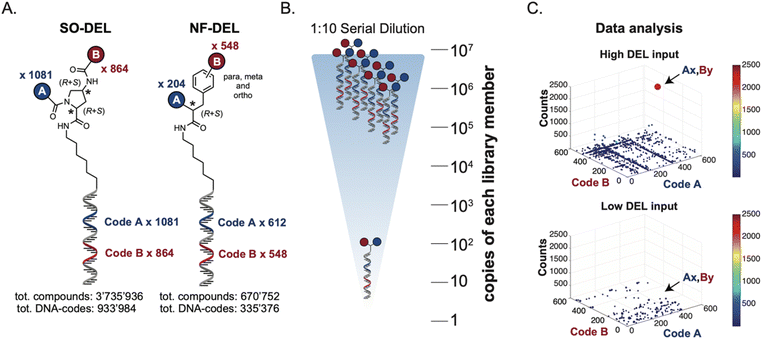
Library design and experimental strategy
The chemical structures of members of the SO-DEL26 and NF-DEL25 libraries, which were used to study the correlation between selection inputs and hit discovery rate and were constructed based on the combinatorial assembly of two sets of building blocks, are depicted in Fig. 1A. The synthesis and validation of the two libraries has been previously reported.25,26 SO-DEL and NF-DEL were constructed using chiral 4-amino-proline or iodo-phenylalanine as central scaffolds, comprising 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735
735![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 936 and 670
936 and 670![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 752 compounds, respectively. Previous screening campaigns performed with both libraries had resulted in the identification of hit compounds for a variety of pharmaceutically relevant target proteins (Table 1).25,26
752 compounds, respectively. Previous screening campaigns performed with both libraries had resulted in the identification of hit compounds for a variety of pharmaceutically relevant target proteins (Table 1).25,26
The two DELs were diluted to a concentration of 10 million copies per compound dissolved in 10 μL (the “selection input”, see also the ESI, Section 3†), and serially diluted to a final concentration of approximately 100 copies of each compound per selection (Fig. 1B).
Subsequently, libraries were screened at different selection inputs (from 107 to 102 copies per compound) in duplicate experiments (ESI, Section 5†) performed against a panel of immobilized target proteins, such as Carbonic Anhydrase IX (CAIX), Human Serum Albumin (HSA) and the SARS-CoV-2 Non-Structural Protein 14 (NSP14).27 The high-throughput sequencing results are presented as three-dimensional matrices, referred to as “fingerprints” (Fig. 1C). In such fingerprints, two dimensions are used to represent the pairs of building blocks which unambiguously determine the chemical structure of each library member, while the third dimension indicates the number of counts for each compound at the end of the DNA-sequencing procedure.
Selections performed using the SO-DEL
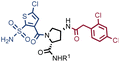
Fig. 2 shows the results of high-throughput sequencing for affinity selections performed with different serial dilutions of the SO-DEL library against three target proteins: CAIX (Fig. 2A), HSA (Fig. 2B), and NSP14 (Fig. 2C). When using 107 copies of SO-DEL for selections against CAIX, the combination A173/B667 (which corresponds to a ligand with KD = 6 ± 2 nM towards CAIX as measured by fluorescence polarization) was highly enriched in comparison to the background (1000 counts for the A173/B667 combination against a background of approximately 1 count; Table 1 and ESI Table S4†). Additionally, several aromatic and heteroaromatic sulfonamides included in the library as building blocks A or B (binding fragments) were enriched, yielding characteristic lines in the fingerprint. As the number of input copies of SO-DEL members used in the selections decreased to 106 and 105, the number of counts for the top enriched combination A173/B667 and other sulfonamides progressively decreased, with the top-enriched combination becoming barely visible at 105 copies. When selections were performed with 104 copies or less, no preferential enrichment of specific building block combinations (singletons) or individual building blocks (lines) was detected (Fig. 2A).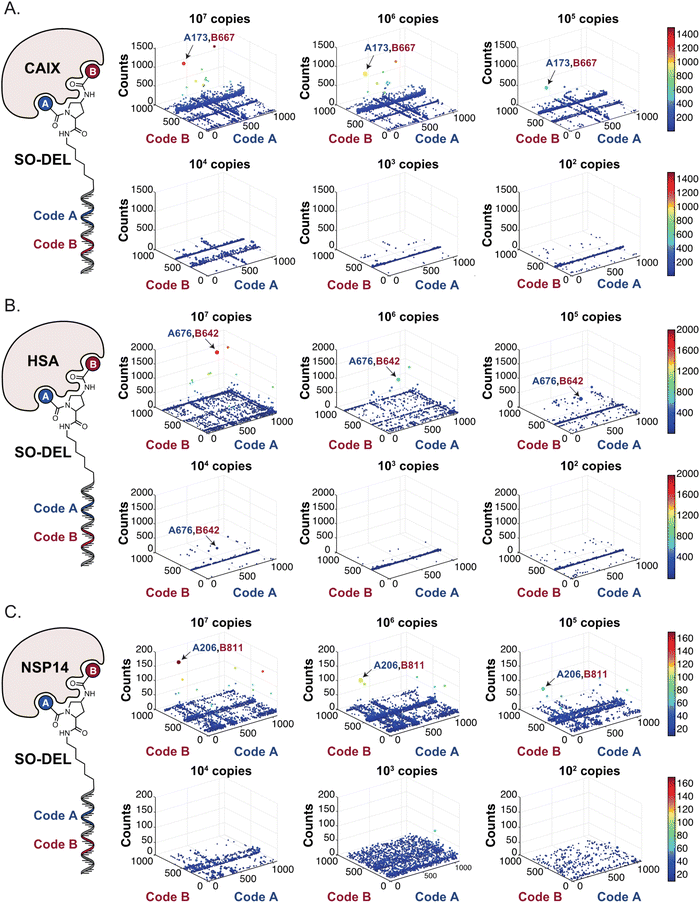 | ||
| Fig. 2 High-throughput sequencing results of affinity selections performed with different inputs (from 107 copies to 100 copies) of SO-DEL against (A) Carbonic Anhydrase IX (CAIX), (B) Human Serum Albumin (HSA) and (C) Non-Structural Protein 14 (NSP14). The data are presented as three-dimensional matrices (fingerprints) as described in Fig. 1. Enriched combinations which have been validated are highlighted with an arrow. Average counts for each selection as well as the number of counts for each enriched combination are reported in the ESI (Section 5†), cut-off = 30 counts. | ||
In selection campaigns against HSA (Fig. 2B), the highest enriched combination corresponded to A676/B642. This compound displayed a dissociation constant of 3 ± 1 nM against the target (Table 1 and ESI Table S4†). HSA fingerprints were characterized by the enrichment of singletons (e.g., A676/B642), indicating the requirement for the presence of both building blocks in the molecule, in order to yield a high-affinity interaction with the cognate target. Also in this case, a selection input of at least 105 copies of SO-DEL members was required in order to obtain high-quality fingerprints.
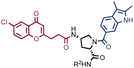
The results of SO-DEL screening on NSP14 (depicted in Fig. 2C) yielded one singleton (building block combination A206/B811) at selection inputs of 105 copies per compound or higher (Table 1 and ESI Table S4†). The compound had been previously validated for its high-affinity binding to NSP14 (KD = 25 ± 3 nM).26 When DEL selections were performed with inputs of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies of compound or lower, no distinct enrichment patterns could be detected over the background signal.
000 copies of compound or lower, no distinct enrichment patterns could be detected over the background signal.
For all three targets (CAIX, HSA and NSP14), additional building block combinations were visible when higher selection inputs were used (i.e., 107 or 106 million copies of SO-DEL).
Selections performed using the NF-DEL
In order to study whether the dependence of selection performance on the number of copies of input changed with different libraries, we repeated CAIX and HSA selections using the NF-DEL library. Ligands to NSP14 could not be found when screening this compound collection. Fig. 3 illustrates the screening results against CAIX (Fig. 3A) and HSA (Fig. 3B). The library contained fragments (several aromatic sulfonamides as building blocks A or B) capable of specific interaction with target protein, with KD values in the single-digit micromolar or even nanomolar range, as well as high-affinity combinations (e.g., A160/B475, Table 1 and ESI Table S4†). When NF-DEL selections were performed against immobilized CAIX at high inputs (107 and 106 copies of the library), the A160/B475 combination was clearly enriched, but the structure–activity information could also be seen in fingerprints corresponding to a selection input of 105 copies of the NF-DEL library. Experiments conducted with selection inputs of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per selection or lower did not produce informative fingerprints (i.e., no hit detected with a sufficiently high enrichment over the background).
000 copies per selection or lower did not produce informative fingerprints (i.e., no hit detected with a sufficiently high enrichment over the background).
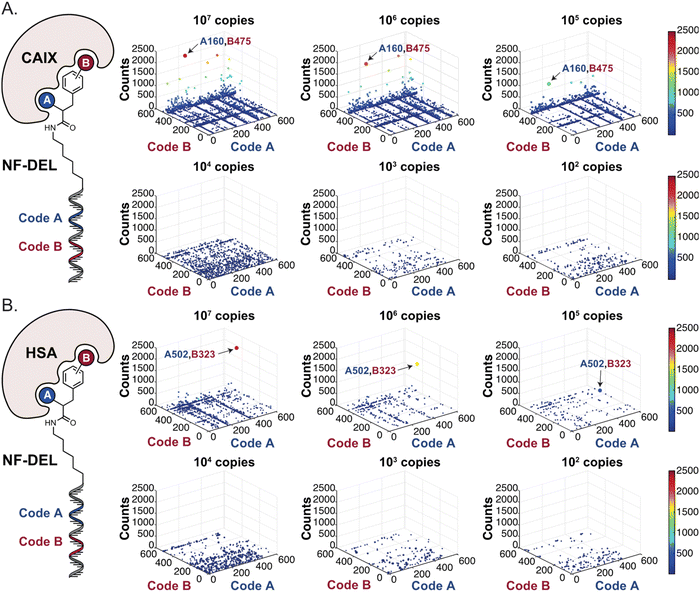 | ||
| Fig. 3 High-throughput sequencing results of affinity selections performed with different inputs of NF-DEL (ranging from 107 to 100 copies) against (A) CAIX and (B) HSA. The data are presented as three-dimensional matrices (fingerprints) as described in Fig. 1. The top enriched combinations are indicated with an arrow. Average counts of each selection and counts for enriched combinations can be found in the ESI (Section 5†), cut-off = 30 counts. | ||
In HSA selections with NF-DEL (Fig. 3B), the A502/B323 singleton was highly enriched at selection inputs of 10 million and 1 million copies of libraries (Table 1 and ESI Table S4†). When screening experiments were performed at 105 copies, the singleton was still detectable, but with lower counts.
Thus, in various selections performed using two libraries (NF-DEL and SO-DEL) against different targets, a minimum threshold of approximately 105 copies of each library member appeared to be required for a reliable detection of hits in affinity capture experiments. When using lower amounts of library copies (i.e., lower than the 105 threshold), singletons (dots) and binding fragments (lines) start to be indistinguishable from the background noise.
Oligonucleotide elongation and selection results
In order to study the impact of the DNA length on the selection output, the annealing region in SO-DEL was extended from 12 to 21 base pairs (bp) at the 3′ extremity (ESI, Section 3.2†). This modification led to a substantial increase in melting temperature (TM,3′, from 42 °C to 63 °C that matches the corresponding TM at the 5′ extremity, TM,5′ = 65 °C) and in an enhanced PCR1 amplifiability of the library (ESI, Fig. S3†). The resulting SO-DEL-long was screened against CAIX (Fig. 4A) and NSP14 (Fig. 4B), following the selection protocol already employed for the experiments presented in Fig. 2. The selection fingerprints did not show significant differences compared to the selection results obtained with the short version of SO-DEL. Also in this case, the previously described hit compound for CAIX (A173/B667) was only identified when a selection input of 105 or more DEL copies was utilized. The NSP14 hit (A206/B881) was slightly enriched in the selection experiment performed at 104 DEL copies, but a higher background noise was observed for selections performed with 103 or lower library copies.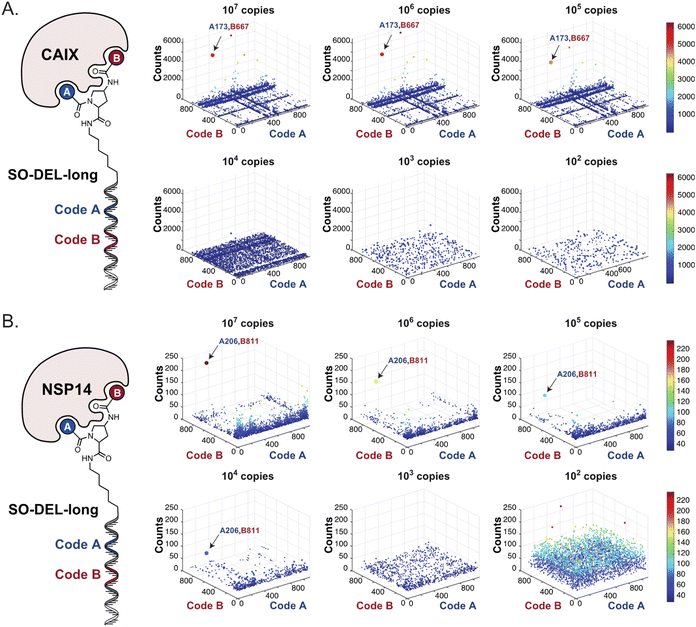 | ||
| Fig. 4 High-throughput sequencing results of affinity selections performed with different inputs of SO-DEL-long (ranging from 107 to 100 copies of library per selection) against (A) CAIX and (B) NSP14. The data are presented as three-dimensional matrices (fingerprints) as described in Fig. 1. The top enriched combinations are indicated with an arrow. Average counts of each selection and counts for enriched combinations can be found in the ESI (Section 5†), cut-off = 30 counts. | ||
Conclusion
DEL technology has become a frequently used methodology for ligand discovery both in industry and in academia, but the impact of experimental procedures on selection success is rarely described in the literature. It has previously been noted that several aspects of the affinity capture procedure (e.g., method of protein immobilization, number of compound copies) and of the subsequent DNA amplification and sequencing may have a substantial impact on the ability to reproducibly detect binding compounds, which are then confirmed as “real” binders upon resynthesis.11,22,24Even within well-established technology such as Phage Display, the achievement of 100% recovery efficiency remains a challenge.28 The success rate of hit discovery in DEL screenings is highly influenced by affinity capture, PCR, and Next Generation Sequencing (NGS) procedures. We have recently demonstrated that high-affinity binders (e.g., acetazolamide against Carbonic Anhydrase IX) can be efficiently captured with a yield close to 30%, unlike micromolar binders (e.g., m-SABA).22,24 The PCR efficiency of the first PCR amplification step after affinity capture is crucial, especially for selections involving library inputs lower than 105 copies.29 Indeed, results of model PCR experiments performed with growing library inputs (using NF-DEL, SO-DEL and SO-DEL-long) show the need for approximately 106–107 DNA molecules to achieve successful barcode amplification (see the ESI, Section 8†). Further worsening of the hit discovery rate happens during the final next-generation sequencing (NGS) step. Modern NGS procedures still suffer from a “count loss” effect, which however equally affects all library members due to PCR normalization normally performed prior to sequencing.
We investigated the impact of library input on the success rate of DEL selections using two high-quality libraries (SO-DEL and NF-DEL) previously described by our laboratory, which had yielded high-affinity hits against various proteins of pharmaceutical interest.25,26 Parallel selection experiments were performed with serially diluted libraries, ranging from 10 million copies to 100 copies per library member. The resulting fingerprints unambiguously revealed that an input threshold of approximately 105 copies of each compound were needed in order to confidently identify potent ligands. Higher input values (e.g., more than 106 copies per library member) may allow the identification of additional binders and further improvement of the signal-to-noise ratio, but this choice depends on the amount of the library which is available for selection experiments. The length of oligonucleotides used for PCR amplification did not appear to have a strong impact on selection performance. In theory, large amounts of DELs can be synthesized, but at some stage the costs for oligonucleotides and building blocks become prohibitively expensive. This limitation underlines the need for a quantitative characterization of input threshold for efficient selections. The correlation between discovery rates at distinct DEL inputs is further discussed in the ESI, Section 7.†
The findings of this study may not only be relevant for the correct execution of DEL selections, but also for the design and use of very large libraries (i.e., those containing billions of compounds). An effective screening (using 106 copies per DEL as selection input) of a library containing 10 billion compounds would require the total use of 16.6 nmoles of total DEL per selection.22,24
In practical terms, this implies that micromoles of final library DNA are needed for realistic screening campaigns, which are performed in at least 100 experiments. Considering a total yield in the range of 1–4% for 2 building blocks DELs25,26 and 0.1–0.2% for 3 building blocks DELs30,31 in relation to the starting DNA material for DEL construction, millimoles of total DNA would be needed, leading to costs which are excessive for most laboratories. In addition, the cost of expensive building blocks would also have to be considered.
As an illustrative example, considering a budget of approximately 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 euros equally distributed between the purchase of DNA codes (oligonucleotides) and small organic building blocks, we can secure approximately 16 nmol of a 3 building-blocks library (factoring in a 20% yield for each coupling step, with a final yield of ∼0.2%
000 euros equally distributed between the purchase of DNA codes (oligonucleotides) and small organic building blocks, we can secure approximately 16 nmol of a 3 building-blocks library (factoring in a 20% yield for each coupling step, with a final yield of ∼0.2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30,31 (see also the ESI, Section 9†). In the context of 100 selections, each performed with 105 DEL copies (considered as the limit of detection based on the finding reported in this article), our calculations suggest that the library could theoretically comprise up to 109 compounds.
30,31 (see also the ESI, Section 9†). In the context of 100 selections, each performed with 105 DEL copies (considered as the limit of detection based on the finding reported in this article), our calculations suggest that the library could theoretically comprise up to 109 compounds.
Since library size has been shown to substantially impact on the probability that high-affinity ligands are discovered in selection campaigns,32–35 the findings described in this article highlight the importance to accurately document experimental parameters in DEL publications and to continue performing research in this area. Discoveries leading to the use of lower copy numbers for individual library members will not only facilitate the efficient use of laboratory resources, but also enable the productive screening of very large DELs.
Our findings indicate that a careful optimization of affinity-capture conditions22 and of decoding methodologies is crucially required if DEL technology is to be productively applied with libraries containing billions of compounds. This aspect is particularly important for the confident detection of singletons (i.e., library members that are found to be enriched only when all building blocks simultaneously contribute to a productive binding interaction). The use of very large libraries, yielding only lines or planes in the selection fingerprints (corresponding to fragments of a molecular structure), is de facto equivalent to the screening of much smaller compound collections and may not allow the full advantage of the potential of DEL technology to be taken.
Data availability
The authors declare that all data supporting the findings of this study are available in the ESI† or from the corresponding authors upon reasonable request.Author contributions
D. N. and N. F. designed the project. S. P. performed the serial dilutions and the affinity selection experiments. S. P., N. F., S. O., G. B., L. P. and J. S. analyzed the HTDS results. S. P. carried out the biotinylation, purification and the quality control of the proteins. The manuscript was written by D. N., N. F., S. P., S. C., and S. O. and the final version was revised and approved by all the authors.Conflicts of interest
D.N. is a cofounder and shareholder of Philogen, a Swiss-Italian Biotech company that operates in the field of ligand-based pharmacodelivery. S. P., L. P., S. O., G. B., S. C., and N. F. are employees of Philochem AG, the daughter company of Philogen acting as the discovery unit of the group. All the other authors declare no competing interests.Acknowledgements
We kindly acknowledge Kristina Schira for providing the NSP14 plasmid and Marco Müller for the production of NSP14.References
- D. A. Erlanson, S. W. Fesik, R. E. Hubbard, W. Jahnke and H. Jhoti, Nat. Rev. Drug Discov., 2016, 15, 605–619 CrossRef CAS PubMed.
- T. E. Nielsen and S. L. Schreiber, Angew. Chem., Int. Ed., 2008, 47, 48–56 CrossRef CAS PubMed.
- S. M. Paul, D. S. Mytelka, C. T. Dunwiddie, C. C. Persinger, B. H. Munos, S. R. Lindborg and A. L. Schacht, Nat. Rev. Drug Discov., 2010, 9, 203–214 CrossRef CAS PubMed.
- E. C. Y. Lee, A. J. McRiner, K. E. Georgiadis, J. Liu, Z. Wang, A. D. Ferguson, B. Levin, M. von Rechenberg, C. D. Hupp, M. I. Monteiro, A. D. Keefe, A. Olszewski, C. J. Eyermann, P. Centrella, Y. Liu, S. Arora, J. W. Cuozzo, Y. Zhang, M. A. Clark, C. Huguet and A. Kohlmann, J. Med. Chem., 2021, 64, 6730–6744 CrossRef CAS PubMed.
- H. Richter, A. L. Satz, M. Bedoucha, B. Buettelmann, A. C. Petersen, A. Harmeier, R. Hermosilla, R. Hochstrasser, D. Burger, B. Gsell, R. Gasser, S. Huber, M. N. Hug, B. Kocer, B. Kuhn, M. Ritter, M. G. Rudolph, F. Weibel, J. Molina-David, J. J. Kim, J. V. Santos, M. Stihle, G. J. Georges, R. D. Bonfil, R. Fridman, S. Uhles, S. Moll, C. Faul, A. Fornoni and M. Prunotto, ACS Chem. Biol., 2019, 14, 37–49 CrossRef CAS.
- A. J. Kennedy, L. Sundström, S. Geschwindner, E. K. Y. Poon, Y. Jiang, R. Chen, R. Cooke, S. Johnstone, A. Madin, J. Lim, Q. Liu, R. J. Lohman, A. Nordqvist, M. Fridén-Saxin, W. Yang, D. G. Brown, D. P. Fairlie and N. Dekker, Commun. Biol., 2020, 3, 782 CrossRef CAS PubMed.
- S. Puglioli, E. Schmidt, C. Pellegrino, L. Prati, S. Oehler, R. de Luca, A. Galbiati, C. Comacchio, L. Nadal, J. Scheuermann, M. G. Manz, D. Neri, S. Cazzamalli, G. Bassi and N. Favalli, Chem, 2023, 9, 411–429 CAS.
- Z. Wu, T. L. Graybill, X. Zeng, M. Platchek, J. Zhang, V. Q. Bodmer, D. D. Wisnoski, J. Deng, F. T. Coppo, G. Yao, A. Tamburino, G. Scavello, G. J. Franklin, S. Mataruse, K. L. Bedard, Y. Ding, J. Chai, J. Summerfield, P. A. Centrella, J. A. Messer, A. J. Pope and D. I. Israel, ACS Comb. Sci., 2015, 17, 722–731 CrossRef CAS PubMed.
- R. A. Goodnow, C. E. Dumelin and A. D. Keefe, Nat. Rev. Drug Discov., 2017, 16, 131–147 CrossRef CAS PubMed.
- R. A. Lerner and S. Brenner, Angew. Chem., 2017, 129, 1184–1185 CrossRef.
- D. Neri and R. A. Lerner, Annu. Rev. Biochem., 2018, 87, 479–502 CrossRef CAS PubMed.
- A. Gironda-Martínez, E. J. Donckele, F. Samain and D. Neri, ACS Pharmacol. Transl. Sci., 2021, 4, 1265–1279 CrossRef PubMed.
- P. A. Harris, S. B. Berger, J. U. Jeong, R. Nagilla, D. Bandyopadhyay, N. Campobasso, C. A. Capriotti, J. A. Cox, L. Dare, X. Dong, P. M. Eidam, J. N. Finger, S. J. Hoffman, J. Kang, V. Kasparcova, B. W. King, R. Lehr, Y. Lan, L. K. Leister, J. D. Lich, T. T. MacDonald, N. A. Miller, M. T. Ouellette, C. S. Pao, A. Rahman, M. A. Reilly, A. R. Rendina, E. J. Rivera, M. C. Schaeffer, C. A. Sehon, R. R. Singhaus, H. H. Sun, B. A. Swift, R. D. Totoritis, A. Vossenkämper, P. Ward, D. D. Wisnoski, D. Zhang, R. W. Marquis, P. J. Gough and J. Bertin, J. Med. Chem., 2017, 60, 1247–1261 CrossRef CAS PubMed.
- S. L. Belyanskaya, Y. Ing, J. F. Callahan, A. L. Lazaar and D. I. Israel, ChemBioChem, 2017, 18(9), 837–842 CrossRef CAS PubMed.
- J. W. Cuozzo, M. A. Clark, A. D. Keefe, A. Kohlmann, M. Mulvihill, H. Ni, L. M. Renzetti, D. I. Resnicow, F. Ruebsam, E. A. Sigel, H. A. Thomson, C. Wang, Z. Xie and Y. Zhang, J. Med. Chem., 2020, 63, 7840–7856 CrossRef CAS PubMed.
- A. G. Gilmartin, T. H. Faitg, M. Richter, A. Groy, M. A. Seefeld, M. G. Darcy, X. Peng, K. Federowicz, J. Yang, S. Y. Zhang, E. Minthorn, J. P. Jaworski, M. Schaber, S. Martens, D. E. McNulty, R. H. Sinnamon, H. Zhang, R. B. Kirkpatrick, N. Nevins, G. Cui, B. Pietrak, E. Diaz, A. Jones, M. Brandt, B. Schwartz, D. A. Heerding and R. Kumar, Nat. Chem. Biol., 2014, 10, 181–187 CrossRef CAS PubMed.
- H. Deng, J. Zhou, F. Sundersingh, J. A. Messer, D. O. Somers, M. Ajakane, C. C. Arico-Muendel, A. Beljean, S. L. Belyanskaya, R. Bingham, E. Blazensky, A. B. Boullay, E. Boursier, J. Chai, P. Carter, C. W. Chung, A. Daugan, Y. Ding, K. Herry, C. Hobbs, E. Humphries, C. Kollmann, V. L. Nguyen, E. Nicodeme, S. E. Smith, N. Dodic and N. Ancellin, ACS Med. Chem. Lett., 2016, 7, 379–384 CrossRef CAS PubMed.
- H. Richter, A. L. Satz, M. Bedoucha, B. Buettelmann, A. C. Petersen, A. Harmeier, R. Hermosilla, R. Hochstrasser, D. Burger, B. Gsell, R. Gasser, S. Huber, M. N. Hug, B. Kocer, B. Kuhn, M. Ritter, M. G. Rudolph, F. Weibel, J. Molina-David, J. J. Kim, J. V. Santos, M. Stihle, G. J. Georges, R. D. Bonfil, R. Fridman, S. Uhles, S. Moll, C. Faul, A. Fornoni and M. Prunotto, ACS Chem. Biol., 2019, 14, 37–49 CrossRef CAS PubMed.
- A. S. Ratnayake, M. E. Flanagan, T. L. Foley, J. D. Smith, J. G. Johnson, J. Bellenger, J. I. Montgomery and B. M. Paegel, ACS Comb. Sci., 2019, 21, 650–655 CrossRef CAS PubMed.
- B. Sauter, L. Schneider, C. Stress and D. Gillingham, Bioorg. Med. Chem., 2021, 52, 116508 CrossRef CAS PubMed.
- A. L. Satz, J. Cai, Y. Chen, R. Goodnow, F. Gruber, A. Kowalczyk, A. Petersen, G. Naderi-Oboodi, L. Orzechowski and Q. Strebel, Bioconjug Chem, 2015, 26, 1623–1632 CrossRef CAS PubMed.
- A. Sannino, E. Gabriele, M. Bigatti, S. Mulatto, J. Piazzi, J. Scheuermann, D. Neri, E. J. Donckele and F. Samain, ChemBioChem, 2019, 20, 955–962 CrossRef CAS PubMed.
- Q. Chen, X. Cheng, L. Zhang, X. Li, P. Chen, J. Liu, L. Zhang, H. Wei, Z. Li and D. Dou, SLAS Discov., 2020, 25, 523–529 CrossRef.
- Y. Li, G. Zimmermann, J. Scheuermann and D. Neri, ChemBioChem, 2017, 18, 848–852 CrossRef CAS PubMed.
- N. Favalli, G. Bassi, C. Pellegrino, J. Millul, R. de Luca, S. Cazzamalli, S. Yang, A. Trenner, N. L. Mozaffari, R. Myburgh, M. Moroglu, S. J. Conway, A. A. Sartori, M. G. Manz, R. A. Lerner, P. K. Vogt, J. Scheuermann and D. Neri, Nat. Chem., 2021, 13, 540–548 CrossRef CAS PubMed.
- S. Oehler, L. Lucaroni, F. Migliorini, A. Elsayed, L. Prati, S. Puglioli, M. Matasci, K. Schira, J. Scheuermann, D. Yudin, M. Jia, N. Ban, D. Bushell, R. Kornberg, S. Cazzamalli, D. Neri, N. Favalli and G. Bassi, Nat. Chem., 2023, 1–13 Search PubMed.
- W. Decurtins, M. Wichert, R. M. Franzini, F. Buller, M. A. Stravs, Y. Zhang, D. Neri and J. Scheuermann, Nat. Protoc., 2016, 11, 764–780 CrossRef CAS PubMed.
- J. McCafferty, A. D. Griffiths, G. Winter and D. J. Chriswell, Nature, 1990, 348(6301), 552–554 CrossRef CAS PubMed.
- L. Plais, A. Lessing, M. Keller, A. Martinelli, S. Oehler, G. Bassi, D. Neri and J. Scheuermann, Chem. Sci., 2022, 13, 967–974 RSC.
- Y. Li, R. De Luca, S. Cazzamalli, F. Pretto, D. Bajic, J. Scheuermann and D. Neri, Nat. Chem., 2018, 10, 441–448 CrossRef CAS PubMed.
- Y. Onda, G. Bassi, A. Elsayed, F. Ulrich, S. Oehler, L. Plais, J. Scheuermann and D. Neri, Chem.–Eur. J., 2021, 27, 7160–7167 CrossRef CAS PubMed.
- A. D. Griffiths, S. C. Williams, O. Hartley, I. M. Tomlinson, P. Waterhouse, W. L. Crosby, R. E. Kontermann, P. T. Jones, N. M. Low and T. A. Allison, EMBO J., 1994, 13, 3245–3260 CrossRef CAS PubMed.
- S. Imanishi, T. Katoh, Y. Yin, M. Yamada, M. Kawai and H. Suga, J. Am. Chem. Soc., 2021, 143, 5680–5684 CrossRef CAS PubMed.
- P. Waterhouse, A. D. Griffiths, K. S. Johnson and G. Winter, Nucleic Acids Res., 1993, 21, 2265 CrossRef CAS PubMed.
- A. S. Perelson and G. F. Oster, J. Theor. Biol., 1979, 81, 645–670 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc03688j |
| This journal is © The Royal Society of Chemistry 2023 |