Open Access Article
Open Access ArticleMicrofluidic-based colorimetric nucleic acid detection of pathogens
Sripadh Guptha
Yedire†
 ,
Haleema
Khan†
,
Tamer
AbdelFatah
,
Roozbeh
Siavash Moakhar
,
Haleema
Khan†
,
Tamer
AbdelFatah
,
Roozbeh
Siavash Moakhar
 and
Sara
Mahshid
and
Sara
Mahshid
 *
*
Department of Bioengineering, McGill University, Montréal, Quebec H3A 0C3, Canada. E-mail: sara.mahshid@mcgill.ca
First published on 27th March 2023
Abstract
Infectious diseases caused by pathogens put a significant burden on global health, as exemplified by the COVID-19 pandemic. There is a need for cost-effective detection techniques that ensure high sensitivity and specificity, comparable to standard methods. Point-of-care (POC) nucleic acid detection techniques provide low-cost, rapid solutions for congregate and remote settings. Microfluidic devices combined with colorimetric read-outs, as one of the portable and easy-to-interpret detection techniques, are apt for POC diagnostics. This paper reviews the most recent advances in colorimetric-based microfluidic devices for the nucleic-acid detection of viruses, bacteria, fungi, and protozoa including influenza, SARS-CoV-2, Listeria, Pseudomonas, Candida, and malaria. The characteristic features of devices, effectiveness of pathogen detection, analysis time, sensitivity and specificity of the results are discussed here. In addition, this paper offers an insight on the future avenues of microfluidic-based colorimetric detection, highlighting the necessary steps for achieving the high caliber set by gold standard techniques. This article suggests that the integration of plasmonic nanostructures with microfluidic devices will address the issue of sensitivity in today's colorimetric-based devices. Future work should also focus on addressing the need for an all-encompassing device, as well as the commercialization of devices to augment translation in clinical settings.
1 Introduction
Infectious diseases are driven by pathogens including bacteria, viruses, fungi, and protozoa. Pathogens create a healthcare challenge as they can rapidly propel the spread of infectious diseases through quick transmission from one host to another. A primary example is the COVID-19 pandemic (the disease caused by novel SARS-CoV-2, that emerged in December 2019),1 which has taken millions of lives, damaged economies worldwide, and tremendously disrupted social operations. As such, effective pathogen diagnosis is a crucial step in reducing the load of deadly infectious diseases. Combatting the COVID-19 pandemic has been a hotspot of recent research and there have been strong efforts worldwide for rapid disease diagnosis and isolation.2 While polymerase chain reaction (PCR) remains the gold standard technique, next generation and advanced pathogen sensing platforms have been recently reported.3,4 Among these, colorimetric detection has gained attention for its ability to deliver results on the order of minutes, as well as for its user-friendly and cost-effective methods. Colorimetric detection can be combined with nucleic acid assays for rapid and accurate detection of disease biomarkers.Nucleic acid amplification techniques (NAATs) for the diagnosis of infectious diseases have been widely implemented, some of which include PCR,5 isothermal amplification assays (IA),6 and CRISPR/Cas.7 While, centralized laboratories remain the dominant hub of pathogen testing worldwide,8 they are limited by the delay in results, need for bench-top analyzers, and highly trained personnel.9 Therefore, it is necessary to employ rapid, sensitive, and economic detection techniques when diagnosing pathogens. Point-of-care (POC) diagnostic systems aim to overcome the challenges presented by standard detection techniques as they offer prompt results for on-site diagnosis and treatment. Some features of POC diagnostics are rapid tests to allow patients to receive immediate treatment plans, sensitive and specific results comparable to those of standard methods, and user-friendly systems.10 This enables healthcare practitioners to make quick medical decisions leading to improved health outcomes for patients, due to disease diagnosis at the earliest stage.
To aid in the development of POC devices, the World Health Organization (WHO) has proposed a gold standard framework dubbed, ASSURED to evaluate and better design POC diagnostic tools.11 This framework expects a POC diagnostic tool to be, affordable, sensitive, specific, user-friendly, rapid/robust, equipment-free, and deliverable (ASSURED).11 In compliance with ASSURED criteria, there are three defining features of a POC system, 1) biochemical assay of detection, 2) sample handling, and 3) signal transduction.11 The choice of the assay for pathogen detection plays an important role in designing a POC diagnostic test. The desired features include cost-effectiveness, high sensitivity and specificity, ease of integration with POC tools like microfluidics, and digital signal transduction techniques. Although the ASSURED criteria are not a necessary criteria for designing POC systems, they aid in discussing and evaluating the POC technologies reported, analogous to a flexible yard stick. In this context, POC systems have been proposed integrating NAATs with microfluidic systems.
Microfluidics have been crucial in designing POC devices for the past decade. Microfluidic platforms are miniature devices which enable the precise manipulation of flow and reaction conditions of fluids on a sub milliliter scale. A crucial feature of microfluidic platforms for pathogen detection is that they can enable the integration of sample processing, extraction, detection, and analysis onto a single platform.12–14 Moreover, confined reaction volumes allow for fast and high throughput analysis.15,16 Microfluidic platforms also carry with them features like portability, automation, and precise handling of small reaction volumes.17 These factors make them ideal candidates for POC applications. Moreover, microfluidics can potentially be integrated with different read-out systems including electrical, optical, and colorimetric.18 Colorimetric methods mainly encompass fluorescent and visual color analysis techniques which are at the heart of this review article. The central idea of colorimetric analysis is the use of specific responsive dyes or nanomaterials that elicit color or fluorescent intensity changes upon variations in local reaction conditions such as the amplification of nucleic acids.19 In the case of the nanomaterial based colorimetric assays, the color change is elicited by the coupling of localized surface plasmon resonance of the nanomaterial, as opposed to colorimetric dyes.20 In both cases, colorimetric (and fluorometric) changes can be assessed by the naked eye or with simple and inexpensive brightfield or fluorescence analysis setups, making colorimetric platforms inexpensive, portable, fast, and highly suitable for POC and low-resource settings.21,22
In recent years and with the advent of the COVID-19 pandemic, the need for economic, sensitive, and portable nucleic acid detection platforms is in the spotlight. Colorimetric-readout platforms offer flexibility in output analysis and give a simplistic readout, which are especially crucial for resource-limited settings. These features can be nicely complemented by microfluidics due to their portability and high throughput. Thus, the integration of colorimetric nucleic acid readout systems onto microfluidic platforms could possibly address the current need for rapid, simple, and cost-effective diagnostic platforms. To capture the state of current research in this area, this review article highlights recent microfluidic colorimetric nucleic acid-based pathogen detection platforms and their assay parameters, discussing important aspects that point to the potential scope for future research. Past review papers have focused on colorimetric assays for pathogen detection.23 To the best of our knowledge there are no other existing reviews that focus on colorimetric nucleic acid detection of all pathogens including viruses, bacteria, fungi, and protozoa using microfluidic technology. This paper aims to address this gap in the literature by centering on microfluidic devices reported in the past 5 years which focus on the detection of viruses, bacteria, fungi, and protozoa. Fig. 1 illustrates the differences between conventional NAATs carried out in the laboratory and those integrated in microfluidic platforms. As depicted in Fig. 1, the former is disadvantaged by requiring trained personnel, long wait times for results, and expensive equipment. In contrast, microfluidic platforms offer POC capabilities including faster results, ease of operation, cost-effectiveness, and portability.
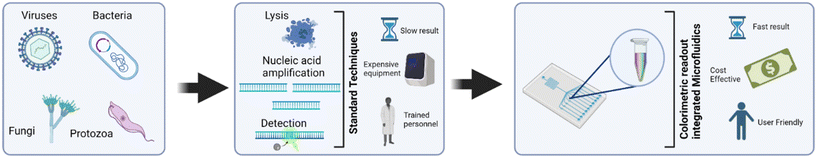 | ||
| Fig. 1 Schematic overview of the review article depicting techniques and pathogens reviewed for diagnosis. | ||
2 Nucleic acid detection techniques
Nucleic acid assays are widely employed for the detection of viruses, bacteria, and fungi.24 Manual microscopy examination, dubbed as microscopic ova and parasite examination (O&P), is still the gold standard for clinical detection of protozoan.25 PCR and LAMP are two nucleic acid detection techniques that provide results within a few hours, making them favorable for POC applications.26 PCR is currently the gold standard technique for the amplification of nucleic acids in vitro due to its rapid amplification of target DNA sequences resulting in fast and high-throughput detection.27 This has led to its prominence as a clinical tool in the diagnosis of bacterial and viral pathogens.28 Similarly, isothermal amplification techniques such as loop-mediated isothermal amplification (LAMP) and recombinase-based amplification, are beneficial for rapid detection of nucleic acids.29 Constant temperatures of isothermal amplification provide a less complicated alternative to PCR, making them suitable for POC applications.30 More recently, CRISPR/Cas technology has emerged as a genome editing tool, based on the ability to recognize and cleave specific DNA or RNA sequences.31 Catalytic activities with CRISPR/Cas can be implemented for nucleic acid detection in biosensors, for example by the degradation of labelled sequences which can be monitored by fluorescent signals.31 A strong advantage of CRISPR/Cas is that multiplexing is very palpable, enabling a single test to identify multiple targets.312.1 Colorimetric indicators-an optical perspective
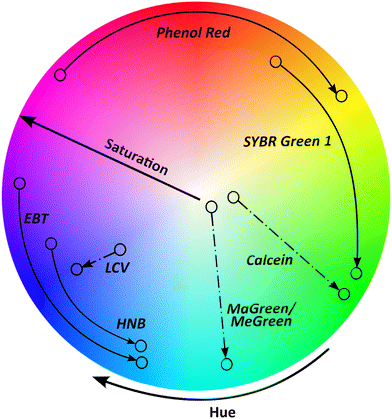
Colorimetric readout is gaining traction and is applied in conjugation with several pathogenic nucleic acid detection techniques.32 Some significant advantages of colorimetric readouts include ease of interpretation, label-free signal transduction, cost-effectiveness, and simplicity.33 The color sensing schemes when coupled with nucleic acid detection assays for pathogens, are broadly categorized into (a) pH responsive colorimetric change,34 (b) color-change via chromogenic indicators,35 (c) colorimetric or florescence change via intercalating dyes,36 (d) nanoparticle mediated colorimetric change.37 One notable obstacle with colorimetric readout is that the human eye is insensitive to low level color contrasts; thus, discriminating outputs between distinct samples is difficult with the naked eye since most conventional colorimetric approaches primarily depend on the colored output's optical density variation. Different colorimetric and fluorescent indicators engender change in optical properties in presence of the target, predominantly in hue and saturation values (Fig. 2). It is evident from Fig. 2, that phenol red offers biggest change in hue upon amplification of the target, enabling quantification. Several systemic studies in the past, compared different colorimetric dyes nucleic acid detection, especially NAATs.38,39 In a recent user-centric study from Scott et al.,40 the authors evaluated six colorimetric LAMP indicators in terms of ease and accuracy of user-interpretation and inter-user variability. The study found that intercalating dyes like leuco crystal violet (LCV), malachite green (MG), that change color in the saturation scale, achieved highest accuracy and agreement between users. Whereas pH dependent phenol red dye was ranked highest for the ease of interpretation. Phenol red posits a high color contrast due to a stark transition from fuchsia to yellow compared to as hydroxynaphthol blue and Eriochrome Black T change to colors with similar hues effecting the sensitivity and ease of interpretation.41,42 It is worth noting that these previous user studies (naked eye) analysis were performed in Eppendorf tubes. Whereas, in the case of microfluidic systems coupled with colorimetric readouts the path length (channel height) of the sensing area, where the strength of the colorimetric signal (related to absorbance) is directly proportional to the path length, according to Beer–Lambert law.43,44 In contrast, for fluorescence readouts, the emission intensity varies inversely with the square of the path length of the sensing area.45 These empirical relations become crucial design considerations when designing capillary or liquid channel based microfluidic setups.46,47 In their study, Scott et al.,40 demonstrated quantitative analysis of LAMP-on-chip reaction with HNB as the indicator, using a digital analyzer. Interestingly, they reported that the short path length did not hamper the quantitative analysis when an image analysis algorithm is employed. Evidently, there has been an increasing the adoption of smartphones and digital cameras coupled with advanced data processing algorithms for color capture and quantification.48–50 Another important consideration when comparing different colorimetric indicators in the context of POC applicability is the requirement of an extra assay step for addition of intercalating dye post-amplification, as opposed to pH sensitive, intercalating, metal ion chromogenic indicators.513 Integration of nucleic acid detection into state-of-the-art portable technologies
Employing colorimetric techniques for pathogen detection have been well researched in the past decade. Interestingly, microfluidic platforms offer advantages like high throughput, requirement of low reaction volumes, and portability, which are largely complementary to the advantages of colorimetric assays. Thus, integration of the above-noted nucleic acid colorimetric assays into microfluidics makes them more conducive to POC. We discuss research exploring this conjunction in the past years for detection of virus, bacteria, protozoa, and fungi, in the following sections.3.1 Colorimetric detection of viruses
Viruses are one of the most serious threats to human life due to their highly transmissive capabilities, as exemplified by the COVID-19 pandemic.52 Within the past two years, the SARS-CoV-2 virus has led to over 6 million mortalities worldwide.53 In addition to COVID-19, each year almost 1 million people die from HIV/AIDS,54 and the WHO has reported between 290![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 650
000 to 650![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths from the seasonal flu,55 as well as up to 887
000 deaths from the seasonal flu,55 as well as up to 887![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 annual deaths from hepatitis B related liver diseases.56 Effective identification of viral pathogens is crucial in the pipeline of disease mitigation as this helps with testing and tracing, appropriate therapeutic recommendations for patients, and robust disease control.57 Many infectious viral diseases are concentrated in low-resource settings; thus, microfluidic-based nucleic acid tests using colorimetric detection may be advantageous in providing a cost-effective, user-friendly alternative to ameliorate existing health disparities.58 Colorimetric virus NAATs depend on the amplification of viral nucleic acids through virus specific primers.59 Genetic material can be directly detected from samples or DNA/RNA extraction can be carried out depending on the virus of interest.59 Notably, great effort has been made in maintaining the accuracy of PCR methods while integrating with colorimetric detection of viruses in microfluidics which is discussed here.
000 annual deaths from hepatitis B related liver diseases.56 Effective identification of viral pathogens is crucial in the pipeline of disease mitigation as this helps with testing and tracing, appropriate therapeutic recommendations for patients, and robust disease control.57 Many infectious viral diseases are concentrated in low-resource settings; thus, microfluidic-based nucleic acid tests using colorimetric detection may be advantageous in providing a cost-effective, user-friendly alternative to ameliorate existing health disparities.58 Colorimetric virus NAATs depend on the amplification of viral nucleic acids through virus specific primers.59 Genetic material can be directly detected from samples or DNA/RNA extraction can be carried out depending on the virus of interest.59 Notably, great effort has been made in maintaining the accuracy of PCR methods while integrating with colorimetric detection of viruses in microfluidics which is discussed here.
A study by Kim and colleagues reported an integrated microdevice incorporating a reverse transcription PCR (RT-PCR) with an immunochromatographic strip (ICS) for rapid gene expression of the influenza A H1N1 virus.60 The microdevice consisted of a five-layered glass polydimethylsiloxane with a functional unit of a pneumatic micropump, RT-PCR chamber, a microvalve, and an ICS. The portable system identified the target H1 gene of the H1N1 virus within 2.5 h. Their work made a valiant effort to scale down the scale of a standard PCR assay into a portable, sensitive microfluidic device. However, the response time can be improved upon for enhanced results in POC settings.
The advent of isothermal amplification techniques for molecular detection of pathogenic DNA, provides a less complicated alternative to PCR, making them more suitable for integration onto microfluidic platforms.61 The main factors contributing to this are, 1) requirement of constant temperature cycles for the denaturation and amplification steps compared to conventional PCR assays, thereby reducing the complexity of the components and set-up required, making it more adaptable to POC settings;62 2) higher specificity and sensitivity compared to conventional techniques;63 and 3) stability against some amplification inhibitors.21 Moreover, isothermal amplification can be combined with gene editing through CRISPR/Cas for detecting target sequences with a colorimetric response.64 LAMP is most widely researched among these isothermal techniques for pathogen detection in microfluidic platforms.62 Microfluidic platforms integrated with LAMP based assays for pathogenic nucleic acid detection have demonstrated POC capabilities in previous works.62 The reduced reagent volumes compared to laboratory techniques make them conducive to use in resource limited settings, facilitating multiplexed detection assays and reduced assay costs.65
With the onset of the COVID-19 pandemic, SARS-CoV-2 detection has been a research hotspot in the diagnostic community. To this end, Yin et al. proposed a smartphone-enabled microfluidic device with integrated nucleic acid extraction and colorimetric LAMP assay for the detection of SARS-CoV-2 and human enteric pathogens in wastewater (Fig. 3a).66 Nucleic acid extraction was facilitated by a Flinders Technology Associates (FTA) membrane, which served as a template for on-chip nucleic acid amplification. A combined recombinase polymerase amplification (RPA) and synergistic enhanced colorimetric LAMP technique was employed for nucleic acid amplification in a sensitive and multiplexed fashion, using Eriochrome Black T (EBT). They reported a sensitivity for SARS-CoV-2 of 100 genome equivalent (GE)/mL within 1 h.
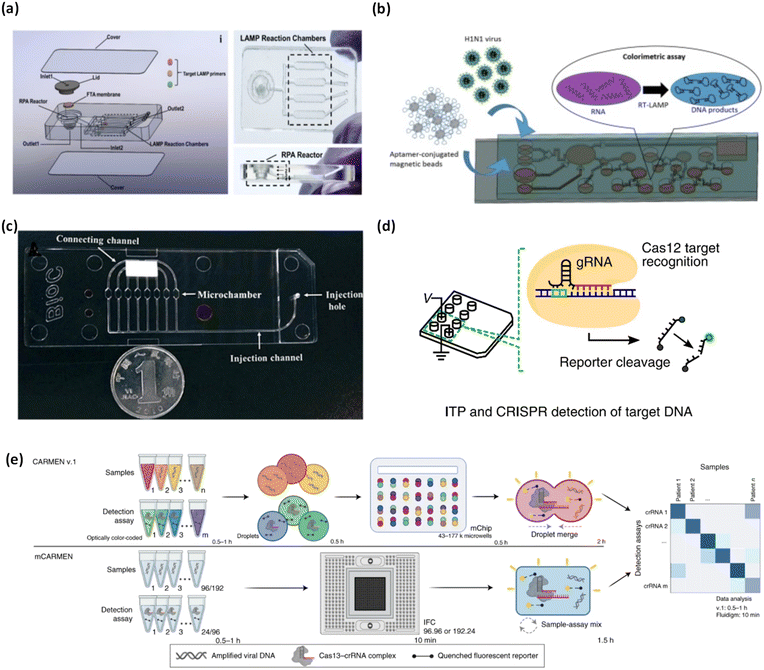 | ||
| Fig. 3 Matrix depicting microfluidic platforms for colorimetric nucleic acid detection of viruses. (a) Integrated microfluidic chip with exploded view, top view, and side view for detection of SARS-CoV-2. Adapted with permission from ref. 66. Copyright 2021, Elsevier. (b) Schematic illustrating self-driven microfluidic device combined with an RT-LAMP assay for colorimetric detection of H1N1. Adapted with permission from ref. 67. Copyright 2019, Elsevier. (c) Graphic depicting developed microfluidic chip for the detection of influenza A. reproduced with permission from ref. 68. Copyright 2018, Royal Society of Chemistry. (d) Schematic of ITP-CRISPR integrated microfluidic device for SARS-CoV-2 detection. Reproduced with permission from ref. 64. Copyright 2020, National Academy of Sciences. (e) Comparison of CARMEN v.1 and mCARMEN for CRISPR-based diagnosis of SARS-CoV-2 and other viruses/variants. Adapted with permission from ref. 69. Copyright 2022, Nature Publishing Group. | ||
Influenza A subtypes are some of the most studied viruses using RT-LAMP based colorimetric detection through microfluidic devices. A novel, self-driven microfluidic device developed by Ma et al. was used to detect H1N1 in just 40 min (Fig. 3b).67 H1N1 virus samples and H1N1 specific aptamer conjugated beads were loaded into corresponding chambers in the microfluidic chip. Capillary forces self-transported the liquid flow and hydrophobic soft valves stopped the transport. The device performed virus isolation via aptamer conjugated beads, virus lysis, RT-LAMP, and a colorimetric reaction with hydroxynaphthol blue. The limit of detection of this device was 3 × 10−4 hemagglutinating units/reaction, which is sufficient sensitivity for clinical applications. This device is the first of its kind due to its self-driven, passive microfluidic action.
Influenza A was also investigated by Wang and colleagues as they integrated RT-LAMP with a microfluidic device for viral colorimetric detection using hydroxynaphthol blue within 1 h (Fig. 3c).68 Initially, the detection chamber had a violet color, the presence of viral RNA of interest and subsequent reaction changed the color to sky blue. Using the developed device, they were able to identify influenza A virus subtypes (H1N1, H3N2, H5N1, and H7N9), influenza B virus, and human adenoviruses. They further evaluated their device using 109 clinical samples achieving high 100% specificity (confidence interval 94.9–100.0) and 96% sensitivity (confidence interval 78.1–99.9). The device notably reported a limit of detection which was 10–100 times better than conventional PCR.
Ideally, diagnostic tests would have the capacity to surveil multiple viruses and variants. CRISPR-based diagnostics offer a highly effective approach to screening numerous viruses and variants simultaneously. In an effort to scale up the capabilities of CRISPR-based diagnostics, Welch and colleagues developed mCARMEN, a microfluidic device with streamlined workflow to rapidly detect multiple viruses and variants simultaneously, based on a quenched fluorescent reporter (Fig. 3e).69 They tested up to 21 viruses including SARS-CoV-2, other coronavirus variants, and influenza, which was bolstered by testing 691 patient samples and 2088 patient samples, for viruses and SARS-CoV-2 variants, respectively. mCARMEN employed integrated fluidic circuits (IFCs) on the commercially available Fluidigm Biomark which uses a particular number of assay combinations that are spatially separated. After manual loading of the IFCs, samples were moved through individual IFC channels until they reached the chip reaction chamber for mixing. Fluorescence was measured with custom automated protocols that captured images of the IFC chip at regular intervals of 5 min for 1–3 h. mCARMEN had 100% analytical specificity and an LOD of 1000 copies per μL for SARS-CoV-2 in a respiratory virus panel (RVP), with a total workflow time of <3 h. Although the workflow time may be deemed as lengthy for POC settings, the high throughput enables broad screening of patients which serves as a huge advantage.
Another work using CRISPR-based diagnostic was by Ramachandran et al., whom developed a device which detected SARS-CoV-2 RNA in ∼30 min (Fig. 3d).64 The workflow of the device involved on-chip isotachophoresis (ITP) extraction of nucleic acids from raw nasal swab samples in viral transport medium. Following this, RT-LAMP was performed off-chip to amplify the viral N and E genes. In the last step, ITP was used to perform on-chip CRISPR-Cas12 based enzymatic reactions for target detection where ITP enabled target DNA recognition and target-activated cleavage of ssDNA reporters. A fluorescent readout was used to verify the presence of target nucleic acids. The device was validated using 64 patient samples where the ITP-CRISPR method correctly detected 30/32 positive samples and showed no false positives. Further, the device showed a robust sensitivity, with an estimated LOD of 10 copies per μL. While their method shows strong sensitivity and fast response time, it is limited by the off-chip RT-LAMP assay, which to some extent, limits full application in POC settings.
Other viruses of interest using LAMP based colorimetric detection include the Ebola virus,70 Zika virus,71 and HPV72 (Table 1). These have shown highly specific results of 100% (ref. 65) or like that of conventional RT-LAMP.71 Additionally, these assays have demonstrated a rapid analysis time of <30 min.65,71 These assays also demonstrated high sensitivity,65,71 where clinical samples have notably shown 100% sensitivity.72
| Pathogen | Indicator | Signal analysis method | Chip material | Sample processed | Limit of detection (LOD)/sensitivity | Specificity | Analysis time | Ref. |
|---|---|---|---|---|---|---|---|---|
| Viruses | ||||||||
| PCR | ||||||||
| H1N1 | Gold nanoparticle-based violet color | Naked eye | Glass-PDMS hybrid | Unspecified | 14.1 pg RNA templates | 100-fold improved detection sensitivity compared to agarose gel | 2.5 h | 60 |
| H1N1 | Taqman florescence probe | Handheld analysis module | Polycarbonate | RNA spiked buffer | 1.0 TCID50/ml | Not specified | 50 min | 105 |
| LAMP | ||||||||
|---|---|---|---|---|---|---|---|---|
| H1N1 | pH indicator (hydroxynaphthol blue) | Naked eye | Hydrophilic film, PDMS, and glass slide | Diluted lab provided samples | 3 × 10−4 hemagglutinating units/reaction | High specificity, confirmed with H3N2, influenza B, and E. coli | 40 min | 67 |
| 10 RNA copies/reaction | ||||||||
| H1N1, H3N2, H5N1, H7N9, influenza B, and human adenoviruses | pH indicator (hydroxynaphthol blue) | Real-time detection instrument | Polycarbonate | Throat swab | 10–100 times better than PCR, high sensitivity (96%) | High specificity (100%) | 1 h | 68 |
| Zika virus | pH indicator (phenol red) | Smartphone | Paper | Tap water, urine, plasma | 1 copy RNA/μL | Similar to conventional RT-LAMP | 15 min | 71 |
| SARS-CoV-2 | Fluorescence analysis (EvaGreen intercalating dye) | Smartphone | Polymer | Nasopharyngeal swab | 50 RNA copies per μL in viral transport medium solution within 30 min, sensitivity of 100% | Specificity of 100% | 30 min | 65 |
| Cutaneous human papillomaviruses | pH indicator (hydroxynaphthol blue) | Naked eye | Not reported | Skin scrapes | 107 viral DNA copies per μL, sensitivity was 100% | Specificity was 34.55–97.62% depending on virus type | Not reported | 72 |
| Vector-borne viruses: Rift Valley fever virus (RVFV), chikungunya virus (CHIKV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), and dengue virus (DENV) subtype I, II, III, and IV | Fluorescence analysis (SYBR green I or calcein dye) | Naked eye or real-time fluorescent analysis device | PDMS | Serum from patients and mosquito samples | 500 copies for RVFV and YFV, 100 copies for CHIKV and DENV-IV, 50 copies for JEV, DENV-I, DENV-II, and DENV-III | Specificity of colorimetric LAMP was the same as real-time LAMP | 50 min | 106 |
| SARS-CoV-2 and human enteric pathogens | pH indicator (Eriochrome Black T) | Smartphone | Methacrylate-based resin | Wastewater samples | 100 GE/mL for SARS-CoV-2 and 500 CFU mL−1 for enteric pathogens | High specificity demonstrated samples tested | 1 h | 66 |
| Dengue virus (DENV) | pH indicator (phenol red) | Naked eye | Paper/polymer based strip | Human blood samples | 5 RNA copies | High specificity among different DENV serotypes | 30 min | 107 |
| SARS-CoV-2 | SYBR green I fluorescence reporter | Smartphone camera | PDMS | Heat inactivated nasopharyngeal swab samples | 100 RNA copies | 100% specificity reported | 1 h | 108 |
| CRISPR | ||||||||
|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2, SARS-CoV-2 variants (including delta and omicron), and influenza | Fluorescence analysis (quenched synthetic fluorescent RNA reporter or RNaseAlert v2) | Fluorescence on Fluidigm Biomark | Commercially available Fluidigm Biomark | Nasopharyngeal swab | LOD range was between 500–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per μL depending on target (SARS-CoV-2 LOD was 1000 copies per μL) 000 copies per μL depending on target (SARS-CoV-2 LOD was 1000 copies per μL) |
100% specificity in respiratory virus panel | <3 h | 69 |
| SARS-CoV-2 | Fluorescence analysis (ssDNA fluorescence quencher reporter) | Fluorescent imaging and analysis | Glass | Nasal swab samples in viral transport medium | 10 copies per μL | 30/32 positive targets correctly detected, no false positives | ∼30 min | 64 |
| SARS-CoV-2, influenza A, and influenza B | Fluorescence analysis (ssDNA reporter) | Naked eye and portable devices | PMMA | Nasopharyngeal swab | 10 copies | Exhibited no cross-reactivity against other coronaviruses | ∼1 h | 109 |
| Bacteria | ||||||||
|---|---|---|---|---|---|---|---|---|
| PCR | ||||||||
| Escherichia coli, Staphylococcus epidermidis, and Staphylococcus saprophyticus | Fluorescence analysis (TaqMan probe) | Photomultiplier tube attached to microscope | PDMS | Blood | 5 CFU ml−1 | None | 4 h | 77 |
| LAMP | ||||||||
|---|---|---|---|---|---|---|---|---|
| P. hauseri, Salmonella and E. coli | Fluorescence analysis (calcein) | Portable fluorescence imaging system | PDMS | In vitro spiked samples for analysis | 1.6 copy number for Salmonella | None | 110 min | 110 |
| Salmonella typhimurium | Magnesium sulfate (MgSO4) mediated turbidity analysis | Smartphone | PDMS | Spiked samples and proof of concept established with meat samples | 14 CFU ml−1 | Listeria monocytogenes, E. coli O157:H7 and Vibrio parahaemolyticus | 90 min | 111 |
| Simultaneous detection of Staphylococcus aureus, Salmonella, Shigella, enterotoxigenic Escherichia coli, and Pseudomonas aeruginosa | Visual color change of LAMP mixture purchased commercially | Portable analyzer | PMMA | Spiked samples and contaminated water | Sau and Sal were both 102 copies per μL, whereas the LODs for Sty, Pae, and Eco were as low as 101 copies per μL | None | 70 min | 112 |
| Salmonella typhimurium | Fluorescence analysis (SYBR green dye) | Fluorescent imaging setup | PDMS and glass | Salmonella typhimurium in milk samples | 5000 CFU mL−1 in the sample or 25 RNA template/25 μL LAMP reaction cocktail | LAMP reaction with S. flexneri (Gram-negative) and S. aureus (Gram-positive) | Not reported | 78 |
| Multiplex detection of P. aeruginosa, S. typhimurium, V. parahaemolyticus, V. vulnificus, S. iniae, or V. alginolyticus | Fluorescent analysis (calcein) | Naked eye or smartphone camera using UV flashlight | PMMA | In vitro bacteria spiked samples | 2 × 102 cells per μL | None | ∼1 h | 82 |
| Vancomycin-resistant Enterococcus (VRE) bacteria | Florescent analysis (calcein) | Optical microscope imaging and analyzed on computer | PDMS and glass | DNA spiked in vitro samples | 1 copy of DNA or 50 copies per μl | Not reported | ∼40 min | 86 |
| Bacterial meningitis DNA, that include Streptococcus agalactiae, Streptococcus pneumoniae, Staphylococcus aureus | Fluorescence analysis (hydroxynaphthol blue) | Fluorescence imaging setup | Paper | Clinical patient DNA samples extracted from culture bacterial meningitis | 4.1 × 102 copies of genomic DNA for Streptococcus pneumoniae | Reported no cross-reactivity for multiple pathogenic DNA | ∼60 min | 79 |
| Escherichia coli O157:H7, Salmonella typhimurium and Vibrio parahaemolyticus | pH indicator (Eriochrome Black T) | Naked eye | PMMA and pressure sensitive adhesive foil | Cultured bacterial cells and extracted genomic DNA | 500 copies when extracted DNA is sample input and 100 copies when bacterial cells are directly used as sample input | None | 60 min | 81 |
| CRISPR | ||||||||
|---|---|---|---|---|---|---|---|---|
| Listeria | Fluorescence analysis (quenched fluorescent ssRNA reporter) | Automated analysis device for fluorescent reading analysis | PMMA | Flammulina velutipes | aM level for each pathogen or 26–96 CFU mL−1 | Strong specificity when comparing fluorescent signals of target and non-target strains | 60 min | 83 |
| Pseudomonas aeruginosa | Fluorescence analysis (tag ssDNA) | Automated analytical equipment with connected to computer | PMMA | Unspecified | 103 CFU mL−1 | High specificity for P. aeruginosa compared to 8 different kinds of bacteria | 1.5 h | 84 |
| Escherichia coli O157:H7 | Fluorescence reported | Automated analysis via smartphone | SU-8-PDMS | Contaminated food samples | 10 CFU mL−1 | Specificity shown against seven other pathogens | 2.5 h | 22 |
| Fungi | ||||||||
|---|---|---|---|---|---|---|---|---|
| LAMP | ||||||||
| Aspergillus spp. | Blue silver nanoplates | Naked eye | Paper | Herbal samples | 100 aflR copies | Specificity was 94.47% | 40 min | 94 |
| Cryptococcus | Fluorescence analysis (SYBR green) | Gel image analysis system | PDMS | Cerebro-spinal fluid | 100 CFU mL−1 | Primers showed 92% when tested against the DNA of 50 pure culture strains | 45 min | 95 |
| Aspergillus fumigatus, Aspergillus flavus, and Cryptococcus neoformans | Hydroxyl naphthol blue (HNB) | Naked eye | PMMA | Spiked aerosol samples | 4 × 104 spores/sample | Specificity tested against six common fungi | 90 min | 113 |
| CRISPR | ||||||||
|---|---|---|---|---|---|---|---|---|
| Candida and Aspergillus | Brown poly(3,3′-diaminobenzidine (DAB)) stripe | Naked eye | Paper | Spiked urine or sputum samples | ∼5 CFU mL−1 | Reasonable specificity when compared with qRT-PCR | 90 min | 96 |
| Protozoa | ||||||||
|---|---|---|---|---|---|---|---|---|
| LAMP | ||||||||
| Plasmodium falciparum (malaria) | Streptavidin labelled red beads | Naked eye | Paper | Blood samples | 105 IU mL−1 after 45 min amplification | None | 45 min | 99 |
| P. falciparum and P. vivax | Fluorescence analysis (calcein) | Mobile analyzer with smartphone readout | PMMA | Spiked human blood samples | 0.5parasites per μl | None. But no interference reported from cellular material in blood sample | 50 min | 100 |
Overall, colorimetric virus detection in microfluidics has been a crucial area of research in finding innovative detection techniques for some of the world's deadliest viruses including SARS-CoV-2, HIV, H1N1, hepatitis B, and the avian flu virus. The quickest analysis time was 30 min from the papers reported which is a drastic improvement from standard methods. For several devices the specificity and sensitivity were close to 100% or comparable to conventional PCR. Moreover, Wang and colleagues' microfluidic device reported a limit of detection which was 10–100 times better than conventional PCR.68Table 1 further summarizes the specks of each device widely. While the devices reported show a proof-of-concept with promising results, further efforts are needed to mobilize these devices for wide clinical applications as exemplified by the SARS-CoV-2 rapid testing kits. Research ought to focus on the integration of all components of analysis on-chip including sample pre-treatment, voltage supplies, and pumps, to enhance user-friendliness and to improve marketability for customers.
3.2 Colorimetric diagnosis of bacteria
Bacteria are one of the major pathogens that regularly engender public health concerns across many parts of the world.73 In clinical settings rapid detection of bacteria would ensure timely diagnosis and treatment of patients.74 Microfluidic technologies with POC capabilities can come to the rescue and prove to be invaluable given their portability, low cost, and rapidity. Research in previous decades has mainly focused on bacterial pathogens in sources that facilitate pathogen transmission like food, material, and water sources, which demand rapid diagnosis and surveillance to curb any potential outbreak at source.75 Similarly, multifunctional microfluidic technologies are being developed to carry out bacteria identification and antibiotic susceptibility testing to accelerate assays.76 Traditionally, culture-based techniques have been used for clinical diagnosis of bacteria, but it is a time intensive process. As an alternative, nucleic acid-based PCR testing has emerged as a rapid and relatively more sensitive alternative. Integrating PCR onto a microfluidic platform brings advantages with the requirement of small sample volumes, enabling ‘sample to results’ in real time. Recently developed colorimetric PCR-coupled microfluidic platforms for bacterial pathogen detection integrating processes from sample preparation to analysis are listed in Table 1. A good example of this is the recent work from Fang et al. which reported a microfluidic-integrated platform for the detection of sepsis induced bacteria.77 The PCR products were detected by the fluorescent analysis of the TaqMan amplicon probe. They reported a good sensitivity of 5 CFU ml−1 which showed minimum interference with blood cellular material facilitated by the enhanced filtration system.LAMP is an excellent alternative to PCR, as evident from the growing interest in this technique. Previously, Azizi et al. reported an application of droplet microfluidics in optimizing the LAMP-based detection of Salmonella typhimurium shown in Fig. 4a.78 The microfluidic platform was employed to encapsulate the bacterial RNA and LAMP cocktail in microdroplets resulting from water-in-oil emulsions, essentially creating microdroplet reactors for the amplification to proceed. The amplification reaction and thus the presence of bacterial RNA in cultured samples and contaminated milk samples, was confirmed by the green fluorescence of SYBR dye in positive samples.78 A mathematical model was also proposed to evaluate the number of droplets required for screening to detect a positive microdroplet, this sped up the process of evaluating detection limits. This approach also resulted in high specificity against other bacterial pathogens. A detection limit of 5000 CFU ml−1 in spiked solutions for cultured samples was reported. However, the demonstration to some extent lacks applicability at POC, owing to the requirement of fluorescent imaging, the laboratory setup for incubation and amplification, and an RNA extraction step off-chip.
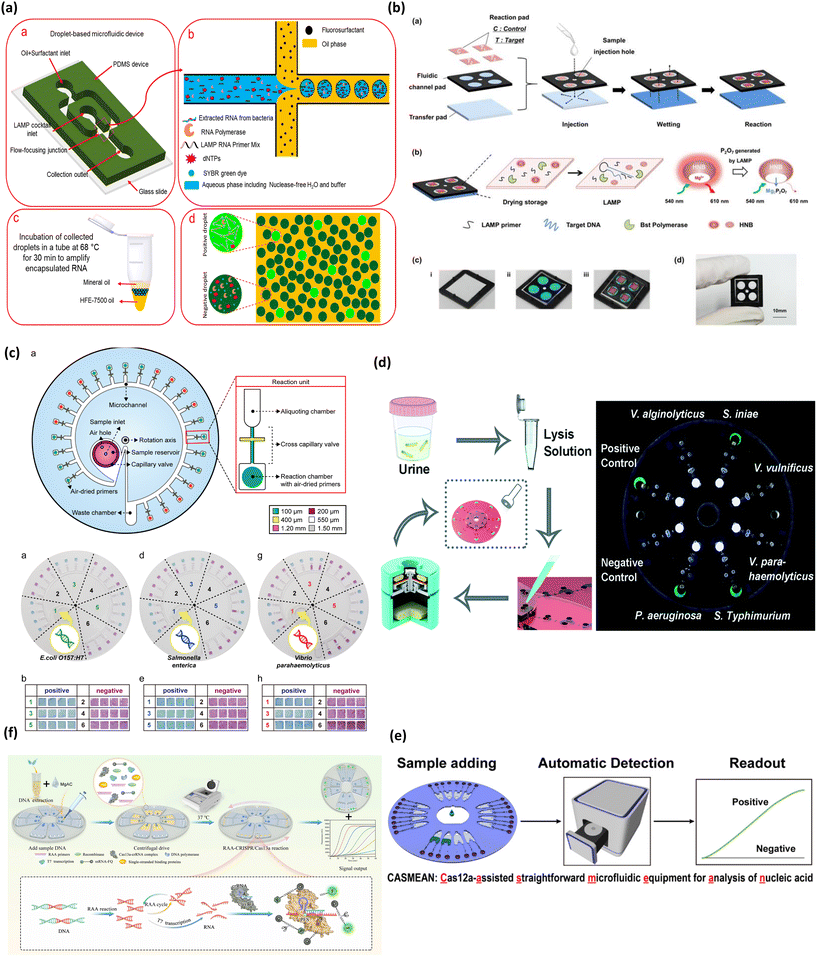 | ||
| Fig. 4 Matrix depicting colorimetric microfluidic platforms for bacteria detection. (a) Schematic depicting detection of pathogenic bacteria utilizing LAMP mediated by droplet microfluidics (a–d). Adapted with permission from ref. 78. Copyright 2019, American Chemical Society. (b) Schematic showing process flow of paper based colorimetric platform for real-time detection of multiple bacterial meningitis DNA via LAMP-based assay resulting in florescence emission of hydroxynaphthol blue (a–d). Reproduced with permission from ref. 79. Copyright 2017, Ivyspring International. (c) Multiplex detection of food pathogens that elicits colorimetric change from purple to sky blue mediated by Eriochrome Black T upon carrying out a LAMP reaction on centrifugal based microfluidic device (a, b, d, e, g and h). Adapted with permission from ref. 81. Copyright 2017, Elsevier. (d) Centrifugal based platform for multiplexed detection of bacterial pathogens based on LAMP assay eliciting a fluorescent signal upon pathogen detection. Reproduced with permission from ref. 82. Copyright 2018, Royal Society of Chemistry. (e) Workflow of microfluidic device combining RAA and CRISPR for detection of Listeria. Adapted with permission from ref. 83. Copyright 2022, Elsevier. (f) Schematic demonstrating RAA-CRISPR detection of Pseudomonas aeruginosa. Adapted with permission from ref. 84. Copyright 2020, American Chemical Society. | ||
In another work, Seok et al. developed a paper-based fluidic device for diagnosis of three types of bacterial meningitis DNA, which include Streptococcus agalactiae, Streptococcus pneumonia and Staphylococcus aureus in ∼60 min, as shown in Fig. 4b.79 The device encompassed three fluidic layers, namely, a reaction pad, fluidic channel pad, and transfer pad. These layers were stacked up in the order mentioned, with a sample injection hole in the center. The sample entering from this hole wetted the reaction pad through the fluidic layer. This system used dried LAMP reagents immobilized on the reaction pad which were activated upon interaction with the wet sample. The amplification and thus the amount of bacterial nuclear material was quantified by monitoring the fluorescence of hydroxynaphthol blue whose real time fluorescence activity reduced with the progression of amplification. The device reported a good sensitivity of 4.1 × 102 copies of genomic DNA for Streptococcus pneumonia which is better than previously reported traditional LAMP assay80 and the specificity of the assay was carried out with other multiple pathogenic DNA which observed no cross reactivity. The device also showed proof-of-concept for DNA extracted from clinical bacterial meningitis cultured in the laboratory.
Another work from Seo et al. in Fig. 4c demonstrated a centrifugal microfluidic platform integrated with LAMP for detection of foodborne pathogens.81 Herein the authors designed a circular microdevice consisting of 24 sample chambers, aliquoting chambers, and amplification reaction chambers. The reaction chambers contained air dried LAMP primer sets while the other components of the LAMP cocktail were loaded with bacterial cells into the sample chambers. The colorimetric detection was mediated by the change of Eriochrome Black T from purple to sky blue for positive samples. The device also exhibited high specificity, with no report of false positives. Interestingly, the device achieved a lower limit of detection at the 100-cell level compared to the 500-copy level when extracted pathogen DNA was used as the input sample. Overall, this research is one of the first works demonstrating on-chip lysis capability indirectly through direct-LAMP with bacterial cells as the input sample. Consequently, this work demonstrated little to no interference of cellular debris on the LAMP reaction, further bolstering the robustness of the LAMP cocktail and proving the applicability of direct-LAMP on microfluidic systems.
Multiplex detection of bacteria is important for applicability in low resource settings, in connection to this, Zhang et al. reported a centrifugal-based microfluidic platform for LAMP-based detection of six types of bacterial pathogens, depicted in Fig. 4d.82 The colorimetric change was mediated by calcein that engendered a fluorescent signal as the LAMP reaction proceeded, which was detected by the naked eye under a hand-held UV light. The device majorly consisted of four compartments, one each for removal of contaminants using zeolite, addition of primer mix, mixing of LAMP cocktail, and the last one for the LAMP reaction and colorimetric detection. The systematic flow between the compartments was controlled by the RPM of the centrifugal device. Although the device consolidates the features of sample purification, amplification, and detection onto a single platform, it requires a separate off-chip bacterial cell lysis step. Overall, the device stands out for the inexpensive setup and potential for POC applications.
Recently, CRISPR-Cas systems have been integrated into microfluidic devices for detection of bacteria pathogens. In a work by Xiang et al. CRISPR-Cas demonstrated the effective detection of Listeria (Fig. 4e).83 Their method was a one-step detection method using recombinase aided amplification (RAA) and CRISPR on a high-throughput centrifugal microfluidic chip, with the capacity to detect 8 samples simultaneously. Their strategy achieved a sample-to-result time in 60 min based on the response of a real-time fluorescence curve. Another work by Chen and colleagues reported a device dubbed CASMEAN which integrated an RAA-CRISPR system in a centrifugal microfluidic POC test (Fig. 4f).84 They applied this device in the detection of Pseudomonas aeruginosa whose presence was confirmed by a fluorescent readout within 1.5 h. Moreover, their results showed that CASMEAN demonstrated an LOD of 103 CFU mL−1 for Pseudomonas aeruginosa samples. These CRISPR-based devices shed light on the capability of high throughput detection of analytes which can dramatically shift the scope of monitoring bacterial infections broadly.
In addition to these, new techniques like droplet-based digital LAMP and digital PCR techniques may offer enhanced sensitivity.85 A recent work by Ma et al. demonstrated a microdroplet based digital LAMP platform for the diagnosis of vancomycin-resistant Enterococcus (VRE) bacteria.86 This is the first work demonstrating the application of emulsion microdroplets for array digital LAMP diagnosis for pathogens. The fluorescence of calcein dye was quantified to evaluate the concentration of pathogenic DNA present. Hence the limit of detection of the platform was deduced to be 1 copy of DNA or 50 copies per μl, which is superior compared to other microfluidic platforms. This work motivated further exploration of these digital nucleic acid amplification platforms, since they confine reactions to small micro volumes which results in a low signal-to-noise ratio and better detection even at low-nucleic acid concentrations.87 Several works have been reported in the past 5–6 years on bacterial nucleic acid detection, some of which are discussed above; the core idea being the integration of colorimetric assays onto microfluidic platforms. The recent works revolving around this idea are reviewed in Table 1.
Overall, nucleic acid integrated microfluidic platforms have an average assay time of ∼90 min, which is a significant improvement from traditional techniques. The platforms moreover achieved a comparable if not better sensitivity/detection limits than traditional off-chip techniques, with the lowest being ∼3 CFU μl−1. Whereas, traditional PCR has a sensitivity of 10–102 CFU μL−1.88 This rapidity in detection and high sensitivity can be attributed to requiring low sample volumes, high reaction rates at confined volumes in microfluidics, and reliable colorimetric agents that are sensitive to reaction products. Bacterial infections in general are highly communicable and the common mode of transmission is contaminated water and food.89 There are devices commercialized mainly for bacteria detection in water, air, and food material and going forward emphasis will be put on commercializing tests for antibiotic resistant bacterial strains. Other works pertaining to nucleic acid integrated microfluidics are reported extensively in Table 1.
3.3 Colorimetric detection of fungi
Fungal infections driven by Candida spp., Cryptococcus spp., and Aspergillus spp., can be life-threatening for patients with profound immunosuppression, due to the destruction of tissue and the stimulation of an inflammatory immune response.90 Moreover, detection of fungal toxins is also important in food as product safety can be compromised which is detrimental to human health.91 With the increased globalization of food, maintaining the quality of food from distant locations remains a problem, as the product may deteriorate in collection and transportation due to microorganisms.92 Several instances of fungi and fungal toxin contamination have been reported with Aspergillus being the dominant agent.93 This has led to strict product regulations and food safety measures. PCR is the standard detection method for fungi and fungal toxins but is limited by its operation cost, requirement for technical expertise, and comprehensive laboratory facilities.94 Consequently, microfluidic paper-based analytical devices using colorimetric detection have demonstrated great advantages for food safety testing.94Colorimetric detection of fungal pathogen Aspergillus flaxus, was demonstrated using a simple, paper-based lab-on-a-chip (Fig. 5a and b).94 This fungus is responsible for postharvest herbal product contamination by releasing mycotoxins, which represents a potentially carcinogenic hazard. In their study, Chaumpluk et al. developed a bendable, paper-based microfluidic platform to rapidly detect Aspergillus spp.94 DNA within 40 min with a limit of detection of 100 aflR copies. The assay amplified the aflatoxin biosynthesis gene aflR using LAMP, followed by target DNA hybridization with probes on blue silver nanoplates, resulting in a colorimetric reaction (Fig. 5a). Dispersed blue silver nanoplates on the detection pad indicated positive results while the colorless or pale-yellow pad represented negative results (Fig. 5b). The bendable chip had two sections: (i) a DNA amplification part and (ii) a detection area connected through channels made of fishing line (Fig. 5b). The LAMP reagent and DNA sample were introduced via inlet 1 and the probe along with silver nanoplate solution was introduced through inlet 2 (Fig. 5b). DNA amplification occurred in amplification pad (1) and squeezed up to the absorption pad (2) (Fig. 5b). Following this, the DNA and probe/silver nanoplate mixture was squeezed down, mixed, passing through the PBS pad (3), before arriving at the paper detection pad (4), where blue color showed presence of Aspergillus samples (Fig. 5b). The assay demonstrated high specificity of 94.47% and high sensitivity of 100%, demonstrating the fabrication of a simple, effective, rapid detection platform for Aspergillus spp.
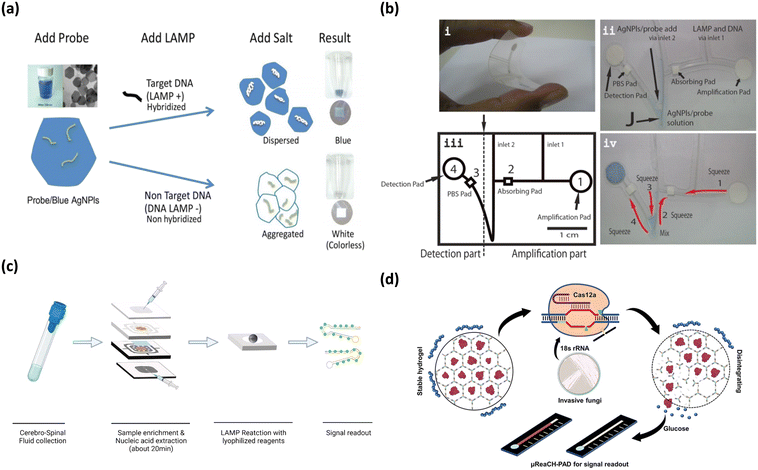 | ||
| Fig. 5 Matrix depicting colorimetric fungi detection. (a) Graphic depicting microfluidic-based colorimetric detection of Aspergillus sp. in herbal specimens. Adapted with permission from ref. 94. Copyright 2016, Wiley. (b) Shows components (i–iv) of microfluidic chip in Chaumpluk and colleague's study. Reproduced with permission from ref. 94. Copyright 2016, Wiley. (c) Workflow of multifunctional microfluidic module for diagnosis of Cryptococcus among patients. Adapted with permission from ref. 95. Copyright 2022, Elsevier. (d) Graphical overview of CRISPR-Cas12 based paper analytical device for detection of Candida and Aspergillus. Adapted with permission from ref. 96. Copyright 2021, American Chemical Society. | ||
Colorimetric LAMP has also been applied to detect Cryptococcus from patients suffering from cryptococcal meningitis.95 In a study by Tian and colleagues, a multifunctional microfluidic module integrating on-chip filtration and nucleic acid extraction was employed based on a SYBR Green fluorescence readout (Fig. 5c).95 The module reported a sensitivity of 100 CFU mL−1 and specificity of the primers was 92% when tested against the DNA of 50 pure culture strains. Moreover, the device reported a detection time of 45 min, which is on the lower end. The major advantage of this study was that the entire process from end-to-end was completed on the module, proposing feasibility for POC applications.
Invasive fungal diseases pose a grave risk to human health which can trigger inflammatory reactions, tissue damage, and organ dysfunction. In an effort to mitigate this threat, Huang et al. developed a paper-based analytical device integrating a microfluidic readout and CRISPR-Cas12a to target Candida and Aspergillus (Fig. 5d).96 The method was comprised of three parts; first, reverse transcribed recombinase aided amplification produced target sequences in high copy numbers. Next, Cas12a identified target DNA hydrogel and initiated DNase activity, subsequently disintegrating the DNA hydrogel and releasing signal molecules which produced glucose in the hydrogel supernatant to trigger signal transduction. The hydrogel supernatant was transferred to the circular reservoir of the paper-based analytical device and flowed in the straight channel, triggering a cascade of reactions enabling the monitoring of a glucose signal in a portable fashion. The specificity of the device was comparable to qPCR and the calculated detection limits were ∼5 CFU mL−1 which is a highly sensitive response, paving the way for future applications of CRISPR diagnostic methods for fungi detection.
The detection of fungi and fungal toxins has been augmented by colorimetric microfluidic methods. The devices discussed here are low cost with high specificity and sensitivity and offer a promising avenue for fungi and fungal toxin detection compared to conventional PCR. Sample detection has shown to be as fast as 40 minutes. Huang et al. reported superb sensitivity with a low LOD of ∼5 CFU mL−1.96 Moreover, the specificity for Aspergillus spp. samples was close to perfect. Table 1 further summarizes the results of each study. Colorimetric testing of food quality would enable high throughput screening of imported produce in order to meet food safety requirements in local communities. Moving forward, the devices reported in this section need to be approved and commercialized to assist in monitoring food safety requirements on a wide scale.
3.4 Colorimetric detection of protozoa
Pathogenic protozoa cause serious gastrointestinal illnesses and infections like malaria. These protozoan infections are rapidly transmissible among human and cattle via environmental and insecticidal carriers,97 making it important to develop rapid diagnostic tools that can help curb the spread of these infections. Several rapid detection tests have been developed proving POC capability and have been reported extensively in the past.98 With already proven success of LAMP integrated microfluidic platforms for other pathogens and even malarial parasites, Reboud et al. fabricated an origami-based paper microfluidic platform for detection of Plasmodium falciparum and Plasmodium pan malarial pathogens, depicted in Fig. 6a.99 The device is unique as it integrates pathogen DNA extraction, LAMP amplification and amplicon detection onto a single platform by employing paper origami techniques and provides the test results in 45 min. The colorimetric change for positive samples was elicited by FITC labelled amplicon upon conjugation with anti-FITC antibodies on the lateral flow strip. The platform showed superior sensitivity and a coincidence rate with gold-standard PCR tests, compared to other commercial and microscopy techniques when tested with real time patient samples. All these factors indicate potential for POC applications in low resource settings which the authors envisioned to demonstrate.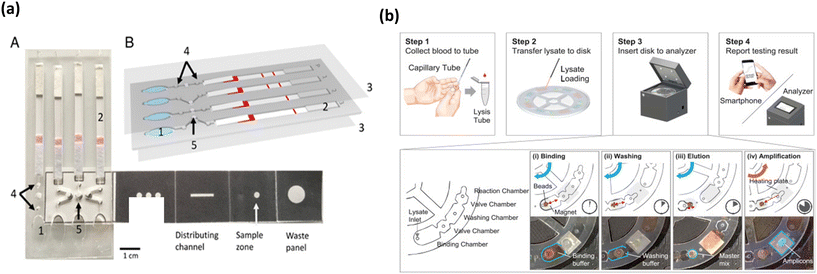 | ||
| Fig. 6 Colorimetric based malarial parasite/biomarker detection platforms. (a) Colorimetric based malaria pathogen detection uses blood as the sample of analysis/detection that elicits color change of the lateral flow assay in presence of amplified bacterial DNA (A and B). Adapted with permission from ref. 99. Copyright 2018, National Academy of Sciences. (b) Workflow of malarial parasite detection microfluidic platform to integrate amplification and detection steps onto a portable device wherein the output is analyzed by fluorescence analysis. Reproduced with permission from ref. 100. Copyright 2018, Elsevier. | ||
In an effort to present a platform that can provide direct sample to result, Choi et al. developed a palm-sized device for the detection of P. falciparum and P. vivax using LAMP.100 The workflow of the assay is shown in Fig. 6b. The device employed a non-centrifugal approach for sample preparation, which used pH switchable magnetic beads. The device was able to perform four tests simultaneously and had four chambers including the binding, valve, washing and reaction chambers. The LAMP output was monitored by fluorescence emission by calcein, which could be directly analyzed via a smartphone, thus reducing any user induced interpretation errors. Moreover, the device presented a good sensitivity of 0.5 parasites per μl and with a turnaround time of 50 min and at low cost. Furthermore, the work showed a proof-of-concept in whole blood spiked with bacterial DNA and this showed no interference or cross reactivity with biological components in blood, further bolstering its stance as a POC malarial diagnosis tool.
Majority of the tests reviewed here are detection tests for the major protozoan which is malarial pathogen (Plasmodium falciparum) or malarial pathogen biomarker (Plasmodium falciparum lactate dehydrogenase). The World Health Organization (WHO) prescribes a sensitivity of 200 parasites per μl for malarial rapid detection tests.101 Almost all the work reported here with only one exception, reported a detection limit greater than the WHO guidelines, which can be observed in Table 1. To suit rapid detection in low resource settings, a majority of the work in the last half decade or so has focused on developing paper microfluidic based devices that elicit a colorimetric output, which is evident from Table 1. Majority of the tests presented in Table 1 use blood as the direct analyte and did not report any interference for biological material in blood matrix, proving true POC capability. In the past, several rapid tests for detection of malaria have been approved by the Center for Disease Control (CDC)102 and/or regulated by WHO.103 Though POC diagnostics for malaria are extensively researched and approved, going forward there may be scope to improve these systems to have lower false positives and false negatives, by integrating camera based digital color analysis techniques104 thereby reducing any user induced reading errors.
4 Conclusion and future perspectives
While traditional pathogen detection approaches, such as PCR and LAMP, are sensitive enough, they suffer from lengthy response times, lack adequate adaptability for use in remote and isolated settings, and are too costly to support efforts against a major pandemic such as COVID-19. The direct impact of these long waiting times and assay complexity is prominent in remote, isolated communities around the globe. Moreover, the requirement of trained personnel further worsens the applicability of these tests for testing the masses. Colorimetric approaches can rightly address this pressing need. Colorimetric platforms for pathogen detection can potentially be an optimal, affordable, accessible, user-friendly, and time-effective alternative to traditional approaches. The average analysis time for colorimetric detection is between 10 minutes and one hour which is rapid compared to traditional approaches. They also offer room for seamless integration of sample-to-result automation, which can avoid any user induced errors. All these features allow for decentralizing testing efforts and pave the way for development of home-based and bed-side tests which can be crucial to curb any pathogen outbreak.Microfluidics offer several advantageous characteristics, including portability, reduced analysis cost, and quicker reaction times at microscales, favoring their translation into POC. These characteristics also make microfluidic colorimetric-based detection beneficial to curb infectious diseases which call for swift action on testing and isolation, such as SARS-CoV-2 and H1N1. The present article discusses recent works in colorimetric-based microfluidic devices for nucleic acid diagnosis of bacteria, viruses, protozoa, and fungi. Despite significant advances in colorimetric-based microfluidic methods for detection of pathogens, this field still faces many challenges like low specificity and sensitivity when integrating nucleic acid detection techniques like PCR, LAMP, and CRISPR into a microfluidic device with a colorimetric readout. Slow but firm progress has been achieved owing to the efforts put in to automate and integrate these platforms with on-chip analytical systems, progressing towards sample-to-answer systems. Going forward, more rigorous research on sensing materials that offer specificity and sensitivity will further push the boundaries of the current sensing caliber towards realizing gold standards.
Specifically, for widespread use of colorimetric microfluidic devices, detection sensitivity has been a major hurdle. Numerous factors influence this sensitivity, but three factors are the most prominent. The first factor is the organic chromogen's extinction coefficient that limits the conventional colorimetric sensor's sensitivity, dictated by the Beer–Lambert law. Further, Beer–Lambert law also dictates the microfluidic channel dimensions (path length) that directly affects the absorbance of the colorimetric changes.114 Colorimetric dyes have varying extinction coefficients which can impact their ability to absorb and reflect light at certain wavelengths.115 The second factor is that the human eye cannot distinguish between low level color contrasts. Particular color changing dyes such as SYBR green, hydroxynaphthol blue, and Eriochrome Black T change to colors in similar hues, making the color change difficult to recognize (Fig. 2).40 Dyes like phenol red that exhibit color change over bigger spectrum of hues would an ideal choice (Fig. 2). The third limiting factor is to keep the colorimetric reagent enzyme active during prolonged storage periods and during transportation since these reagents are crucial to avoid any occurrences of false positives and/or negatives. Temperature changes in the environment and exposure to light are two variables that can alter enzyme activity in chemical reactions. Different strategies have been employed to address these challenges like single step amplification techniques, sensing platforms employing nanomaterials with surface reaction dependent physicochemical characteristics, and specialized nanomaterials. Previously, nanomaterials were explored for their strong colorimetric and/or fluorometric responses,116 often integrated with lateral flow microfluidic assays.117 Although gold nanoparticles were the most commonly used for colorimetric readouts, there have been continued efforts towards developing new nanomaterials to enhance the sensitivity of colorimetric readouts.118 It is interesting to point out a class of nanostructures, that elicit different reflectance spectra in relation to specialized metal nanostructure arrangements at various dimensions with an incident light source.119 More importantly, a change in the surface analyte's refractive index brings about change in colorimetric characteristics of these nanostructures which can simply be analyzed by the naked eye enabled by a portable optical setup with an inbuilt light source. Moreover, the optical properties of these plasmonic nanostructures can be manipulated by varying either the size/morphology and/or interparticle distances, making them worth exploring for colorimetric detection applications. These inherent features of plasmonic nanostructures offer amazing advantages, particularly high sensitivity in response to external stimuli in comparison to conventional colorimetric assays using organic chromophores. With growing interest in making existing microfluidic technologies truly POC with high sensitivity and throughput, the use of these plasmonic structures can play a crucial role in bridging the gap between colorimetric detection and excellent sensitivity.
The noteworthy progress in colorimetric-based microfluidic research in pathogen detection and their ideas and innovation are detailed in this review. With respect to pathogen diagnosis, major attempts have been made in the area of integrating extremely sensitive nucleic acid methods-PCR, LAMP, and CRISPR-into microfluidic platforms, which is evident from the high volume of works pertaining to this in recent years. With the advent of centrifugation-based platforms, high sensitivity and throughput have been achieved. In an analogous way, paper-based microfluidic platforms have enabled acceptable sensitivity, portability, and cost-effectiveness. Despite considerable advances in POC diagnostics by these techniques, an inclusive and comprehensive system with the ability to perform all detection steps on-chip is still lacking. In most of the previously conducted studies, sample pre-processing steps are conducted off-chip, that is, cellular lysis, nuclear material extraction, and/or need specialized equipment such as a thermocycler, microscope, bench-top centrifugation setup, and heating oven, among others. Thus, they are not encouraging for application at POC. Nevertheless, earlier works can be improved for integrating extraction, cellular lysis, and detection steps onto a single platform, while being modular for the inclusion/exclusion of other detection assay features, if required. Similarly, pursuing the integration of digital droplet microfluidics with colorimetric detection assays would be interesting since it has been recently shown that they can detect pathogens in vitro using LAMP in limited microdroplets and provide great throughput and sensitivity.
Another serious challenge is how to quantitatively detect with personalized equipment (e.g., smartphones, wearable devices) and transfer the technique into the clinic, which will be the future goal for colorimetric detection. It is possible to perform imaging by a personalized smartphone and process the results to digital information. Through this, the quantitative analysis of the analytes is realized. For improving the quantitative assessment capacity, we should adopt a robust automated imaging and subsequent analysis approach by integrating with smart gadgets. It is required to extend this concept into colorimetric-based microfluidic sensing platforms. If not, many of the existing colorimetric sensing systems are only proof-of-concept demonstrations and are challenging for clinical translation.
Considering the future of such assays related to POC/clinical diagnostics, there is still a need to focus on the fundamental research and its clinical translation, with the integration of advanced nanotechnology, biotechnology, and other emerging technologies such as colorimetric-based microfluidic chips and personalized equipment. For accelerating clinical translation, there is a need for collaboration between front-line medical systems and the academic arena. As colorimetric-based microfluidics has a multidisciplinary nature, there is a need for ongoing coordination between all related parties, such as scientists, engineers, end-users (e.g., physicians and medical examiners), and commercial partners (e.g., marketing experts and investors) in a unified and collaborative manner for successful realization of microfluidic tools/technology to the market. Clinical trials should pursue proof-of-concept experiments along with volume-manufacturing considerations so that both commercial and performance success can be obtained.
Integration and standardization are two other challenges with device commercialization. Unreproducible results can appear due to the variability in outcomes of fabricated devices. In addition, many works have shown a failure to integrate all sample analysis steps in an all-inclusive device. It is important to allocate efforts for device integration and standardization improvement for augmenting the scale of pathogen diagnosis. Currently, there are many opportunities for the development and application of colorimetric-based microfluidic technologies for addressing different pathogen-related challenges that are globally emerging. With the entry of microfluidics to its third decade, it is expected that growth and expansion will be extended beyond simple proof-of-concept systems into broad, commercial real-world applications, particularly for nucleic acid detection using colorimetric-based microfluidic systems.
Author contributions
SGY, HK contributed to the conceptualization, planning, execution and drafting of the article. TA and RSM contributed to discussing standard detection techniques and future perspectives. SM contributed with supervision, resources, review and editing of the article.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank the Faculty of Engineering at McGill University, the Natural Science and Engineering Research Council of Canada (NSERC, G247765), Canadian Institutes of Health Research (CIHR, 257352), the Canada Foundation for Innovation (CFI, G248924), MI4 (McGill Interdisciplinary Initiative in Infection and Immunity) for financial support. SM is thankful to Canada Research Chair program for the financial support. SGY is thankful to McGill Faculty of Engineering for financial support.References
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F. Gao and W. Tan, N. Engl. J. Med., 2020, 382, 727–733 CrossRef CAS PubMed.
- Y. Orooji, H. Sohrabi, N. Hemmat, F. Oroojalian, B. Baradaran, A. Mokhtarzadeh, M. Mohaghegh and H. Karimi-Maleh, Nano-Micro Lett., 2021, 13, 1–30 CrossRef CAS PubMed.
- R. Siavash Moakhar, C. del Real Mata, M. Jalali, H. Shafique, A. Sanati, J. de Vries, J. Strauss, F. Ghasemi, M. McLean, I. I. Hosseini, Y. Lu, S. G. Yedire, S. S. Mahshid, M. A. Tabatabaiefar, C. Liang and S. Mahshid, Adv. Sci., 2022, 220424 Search PubMed.
- W. Gu, X. Deng, M. Lee, Y. D. Sucu, S. Arevalo, D. Stryke, S. Federman, A. Gopez, K. Reyes, K. Zorn, H. Sample, G. Yu, G. Ishpuniani, B. Briggs, E. D. Chow, A. Berger, M. R. Wilson, C. Wang, E. Hsu, S. Miller, J. L. DeRisi and C. Y. Chiu, Nat. Med., 2021, 27, 115–124 CrossRef CAS PubMed.
- B. L. Fernández-Carballo, I. McGuiness, C. McBeth, M. Kalashnikov, S. Borrós, A. Sharon and A. F. Sauer-Budge, Biomed. Microdevices, 2016, 18(2) DOI:10.1007/s10544-016-0060-4.
- J. Zhong and X. Zhao, Food Anal. Methods, 2018, 11, 1543–1560 CrossRef.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38(7), 870–874 CrossRef CAS PubMed.
- E. Nunez-Bajo, A. S. P. Collins, M. Kasimatis, Y. Cotur, T. Asfour, U. Tanriverdi, M. Grell, M. Kaisti, G. Senesi, K. Stevenson and F. Güder, Nat. Commun., 2020, 11(1), 1–10 CrossRef PubMed.
- W. Jung, J. Han, J. W. Choi and C. H. Ahn, Microelectron. Eng., 2015, 132, 46–57 CrossRef CAS.
- S. K. Vashist, Biosensors, 2017, 7(4), 62 CrossRef PubMed.
- W. Y. Lim, B. L. Lan and N. Ramakrishnan, Biosensors, 2021, 11(11), 434 CrossRef CAS PubMed.
- M. Jalali, T. Abdel Fatah, S. S. Mahshid, M. Labib, A. S. Perumal and S. Mahshid, Small, 2018, 14(35), 180189 Search PubMed.
- K. Tsougeni, G. Kaprou, C. M. Loukas, G. Papadakis, A. Hamiot, M. Eck, D. Rabus, G. Kokkoris, S. Chatzandroulis, V. Papadopoulos, B. Dupuy, G. Jobst, E. Gizeli, A. Tserepi and E. Gogolides, Sens. Actuators, B, 2020, 320, 128345 CrossRef CAS.
- T. Abdelfatah, M. Jalali and S. Mahshid, Biomicrofluidics, 2018, 12, 64103 CrossRef PubMed.
- R. Siavash Moakhar, T. Abdelfatah, A. Sanati, M. Jalali, S. E. Flynn, S. S. Mahshid and S. Mahshid, ACS Appl. Mater. Interfaces, 2020, 12(20), 23298–23310 CrossRef CAS PubMed.
- E. J. Walsh, A. Feuerborn, J. H. R. Wheeler, A. N. Tan, W. M. Durham, K. R. Foster and P. R. Cook, Nat. Commun., 2017, 8(1), 1–9 CrossRef CAS PubMed.
- S. Panesar and S. Neethirajan, Nano-Micro Lett., 2016, 8, 204–220 CrossRef CAS PubMed.
- A. Sanati, M. Jalali, K. Raeissi, F. Karimzadeh, M. Kharaziha, S. S. Mahshid and S. Mahshid, Microchim. Acta, 2019, 186, 1–22 CrossRef PubMed.
- N. A. Tanner, Y. Zhang and T. C. Evans, BioTechniques, 2015, 58, 59–68 CrossRef CAS PubMed.
- A. Rostami, A. Hadjizadeh and S. Mahshid, J. Mater. Sci., 2020, 55(18), 7969–7980 CrossRef CAS.
- Y. Shang, J. Sun, Y. Ye, J. Zhang, Y. Zhang and X. Sun, Crit. Rev. Food Sci. Nutr., 2020, 60, 201–224 CrossRef CAS PubMed.
- Y. Shang, G. Xing, X. Liu, H. Lin and J. M. Lin, Anal. Chem., 2022, 94, 16787–16795 CrossRef CAS PubMed.
- V. X. T. Zhao, T. I. Wong, X. T. Zheng, Y. N. Tan and X. Zhou, Mater. Sci. Energy Technol., 2020, 3, 237–249 CAS.
- L. Zhang, B. Ding, Q. Chen, Q. Feng, L. Lin and J. Sun, TrAC, Trends Anal. Chem., 2017, 94, 106–116 CrossRef CAS.
- T. R. Kozel and B. Wickes, Cold Spring Harbor Perspect. Med., 2014, 4(4) DOI:10.1101/cshperspect.a019299.
- X. Tan, M. K. Khaing Oo, Y. Gong, Y. Li, H. Zhu and X. Fan, Analyst, 2017, 142, 2378–2385 RSC.
- D. Brinati, A. Campagner, D. Ferrari, M. Locatelli, G. Banfi and F. Cabitza, J. Med. Syst., 2020, 44, 1–12 CrossRef PubMed.
- H. S. Huang, C. L. Tsai, J. Chang, T. C. Hsu, S. Lin and C. C. Lee, Clin. Microbiol. Infect., 2018, 24, 1055–1063 CrossRef CAS PubMed.
- J. Kashir and A. Yaqinuddin, Med. Hypotheses, 2020, 141, 109786 CrossRef CAS PubMed.
- K. U. Ludwig, R. M. Schmithausen, D. Li, M. L. Jacobs, R. Hollstein, K. Blumenstock, J. Liebing, M. Słabicki, A. Ben-Shmuel, O. Israeli, S. Weiss, T. S. Ebert, N. Paran, W. Rüdiger, G. Wilbring, D. Feldman, B. Lippke, N. Ishorst, L. M. Hochfeld, E. C. Beins, I. H. Kaltheuner, M. Schmitz, A. Wöhler, M. Döhla, E. Sib, M. Jentzsch, J. D. Borrajo, J. Strecker, J. Reinhardt, B. Cleary, M. Geyer, M. Hölzel, R. Macrae, M. M. Nöthen, P. Hoffmann, M. Exner, A. Regev, F. Zhang and J. L. Schmid-Burgk, Nat. Biotechnol., 2021, 39(12), 1556–1562 CrossRef CAS PubMed.
- R. Aman, A. Mahas and M. Mahfouz, ACS Synth. Biol., 2020, 9, 1226–1233 CrossRef CAS PubMed.
- Q. H. Nguyen and M. Il Kim, TrAC, Trends Anal. Chem., 2020, 132, 116038 CrossRef CAS PubMed.
- D. M. Kim and S. M. Yoo, Biosensors, 2022, 12(7), 532 CrossRef CAS PubMed.
- P. Singh, S. Kakkar, Bharti, R. Kumar and V. Bhalla, Chem. Commun., 2019, 55(33), 4765–4768 RSC.
- M. Yang, X. Liu, Y. Luo, A. J. Pearlstein, S. Wang, H. Dillow, K. Reed, Z. Jia, A. Sharma, B. Zhou, D. Pearlstein, H. Yu and B. Zhang, Nat. Food, 2021, 2, 110–117 CrossRef.
- A. Garrido-Maestu and M. Prado, Compr. Rev. Food Sci. Food Saf., 2022, 21, 1913–1939 CrossRef PubMed.
- C. Yan, Y. Sun, M. Yao, X. Jin, Q. Yang and W. Wu, Sens. Actuators, B, 2022, 354, 131117 CrossRef CAS.
- M. Chahar, A. Anvikar, R. Dixit and N. Valecha, Exp. Parasitol., 2018, 190, 1–9 CrossRef CAS PubMed.
- J. Fischbach, N. C. Xander, M. Frohme and J. F. Glökler, BioTechniques, 2015, 58, 189–194 CrossRef CAS PubMed.
- A. T. Scott, T. R. Layne, K. C. O'Connell, N. A. Tanner and J. P. Landers, Anal. Chem., 2020, 92, 13343–13353 CrossRef CAS PubMed.
- A. Neshani, H. Zare, H. Sadeghian, H. Safdari, B. Riahi-Zanjani and E. Aryan, Diagnostics, 2023, 13(1), 155 CrossRef CAS PubMed.
- S. L. Wastling, K. Picozzi, A. S. L. Kakembo and S. C. Welburn, PLoS Neglected Trop. Dis., 2010, 4, e865 CrossRef PubMed.
- K. E. McCracken and J. Y. Yoon, Anal. Methods, 2016, 8, 6591–6601 RSC.
- Z. Li, H. Liu, D. Wang, M. Zhang, Y. Yang and T. L. Ren, TrAC, Trends Anal. Chem., 2023, 158, 116790 CrossRef CAS.
- L. Gervais, N. De Rooij and E. Delamarche, Adv. Mater., 2011, 23, H151–H176 CrossRef CAS PubMed.
- J. J. Creelman, E. A. Luy, G. C. H. Beland, C. Sonnichsen and V. J. Sieben, Sensors, 2021, 21(18), 6250 CrossRef CAS PubMed.
- M. H. V. Werts, V. Raimbault, R. Texier-Picard, R. Poizat, O. Franais, L. Griscom and J. R. G. Navarro, Lab Chip, 2012, 12, 808–820 RSC.
- M. S. Woolf, L. M. Dignan, A. T. Scott and J. P. Landers, Nat. Protoc., 2021, 16, 218–238 CrossRef CAS PubMed.
- G. Xing, J. Ai, N. Wang and Q. Pu, TrAC, Trends Anal. Chem., 2022, 157, 116792 CrossRef CAS.
- T. Kaneta, W. Alahmad and P. Varanusupakul, Appl. Spectrosc. Rev., 2019, 54, 117–141 CrossRef.
- M. Zhang, J. Ye, J. He, F. Zhang, J. Ping, C. Qian and J. Wu, Anal. Chim. Acta, 2020, 1099, 1–15 CrossRef CAS PubMed.
- S. Sanche, Y. T. Lin, C. Xu, E. Romero-Severson, N. Hengartner and R. Ke, Emerging Infect. Dis., 2020, 26, 1470–1477 CrossRef CAS PubMed.
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU).
- Our world in data, https://ourworldindata.org/hiv-aids, (accessed 2022).
- World Health Organization, https://www.who.int/news/item/11-03-2019-who-launches-new-global-influenza-strategy#:~:text=WHO%20will%20expand%20partnerships%<?pdb_no 20to?>20to<?pdb END?>,to%20prevent%20and%20control%20influenza, (accessed 2021).
- World Health Organization, https://www.who.int/news-room/fact-sheets/detail/hepatitis-b, (accessed 2021).
- A. J. Kucharski, P. Klepac, A. J. K. Conlan, S. M. Kissler, M. L. Tang, H. Fry, J. R. Gog, W. J. Edmunds, J. C. Emery, G. Medley, J. D. Munday, T. W. Russell, Q. J. Leclerc, C. Diamond, S. R. Procter, A. Gimma, F. Y. Sun, H. P. Gibbs, A. Rosello, K. van Zandvoort, S. Hué, S. R. Meakin, A. K. Deol, G. Knight, T. Jombart, A. M. Foss, N. I. Bosse, K. E. Atkins, B. J. Quilty, R. Lowe, K. Prem, S. Flasche, C. A. B. Pearson, R. M. G. J. Houben, E. S. Nightingale, A. Endo, D. C. Tully, Y. Liu, J. Villabona-Arenas, K. O'Reilly, S. Funk, R. M. Eggo, M. Jit, E. M. Rees, J. Hellewell, S. Clifford, C. I. Jarvis, S. Abbott, M. Auzenbergs, N. G. Davies and D. Simons, Lancet Infect. Dis., 2020, 20, 1151–1160 CrossRef CAS PubMed.
- S. Sharma, J. Zapatero-Rodríguez, P. Estrela and R. O'Kennedy, Biosensors, 2015, 5, 577–601 CrossRef PubMed.
- L. Bokelmann, O. Nickel, T. Maricic, S. Pääbo, M. Meyer, S. Borte and S. Riesenberg, Nat. Commun., 2021, 12, 1467 CrossRef CAS PubMed.
- Y. T. Kim, Y. Chen, J. Y. Choi, W. J. Kim, H. M. Dae, J. Jung and T. S. Seo, Biosens. Bioelectron., 2012, 33(1), 88–94 CrossRef CAS PubMed.
- L. Gorgannezhad, H. Stratton and N. T. Nguyen, Micromachines, 2019, 10(6), 408 CrossRef PubMed.
- B. A. Lakshmi and S. Kim, in Recent Developments in Applied Microbiology and Biochemistry, Elsevier, 2021, pp. 291–297 Search PubMed.
- M. N. Aoki, B. de Oliveira Coelho, L. G. B. Góes, P. Minoprio, E. L. Durigon, L. G. Morello, F. K. Marchini, I. N. Riediger, M. do Carmo Debur, H. I. Nakaya and L. Blanes, Sci. Rep., 2021, 11(1), 1–10 CrossRef PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- A. Ganguli, A. Mostafa, J. Berger, M. Y. Aydin, F. Sun, S. A. Stewart de Ramirez, E. Valera, B. T. Cunningham, W. P. King and R. Bashir, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 22727–22735 CrossRef CAS PubMed.
- K. Yin, X. Ding, Z. Xu, Z. Li, X. Wang, H. Zhao, C. Otis, B. Li and C. Liu, Sens. Actuators, B, 2021, 344, 130242 CrossRef CAS PubMed.
- Y. D. Ma, Y. S. Chen and G. Bin Lee, Sens. Actuators, B, 2019, 296, 126647 CrossRef CAS.
- R. Wang, R. Zhao, Y. Li, W. Kong, X. Guo, Y. Yang, F. Wu, W. Liu, H. Song and R. Hao, Lab Chip, 2018, 18, 3507–3515 RSC.
- N. L. Welch, M. Zhu, C. Hua, J. Weller, M. E. Mirhashemi, T. G. Nguyen, S. Mantena, M. R. Bauer, B. M. Shaw, C. M. Ackerman, S. G. Thakku, M. W. Tse, J. Kehe, M.-M. Uwera, J. S. Eversley, D. A. Bielwaski, G. McGrath, J. Braidt, J. Johnson, F. Cerrato, G. K. Moreno, L. A. Krasilnikova, B. A. Petros, G. L. Gionet, E. King, R. C. Huard, S. K. Jalbert, M. L. Cleary, N. A. Fitzgerald, S. B. Gabriel, G. R. Gallagher, S. C. Smole, L. C. Madoff, C. M. Brown, M. W. Keller, M. M. Wilson, M. K. Kirby, J. R. Barnes, D. J. Park, K. J. Siddle, C. T. Happi, D. T. Hung, M. Springer, B. L. MacInnis, J. E. Lemieux, E. Rosenberg, J. A. Branda, P. C. Blainey, P. C. Sabeti and C. Myhrvold, Nat. Med., 2022, 28, 1083–1094 CrossRef CAS PubMed.
- X. Lin, X. Jin, B. Xu, R. Wang, R. Fu, Y. Su, K. Jiang, H. Yang, Y. Lu, Y. Guo and G. Huang, Micromachines, 2019, 10(11), 777 CrossRef PubMed.
- K. Kaarj, P. Akarapipad and J.-Y. Yoon, Sci. Rep., 2018, 8, 12438 CrossRef PubMed.
- Y. Wang, G. Ge, R. Mao, Z. Wang, Y. Z. Sun, Y. G. Du, X. H. Gao, R. Q. Qi and H. D. Chen, Virol. J., 2020, 17, 99 CrossRef CAS PubMed.
- B. M. Andersen, in Prevention and Control of Infections in Hospitals, Springer International Publishing, 2019, pp. 1021–1028 Search PubMed.
- H. Wang, H. Ceylan Koydemir, Y. Qiu, B. Bai, Y. Zhang, Y. Jin, S. Tok, E. C. Yilmaz, E. Gumustekin, Y. Rivenson and A. Ozcan, Light: Sci. Appl., 2020, 9, 2047–7538 Search PubMed.
- N. Shauloff, A. Morag, K. Yaniv, S. Singh, R. Malishev, O. Paz-Tal, L. Rokach and R. Jelinek, Nano-Micro Lett., 2021, 13, 1–15 CrossRef PubMed.
- D. J. Shin, N. Andini, K. Hsieh, S. Yang and T. H. Wang, Annu. Rev. Anal. Chem., 2019, 12, 41–67 CrossRef CAS PubMed.
- Y.-L. Fang, C.-H. Wang, Y.-S. Chen, C.-C. Chien, F.-C. Kuo, H.-L. You, M. S. Lee and G.-B. Lee, Lab Chip, 2021, 21, 113–121 RSC.
- M. Azizi, M. Zaferani, S. H. Cheong and A. Abbaspourrad, ACS Sens., 2019, 4, 841–848 CrossRef CAS PubMed.
- Y. Seok, H.-A. Joung, J.-Y. Byun, H.-S. Jeon, S. J. Shin, S. Kim, Y.-B. Shin, H. Soo and M.-G. Kim, Theranostics, 2017, 7(8), 2220–2230 CrossRef CAS PubMed.
- Y. Xia, X. G. Guo and S. Zhou, J. Thorac. Dis., 2014, 6, 1193–1199 Search PubMed.
- J. H. Seo, B. H. Park, S. J. Oh, G. Choi, D. H. Kim, E. Y. Lee and T. S. Seo, Sens. Actuators, B, 2017, 246, 146–153 CrossRef CAS.
- L. Zhang, F. Tian, C. Liu, Q. Feng, T. Ma, Z. Zhao, T. Li, X. Jiang and J. Sun, Lab Chip, 2018, 18, 610–619 RSC.
- X. Xiang, F. Li, Q. Ye, Y. Shang, M. Chen, J. Zhang, B. Zhou, H. Suo, Y. Ding and Q. Wu, Sens. Actuators, B, 2022, 358, 131517 CrossRef CAS.
- Y. Chen, Y. Mei, X. Zhao and X. Jiang, Anal. Chem., 2020, 92, 14846–14852 CrossRef CAS PubMed.
- H. Yuan, Y. Chao and H. C. Shum, Small, 2020, 16, 1904469 CrossRef CAS PubMed.
- Y. D. Ma, K. Luo, W. H. Chang and G. Bin Lee, Lab Chip, 2018, 18, 296–303 RSC.
- Z. Zhang, S. Zhao, F. Hu, G. Yang, J. Li, H. Tian and N. Peng, Micromachines, 2020, 11, 177 CrossRef PubMed.
- J. H. Melendez, Y. M. Frankel, A. T. An, L. Williams, L. B. Price, N. Y. Wang, G. S. Lazarus and J. M. Zenilman, Clin. Microbiol. Infect., 2010, 16, 1762–1769 CrossRef CAS PubMed.
- J. R. Rohr, C. B. Barrett, D. J. Civitello, M. E. Craft, B. Delius, G. A. DeLeo, P. J. Hudson, N. Jouanard, K. H. Nguyen, R. S. Ostfeld, J. V. Remais, G. Riveau, S. H. Sokolow and D. Tilman, Nat. Sustain., 2019, 2(6), 445–456 CrossRef PubMed.
- K. K. Hussain, D. Malavia, E. M. Johnson, J. Littlechild, C. P. Winlove, F. Vollmer and N. A. R. Gow, J. Fungi, 2020, 6, 1–26 Search PubMed.
- World Health Organization, https://www.who.int/publications/i/item/978924159444, (accessed 2007).
- J. R. Hodges and A. M. Kimball, Global. Health, 2005, 1, 4 CrossRef PubMed.
- I. Rizzo, G. Vedoya, S. Maurutto, M. Haidukowski and E. Varsavsky, Microbiol. Res., 2004, 159, 113–120 CrossRef CAS PubMed.
- P. Chaumpluk, P. Plubcharoensook and S. Prasongsuk, Biotechnol. J., 2016, 11, 768–779 CrossRef CAS PubMed.
- Y. Tian, T. Zhang, J. Guo, H. Lu, Y. Yao, X. Chen, X. Zhang, G. Sui and M. Guan, Talanta, 2022, 236, 122827 CrossRef CAS PubMed.
- D. Huang, D. S. Ni, M. Fang, Z. Shi and Z. Xu, Anal. Chem., 2021, 93, 16965–16973 CrossRef CAS PubMed.
- E. P. Mwanga, E. G. Minja, E. Mrimi, M. G. Jiménez, J. K. Swai, S. Abbasi, H. S. Ngowo, D. J. Siria, S. Mapua, C. Stica, M. F. Maia, A. Olotu, M. T. Sikulu-Lord, F. Baldini, H. M. Ferguson, K. Wynne, P. Selvaraj, S. A. Babayan and F. O. Okumu, Malar. J., 2019, 18, 341 CrossRef PubMed.
- N. Kolluri, C. M. Klapperich and M. Cabodi, Lab Chip, 2018, 18, 75–94 RSC.
- J. Reboud, G. Xu, A. Garrett, M. Adriko, Z. Yang, E. M. Tukahebwa, C. Rowell and J. M. Cooper, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4834–4842 CrossRef CAS PubMed.
- G. Choi, T. Prince, J. Miao, L. Cui and W. Guan, Biosens. Bioelectron., 2018, 115, 83–90 CrossRef CAS PubMed.
- World Health Organization, WHO Global Malaria Programme Good practices for selecting and procuring rapid diagnostic tests for malaria, 2011 Search PubMed.
- Centers for Disease control and Prevention, https://www.cdc.gov/malaria/diagnosis_treatment/diagnostic_tools.html, (accessed 2023) Search PubMed.
- W. World Health Organization, WHO list of prequalified in vitro diagnostic products, 2020 Search PubMed.
- L. Saisin, R. Amarit, A. Somboonkaew, O. Gajanandana, O. Himananto and B. Sutapun, Sensors, 2018, 18(11), 4026 CrossRef PubMed.
- D. Xu, X. Jiang, T. Zou, G. Miao, Q. Fu, F. Xiang, L. Feng, X. Ye, L. Zhang and X. Qiu, Microfluid. Nanofluid., 2022, 26, 1–9 CrossRef PubMed.
- Y. Yao, N. Zhao, W. Jing, Q. Liu, H. Lu, W. Zhao, W. Zhao, Z. Yuan, H. Xia and G. Sui, Sens. Actuators, B, 2021, 333, 129521 CrossRef CAS.
- P. Biswas, G. N. Mukunthan Sulochana, T. N. Banuprasad, P. Goyal, D. Modak, A. K. Ghosh and S. Chakraborty, ACS Sens., 2022, 7, 3720–3729 CrossRef CAS PubMed.
- R. R. G. Soares, A. S. Akhtar, I. F. Pinto, N. Lapins, D. Barrett, G. Sandh, X. Yin, V. Pelechano and A. Russom, Lab Chip, 2021, 21, 2932–2944 RSC.
- H. Wang, J. Xu, S. Li, X. Wang, G. Liu, S. Yang, F. Zhao, Q. Liu, X. Chen, C. He and M. Li, Anal. Chim. Acta, 2023, 1242, 340812 CrossRef CAS PubMed.
- P. Chen, C. Chen, H. Su, M. Zhou, S. Li, W. Du, X. Feng and B. F. Liu, Talanta, 2021, 224, 121844 CrossRef CAS PubMed.
- S. Wang, N. Liu, L. Zheng, G. Cai and J. Lin, Lab Chip, 2020, 20, 2296–2305 RSC.
- D. Liu, Y. Zhu, N. Li, Y. Lu, J. Cheng and Y. Xu, Sens. Actuators, B, 2020, 310, 127834 CrossRef CAS.
- H. Lu, J. Zhu, T. Zhang, X. Zhang, X. Chen, W. Zhao, Y. H. Yao, W. Zhao and G. Sui, Talanta, 2022, 246, 123467 CrossRef CAS PubMed.
- V. C. Pinto, C. F. Araújo, P. J. Sousa, L. M. Gonçalves and G. Minas, Sens. Actuators, B, 2019, 290, 285–292 CrossRef CAS.
- H. Wang, H. Rao, M. Luo, X. Xue, Z. Xue and X. Lu, Coord. Chem. Rev., 2019, 398, 113003 CrossRef CAS.
- A. Rubio-Monterde, D. Quesada-González and A. Merkoçi, Anal. Chem., 2023, 95, 468–489 CrossRef CAS PubMed.
- Z. Wang, J. Zhao, X. Xu, L. Guo, L. Xu, M. Sun, S. Hu, H. Kuang, C. Xu and A. Li, Small Methods, 2022, 6(1), 210114 Search PubMed.
- X. Chen, Y. Leng, L. Hao, H. Duan, J. Yuan, W. Zhang, X. Huang and Y. Xiong, Theranostics, 2020, 10, 3737 CrossRef CAS PubMed.
- L. Wang, R. J. H. Ng, S. Safari Dinachali, M. Jalali, Y. Yu and J. K. W. Yang, ACS Photonics, 2016, 3, 627–633 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |