Open Access Article
Open Access ArticleAll-in-one terahertz taste sensor: integrated electronic and bioelectronic tongues
Jin
Wang
 *,
Kenji
Sakai
and
Toshihiko
Kiwa
*,
Kenji
Sakai
and
Toshihiko
Kiwa
Graduate School of Interdisciplinary Science and Engineering in Health Systems, Okayama University, 3-1-1, Tsushima-Naka, Kitaku, Okayama, 700-8530, Japan. E-mail: wangjin@okayama-u.ac.jp
First published on 13th March 2023
Abstract
Taste sensors, also known as electronic tongues or bioelectronic tongues, are designed to evaluate food and beverages, as well as for medical diagnostics. These devices mimic the ability of the human tongue to detect and identify different tastes in liquid samples, such as sweet, sour, salty, bitter, and umami. In this study, a novel all-in-one terahertz taste sensor was proposed, which differs from traditional electrochemical approaches. This sensor utilizes terahertz technology for imaging and sensing chemical reactions on the terahertz semiconductor emitter surface. The surface can be functionalized with ion-sensitive membranes, proteins, DNA aptamers, and organic receptors, enabling the detection of various substances, such as solution pH, physiological ions, sugars, toxic chemicals, drugs, and explosives. Terahertz taste sensors offer several advantages, including being label-free, high sensitivity and selectivity, rapid response, minimal sample consumption, and the ability to detect non-charged chemical substances. By integrating multiple receptors or sensing materials on a single chip, the all-in-one terahertz taste sensor has significant potential for future taste substance detection, nutrition evaluation, metabolite and drug monitoring, and biomarker sensing.
Taste sensors
Taster sensors, also known as electronic or bioelectronic tongues, are devices that mimic the human sense of taste by identifying and measuring different chemical compounds in a sample solution. Researchers began developing taste sensors in the late twentieth century to analyze the chemical compounds found in foods and beverages, which are difficult to measure or identify. These electronic tongues are generally potentiometric or amperometric devices or equipment with ion-selective electrodes,1 metal oxides,2,3 or artificial lipid polymer arrays,4–8 which can detect or measure changes in pH and ion concentration as well as identify a wide range of taste substances or chemical compounds in liquid samples (milk, beer, tea, etc.) by measuring the electrical potential or current. Legin et al. developed a sensor array-based electronic tongue and pattern recognition algorithm for the qualitative measurement of several different beverages.9 Dias et al. successfully built 36 cross-sensibility sensors for the recognition of five basic taste substances.10 Moreover, several sensor devices and equipment have been manufactured and commercialized for laboratory research and industrial applications. For example, the TS-5000Z taste sensing system (Fig. 1A), developed by Professor Toko and Intelligent Sensor Technology Inc., Japan, detects five basic tastes, flavor properties (sharpness and richness), and taste masking properties (Fig. 1B).11,12 Another commercial tongue consists of seven ion-selective field effect transistors (ISFET) developed by Astree II (Alpha MOS, Toulouse, France), which are also featured in taste evaluation, food quality control, and pharmaceutical industry applications.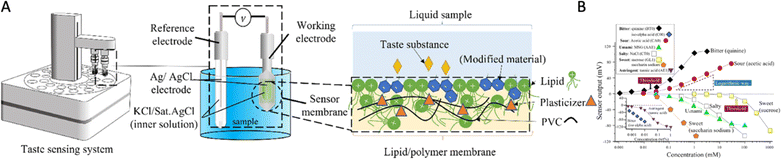 | ||
| Fig. 1 (A) Taste sensor (TS-5000Z) system, configuration, and lipid/polymer membrane. (B) TS-5000Z sensor response versus five basic tastes and astringency substances. Reprinted with permission from ref. 12, copyright 2023, Elsevier. | ||
In the early twenty-first century, another important development of the bioelectronic tongue was presented to analyze complex mixtures of chemical compounds and identify individual compounds that are not available using traditional methods.13–16 Bioelectronic tongues are based on a variety of recognition elements,17–23 such as nucleic acids, proteins, antibodies, cells, and taste receptors. A key feature of bioelectronic tongues is their high selectivity and sensitivity. This enables the identification of specific chemical compounds that may not be detectable using traditional analytical methods. Furthermore, bioelectronic tongues can be used for the detection of biomolecules, such as proteins and nucleic acids, using enzyme-, antibody-, or nucleic acid-based receptors. Qin et al. proposed a biohybrid tongue that combines a microelectrode array chip with mammalian gustatory epithelium to create a highly sensitive taste sensing system. The study quantified the interaction effects of sourness on sweetness, revealing that suppression of sweetness by sourness increases with the concentration of the sour stimulus (Fig. 2). Jeong et al. successfully developed an ultrasensitive artificial sweet taste bioelectronic tongue based on the T1R2 VFT of a human sweet taste receptor, which detects substances down to a limit of 0.1 fM and selectively discriminates sweet substances from other taste substances (Fig. 3).
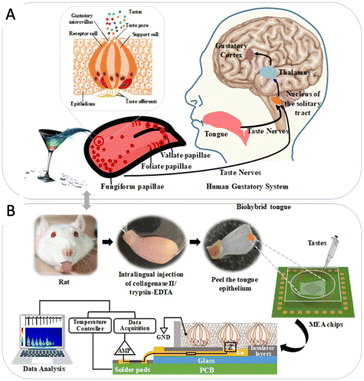 | ||
| Fig. 2 Biohybrid tongue. (A) Human gustatory system. (B) Gustatory epithelium functionalized microelectrode array chip. Reprinted with permission from ref. 19, copyright 2022, American Chemical Society. | ||
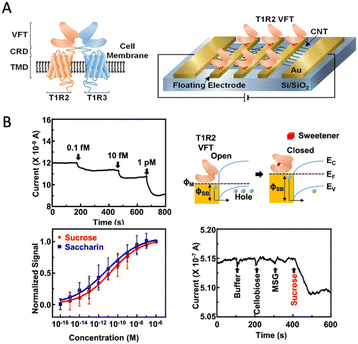 | ||
| Fig. 3 Bioelectronic tongues. (A) Sweet taste receptor-based CNT-FET tongue. (B) Real-time sensor response corresponding to different concentrations of sucrose solutions. Reprinted with permission from ref. 23, copyright 2022, American Chemical Society. | ||
Recent advancements in technology, materials science, computational biology, and artificial intelligence have enabled the incorporation of novel receptors and material structures into taste sensors. These include metallic nanostructure-based plasmonic sensors,24,25 voltammetric electronic tongue with machine learning algorithms,26 and electrochemical sensors.27,28 These advancements have significantly enhanced the performance of electronic and bioelectronic tongues, thereby offering more accurate and robust measurements.
Current challenges
Taste sensors are still considered a research area and are not widely employed in commercial applications. They have been used in laboratory settings to analyze a wide range of samples, including foods and beverages and medical and environmental samples. They have also been used for quality control in the food and beverage industry, as well as in medical diagnostics.However, they have difficulties in detecting and selectively identifying non-charged taste substances such as glucose and caffeine, lack applicability for detecting nutrients, drugs, metabolites, and heavy ions at the same time, and have limitations in the design of bioreceptors for multiplex sensing.
There is still a need for further development and validation to make them more practical for commercial use. Scientists are still working on developing and improving electronic and bioelectronic tongues with the goal of making them more accurate, reliable, and practical for commercial use. This includes developing new sensors, improving signal processing and data analysis methods, and reducing the costs of the devices. The main challenges in the development and practical use of electronic and bioelectronic tongues include the following:
• Require more receptors or sensing materials for multi-analyte measurement.
• Improve the accuracy and reliability with machine learning algorithms and data analysis approaches.
• Reduce the cost of the sensor system or device equipment for commercialization.
• Simplify the sample preparation process and operation of the sensor system.
• Integrate multiple sensors or obtain a more precise and comprehensive view.
• Conduct standardization when using the taste sensors.
Sensing materials and receptors play a critical role in the development of advanced taste sensors. In recent years, there have been several widely used sensing materials or receptors for sensing taste chemical compounds in electronic and bioelectronic tongues. These include ion-selective membranes, metal oxides, artificial lipid polymer membranes, and bioreceptors, including enzymes, antibodies, nucleic acids, and cells. For example, cell-based bioelectronic tongues can be highly selective but are also more expensive to produce and maintain, require more time to prepare, and have limited selectivity and specificity in detecting certain chemical compounds compared to other types of sensors.
As is well-known, thousands of chemical compounds contribute to taste, including sweet, sour, salty, bitter, and umami tastes. Table 1 shows a summary of the main substances and properties of the five basic tastes. Specific compounds that contribute to each taste can vary depending on the food or beverage being tasted. Measuring certain taste substances, such as noncharged glucose and caffeine, using potentiometric electronic tongues can be challenging. Therefore, further research is needed to develop new taste receptors or sensing materials coupled with nanotechnology and microfabrication techniques for advanced taste sensor development. For example, conductive polymers, organic and inorganic materials, nanomaterials, and artificial receptor molecularly imprinted polymers (MIP) have been widely developed for electronic tongue development.29–31 These sensing materials can be integrated into electronic devices such as field-effect transistors (FET), quartz crystal microbalances (QCM), surface plasmon resonance sensors (SPR), or cantilever sensors through an array-based approach and machine learning algorithms to detect and identify the target chemical compounds with high-throughput, simple, and low-cost measurements, overcoming the current challenges.32 Additionally, nanomaterials such as carbon nanotubes can be combined with bioreceptors, enzymes, antibodies, cells, or other biological molecules to develop more sensitive and specific taste sensors.
| Taste | Chemical compounds | Properties |
|---|---|---|
| Sweet | Glucose, fructose, sucrose | Source of natural sugar and vitamins |
| Sour | Citric acid, lactic acid, acetic acid | Source of vitamin C |
| Salty | Sodium, potassium, other minerals | Maintain electrolyte balance |
| Bitter | Caffeine, quinine, alkaloids | Noxious compounds but some have beneficial effects |
| Umami | Amino acids, glutamate, aspartate | Provide essential amino acids and other nutrients |
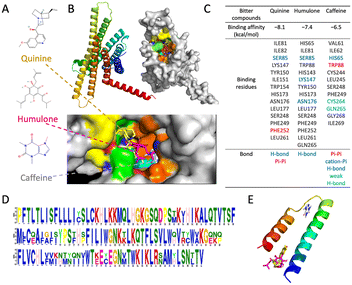
For bioelectronic tongue development, small receptors like peptides or aptamers over antibodies, cells, or enzymes have several advantages such as the following: they can be engineered to have specific binding properties and are versatile and easier to synthesize with low cost, which makes them ideal for use in biosensors and diagnostic assays. To better understand taste receptors for taste substance perception, in silico investigation is required. AlphaFold2 is a novel computational method that utilizes deep learning and neural networks to predict the structures of proteins from the amino acid sequences.33–35 It is a valuable tool for researchers in the fields of structural biology and bioinformatics because it eliminates the need for costly and time-consuming experimental methods to determine the protein structure. As shown in Fig. 4A and B, a new bitter taste receptor, TAS2R20, which is a G protein-coupled receptor (GPCR), was predicted and analyzed by molecular docking.36 Three typical bitter chemicals – quinine, humulone, and caffeine – interact with TAS2R20 with different binding affinities, and the binding residues are shown in Fig. 4C. This indicates how bitter molecules interact with taste proteins. Additionally, the binding motif was predicted, which can aid in the future design of short peptides for binding bitter substances using artificial neural networks (Fig. 4D and E).
Many studies have used computational algorithms to predict the structure and binding properties of peptides, including computational design of odorant-binding peptides or rational design of binding peptides from odorant-binding proteins (OBPs),37–40 which can be a very efficient way to identify potential candidates for further experimentation. In our previous study, we rationally designed a small receptor peptide from an anti-TNT monoclonal antibody in the complementarity determination region (CDR) (Fig. 5A and B), which is the antigen-binding region. Based on the molecular docking simulation and array-based fluorescence measurements (Fig. 5C), 15-mer amino acid peptides were selected as the TNT-binding peptides.41,42 The small receptor exhibited high affinity (Kd = 10.14 nM) for targeting TNT molecules and excellent selectivity.43 These studies revealed a novel approach for the development of small receptors.
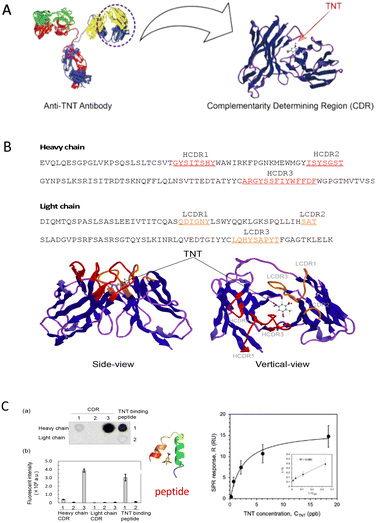 | ||
| Fig. 5 Rational design of a TNT binding peptide from an anti-TNT monoclonal antibody. (A) Structure of the TNT antibody, CDR and designed binding peptide. Reprinted with permission from ref. 42, copyright 2018, Elsevier. (B) Homology modelling and amino acid sequence analysis. (C) Experimental fluorescence results of the array-based peptide measurements (left), docking simulation (middle), and surface plasmon resonance kinetic analysis (right). Reprinted with permission from ref. 41 and 43, copyright 2017, American Chemical Society. Copyright 2019, Elsevier. | ||
All-in-one terahertz taste sensor
Terahertz (THz) waves are electromagnetic radiation that fall between the microwave and infrared frequencies in the electromagnetic spectrum. They have a frequency range of approximately 0.1 to 10 THz. THz waves have unique properties that make them useful for a variety of applications in different fields, such as spectroscopy, imaging, sensing, communications, materials science, etc.44 Terahertz technology has been considered as a potential method for taste sensing, environmental monitoring, and medical diagnosis, as it offers a non-invasive, highly sensitive, selective, and label-free method for detecting chemical compounds and identifying their concentrations in a sample.Our group developed an advanced terahertz chemical microscope (TCM). The TCM utilizes a semiconductor device, known as the sensing plate, to visualize the electrical potential distribution.45–49 The sensing plate was constructed by depositing a thin silicon film on a sapphire substrate and then adding a silicon oxide layer on the top (Fig. 6A). The thickness of the silicon film was 500 nm and that of the silicon oxide film was several nanometers (Fig. 6B and C). The sensing plate was 10 mm square for easy handling; however, it can be fabricated to wafer size depending on the sensing area that needs to be measured. The defects near the boundary between the silicon and silicon oxide films caused the energy band to bend towards the boundary surface, creating a depletion layer electric field. The direction of the bend depended on the doping type of the silicon film. When a femtosecond laser pulse with photon energy equal to or greater than the silicon bandgap was applied to the sensor plate from the substrate side, the charge carriers within the silicon film were excited and accelerated by the depletion layer electric field. As shown in eqn (1), the amplitude intensity of the generated terahertz wave (ETHz) is proportional to the square root of the electric potential (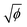 ). In this equation, El represents the depletion layer, e represents the electric elementary charge, N represents the electric charge density per unit area, and ε0 denotes the dielectric constant of vacuum. Therefore, various types of chemical reactions occurring on the sensing plate can be measured.
). In this equation, El represents the depletion layer, e represents the electric elementary charge, N represents the electric charge density per unit area, and ε0 denotes the dielectric constant of vacuum. Therefore, various types of chemical reactions occurring on the sensing plate can be measured.
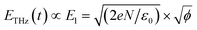 | (1) |
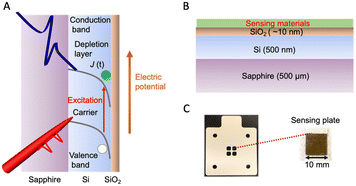 | ||
| Fig. 6 (A) Schematic of the energy band of the terahertz sensing plate. (B) Components of the terahertz sensing plate. (C) A photograph of the plastic holder for microwell formation and the sensing plate. Reprinted with permission from ref. 53, copyright 2023, Elsevier. | ||
To address the challenges, an innovative solution – an all-in-one terahertz taste sensor – has been proposed. This terahertz taste sensor can detect and quantify a diverse range of chemicals and biochemicals using multi-well arrays on a single semiconductor chip (sensing plate). It can visualize chemical and biochemical reactions with minimal sample volume consumption by using a terahertz chemical microscope system. This technology has shown a wide range of potential applications (Fig. 7A and B), including measuring physiological ions, the solution pH, non-charged molecules (Fig. 7C and D), sugars (Fig. 7E), toxic chemicals and drugs (Fig. 7F), enzyme kinetics, detecting cancer cells, analyzing the binding of antibodies and antigens in immunoassays, and evaluating cosmetic products and lithium-ion batteries.50–54 The terahertz sensing plate can be functionalized with ion-sensitive membranes, DNA aptamers, organic molecules, proteins, and antibodies.53,55 By measuring and visualizing the distribution of the electrochemical potential on the semiconductor sensing plate, the TCM can provide detailed information about chemical reactions. Furthermore, the TCM is a label-free, highly sensitive technology that can detect chemical and physical changes in the terahertz sensing plate. Unlike other established techniques such as FETs, SPR, QCMs, light-addressable potentiometric sensors (LAPS), and electrochemical sensors, the TCM does not have the limitations of Debye length design, molecular mass weight and charged molecule requirements or three electrodes requirements.56–59 For example, non-charged toxic and drug compounds have been successfully measured by simply modifying the semiconductor chip with a polymer and a DNA aptamer based on charge-transfer complex formation and conformational changes of the negatively charged DNA structure.53 Moreover, as previously discussed, the development of a wide range of bioreceptors and sensing materials through in silico or rational design, combined with the all-in-one terahertz taste sensor, could accelerate the development of multi-receptor arrays for detecting various types of chemicals.
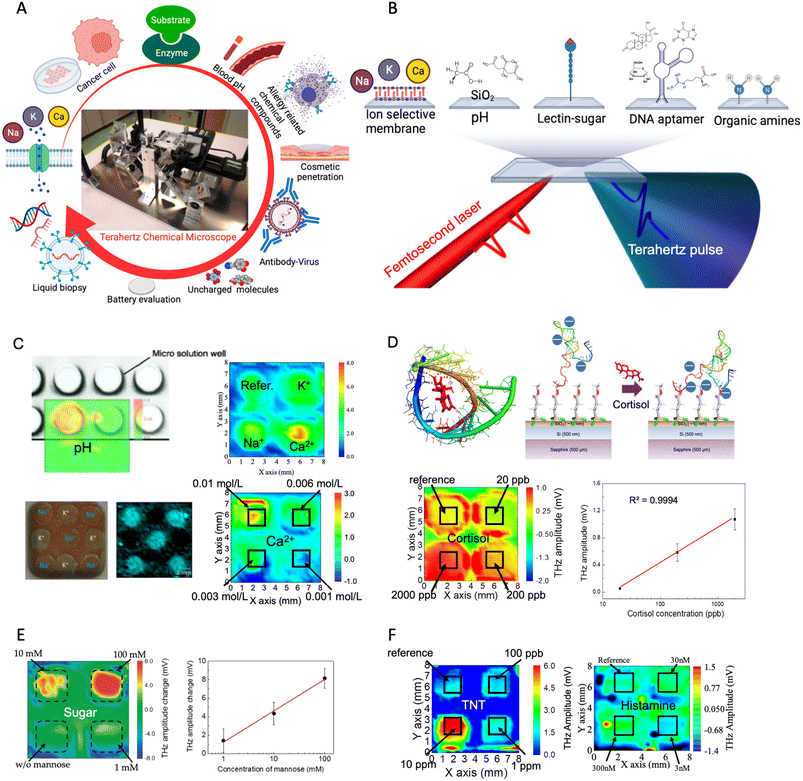 | ||
| Fig. 7 (A) Multifunctional terahertz chemical microscope in a variety of applications. Reprinted with permission from ref. 54. Copyright 2022 MDPI (Basel, Switzerland). (B) Sensing plate functionalization methods. Reprinted with permission from ref. 54. Copyright 2021 MDPI (Basel, Switzerland). (C) Visualization of pH, Na+, K+, and Ca2+. Reprinted with permission from ref. 45 and 54. Copyright 2018 Optical Society of America and SPIE. Reprinted with permission from ref. 53. Copyright 2023, Elsevier. (D) DNA aptamer-based terahertz chip for non-charge cortisol detection with high sensitivity. Reprinted with permission from ref. 53, copyright 2023, Elsevier. (E) Sensitive detection of the sugar compound mannose. Reprinted with permission from ref. 45. Copyright 2019 MDPI (Basel, Switzerland). (F) TNT explosive (left) and toxic histamine detection (right). Reprinted with permission from ref. 53. Copyright 2023, Elsevier and reprinted with permission from ref. 55. Copyright 2022 MDPI (Basel, Switzerland). | ||
However, the all-in-one terahertz taste sensor has some limitations or drawbacks, such as long-term stability problems of the laser power output and the non-uniform initial potential of the terahertz semiconductor chip caused by the manufacturing process. It requires specialized equipment and trained personnel to operate, which may limit its use under certain conditions. Therefore, future research and development could focus on maintaining the stability of the laser power output, improving the manufacturing process of the terahertz semiconductor chip, reducing the cost of the optical setup to make it more accessible, and integrating the sensor for ease of use. In addition, the development of advanced data processing or machine learning algorithms is necessary to achieve more reliable and accurate measurements.
In summary, the all-in-one terahertz taste sensor shows great potential for both academic research and industrial applications, and its non-invasive, label-free, and multi-analyte sensing approach will be a major advancement in taste sensing technology. It also has potential for use in environmental monitoring, nutrition evaluation, metabolite and drug measurement, biomarker sensing, and even for odorant identification, which could provide a comprehensive view and status and contribute to the future Food Data Bank (FDB).
Author contributions
J. W. wrote the manuscript. K. S. and T. K. reviewed the manuscript. All the authors provided permission for submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by a JSPS KAKENHI Grant-in-Aid for Early-Career Scientists (grant number: 21K14169).References
- L. Moreno, A. Merlos, N. Abramova, C. Jiménez and A. Bratov, Sens. Actuators, B, 2006, 116, 130–134 CrossRef CAS.
- Y. Kojima and Y. Hasegawa, AIP Conf. Proc., 2011, 1362, 195–196 CrossRef CAS.
- H.-M. Jeong, H.-C. Kwon, B. Xu, D. Jung, M. Han, D.-H. Kwon and S.-W. Kang, Sens. Actuators, B, 2020, 308, 127661 CrossRef CAS.
- K. Toko, Mater. Sci. Eng., C, 1996, 4, 69–82 CrossRef.
- K. Hayashi, M. Yamanaka, K. Toko and K. Yamafuji, Sens. Actuators, B, 1990, 2, 205–213 CrossRef CAS.
- Y. Kobayashi, M. Habara, H. Ikezazki, R. Chen, Y. Naito and K. Toko, Sensors, 2010, 10, 3411–3443 CrossRef CAS PubMed.
- Y. Tahara, A. Ikeda, Y. Maehara, M. Habara and K. Toko, Sensors, 2011, 11, 9878–9886 CrossRef CAS PubMed.
- J. Yoshimatsu, K. Toko, Y. Tahara, M. Ishida, M. Habara, H. Ikezaki, H. Kojima, S. Ikegami, M. Yoshida and T. Uchida, Sensors, 2020, 20, 1–13 CrossRef PubMed.
- A. Legin, A. Rudnitskaya, Y. G. Vlasov, C. Di Natale, F. Davide and A. D'Amico, Sens. Actuators, B, 1997, 44, 291–296 CrossRef CAS.
- L. A. Dias, A. M. Peres, A. C. A. Veloso, F. S. Reis, M. Vilas-Boas and A. A. S. C. Machado, Sens. Actuators, B, 2009, 136, 209–217 CrossRef CAS.
- X. Wu, Y. Tahara, R. Yatabe and K. Toko, Anal. Sci., 2020, 36, 147–159 CrossRef CAS PubMed.
- X. Wu and K. Toko, TrAC, Trends Anal. Chem., 2023, 158, 116874 CrossRef CAS.
- G. Valdés-Ramírez, M. Gutiérrez, M. del Valle, M. T. Ramírez-Silva, D. Fournier and J. L. Marty, Biosens. Bioelectron., 2009, 24, 1103–1108 CrossRef PubMed.
- M. Gutiérrez, S. Alegret and M. del Valle, Biosens. Bioelectron., 2008, 23, 795–802 CrossRef PubMed.
- M. Gutiérrez, S. Alegret and M. del Valle, Biosens. Bioelectron., 2007, 22, 2171–2178 CrossRef PubMed.
- J. Zeravik, A. Hlavacek, K. Lacina and P. Skládal, Electroanalysis, 2009, 21, 2509–2520 CrossRef CAS.
- D. Ha, Q. Sun, K. Su, H. Wan, H. Li, N. Xu, F. Sun, L. Zhuang, N. Hu and P. Wang, Sens. Actuators, B, 2015, 207, 1136–1146 CrossRef CAS.
- Z. Qin, B. Zhang, L. Hu, L. Zhuang, N. Hu and P. Wang, Biosens. Bioelectron., 2016, 78, 374–380 CrossRef CAS PubMed.
- C. Qin, C. Chen, Q. Yuan, N. Jiang, M. Liu, Y. Duan, H. Wan, R. Li, L. Zhuang and P. Wang, Anal. Chem., 2022, 94(19), 6976–6985 CrossRef CAS PubMed.
- Y. Hou, M. Genua, D. Tada Batista, R. Calemczuk, A. Buhot, P. Fornarelli, J. Koubachi, D. Bonnaffé, E. Saesen, C. Laguri, H. Lortat-Jacob and T. Livache, Angew. Chem., Int. Ed., 2012, 51, 10394–10398 CrossRef CAS PubMed.
- H. S. Song, O. S. Kwon, S. H. Lee, S. J. Park, U. K. Kim, J. Jang and T. H. Park, Nano Lett., 2013, 13, 172–178 CrossRef CAS PubMed.
- H. S. Song, H. J. Jin, S. R. Ahn, D. Kim, S. H. Lee, U. K. Kim, C. T. Simons, S. Hong and T. H. Park, ACS Nano, 2014, 8, 9781–9789 CrossRef CAS PubMed.
- J. Y. Jeong, Y. K. Cha, S. R. Ahn, J. Shin, Y. Choi, T. H. Park and S. Hong, ACS Appl. Mater. Interfaces, 2022, 14, 2478–2487 CrossRef CAS PubMed.
- G. Macias, J. R. Sperling, W. J. Peveler, G. A. Burley, S. L. Neale and A. W. Clark, Nanoscale, 2019, 11, 15216–15223 RSC.
- S. Forest, T. Theórêt, J. Coutu and J. F. Masson, Anal. Methods, 2020, 12, 2460–2468 RSC.
- Z. Yang, N. Miao, X. Zhang, Q. Li, Z. Wang, C. Li, X. Sun and Y. Lan, Food Control, 2021, 121, 107608 CrossRef CAS.
- Z. Chen, Q. Zhang, J. Shan, Y. Lu and Q. Liu, ACS Omega, 2020, 5, 27536–27545 CrossRef CAS PubMed.
- D. C. Braz, M. P. Neto, F. M. Shimizu, A. C. Sá, R. S. Lima, A. L. Gobbi, M. E. Melendez, L. M. R. B. Arantes, A. L. Carvalho, F. V. Paulovich and O. N. Oliveira Jr, Talanta, 2022, 243, 123327 CrossRef CAS PubMed.
- H. Sun, Z. H. Mo, J. T. S. Choy, D. R. Zhu and Y. S. Fung, Sens. Actuators, B, 2008, 131, 148–158 CrossRef CAS.
- A. Herrera-Chacon, A. González-Calabuig, I. Campos and M. del Valle, Sens. Actuators, B, 2018, 258, 665–671 CrossRef CAS.
- M. Podrazka, E. Báczyńska, M. Kundys, P. S. Jeleń and E. W. Nery, Biosensors, 2017, 8, 1–24 CrossRef PubMed.
- M. Wang, X. Cetó and M. del Valle, Biosens. Bioelectron., 2022, 198, 113807 CrossRef CAS PubMed.
- M. Varadi, S. Anyango, M. Deshpande, S. Nair, C. Natassia, G. Yordanova, D. Yuan, O. Stroe, G. Wood, A. Laydon, A. Zídek, T. Green, K. Tunyasuvunakool, S. Petersen, J. Jumper, E. Clancy, R. Green, A. Vora, M. Lutfi, M. Figurnov, A. Cowie, N. Hobbs, P. Kohli, G. Kleywegt, E. Birney, D. Hassabis and S. Velankar, Nucleic Acids Res., 2022, 50, D439–D444 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- K. Tunyasuvunakool, J. Adler, Z. Wu, T. Green, M. Zielinski, A. Žídek, A. Bridgland, A. Cowie, C. Meyer, A. Laydon, S. Velankar, G. J. Kleywegt, A. Bateman, R. Evans, A. Pritzel, M. Figurnov, O. Ronneberger, R. Bates, S. A. A. Kohl, A. Potapenko, A. J. Ballard, B. Romera-Paredes, S. Nikolov, R. Jain, E. Clancy, D. Reiman, S. Petersen, A. W. Senior, K. Kavukcuoglu, E. Birney, P. Kohli, J. Jumper and D. Hassabis, Nature, 2021, 596, 590–596 CrossRef CAS PubMed.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, Nat. Methods, 2022, 19, 679–682 CrossRef CAS PubMed.
- T. Wasilewski, B. Szulczyński, M. Wojciechowski, W. Kamysz and J. Gębicki, Sensors, 2019, 19(19), 4284 CrossRef CAS PubMed.
- A. J. M. Barbosa, A. R. Oliveira and A. C. A. Roque, Trends Biotechnol., 2018, 36, 1244–1258 CrossRef CAS PubMed.
- J. Wang, K. Sakai and T. Kiwa, Molecules, 2022, 27, 3917 CrossRef CAS PubMed.
- Y. Lu and Q. Liu, Sens. Diagn., 2022, 1, 1126–1142 RSC.
- M. Okochi, M. Muto, K. Yanai, M. Tanaka, T. Onodera, J. Wang, H. Ueda and K. Toko, ACS Comb. Sci., 2017, 19, 625–632 CrossRef CAS PubMed.
- J. Wang, M. Muto, R. Yatabe, Y. Tahara, T. Onodera, M. Tanaka, M. Okochi and K. Toko, Sens. Actuators, B, 2018, 264, 279–284 CrossRef CAS.
- T. Komikawa, M. Tanaka, K. Yanai, B. R. G. Johnson, K. Critchley, T. Onodera, S. D. Evans, K. Toko and M. Okochi, Biosens. Bioelectron., 2020, 153, 112030 CrossRef CAS PubMed.
- M. Tonouchi, Nat. Photonics, 2007, 1, 97–105 CrossRef CAS.
- T. Kiwa, T. Kamiya, T. Morimoto, K. Fujiwara, Y. Maeno, Y. Akiwa, M. Iida, T. Kuroda, K. Sakai, H. Nose, M. Kobayashi and K. Tsukada, Photonics, 2019, 6(1), 10 CrossRef CAS.
- H. Murakami, N. Uchida, R. Inoue, S. Kim, T. Kiwa and M. Tonouchi, Proc. IEEE, 2007, 95, 1646–1657 Search PubMed.
- K. Akimune, Y. Okawa, K. Sakai, T. Kiwa and K. Tsukada, Appl. Phys. Express, 2014, 7, 122401 CrossRef.
- T. Kiwa, T. Kamiya, T. Morimoto, K. Sakai and K. Tsukada, Opt. Express, 2018, 26, 8232 CrossRef CAS PubMed.
- T. Kiwa, A. Tenma, S. Takahashi, K. Sakai and K. Tsukada, Sens. Actuators, B, 2013, 187, 8–11 CrossRef CAS.
- T. Kiwa, Y. Kondo, Y. Minami, I. Kawayama, M. Tonouchi and K. Tsukada, Appl. Phys. Lett., 2010, 96, 1–4 CrossRef.
- F. Ahmed, A. Mahana, K. Taniizumi, J. Wang, K. Sakai and T. Kiwa, Jpn. J. Appl. Phys., 2021, 60, 027003 CrossRef CAS.
- M. Shimizu, R. Yamanaka, T. Teranishi, J. Wang, K. Sakai, K. Tsukada and T. Kiwa, Electr. Eng. Japan, 2021, 214, 1–7 CrossRef.
- J. Wang, M. Ando, H. Nagata, S. Yoshida, K. Sakai and T. Kiwa, Biosens. Bioelectron., 2023, 220, 114901 CrossRef CAS PubMed.
- T. Kiwa, K. Sakai and K. Tsukada, SPIE Newsroom, 2013, 2–5 Search PubMed.
- J. Wang, K. Sato, Y. Yoshida, K. Sakai and T. Kiwa, Photonics, 2022, 9(1), 26 CrossRef CAS.
- M. del Valle, Electroanalysis, 2010, 22, 1539–1555 CAS.
- P. Vahdatiyekta, M. Zniber, J. Bobacka and T. P. Huynh, Anal. Chim. Acta, 2022, 1221, 340114 CrossRef CAS PubMed.
- L. Du, J. Wang, W. Chen, L. Zhao, C. Wu and P. Wang, Anal. Chim. Acta, 2018, 1022, 106–112 CrossRef CAS PubMed.
- T. Wagner, W. Vornholt, C. F. Werner, T. Yoshinobu, K. I. Miyamoto, M. Keusgen and M. J. Schöning, Phys. Med., 2016, 1, 2–7 CrossRef.
| This journal is © The Royal Society of Chemistry 2023 |