Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A cost-effective aptasensor capable of early diagnosis and monitoring of Alzheimer's disease with the rapid analysis of beta-amyloid peptide 1–40†
Ashley
Khang‡
,
Nnebuefe
Idegwu‡
and
Ji Hoon
Lee
 *
*
Luminescent MD, LLC, Hagerstown, MD 21742, USA. E-mail: jhlee@luminescentmd.com
First published on 17th January 2023
Abstract
A cost-effective biosensor with guanine chemiluminescence detection capable of sensing trace levels of beta-amyloid peptide 1–40 was developed for the early diagnosis and accurate monitoring of Alzheimer's disease (AD), which is the biggest cause of dementia-related memory loss, and other cognitive abilities. A specific DNA aptamer binds rapidly with beta-amyloid peptide 1–40 within 1 hour at room temperature. Free DNA aptamers remaining after the procedure to capture beta-amyloid peptide 1–40 in a sample electrostatically bind with luminescent dye within 5 min at room temperature. After the procedure, 3-methoxylphenylglyoxal hydrate in dimethyl furan and tetra-n-propylammonium hydroxide in deionized water were added to the solution to generate green chemiluminescence. Bright chemiluminescence, created by the internal chemiluminescence resonance energy transfer (inter-CRET) of the luminescent dye bound with the DNA aptamer and the high-energy intermediate formed in guanine chemiluminescence, was measured immediately for 20 s. The relative CL intensity decreased proportionally with the increase of the beta-amyloid peptide 1–40. The limit of detection (LOD) of the biosensor having a wide linear dynamic range (5–500 ng ml−1) was as low as 2 ng ml−1. In addition, it was confirmed that the biosensor can quantify beta-amyloid peptide 1–40 with good accuracy, precision, recovery, and selectivity. We expect the principle and concepts confirmed here to be developed to be applied as a new method capable of early diagnosis and prognosis of human diseases with good reliability.
Introduction
The United Nations (UN) has announced that the global population of those aged 65 and over is growing more rapidly than that of any other age group (https://www.un.org/en/sections/issues-depth/ageing/). With this trend, the number of patients suffering from Alzheimer's disease, the most common cause of dementia among adults aged 65 years and older, is increasing continuously even though Alzheimer's disease is not a normal part of aging. However, even among carriers of Alzheimer's disease genes, a few people reach age 100 and older without ever developing the disease – demonstrating that environmental risk contributes to the disease.1Symptoms of Alzheimer's disease usually appear after age 60 even though it starts 10 or more years before. Thus, it is important to diagnose Alzheimer's disease early to prevent and delay the appearance of the symptoms. Beta-amyloid peptides (e.g., 1–40, 1–42) and tau levels in human fluid samples are widely applied as biomarkers to indicate the early stages of Alzheimer's disease.2–4 They are quantified and monitored with various analytical methods such as time-consuming and expensive immunoassays with a specific sensor such as chemiluminescent, colorimetric, electrochemical, or fluorescent.5–8 In order to improve on immunoassays operated with intractable antibodies produced with the sacrifice of small animals, recently, cost-effective and easy-to-use single-strand DNA (ssDNA) and ribonucleic acid (RNA) aptamers synthesized with appropriate chemicals were designed and applied to the detection of beta-amyloid peptides.9–11
Guanines of a free DNA aptamer, not bound with a biomarker, react with 3,4,5-trimethoxylphenylglyoxal (TMPG) to produce a high-energy intermediate as shown in Scheme 1.12,13 The high-energy intermediate formed from the reaction of guanine and TMPG in the presence of tetra-n-propylammonium hydroxide (TPA) and dimethylformamide (DMF) can emit dim light by itself14,15 (see Scheme 1A) as well as transfer energy to a luminescent dye conjugated with the DNA aptamer based on the principle of intra chemiluminescence resonance energy transfer (intra-CRET) as shown in Scheme 1B. After the intra-CRET, the luminescent dye emits bright visible light. The colour of the light depends on the chemical and physical properties of the fluorescent dye as shown in Scheme 1B.13,16–20
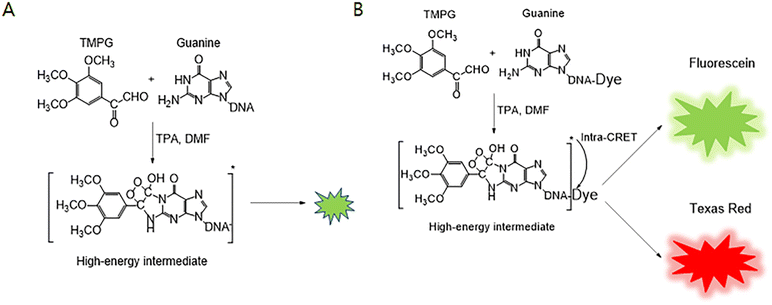 | ||
| Scheme 1 (A) Guanine chemiluminescence in the absence of a luminescent dye,14,15 (B) guanine chemiluminescence reaction in the presence of a luminescent dye such as fluorescein or Texas Red.13,16–20 | ||
Commercially available dyes, such as QuantiFluor, SFC Green, and Chamel Green, electrostatically bound with ssDNA emit bright fluorescence even though they cannot emit light in the absence of ssDNA.21,22 Based on the specific properties of QuantiFluor, some biosensors have been developed using the interaction of the dye and free DNA aptamers not bound with a target marker using fluorescence detection.21
In this research, we developed for the first time an aptasensor with chemiluminescence detection for the early diagnosis of Alzheimer's disease using a cost-effective DNA aptamer of beta-amyloid peptide 1–40, instead of expensive and intractable antibodies,23,24 and Chamel Green based on two hypotheses. First, Chamel Green electrostatically bound with free DNA aptamers emits bright chemiluminescence if the high-energy intermediate formed from the reaction between guanines of the free DNA aptamer and a phenylglyoxal hydrate derivative, as shown in Scheme 1(A), can transfer energy to Chamel Green intercalated with the DNA aptamer based on internal-CRET instead of conventional intra-CRET as shown in Scheme 1(B).13,16–20 Schemes S1 and S2† show the differences between intra- and inter-CRET in the guanine chemiluminescence reaction. Intra-CRET between the high-energy intermediate and a fluorescent dye occurs when the fluorescence dye covalently binds with the high-energy intermediate formed in the guanine chemiluminescence reaction as shown in Schemes 1 and S1.†13,16,17 As shown in Scheme S2,† the inter-CRET between the high-energy intermediate and a fluorescent dye occurs when the distance between the high-energy intermediate and the fluorescent dye is shorter than 10 nm.25–29 Second, if the brightness of the light emitted from the aptasensor decreases with the increase of beta-amyloid peptide 1–40 in a sample, it is possible to develop an easy-to-use aptasensor operated without time-consuming and tedious washing procedures. The research is described in detail in this manuscript.
Materials and methods
Chemicals and materials
Beta-amyloid peptide 1–40 aptamer (5′-TGG GGG GCG GAC GAT AGG GGC CCC CCG GTA GGA TGG ACG-3′)30 was purchased from Alpha DNA (Montreal, Quebec, Canada). 3,4,5-Trimethoxylphenylglyoxal hydrate (TMPG, 97%) and 3-methoxylphenylglyoxal hydrate (3-MPG) were purchased from Matrix Scientific (Columbia, SC, USA). Tetra-n-propylammonium (TPA, HPLC grade) was purchased from Alfa Aesar (Ward Hill, MA, USA). N,N-Dimethylformamide (DMF) and various biomaterials were purchased from EMD (Billerica, MA, USA). Beta-amyloid peptides 1–40 and 1–42 were purchased from Peptide International (Louisville, KY, USA). Pooled human cerebrospinal fluid (CSF) was purchased from Innovative Research (Novi, MI, USA). Artificial CSF A and B (1000 mL of each, ready to use for electrophysiology) were purchased from Ecocyte Bioscience (Austin, TX, USA). Several types of buffer solution (pH 7.0–8.5) were purchased from Teknova (Hollister, CA, USA). SFC Green and Chamel Green, which are derivatives of a merocyanine-based compound (see Scheme S1†), were provided by Sun Fine Chem (Cheongju, Chungbuk, Republic of Korea).Methods
Results and discussion
Electrostatic binding of luminescent dye and ssDNA
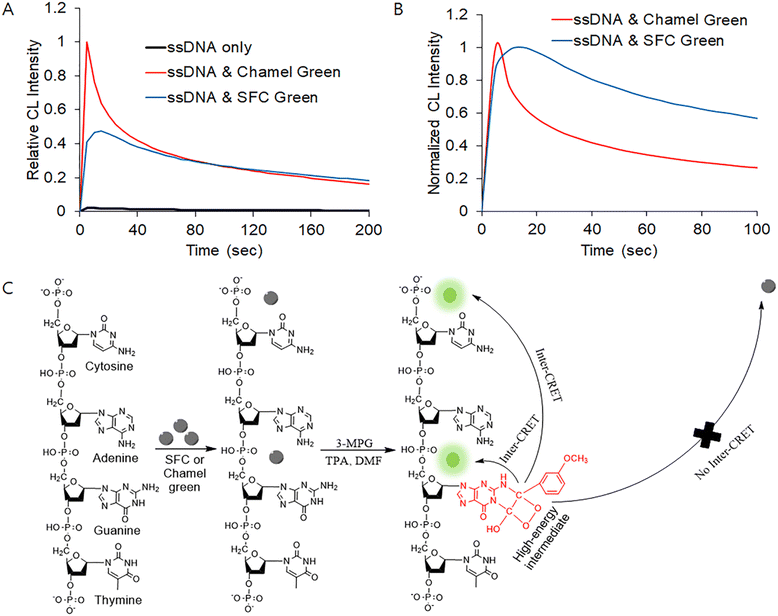
As shown in Fig. 1(A), the brightness of the guanine chemiluminescence of the ssDNA depends on the reaction conditions. With the addition of SFC Green or Chamel Green, the brightness of the guanine chemiluminescence was enhanced. In addition, the CL spectrum in the presence of SFC Green was different from that in the presence of Chamel as shown in Fig. 1(B). The time (τmax: 15 s) necessary for attaining the maximum intensity (Imax) of the CL in the presence of SFC Green was shorter than that (5 s) in the presence of Chamel Green. Also, the half-life time (τ1/e: 190 s) necessary for attaining the half intensity (I1/e) of the former was shorter than that (35 s) of the latter. Fig. 1(B) indicates that the shape of the spectrum depends on the properties of the luminescent dye added in the guanine chemiluminescence reaction.Fig. 1(C) shows the possible guanine chemiluminescence reaction mechanism proposed based on the results shown in Fig. 1(A) and (B). First, it is expected that SFC Green and Chamel Green electrostatically bind with cytosine and guanine of the ssDNA. After the binding interaction, guanine reacts with 3-MPG in the presence of TPA and DMF to produce a high-energy intermediate. Then, the high-energy intermediate transfers energy to the luminescent dye (SFC or Chamel Green) intercalated in the ssDNA based on the principle of inter-CRET. The luminescent dye excited due to the inter-CRET emits bright light, whereas the remaining SFC (or Chamel) Green not bound with ssDNA cannot emit light because the distance (>10 nm) between the high-energy intermediate and free SFC (or Chamel) Green is too great for the inter-CRET to occur. It is well-known that the distance between a donor and a receiver for CRET should be shorter than 10 nm.31,32
Effect of phenylglyoxal hydrate derivative
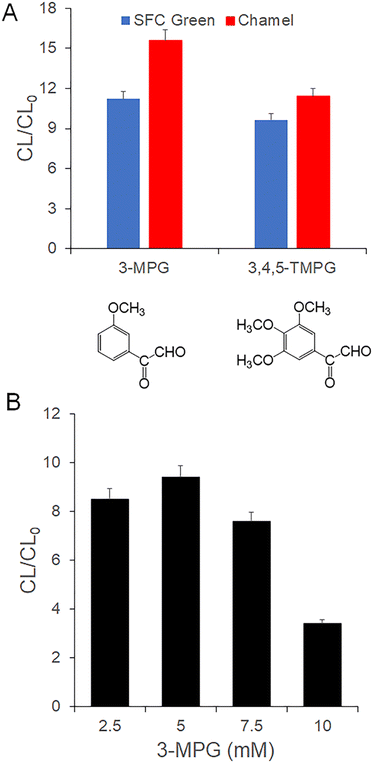
As shown in Fig. 2(A), CL/CL0 determined in the presence of 3-MPG is higher than that in the presence of 3,4,5-TMPG. CL0 is the chemiluminescence intensity of ssDNA measured in the absence of SFC (or Chamel) Green. CL is the chemiluminescence intensity of ssDNA measured in the presence of SFC (or Chamel) Green. The results indicate that the efficiency of 3-MPG is better than TMPG widely used in the guanine chemiluminescence reaction.12,13,16–20 In other words, a high-energy intermediate formed from the reaction between 3-MPG and guanine of the ssDNA in the presence of TPA and DMF can effectively transfer energy to SFC Green and Chamel Green bound with cytosine and guanine of the ssDNA. This is because the CL emission of the Chamel Green and SFC Green is dependent on the energy, which is a self-emission wavelength, of the high-energy intermediate formed from the reaction of 3-MPG (or TMPG) and guanine. Unfortunately, the self-emission of the high-energy intermediate formed with 3-MPG or TMPG (see Scheme 1(A)) was too low to determine the maximum CL emission wavelength with a charge coupled device (CCD, USB 2000, Ocean Insight, Orlando, FL). As indirect evidence, the fluorescence wavelength (470 nm) of the complex formed from the reaction of guanine and 3-MPG is 10 nm longer than that (460 nm) formed from the reaction of guanine and TMPG.33 Based on the results shown in Fig. 2(A) and the indirect evidence,33 the wavelength of the self-emission generated from the high-energy intermediate in the presence of 3-MPG is appropriate for emission of relatively strong CL of SFC Green and Chamel Green with the inter-CRET shown in Fig. 1(C) and Scheme S2.† In conclusion, 3-MPG was selected for the development of an aptasensor in this research.Fig. 2(B) shows that CL/CL0 depends on the concentration of 3-MPG. Based on the results, 5 mM 3-MPG was selected to develop an aptasensor capable of sensing beta-amyloid peptide 1–40 in a sample. CL/CL0 in the presence of 3-MPG at a concentration greater than 5 mM was lower than that in the presence of 5 mM 3-MPG due to the self-quenching effect observed in the presence of the excess high-energy intermediate formed from the reaction of guanine and relatively high concentration of phenylglyoxal hydrate derivatives such as 3-MPG and TMPG.13,16
Effect of TPA
As shown in Fig. 3, CL/CL0 depends on the concentration of TPA in the guanine chemiluminescence reaction in the presence of Chamel Green. The results indicate that the concentration of the high-energy intermediate formed from the reaction of 3-MPG and guanines of the ssDNA is dependent on the concentration of TPA. Fig. 3 indicates that the concentration of the high-energy intermediate formed in the presence of 10 mM TPA is lower than that in the presence of 20 mM TPA. Thus, CL/CL0 under the former conditions is lower than that under the latter conditions. CL/CL0 in the presence of 40 mM TPA was also lower than that in the presence of 20 mM TPA due to the self-quenching effect. In other words, too many of the electrons of Chamel Green excited by the inter-CRET between the high-energy intermediate and Chamel Green were relaxed due to rapid collision (self-quenching) in the excited state before the emission of chemiluminescence. Based on the experimental results shown in Fig. 3, 20 mM TPA was selected for this research.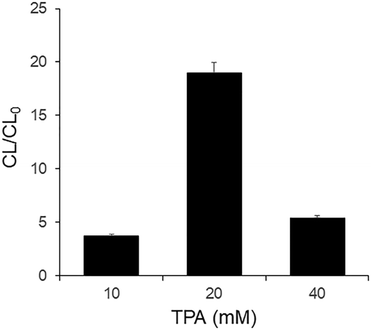 | ||
| Fig. 3 Effect of TPA. [3-MPG] = 5 mM in DMF, [ssDNA] = 500 nM, [Chamel Green] = 5 μM in 10 mM Tris-HCl (pH 8). | ||
Effects of Chamel Green and DNA aptamer
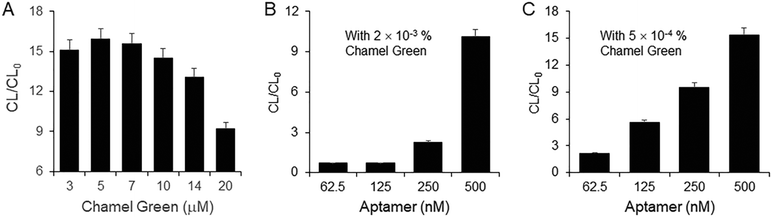
Fig. 4(A) shows that the CL/CL0 depends on the concentration of Chamel Green. Six different concentrations of Chamel Green in 10 mM Tris-HCl (pH 8) were prepared by diluting the 10 mM Chamel Green stock solution. 500 nM aptamer of beta-amyloid peptide 1–40 was used. CL/CL0 was the highest in the presence of 5 μM Chamel Green prepared by 2000-fold dilution of the stock in 10 mM Tris-HCl (pH 8). The CL/CL0 calculated in the presence of higher concentrations than 5 μM of Chamel Green was lower than that in the presence of 5 μM Chamel Green due to the self-quenching effect. Based on the results shown in Fig. 4(A), we selected 5 μM Chamel Green for the development of the label free aptasensor with guanine chemiluminescence detection.As shown in Fig. 4(B), CL/CL0 in the presence of 250 or 500 nM aptamer was higher than 1 because Chamel Green emits bright chemiluminescence due to the inter-CRET between the high-energy intermediate and Chamel Green intercalated in the relatively high concentration of the aptamer. However, CL/CL0 in the presence of 62.5 or 125 nM was lower than 1 due to the self-quenching of excess Chamel Green electrons excited in the relatively low concentration of the aptamer with inter-CRET. Thus, CL was lower than CL0 measured in the presence of the aptamer only. The results in Fig. 4(B) indicate that the Chamel Green should be more dilute to obtain bright chemiluminescence in the presence of a relatively low concentration of aptamer without the self-quenching effect.
As shown in Fig. 4(C), CL/CL0 in the presence of four different concentrations of the aptamer was higher than 1 when the concentration of Chamel Green was 4-fold lower than that used in Fig. 4(B). Thus, CL/CL0 was proportionally enhanced with the increase of aptamer concentration. In addition, CL/CL0 in the presence of 250 nM aptamer in Fig. 4(C) was about 3-fold higher than that in Fig. 4(B) because for the former, the CL was generated without the self-quenching effect. Based on the results, we confirmed again that 5 μM Chamel Green is the optimum concentration.
Effects of the buffer and pH
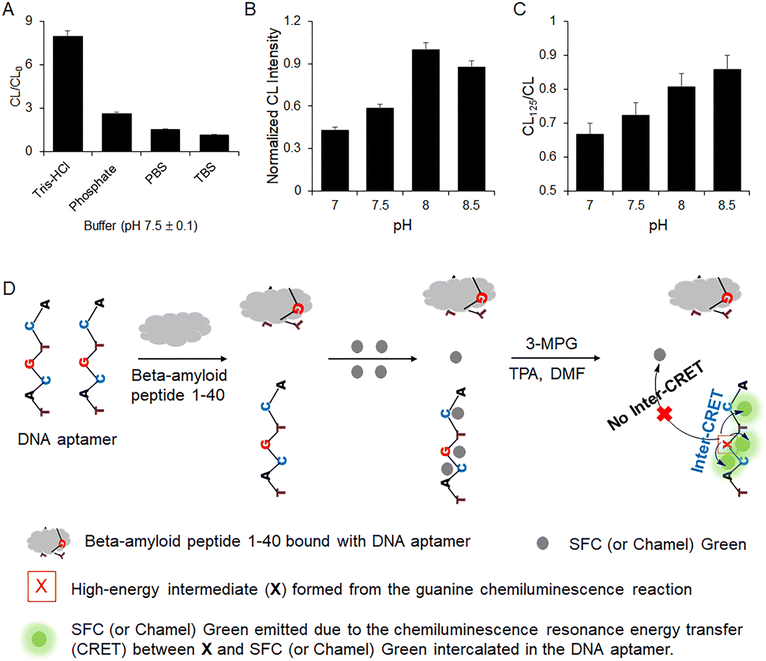
As shown in Fig. 5(A), CL/CL0 was dependent on the composition of the buffer. CL/CL0 in 10 mM Tris-HCl (pH 7.5) was much higher than those in the other buffers. The results indicate that the electrostatic binding interaction of the DNA aptamer and Chamel Green in 10 mM Tris-HCl is faster and more stable than that in other buffers. Fig. 5(A) shows that CL/CL0 in PBS and TBS is slightly higher than 1. The results indicate that the CL in the presence of Chamel Green is as low as CL0. The results indicate that the electrostatic binding of Chamel Green and the aptamer or the inter-CRET between Chamel Green and the high-energy intermediate is very difficult in PBS and TBS. Based on the research results, 10 mM Tris-HCl was selected to prepare the aptamer and Chamel Green working solutions.Fig. 5(B) shows that the CL emission in the presence of the aptamer and Chamel Green was the brightest in Tris-HCl (pH 8). However, Fig. 5(C) indicates that the binding reaction of the aptamer and beta-amyloid peptide 1–40 in pH 8 was slower than those in pH 7 and 7.5, because CL125/CL at pH 8 was higher than those in pH 7 and 7.5. CL125 is the relative CL intensity with the intercalation of Chamel Green and the aptamer remaining after the binding reaction of the aptamer and beta-amyloid peptide 1–40 (125 ng mL−1) for 1 hour at room temperature (21 ± 2 °C). CL is the relative CL intensity emitted in the presence of the aptamer and Chamel Green without beta-amyloid peptide 1–40. Fig. 5(C) indicates that the aptamer bound with beta-amyloid peptide 1–40 cannot emit light because guanines of the aptamer bound with beta-amyloid peptide 1–40 cannot react with 3-MPG to produce the high-energy intermediate. In addition, the intensities of the CL and CL125 under acidic conditions (Tris-HCl: pH < 7) were too low to be applied for the development of the biosensor because the pH for the intra-CRET between the high-energy intermediate and Chamel Green should be higher than 7. For example, the relative CL intensity measured at pH 6 was less than 10% of that at pH 7. The pH dependence of the intra-CRET in guanine chemiluminescence is consistent with that of the inter-CRET because it is difficult for the high-energy intermediate to form from the reaction between 3-MPG and guanine under the acidic conditions.13,16,17 Based on the results shown in Fig. 5(C), we selected Tris-HCl (pH 7) for the development of the biosensor capable of quantifying trace levels of beta-amyloid peptide 1–40 in a sample.
Based on the preliminary research results, we developed an aptasensor operating with the procedure shown in Fig. 5(D). First, the aptamer binds with beta-amyloid peptide 1–40. Then, Chamel Green was added to the solution for the intercalation between Chamel Green and the aptamer remaining after the binding reaction. Finally, the CL of the solution was measured with the addition of chemiluminescence reagents such as 3-MPG, TPA, and DMF as shown in Fig. 5(D).
Analysis of beta-amyloid peptide 1–40
As shown in Fig. 6(A), the relative CL intensity exponentially decreased with the increase of beta-amyloid peptide 1–40 in a sample because the DNA aptamer bound with beta-amyloid peptide 1–40 cannot emit chemiluminescence as shown in Fig. 5(C). Fig. 6(B) is the linear calibration curve for the quantification of beta-amyloid peptide 1–40 in a sample. The wide dynamic range of the calibration curve has a good linearity from 5 to 500 ng mL−1. The limit of detection (LOD = 3σ/slope) was as low as 2 ng mL−1. σ is the standard deviation of the chemiluminescence intensity measured in the absence of beta-amyloid peptide 1–40 (N = 15). The LOD of the easy-to-use aptasensor is similar to those of other analytical methods34–38 that have been recently reported. Table S1† indicates that the aptasensor can rapidly quantify beta-amyloid peptide 1–40, whereas the other methods were operated with complicated and time-consuming procedures. Using the linear calibration curve of the biosensor, it is possible to diagnose Alzheimer's disease (AD) related dementia because the range (12.2–19.9 ng mL−1) of beta-amyloid peptide 1–40 measured from patients with AD related dementia39 is in the dynamic range of the calibration curve of the biosensor developed in this research.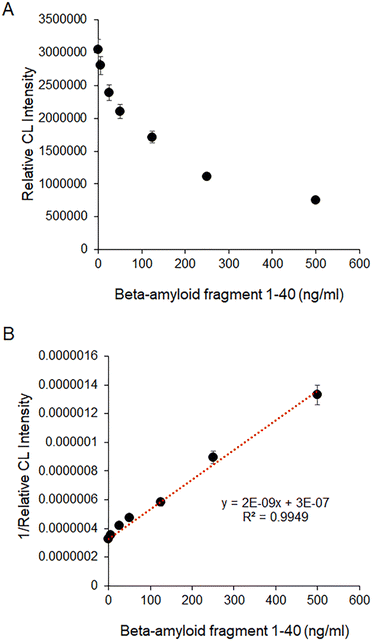 | ||
| Fig. 6 (A) Relative CL intensities in the presence of various concentrations of beta-amyloid peptide 1–40, (B) linear calibration curve capable of quantifying beta-amyloid peptide 1–40 in a sample. | ||
Accuracy, precision, recovery, and selectivity of the biosensor
In order to study the accuracy, precision, and recovery of the aptasensor in CSF samples, three different CSF samples containing certain concentrations of beta-amyloid peptide 1–40 were used. Each sample was prepared with CSF. Then, we spiked the samples with additional beta-amyloid peptide 1–40 (15, 50, and 150 ng mL−1). As shown in Table S2,† the accuracy, precision, and recovery of the aptasensor were within the statistically acceptable error ranges. The accuracy and precision ranges of the biosensor were 7.2–8.3% and 4.8–10%. The recovery range of the biosensor was from 92.7 to 108.3%.Fig. 7 shows that the DNA aptamer used for this research exhibits good selectivity for the analysis of beta-amyloid peptide 1–40. This aptamer did not bind with other biological materials including beta-amyloid peptide 1–42. This result is consistent with the previous report.30
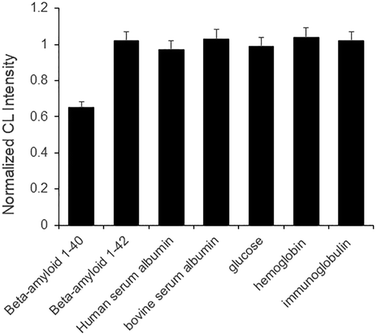 | ||
| Fig. 7 Selectivity of the biosensor operated with DNA aptamer capable of capturing beta-amyloid peptide 1–40. | ||
Conclusions
The electrostatic binding interaction of the DNA aptamer and Chamel Green was studied for the first time to develop a new aptasensor for the analysis of beta-amyloid peptide 1–40 in a sample. It was confirmed that the complex formed from the electrostatic binding between Chamel Green and the aptamer generates bright chemiluminescence in the guanine chemiluminescence reaction due to the inter-CRET between Chamel Green and the high-energy intermediate formed from the reaction of 3-MPG and guanine of the DNA aptamer. Additionally, the DNA aptamer rapidly binds with beta-amyloid peptide 1–40 in CSF. In conclusion, trace levels of beta-amyloid peptide 1–40 were quantified with the aptasensor with guanine chemiluminescence detection. Additionally, we confirmed that the aptasensor could be operated with good accuracy, precision, recovery, and selectivity.It is expected that the analytical method first developed in this research can be applied to develop advanced aptasensors capable of rapidly sensing other biomarkers of Alzheimer's disease such as beta-amyloid peptide 1–42 and tau. We expect that the accurate and early diagnosis of Alzheimer's disease is possible with the consecutive analyses of three biomarkers, including beta-amyloid peptide 1–40 and 1–42, and tau, in a sample. The aptasensor with guanine chemiluminescence detection operated based on the principle of inter-CRET can be applied to diagnose various human diseases such as cardiac ailments, cancers, and infectious diseases because numerous DNA aptamers for the diagnosis of human diseases have been widely developed.40,41 Additionally, it is possible that this technology can be applied in various research fields such as biology, biomedical engineering, chemistry, environmental science and engineering, and toxicology.
Author contributions
Ashley Khang: conceptualization, methodology, validation, data curation, investigation, writing – original draft. Nnebuefe Idegwu: conceptualization, methodology, validation, data curation, investigation, writing – original draft. Ji Hoon Lee: conceptualization, methodology, writing – review & editing, supervision.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This project was performed as part of an intern program (LMD-2001) of Luminescent MD, LLC.Notes and references
- M. G. M. O. Rikkert, J.-P. Teunisse and M. Vernooij-Dassen, Am. J. Alzheimer's Dis. Other Dement., 2005, 20, 269–272 CrossRef.
- A. Mobed and M. Hasanzadeh, Int. J. Biol. Macromol., 2020, 161, 59–71 CrossRef CAS PubMed.
- S. Wang, H. Lindroth, C. Chan, R. Greene, P. Serrano-Andrews, S. Khan, G. Rios, S. Jabbari, J. Lim, A. J. Saykin and B. Khan, J. Am. Geriatr. Soc., 2021, 69, 255–263 CrossRef PubMed.
- S. Rayaprolu, L. Higginbotham, P. Bagchi, C. M. Watson, T. Zhang, A. I. Levey, S. Rangaraju and N. T. Seyfried, Neuropsychopharmacology, 2021, 46, 98–115 CrossRef CAS PubMed.
- D. Diaz-Lucena, G. Escaramis, A. Villar-Pique, P. Hermann, M. Schmitz, D. Varges, I. Santana, J. A. del Rio, E. Marti, I. Ferrer, I. Baldeiras, I. Zerr and F. Llorens, J. Neurol., 2020, 267, 2567–2581 CrossRef CAS PubMed.
- S. B. Sankar, C. Infante-Garcia, L. D. Weinstock, J. J. Ramos-Rodriguez, C. Hierro-Bujalance, C. Fernandez-Ponce, L. B. Wood and M. Garcia-Alloza, J. Neuroinflammation, 2020, 17, 38 CrossRef CAS PubMed.
- C. D. Whelan, N. Mattsson, M. W. Nagle, S. Vijayaraghavan, C. Hyde, S. Janelidze, E. Stomrud, J. Lee, L. Fitz, T. A. Samad, G. Ramaswamy, R. A. Margolin, A. Malarstig and O. Hansson, Acta Neuropathol. Commun., 2019, 7, 14 CrossRef PubMed.
- M. M. Mielke, J. A. Syrjanen, K. Blennow, H. Zetterberg, I. Skoog, P. Vemuri, M. M. Machulda, J. Graff-Radford, D. S. Knopman, C. R. Jack, R. C. Petersen and S. Kern, Alzheimer's Dementia, 2019, 15, 1437–1447 CrossRef.
- H. X. Ren, Y. B. Miao and Y. D. Zhang, Microchim. Acta, 2020, 187, 114 CrossRef CAS PubMed.
- M. Negandary and H. Heli, Talanta, 2019, 198, 510–517 CrossRef PubMed.
- Y. Obata, K. Murakami, T. Kawase, K. Hirose, N. Izuo, T. Shimizu and K. Irie, ACS Omega, 2020, 5, 21531–21537 CrossRef CAS PubMed.
- M. Kai, S. Kishida and K. Sakai, Anal. Chim. Acta, 1999, 381, 155–163 CrossRef CAS.
- S. Cho, L. Park, R. Chong, Y. T. Kim and J. H. Lee, Biosens. Bioelectron., 2014, 52, 310–316 CrossRef CAS PubMed.
- X. Hun, Z. Mei, Z. Wang and Y. He, Spectrochim. Acta, Part A, 2012, 95, 114–119 CrossRef CAS PubMed.
- X. Hun, Y. Xu and C. Chen, Sens. Actuators, B, 2014, 202, 594–599 CrossRef.
- T. Cha, S. Cho, Y. T. Kim and J. H. Lee, Biosens. Bioelectron., 2014, 62, 31–37 CrossRef CAS PubMed.
- J. Chong, H. Chong and J. H. Lee, Anal. Biochem., 2019, 564, 102–107 CrossRef PubMed.
- J. Khang, D. Kim, K. W. Chung and J. H. Lee, Talanta, 2016, 147, 177–183 CrossRef CAS PubMed.
- B. Kim, K. W. Chung and J. H. Lee, J. Pharm. Biomed. Anal., 2018, 152, 315–321 CrossRef CAS PubMed.
- M. Kwon, Y. Park and J. H. Lee, RSC Adv., 2015, 5, 94629–94634 RSC.
- R. M. Bialy, M. M. Ali, Y. F. Li and J. D. Brennan, Chem. – Eur. J., 2020, 26, 5085–5092 CrossRef CAS PubMed.
- N. Ardjomandi, J. Huth, D. R. Stamov, A. Henrich, C. Klein, H. P. Wendel, S. Reinert and D. Alexander, Mater. Sci. Eng., C, 2016, 67, 267–275 CrossRef CAS PubMed.
- A. Santoro, M. Grimaldi, M. Buonocore, I. Stillitano, A. Gloria, M. Santin, F. Bobba, M. Sublimi Saponetti, E. Ciaglia and A. M. D'Ursi, Eur. J. Med. Chem., 2022, 237, 114400 CrossRef CAS PubMed.
- K. Yamashita, S. Watanabe, K. Ishiki, M. Miura, Y. Irino, T. Kubo, J. Matsui, K. Hagino, S. Iwanaga and T. Yoshida, Biochem. Biophys. Res. Commun., 2021, 576, 22–26 CrossRef CAS PubMed.
- J. Jeon, D. G. You, W. Um, J. Lee, C. H. Kim, S. Shin, S. Kwon and J. H. Park, Sci. Adv., 2020, 6, eaaz8400 CrossRef CAS PubMed.
- W. An, R. P. Mason and A. R. Lippert, Org. Biomol. Chem., 2018, 16, 4176–4182 RSC.
- J. Lou, X. Tang, H. Zhang, W. Guan and C. Lu, Angew. Chem., Int. Ed., 2021, 60, 13029–13034 CrossRef CAS PubMed.
- W. Peng, W. Li, H. Han, H. Liu, P. Liu, X. Gong and J. Chang, VIEW, 2022, 3, 20200191 CrossRef CAS.
- Y. Qi and B. Li, Chem. – Eur. J., 2011, 17, 1642–1648 CrossRef CAS PubMed.
- M. Chakravarthy, H. AlShamaileh, H. Huang, R. K. Tannenberg, S. Chen, S. Worrall, P. R. Dodd and R. N. Veedu, Chem. Commun., 2018, 54, 4593–4596 RSC.
- D. Lee, S. Khaja, J. C. Velasquez-Castano, M. Dasari, C. Sun, J. Petros, W. R. Taylor and N. Murthy, Nat. Mater., 2007, 6, 765–769 CrossRef CAS PubMed.
- X. Y. Huang, L. Li, H. F. Qian, C. Q. Dong and J. C. Ren, Angew. Chem., Int. Ed., 2006, 45, 5140–5143 CrossRef CAS PubMed.
- E. Kojima, Y. Ohba, M. Kai and Y. Ohkura, Anal. Chim. Acta, 1993, 280, 157–162 CrossRef CAS.
- X. Zhu, N. N. Zhang, Y. T. Zhang, B. X. Liu, Z. Chang, Y. L. Zhou, Y. Q. Hao, B. X. Ye and M. T. Xu, Anal. Methods, 2018, 10, 641–645 RSC.
- T. Hu, C. X. Chen, G. M. Huang and X. R. Yang, Sens. Actuators, B, 2016, 234, 63–69 CrossRef CAS.
- R. Picou, J. P. Moses, A. D. Wellman, I. Kheterpal and S. D. Gilman, Analyst, 2010, 135, 1631–1635 RSC.
- J. Sethi, M. Van Bulck, A. Suhail, M. Safarzadeh, A. Perez-Castillo and G. H. Pan, Microchim. Acta, 2020, 187, 10 CrossRef.
- N. Zakaria, M. Z. Ramli, K. Ramasamy, L. S. Meng, C. Y. Yean, K. K. B. Singh, Z. M. Zain and K. F. Low, Anal. Biochem., 2018, 555, 12–21 CrossRef CAS PubMed.
- E. Niemantsverdriet, J. Ottoy, C. Somers, E. De Roeck, H. Struyfs, F. Soetewey, J. Verhaeghe, T. Van den Bossche, S. Van Mossevelde, J. Goeman, P. P. De Deyn, P. Marien, J. Versijpt, K. Sleegers, C. Van Broeckhoven, L. Wyffels, A. Albert, S. Ceyssens, S. Stroobants, S. Staelens, M. Bjerke and S. Engelborghs, J. Alzheimer's Dis., 2017, 60, 561–576 CAS.
- H.-M. Meng, H. Liu, H. Kuai, R. Peng, L. Mo and X.-B. Zhang, Chem. Soc. Rev., 2016, 45, 2583–2602 RSC.
- Y. Hang, J. Boryczka and N. Wu, Chem. Soc. Rev., 2022, 51, 329–375 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00162d |
| ‡ Ashley Khang and Nnebuefe Idegwu contributed equally in this research. |
| This journal is © The Royal Society of Chemistry 2023 |