Open Access Article
Open Access ArticleA novel point-of-care diagnostic prototype system for the simultaneous electrochemiluminescent sensing of multiple traumatic brain injury biomarkers†
Milica
Jović
 a,
Denis
Prim
a,
Denis
Prim
 a,
Ophélie
Righini
a,
Ophélie
Righini
 a,
David
Tagan
a,
David
Tagan
 b,
Mélanie
Stäuble
b,
Mélanie
Stäuble
 a,
Marc
Pignat
a,
Marc
Pignat
 b,
Steve
Gallay
b,
Steve
Gallay
 b,
Martial
Geiser
b,
Martial
Geiser
 c and
Marc E.
Pfeifer
c and
Marc E.
Pfeifer
 *a
*a
aDiagnostic Systems Research Group, Institute of Life Technologies, School of Engineering, University of Applied Sciences and Arts Western Switzerland (HES-SO Valais-Wallis), Rue de l'Industrie 19, 1950 Sion, Switzerland. E-mail: marc.pfeifer@hevs.ch
bInstitute of Systems Engineering, School of Engineering, University of Applied Sciences and Arts Western Switzerland (HES-SO Valais-Wallis), Rue de l'Industrie 23, 1950 Sion, Switzerland
cY-YBAR, Rue de la Dixence 10, 1950 Sion, Switzerland
First published on 30th May 2023
Abstract
Traumatic brain injuries (TBI) are typically acquired when a sudden violent event causes damage to the brain tissue. A high percentage (70–85%) of all TBI patients are suffering from mild TBI (mTBI), which is often difficult to detect and diagnose with standard imaging tools (MRI, CT scan) due to the absence of significant lesions and specific symptoms. Recent studies suggest that a screening test based on the measurement of a protein biomarker panel directly from a patient's blood can facilitate mTBI diagnosis. Herein, we report a novel prototype system designed as a precursor of a future hand-held point-of-care (POC) diagnostic device for the simultaneous multi-biomarker sensing, employing a microarray-type spatially resolved electrochemiluminescence immunoassay (SR-ECLIA). The small tabletop prototype consists of a screen-printed electrode compartment to conduct multi-analyte ECL sandwich assays, a potentiostat module and a light collection module, all integrated into a compact 3D-printed housing (18.2 × 16.5 × 5.0 cm), as well as an sCMOS detector. Based on this design concept, further miniaturization, system integration, performance optimization and clinical evaluation shall pave the way towards the development of a portable instrument for use at the site of accident and healthcare. To demonstrate the system's feasibility, current performance and efficiency, the simultaneous detection of three mTBI biomarkers (GFAP, h-FABP, S100β) in 50% serum was achieved in the upper pg mL−1 range. The proposed device is amenable to the detection of other biomarker panels and thus could open new medical diagnostic avenues for sensitive multi-analyte measurements with low-volume biological sample requirements.
Introduction
Each year about 70 million people all over the world, and 2.5 million people in Europe suffer from traumatic brain injuries (TBI). The figures are considerably underestimated partly because it is currently impossible to diagnose TBI in a decentralized way. Especially in the case of mild TBI (mTBI, concussion), a significant number of people are not seeking medical help due to the absence, or presence of only non-specific, light symptoms (headaches, nausea, etc.).1 Typical causes of TBI are sports accidents (e.g., American football, boxing, skiing, mountain biking), vehicle accidents (e.g., car, e-scooter), and violence (e.g., domestic, military, etc.). Short- to long-term consequences from TBI can have a strong correlation with neurodegenerative diseases in later years (Alzheimer's disease, Parkinson's disease, chronic traumatic encephalopathy, etc.). Currently, patients suspected of having suffered TBI are assessed using a Glasgow Coma Scale (GCS), which mainly evaluates their consciousness level and gives an indication of the severity of TBI (mild GCS 13–15; moderate GCS 9–12; severe GCS 3–8). The downside is that the GCS is based on subjective neurological and physical evaluation and can be time-consuming for both patients and doctors.2–4 Neuroimaging tools (CT, MRI) are also used for TBI diagnostics, but their main limitations include long analysis time (3–6 h), cost (2-million-dollar equipment), and radiation exposure (radiation equal to 200 chest X-rays5). Furthermore, according to the study by Korley et al., for 82% of all TBI patients taking a CT scan, there will for less than 10% of them evidence of a traumatic abnormality.6TBI biomarkers can pass the blood–brain barrier (BBB) following the injury and enter the blood circulation, where their typical concentration range in serum or plasma is mostly in the lower picogram range (pg mL−1).7 As recently reported, diagnostic screening tests based on biomarker measurements can significantly improve TBI diagnostics in three ways: (i) for patients: by having easy access to diagnostic tools at the site of the accident, avoiding radiation from medical imaging, and having rapid reassurance and timely discharge; (ii) for hospitals: by improving patient triage and staff productivity, reducing the number of unnecessary CT scans, and reducing the number of patients being discharged without proper diagnosis; (iii) for the healthcare system: by improving long-term patient outcomes, reducing costs of medical analysis and patient waiting times, and overall in reducing 400 billion dollars of global economic burden of TBI.
One of the most important challenges for a TBI diagnostic screening test is to be able to detect multiple biomarkers at the same time. Given the complexity of the human brain, it is very unlikely that any single biomarker would have sufficient clinical sensitivity and specificity,8,9 so there is a need for a test that will be able to detect a biomarker panel and allow decentralized TBI diagnostics. The road has been paved in January 2021 by Abbott's i-STAT™ Alinity™ hand-held device, which amperometrically measures two biomarkers (UCH-L1 and GFAP).10 However, there is still significant room for technologies and devices that would enable sensing of a higher number of biomarker targets and sequential multi-point measurements, which is especially important considering rather different post-injury kinetics.11,12 The conception and development of such a minimally invasive diagnostic device would certainly make a positive impact on patient management and improve the current state of healthcare.
Electrochemiluminescence (ECL) had previously shown to be a powerful method for bioanalytical applications as it possesses high temporal and spatial control of electrochemical techniques, as well as high sensitivities and low backgrounds,13 making it superior to photoluminescence or other optical methods that rely on external light excitation. The advantages over its analogous technique, chemiluminescence (CL), are based on the electrochemical reaction that allows the time and position of the light-emitting reaction to be controlled. Control over position allows confinement of light emission to a region precisely positioned to the detector, improving sensitivity by increasing the signal-to-noise ratio. ECL has been employed for the detection of a wide range of analytes, including small-size molecules,14–16 proteins,17,18 nucleic acids,19,20 and in the assays involving complex biological samples.21,22 In fact, immunoassays based on ECL are well established products for the in vitro diagnostic (IVD) use, run on high-throughput instruments in centralized clinical laboratories (e.g., Roche Cobas analysers). Thus, while ECL appears to be a proven assay and read-out technology for high sensitivity applications in laboratory medicine, to our knowledge its appropriateness for field deployable and reliable POC medical diagnostic use has not yet been shown. In recent years however, various researchers have made significant efforts to develop and establish miniaturized ECL devices suitable for biomedical applications. Recent reviews by Bhaiyya et al.23 and Ying et al.24 give an overview of ECL-based systems/devices developed for POC settings. Even though it is evident that miniaturized ECL devices are gaining attention, there is still a need for miniaturized ECL platforms that will allow a simultaneous multi-analyte sensing.23
ECL assays have been rarely combined with protein microarray approaches, the latter being known for their multi-analyte detection capabilities, higher throughput, and potential for miniaturization.25 Like classical ELISA, microarrays employ a recognition element fixed on the carrier material and various label-free or labelled-probe detection methods (mostly limited to fluorescence and chemiluminescence).26–28 It is not obvious, and limited evidence in the scientific literature show that screen-printed carbon electrodes (SPCE) can meet demanding multiplex high-sensitivity sandwich immunoassay performance and reliability requirements, as is needed for mTBI diagnostic applications. Inadequate electrode solid-surface properties may lead to either non-optimal, irreproducible immobilizations of antibodies, inhomogeneities, high background (blank) signal intensities, or poor electrochemical conductivities. In our previous work, we have combined a microarray approach with an electrochemiluminescence immunoassay (ECLIA) for the simultaneous detection of an mTBI biomarker panel on a single screen-printed carbon electrode (SPCE).29 We observed significantly different ECL signal intensities as a function of electrode material employed and physical and chemical surface modifications had indeed, in some instances, a signal-amplifying effect when working with homogeneous luminophore solutions. Herein, we focused on increasing the technology readiness level (TRL) of our previous laboratory breadboard and improved the usability for possible early evaluations in a more relevant environment (e.g., by clinical partners). We designed and incorporated various hard- and software components into a miniaturized device that could be easily expanded to a prototype mini-series. This tabletop device comprises an SPCE carrying a 3-plex microarray assay, a potentiostat module and a light collection module, everything integrated into a 3D-printed housing to completely shield the signal generation from ambient light and data acquisition process, as well as an sCMOS detector (12.2 × 8.4 × 8.4 cm). As shown below, this prototype instrument can be employed for single biomarker detection (CRP) and multiple biomarkers detection (GFAP, h-FABP, S100β), but the degree of multiplexing can certainly be expanded if required in the future.
Experimental
Materials
Assay protocols
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was used for all detection antibodies, while the labelling incorporation ratio was calculated by measuring the OD455 values using the NanoDrop OneC Microvolume UV-VIS Spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA). The calculated label ratio for detection antibodies was 9
1 was used for all detection antibodies, while the labelling incorporation ratio was calculated by measuring the OD455 values using the NanoDrop OneC Microvolume UV-VIS Spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA). The calculated label ratio for detection antibodies was 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 20 min, then rinsed with MQ water, and dried with argon. Then an S3 contactless nano-spotting device from Scienion AG (Berlin, Germany) equipped with a Piezo Dispense Capillary (PDC-70, coating type 3, p/n: P-2030 S-6051) was used to dispense drops of 350 pL (±10 pL). For the microarray sandwich assays, capture antibody solutions of biomarkers were spotted on pre-defined positions of SPCE to form spots with diameters of ∼450 μm (±50 μm). The source plate temperature was set at 15 °C and the relative humidity in the spotting area at 60%. After the spotting process SPCE were left in the spotting area during 30–60 min before blocking with 50 μL of 2% BSA (w/v) in PBS 1× solution (60 min, RT). The electrodes were then washed with wash buffer (PBS 1× with 0.06% Tween-20 (v/v)) and placed inside the custom-made incubation cells fabricated from Teflon, as previously reported.29 The incubation cells were designed to accommodate 12 SPEs, leaving only the WE areas exposed to the reagents during various incubation/mixing/washing steps of the immunoassay. Mixtures of antigens or blank solutions (50 μL) were added to the wells and incubated (60 min, 500 rpm, RT). After the second washing step (3 × 300 μL), 50 μL of the detection antibody mixture was added and incubated (60 min, 500 rpm, RT). After the final washing step (6 × 300 μL), the electrodes were taken out of the cells, were rinsed with wash buffer, and 75 μL of read buffer T 2× solution was added to initiate the signal generation and read-out process. ImageJ software (v1.53) was used for image processing.
1) for 20 min, then rinsed with MQ water, and dried with argon. Then an S3 contactless nano-spotting device from Scienion AG (Berlin, Germany) equipped with a Piezo Dispense Capillary (PDC-70, coating type 3, p/n: P-2030 S-6051) was used to dispense drops of 350 pL (±10 pL). For the microarray sandwich assays, capture antibody solutions of biomarkers were spotted on pre-defined positions of SPCE to form spots with diameters of ∼450 μm (±50 μm). The source plate temperature was set at 15 °C and the relative humidity in the spotting area at 60%. After the spotting process SPCE were left in the spotting area during 30–60 min before blocking with 50 μL of 2% BSA (w/v) in PBS 1× solution (60 min, RT). The electrodes were then washed with wash buffer (PBS 1× with 0.06% Tween-20 (v/v)) and placed inside the custom-made incubation cells fabricated from Teflon, as previously reported.29 The incubation cells were designed to accommodate 12 SPEs, leaving only the WE areas exposed to the reagents during various incubation/mixing/washing steps of the immunoassay. Mixtures of antigens or blank solutions (50 μL) were added to the wells and incubated (60 min, 500 rpm, RT). After the second washing step (3 × 300 μL), 50 μL of the detection antibody mixture was added and incubated (60 min, 500 rpm, RT). After the final washing step (6 × 300 μL), the electrodes were taken out of the cells, were rinsed with wash buffer, and 75 μL of read buffer T 2× solution was added to initiate the signal generation and read-out process. ImageJ software (v1.53) was used for image processing.
Prototype components
Results and discussion
Prototype development and performance evaluation
The POC diagnostic prototype system is composed of the following carefully selected, pre-existing as well as newly designed elements (Fig. 1a and b, SI-1 and SI-2†):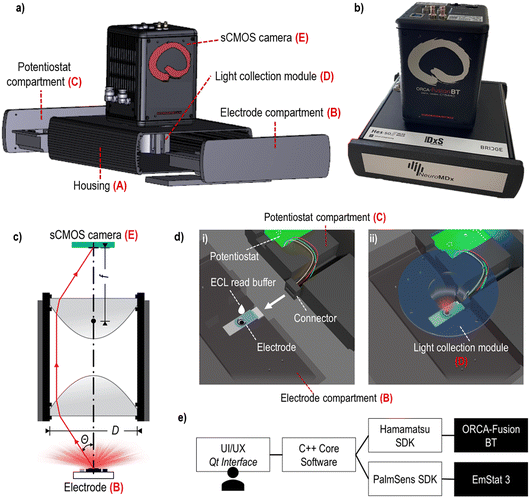 | ||
| Fig. 1 Prototype build-up: a) a layer rendering image of the prototype main components; b) photo of the prototype device; c) schematic representation of the light collection module; d) schematic rendering of the electrode placed inside the electrode compartment: (i) depicts the system when the electrode and potentiostat compartments are disconnected (some parts are made transparent here), and ECL read buffer is manually added; (ii) depicts the system during the measurement step when the compartments are engaged and properly positioned beneath the light collection module (transparent blue); e) software architecture diagram. The graphical interface is shown in the SI-3.† | ||
A. Housing;
B. Electrode compartment;
C. Potentiostat compartment;
D. Light collection module (Fig. 1c);
E. sCMOS camera;
F. Computer and software.
The prototype device housing (A) had a role to shield the ECL and detection process from ambient light. The housing included a 3D-printed case with two compartments (B and C) that contained the electrode and the potentiostat, respectively. The light collection module (D) was accurately positioned on top of the electrode (B) and aligned with the sCMOS camera (E) located on top of the housing. The disposable electrode was placed into a dedicated rectangular groove of the lower part of the electrode compartment (B), which, when assembled with the upper part, incorporated a small reservoir for the addition of the ECL read buffer (SI-2†). The potentiostat module was placed inside the potentiostat compartment (C), opposite to the electrode compartment, and was used to trigger the ECL signal generation by applying a small external potential (Fig. 1d).
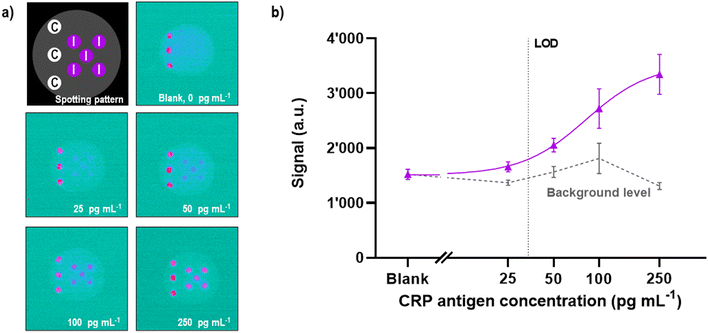
The ECL light emitted from the electrode was collected by an optical system named light collection module (D), consisting of two reversed identical lenses, where the aperture was not clearly defined to prioritize the photon collection efficiency over the image quality (Fig. 1c). We chose aspheric lenses from Edmund Optics (USA) with a 0.66 numerical aperture (cos(θ) = 0.606), a focal length (f) of 18.75 mm and a diameter (D) of 25.00 mm. The photon collection efficiency (η, 19.7%) was calculated using the eqn (1) where Ω is the solid angle sustained by the optical system in the object space:
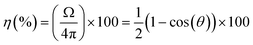 | (1) |
The last component of the prototype system was the software (F) that has been programmed to control and synchronize the potentiostat module (C) with the sCMOS camera (E) (Fig. 1e). The graphical interface allows the user to select the sCMOS exposition duration and binning, as well as the potentiostat stimulus duration and the voltage (SI-3†). After the measurement, an image file and a corresponding text file containing the generated data and the measurement parameters are saved to the hard disk.
Conceptualization of an ECL microarray on SPCE
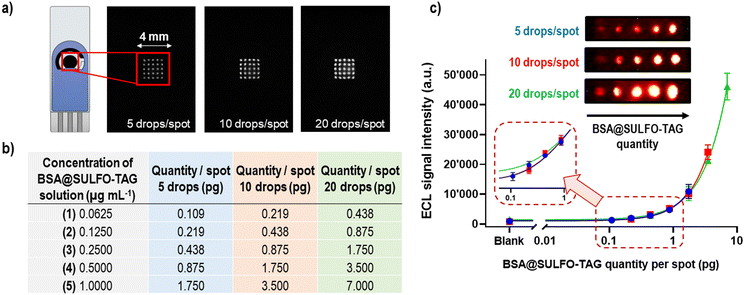
The concept of microarray immunoassays is based on the ambient analyte theory46 and the use of very small amounts of antibodies (or other recognition elements) that are typically orders of magnitude lower than those in classical immunoassays. In microarray methods the signal-to-background ratio will increase with the decrease of the spot area containing recognition elements, leading to higher assay sensitivity (SI-4†).46 Some of the advantages of immunoassays conforming to ambient analyte assay principles include the possibility to detect multiple targets, having minimized diffusion constraints on analytes binding to antibodies, faster reaction kinetics, and higher assay sensitivities compared to the other immunoassay designs.46The first step in the conceptualization of the ECL microarray on SPCE was to spot the carbon working electrode with various amounts of proteins carrying ECL labels (Ru(bpy)32+) to test the detector's sensitivity, spatial resolution, and dynamic range. For this purpose, 1 μg mL−1 of BSA protein labelled with the SULFO-TAG was spotted on the carbon WE in the form of a 5 × 5 spot microarray, using the following approach (Fig. 2a): 5 drops per spot (1.75 pg of BSA@SULFO-TAG protein, ∼250 μm spot diameter), 10 drops per spot (3.50 pg of BSA@SULFO-TAG protein, ∼300 μm spot diameter), and 20 drops per spot (7.00 pg of BSA@SULFO-TAG protein, ∼400 μm spot diameter). To perform the ECL read-out the electrode was placed inside the prototype device and 75 μL of MSD Read Buffer T 2X containing tripropylamine was added on top of it. The obtained ECL images showed excellent spatial resolution obtained with the sCMOS detector and good reproducibility of signals from the different spots of the microarray. In the second step, SPCE were spotted with increasing amounts of BSA@SULFO-TAG protein by using solutions of different concentrations and different spot sizes (Fig. 2b). Obtained ECL images and corresponding signals are shown in Fig. 2c, indicating that BSA@SULFO-TAG quantities as low as 0.1 pg could be successfully detected, with excellent correlation between spotted quantities and detected ECL signals, and with very good spot-to-spot and electrode-to-electrode reproducibility. Based on these findings it can be concluded that the ECL microarray approach combined with the sCMOS based prototype system offers great potential for detection of very low quantities of proteins.
Microarray-type spatially resolved ECL immunoassay (SR-ECLIA) for single biomarker detection
Once the ECL microarray concept has been established on SPCE using POC diagnostic prototype system for the read-out, the next step was to evaluate the performance of such system for single biomarker detection. For that purpose, a microarray-type spatially resolved ECL immunoassay (SR-ECLIA) was developed for C-reactive protein (CRP) biomarker. CRP is well-known and widely used biomarker of inflammation, however, recently there has been growing evidence implicating high sensitivity CRP (hsCRP) as a biomarker for neurological diseases. hsCRP is being particularly interesting as a potential biomarker for TBI47–49 or part of mTBI biomarker panel, due to high sensitivity, specificity, and negative predictive value during the acute mTBI phase.50The SR-ECLIA-type sandwich assay for hsCRP was developed by spotting carbon working electrodes with CRP capture antibodies and BSA@SULFO-TAG protein as control/alignment spots. The spotting pattern was composed of 3 control spots (BSA@SULFO-TAG) and 5 spots for the CRP assay (Fig. 3a). Methods and conditions of the assay are listed in SI-5.† The rest of the protocol is the same as for any immunoassay, with blocking, antigen addition, washing, and detection antibody (carrying ECL labels in form of SULFO-TAG being Ru(bpy)32+) addition steps. ECL detection was accomplished by adding the read buffer containing tripropylamine (TPA). The ECL signal on SPCE was triggered using chronoamperometry at a potential of 1.55 V. Once the detection step was finished, the recorded ECL images were processed using the ImageJ software, where the signal intensity of each spot area was calculated for each spot.
 | ||
| Fig. 3 Microarray-type spatially resolved (SR-ECLIA) CRP singleplex assay performance in “clean target” (buffer solution) using POC prototype detection system: a) capture antibody spotting pattern (letter “C” – BSA@SULFO-TAG control spots; letter “I” – inflammatory (CRP) biomarker spot; and corresponding ECL raw data images generated at various CRP concentrations (0, 25, 50, 100, and 250 pg mL−1) (8 × 8 binning and dark subtraction, but no other postprocessing); b) obtained calibration curve. The error bars represent the standard deviations from two replicates (n = 2) and the fitting was performed with a 4-PL equation; dashed horizontal curve represent the evolution of the background measured on the electrode surface, in the surrounding area of the spots. The signals were not normalized. Dotted vertical line represent the LOD (calculated at 34 pg mL−1). Assay conditions are listed in SI-5.† | ||
The Ru(bpy)32+/TPA co-reactant system was used to generate the ECL signals, as it offers the best ECL performance, it is bio-stable, well-validated and widely applied in commercial diagnostic applications.51 The ECL signal generation of the co-reactant couple is produced by the reaction between deprotonated TPA˙ radical and electrogenerated Ru(bpy)33+, as indicated in the following reactions (eqn (2)–(5)):52
| Ru(bpy)32+ → Ru(bpy)33+ + e− | (2) |
| Ru(bpy)33+ + TPA → Ru(bpy)32+ + TPA˙ | (3a) |
| TPA → TPA˙ + e− | (3b) |
| Ru(bpy)33+ + TPA˙ → Ru(bpy)32+* | (4) |
| Ru(bpy)32+* → Ru(bpy)32+ + hv (λmax = 610 nm) | (5) |
To evaluate the performance of the POC diagnostic prototype system, buffer solutions containing CRP concentrations from 25 to 250 pg mL−1 were analysed and based on the obtained images (Fig. 3a) a calibration curve was established (Fig. 3b). Data was analysed assuming that the ECL signal was proportional to the CRP concentration through a four-parameter dose–response regression fit model (4PL). The LOD value of 34 pg mL−1 [CI 95%, 19–44 pg mL−1] was calculated by interpolating the curve using the average value of the blank plus three times the standard deviation of the blank. The obtained LOD in double-digit pg range is impressive, considering the miniaturize size of the device/detector system, which often presents limitations for reaching high sensitivities. The background is kept at an acceptable level (∼1500 a.u.) at all concentrations, while the variability, knowing the work in progress stage of the demonstrator, is below 20% (ranging from 5% to 15%). The focus was made on the low antigen concentration level to determine the device/assay sensitivity; however, further evaluations are planned at higher concentration levels to determine the dynamic range.
Microarray-type spatially resolved ECL immunoassay (SR-ECLIA) for mTBI biomarker panel detection
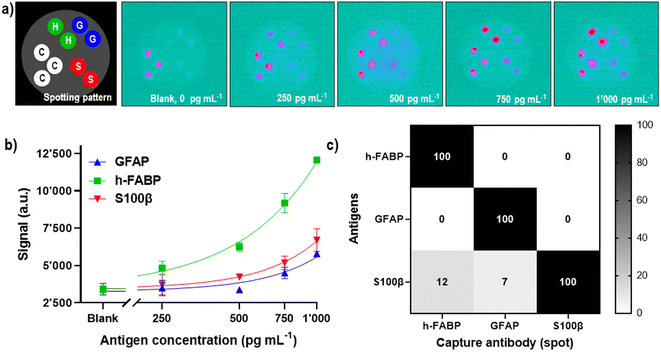
In our previous publication, we have developed three single-plex assays for the mTBI biomarkers GFAP, h-FABP, S100β, and shown the proof-of-concept of how these three assays can be combined into a 3-plex microarray type assay format on a single SPCE using the SR-ECLIA approach (Fig. 4a).29 The protocol is similar to single biomarker detection protocol described in the previous chapter, with the difference that carbon working electrodes were spotted with BSA@SULFO-TAG protein (control/alignment spots, 3 spots) and with capture antibodies of all three biomarkers (2 spots) (Fig. 4a, STEP 1). After the rest of the sandwich assay was performed (Fig. 4a, STEP 2), the ECL detection was done by adding the read buffer solution containing tripropylamine (Fig. 4a, STEP 3) and performing chronoamperometry at a potential of 1.55 V. Recorded ECL images were processed using the ImageJ software, where the signal intensity of each spot area was calculated for each biomarker (Fig. 4a, STEP 4).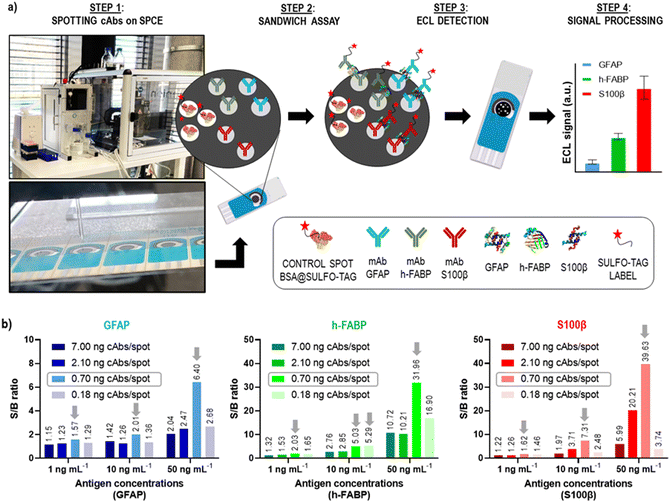 | ||
| Fig. 4 Microarray-type SR-ECLIA 3-plex protocol: a) schematic representation of the assay established for detection of three mTBI biomarkers (GFAP, h-FABP, S100β) on a single SPCE. STEP 1 includes the spotting of small quantities of capture antibodies of each biomarker on the predefined spot positions on the SPCE using a spotter device (BSA labelled with SULFO-TAG was used for control/alignment purposes); STEP 2 includes the typical sandwich assay protocol composed of a blocking step, addition of three antigens, and addition of detection antibodies modified with the SULFO-TAG label carrying Ru(bpy)32+; STEP 3 is the detection step that includes addition of the tripropylamine (TPA) co-reactant and applying a potential on the SPCE to trigger the ECL signal generation from each biomarker spot; STEP 4 comprises the raw data processing; b) optimization of capture antibody amount per spot (cAb amount for all biomarkers: 7.00 ng per spot, 2.10 ng per spot, 0.70 ng per spot, 0.18 ng per spot). The antigen concentrations were 0, 1, 10 and 50 ng mL−1. The other assay conditions are listed in SI-5.† | ||
The obtained 3-plex microarray SR-ECLIA assay was further optimized by fine-tuning the quantity of spotted capture antibodies (cAbs) on the electrode surface. The cAbs in the assay should provide optimal electrode surface coverage and proper orientation for the best recognition of target antigens. For that purpose, SPCE were spotted with four different concentrations/quantities of cAbs of each biomarker: (i) 25 μg mL−1 (0.18 ng per spot); (ii) 100 μg mL−1 (0.70 ng per spot); (iii) 300 μg mL−1 (2.10 ng per spot); and (iv) 1000 μg mL−1 (7.00 ng per spot), according to the pattern shown in Fig. 4a (the assay conditions are listed in SI-5†). For each spotting condition, a complete multi-analyte assay was performed with four different antigen concentrations in triplicate (0, 1, 10, and 50 ng mL−1). Fig. 4b shows obtained S/B ratios for all tested conditions and for all three biomarkers. It was observed that when cAb quantities on the electrodes were higher than >0.70 ng per spot (2.10 and 7.00 ng per spot, respectively) and lower than <0.70 ng per spot (0.18 ng per spot), low S/B ratios were obtained, possibly indicating sub-optimal electrode surface coverage. The best S/B ratios were obtained with spots containing 0.70 ng of cAbs, thus this condition was used for further experiments.
Once the conditions for the 3-plex assay were optimized, its analytical performance was assessed by establishing biomarker calibration curves in 50% human serum (v/v), in the concentration range from 0 to 1 ng mL−1 (Fig. 5). The obtained ECL images (Fig. 5a) were treated using ImageJ software, and calculated signal intensities from the spots were used for the construction of a calibration curve using the four-parameter dose–response regression fit model (4PL) (Fig. 5b). The analytical performance of the 3-plex assay was reasonably good, reaching LODs of 742 pg mL−1 (R2 = 0.78), 237 pg mL−1 (R2 = 0.97) and 583 pg mL−1 (R2 = 0.82) for GFAP, h-FABP and S100β, respectively. When the LODs are compared with biomarkers' cut-off values (physiological concentrations used to distinguish “healthy” vs. “mild TBI cases”), which are 22 pg mL−1 for GFAP,53 2620 pg mL−1 for h-FABP,54 and 42 pg mL−1 for S100β,54 it can be concluded that current sensitivity of the assay/prototype is sufficient to detect h-FABP biomarker (LOD is at 1/10 of biomarker's cut-off value), but for GFAP and S100β further improvements of analytical sensitivities will be required. The differences in analytical sensitivities are presumably due to lower binding affinities of the GFAP and S100β antibodies (compared to the h-FABP ones). Based on the obtained results with single biomarker detection (CRP, LOD 34 pg mL−1), it is conceivable that further optimization of the h-FABP assay part will lead to an LOD in the lower double-digit pg mL−1 range, the sCMOS detector sensitivity thereby not being a limiting factor. Furthermore, higher LODs obtained for the 3-plex assay vs. singleplex assay are certainly not surprising, considering the complexity of multianalyte interactions and cross-reactivities between antigens and/or antibodies.
It should be noted that the SPCE were spotted, with capture antibodies, and blocked, with the appropriate reagents, on the day of the experiments and used for one single measurement due to the adsorption of tripropylamine on the carbon surface. Further experiments are planned to evaluate and, if necessary, improve the shelf-life of the prepared electrodes upon prolonged storage. To show that there was no major 3-plex assay specificity issue, i.e., that the antibodies did not recognize non-target antigens, a set of electrodes were spotted with all three capture antibodies before probing the assays with the individual analytes at a concentration of 2.5 ng mL−1 and subsequent detection with the mixture of GFAP, h-FABP and S100β detection antibodies. Under the not yet fully optimized assay conditions the presence of the S100β antigen showed a 7% non-specific reaction with the GFAP spot position and a 12% non-specific reaction with the h-FABP spot position, respectively. This cross-reaction signal could be further resolved by using more specific Ab clone, adding detergent and/or buffer composition change. With samples containing either h-FABP or GFAP antigens no non-specific signals were observed (0%) (Fig. 5c). The mixture of all three detection antibodies when added to the spotted electrodes in absence of antigens did not reveal any cross-reactivity (see “Blank” in Fig. 5a). The positional accuracy of the spots functionalized with cAbs and BSA@SULFO-TAG across multiple SPCE (inter-electrode spot position variability) was remarkably good. A close-up view on the ECL images also reveals well-defined and reasonably symmetrical spots with no evidence for significant bleeding or spreading. This suggests a certain robustness with regards to both the automated spotting process and the implemented multi-step sandwich immunoassay.
Assay repeatability and preliminary recovery test
The applicability and reliability of the multiplex SR-ECLIA for the quantification of the mTBI biomarkers on the POC diagnostic prototype device was evaluated in a complex physiological matrix by the method of standard addition. Biomarker concentrations were spiked into human serum diluted 2× with assay diluent (“spiked” concentration). Based on the obtained ECL signal intensity, the corresponding biomarker concentrations (“detected” concentration) were calculated according to the regression equation performed using the same matrix (4PL dose–response curve). The recoveries were calculated by the ratio between the “detected” and “spiked” concentrations. The obtained results are summarized in Table 1. The determined recoveries ranged from 95% to 111%, which suggests a reasonable preliminary performance at this stage of development.| Biomarker | Concentration (pg mL−1) | Inter-assay CV (%) | Recovery of controls in serum (%) |
|---|---|---|---|
| h-FABP | 250 | 11 | 95 |
| 500 | 14 | ||
| 750 | 15 | ||
| 1000 | 5 | ||
| GFAP | 250 | 7 | 105 |
| 500 | 6 | ||
| 750 | 9 | ||
| 1000 | 9 | ||
| S100β | 250 | 9 | 111 |
| 500 | 14 | ||
| 750 | 16 | ||
| 1000 | 11 | ||
Future & Perspectives
It is evident that miniaturized ECL systems/devices can be very powerful analytical and medical diagnostic tools that can enable detection of low-level biomarkers in body fluids at the point-of-care. Table SI-6† gives a brief overview of recently published articles on such systems/devices, including details on used reagents, detectors, electrodes, analytes and reported LODs (non-POC diagnostic type device publications were not considered). What most of these systems are lacking, as pointed out in the review by Bhaiyya et al.,23 is the simultaneous sensing of multiple analytes. This feature is particularly important when biological samples with limited volumes need to be analysed (e.g., paediatric patients), and/or when the diagnosis cannot be established based on a single biomarker measurement like for mTBI.Device compactness and lightweight, thus portability of such a detection system is certainly another key advantage considering the need for early post-injury measurements at the site of accident which is clearly beneficial in the case of mTBI diagnosis. The possibility to utilize the device, for instance in the ambulance, or at a general practitioner's cabinet in remote regions, allows the acquisition of multiple measurement values useful for biomarker kinetic assessments. We tried to answer that need by combining a microarray approach with a spatially resolved ECL detection and integrating this methodology into a compact prototype with an sCMOS detector to enable simultaneous multi-analyte detection on a cheap and disposable SPCE. Even though full system integration is not without technical challenges, we have shown that all key components can be successfully integrated into a well-performing and reasonably small-size tabletop read-out device that can serve as a good starting point for further miniaturization, assay optimization and for early-stage pre-clinical evaluations. In fact, valuable feedback for design improvements by early healthcare adopters/end-users is always helpful. The smart integration of already existing and proven components, that are manufacturable at large scale (e.g., SPCE, potentiostat, board-level sCMOS, etc.), may from a product development and fabrication cost point-of-view be seen as economically favourable and de-risking, thus could support a path forward towards ultimate regulatory approval and commercialization.
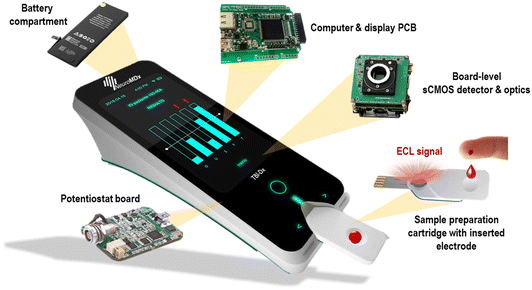
The next development effort towards a comprehensive system prototype (TRL 4–5) focuses on the device docking unit for the sample processing cartridge currently under development. Incidentally, the cartridge is a disposable item that must meet minimal cost-of-goods-manufactured (COGM) and rapid assay time-to-results requirements. Our vision of the future device is shown in Fig. 6. Beyond the mTBI biomarker panel, other multi-analyte medical applications (e.g., cardiovascular, inflammation, oncology, etc.) can be envisioned, however, the advantages of such as POC diagnostic device against existing laboratory platforms would have to be evaluated carefully in terms of economic viability and clinical utility.
Conclusions
In summary, this publication introduces and describes the concept and fabrication of a miniaturized preliminary POC prototype system for the simultaneous sensing of multiple analytes in a single sample based on a microarray-type ECLIA. The motivation to develop such prototype was: 1) to demonstrate that the spatially resolved electrochemiluminescence immunoassay concept (SR-ECLIA) was robust enough and had the potential for good and reliable test performances, and 2) to expedite the construction of a small tabletop instrument to investigate the possibilities of designing and manufacturing in detail a comprehensive advanced prototype solution of a future handheld POC diagnostic device for mTBI. The basic components of the prototype are a screen-printed carbon electrode compartment, an OEM potentiostat module and a specifically designed light collection module, all integrated into a 3D printed housing, as well as an sCMOS detector. This prototype device was evaluated for the detection of single biomarker in clean target (CRP, LOD 34 pg mL−1), as well for simultaneous detection of three mild traumatic brain injury (mTBI) biomarkers (GFAP, h-FABP, S100β), achieving LODs close to 200 pg mL−1 (for h-FABP) in 50% human serum and reasonable recoveries with spiked serum samples.So far, no major obstacles in this endeavour have been encountered. However, the efficient and cost-effective functionalization (spotting) of the SPCE and the scalable assembly into cartridges are challenges on the horizon, although rather engineering than scientific ones. Aside of further assay analytical sensitivity improvements, the next instrument development milestone will include the detailed design of the docking system comprising the sample processing cartridge and the docking port of the reader device. The prototype and assay performances are intended to be evaluated with mTBI patient samples in the next step. In conclusion, the prototype system described in this publication can already serve as an “enabling” tool on the possible path towards industrial product development.
Author contributions
Milica Jović: conceptualization, methodology, formal analysis, investigation, writing – original draft preparation, supervision. Denis Prim: conceptualization, methodology, hardware, formal analysis, investigation, writing – review & editing, visualization, supervision. Ophélie Righini: formal analysis, investigation, writing – review & editing. David Tagan: methodology, hardware. Mélanie Stäuble: formal analysis, investigation. Marc Pignat: software. Steve Gallay: hardware. Martial Geiser: methodology, hardware. Marc E. Pfeifer: conceptualization, methodology, writing – original draft preparation, visualization, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by BRIDGE (joint program conducted by the Swiss National Science Foundation (SNSF) and Innosuisse – the Swiss Innovation Agency) [grant no. 40B2-0_181013], the Axe Santé HES-SO Valais-Wallis [innovation cheque 2022 (TBI POCDx Prototype)], and the Institute of Life Technologies HES-SO Valais-Wallis. The authors would like to thank the mechanical workshop of the HES-SO Valais-Wallis for the fabrication of customized incubation cells for SPEs.References
- S. Mondello, U. Muller, A. Jeromin, J. Streeter, R. L. Hayes and K. K. W. Wang, Expert Rev. Mol. Diagn., 2011, 11, 65–78 CrossRef PubMed.
- R. Mehta, G. P. Trainee1, K. Chinthapalli and C. Neurologist2, BMJ, 2019, 365, l1296 CrossRef PubMed.
- M. C. Dewan, A. Rattani, S. Gupta, R. E. Baticulon, Y.-C. Hung, M. Punchak, A. Agrawal, A. O. Adeleye, M. G. Shrime, A. M. Rubiano, J. V. Rosenfeld and K. B. Park, J. Neurosurg., 2018, 130, 1080–1097 Search PubMed.
- J. M. Powell, J. V. Ferraro, S. S. Dikmen, N. R. Temkin and K. R. Bell, Arch. Phys. Med. Rehabil., 2008, 89, 1550–1555 CrossRef PubMed.
- R. Smith-Bindman, J. Lipson, R. Marcus, K. P. Kim, M. Mahesh, R. Gould, A. B. de Gonzalez and D. L. Miglioretti, Arch. Intern. Med., 2009, 169, 2078 CrossRef PubMed.
- F. K. Korley, G. D. Kelen, C. M. Jones and R. Diaz-Arrastia, J. Head Trauma Rehabil., 2016, 31, 379–387 CrossRef PubMed.
- A. D. Krausz, F. K. Korley and M. A. Burns, Biosensors, 2021, 11, 319 CrossRef CAS PubMed.
- J. P. Posti, R. S. K. Takala, L. Lagerstedt, A. M. Dickens, I. Hossain, M. Mohammadian, H. Ala-Seppälä, J. Frantzén, M. van Gils, P. J. Hutchinson, A. J. Katila, H.-R. Maanpää, D. K. Menon, V. F. Newcombe, J. Tallus, K. Hrusovsky, D. H. Wilson, J. Gill, J.-C. Sanchez, O. Tenovuo, H. Zetterberg and K. Blennow, J. Neurotrauma, 2019, 36, 2178–2189 CrossRef PubMed.
- L. Lagerstedt, J. J. Egea-Guerrero, A. Bustamante, A. Rodríguez-Rodríguez, A. El Rahal, M. Quintana-Diaz, R. García-Armengol, C. M. Prica, E. Andereggen and L. Rinaldi, PLoS One, 2018, 13(7), e0200394 CrossRef PubMed.
- Abbott Receives FDA 510(k) Clearance for the First Rapid Handheld Blood Test for Concussions, https://abbott.mediaroom.com/2021-01-11-Abbott-Receives-FDA-510-k-Clearance-for-the-First-Rapid-Handheld-Blood-Test-for-Concussions, (accessed March 24, 2021).
- S. Azizi, D. B. Hier, B. Allen, T. Obafemi-Ajayi, G. R. Olbricht, M. S. Thimgan and D. C. Wunsch, Front. Neurol., 2021, 12, 668606 CrossRef PubMed.
- H. Adrian, K. Marten, N. Salla and V. Lasse, eNeuro, 2016, 3(6), ENEURO.0294-16.2016 CrossRef PubMed.
- Z. Fu, W. Wei, C. Li and Z. Wang, Zhongguo Kexue: Huaxue, 2011, 41, 773–784 Search PubMed.
- J. Miranda, N. Humphrey, R. Kinney, R. O'Sullivan, B. Thomas, I. E. Mondaca Medina, R. Freedman and E. Fahrenkrug, ACS Sens., 2021, 6, 4136–4144 CrossRef CAS PubMed.
- X. Jiang, H. Wang, R. Yuan and Y. Chai, Anal. Chem., 2018, 90, 8462–8469 CrossRef CAS PubMed.
- J. Liu, J. Liang, C. Wu and Y. Zhao, Anal. Chem., 2019, 91, 6902–6909 CrossRef CAS PubMed.
- Y. Liu, H. Zhang, B. Li, J. Liu, D. Jiang, B. Liu and N. Sojic, J. Am. Chem. Soc., 2021, 143, 17910–17914 CrossRef CAS PubMed.
- L.-Y. Huang, X. Hu, H.-Y. Shan, L. Yu, Y.-X. Gu, A.-J. Wang, D. Shan, P.-X. Yuan and J.-J. Feng, Sens. Actuators, B, 2021, 344, 130300 CrossRef CAS.
- Y. Li, C. Z. Huang and Y. F. Li, Anal. Chem., 2019, 91, 9308–9314 CrossRef CAS PubMed.
- Q. Hao, Q. Xu, S. Niu, C. Ding and X. Luo, Anal. Chem., 2021, 93, 10679–10687 CrossRef CAS PubMed.
- A. J. Stewart, K. Brown and L. Dennany, Anal. Chem., 2018, 90, 12944–12950 CrossRef CAS PubMed.
- K. Brown, M. McMenemy, M. Palmer, M. J. Baker, D. W. Robinson, P. Allan and L. Dennany, Anal. Chem., 2019, 91, 12369–12376 CrossRef CAS PubMed.
- M. Bhaiyya, P. K. Pattnaik and S. Goel, Curr. Opin. Electrochem., 2021, 30, 100800 CrossRef CAS.
- X. Ying, L. Zhou, W. Fu, Y. Wang and B. Su, Sens. Diagn., 2023, 2, 480–491 RSC.
- M. Cretich, F. Damin, G. Pirri and M. Chiari, Biomol. Eng., 2006, 23, 77–88 CrossRef CAS PubMed.
- F. X. R. Sutandy, J. Qian, C. Chen and H. Zhu, Curr. Protoc. Protein Sci., 2013, 72, 2711–27116 Search PubMed.
- L. Berrade, A. E. Garcia and J. A. Camarero, Pharm. Res., 2011, 28, 1480–1499 CrossRef CAS PubMed.
- Developments and Applications of Functional Protein Microarrays*|Elsevier Enhanced Reader, (accessed September 20, 2022).
- M. Jović, D. Prim, E. Saini and M. E. Pfeifer, Biosensors, 2022, 12, 172 CrossRef PubMed.
- L. D'Alton, S. Carrara, G. J. Barbante, D. Hoxley, D. J. Hayne, P. S. Francis and C. F. Hogan, Bioelectrochemistry, 2022, 146, 108107 CrossRef PubMed.
- S. Li, D. Zhang, J. Liu, C. Cheng, L. Zhu, C. Li, Y. Lu, S. S. Low, B. Su and Q. Liu, Biosens. Bioelectron., 2019, 129, 284–291 CrossRef CAS PubMed.
- F. Du, Z. Dong, Y. Guan, A. M. Zeid, D. Ma, J. Feng, D. Yang and G. Xu, Anal. Chem., 2022, 94, 2189–2194 CrossRef CAS PubMed.
- S. Li, J. Liu, Z. Chen, Y. Lu, S. S. Low, L. Zhu, C. Cheng, Y. He, Q. Chen, B. Su and Q. Liu, Sens. Actuators, B, 2019, 297, 126811 CrossRef CAS.
- J. L. Delaney, C. F. Hogan, J. Tian and W. Shen, Anal. Chem., 2011, 83, 1300–1306 CrossRef CAS PubMed.
- S. Li, Y. Lu, L. Liu, S. S. Low, B. Su, J. Wu, L. Zhu, C. Li and Q. Liu, Sens. Actuators, B, 2019, 285, 34–41 CrossRef CAS.
- T. Liu, J. He, Z. Lu, M. Sun, M. Wu, X. Wang, Y. Jiang, P. Zou, H. Rao and Y. Wang, Chem. Eng. J., 2022, 429, 132462 CrossRef CAS.
- H. J. Kwon, E. C. Rivera, M. R. C. Neto, D. Marsh, J. J. Swerdlow, R. L. Summerscales and P. P. Tadi Uppala, Results Chem., 2020, 2, 100029 CrossRef CAS.
- M. M. P. S. Neves, P. Bobes-Limenes, A. Pérez-Junquera, M. B. González-García, D. Hernández-Santos and P. Fanjul-Bolado, Anal. Bioanal. Chem., 2016, 408, 7121–7127 CrossRef CAS PubMed.
- L. Zhu, W. Fu, J. Chen, S. Li, X. Xie, Z. Zhang, J. Liu, L. Zhou, B. Su and X. Chen, Sens. Actuators, B, 2022, 366, 131972 CrossRef CAS.
- R. Tan, Y. Shen, D. Li, Y. Yang and Y. Tu, Electroanalysis, 2022, 34, 423–431 CrossRef CAS.
- Y. Chen, J. Wang, Z. Liu, X. Wang, X. Li and G. Shan, Biochem. Eng. J., 2018, 129, 1–6 CrossRef CAS.
- W. Li, M. Li, S. Ge, M. Yan, J. Huang and J. Yu, Anal. Chim. Acta, 2013, 767, 66–74 CrossRef CAS PubMed.
- M. Bhaiyya, M. B. Kulkarni, P. K. Pattnaik and S. Goel, Luminescence, 2022, 37, 357–365 CrossRef CAS PubMed.
- Y. Fan, J. Li, Y. Guo, L. Xie and G. Zhang, Measurement, 2021, 171, 108829 CrossRef.
- K. G. Shah, V. Singh, P. C. Kauffman, K. Abe and P. Yager, Anal. Chem., 2018, 90, 6967–6974 CrossRef CAS PubMed.
- R. Ekins, in The Immunoassay Handbook, ed. D. Wild, Elsevier, Oxford, 4th edn, 2013, pp. 109–121 Search PubMed.
- L. B. Xu, J. K. Yue, F. Korley, A. M. Puccio, E. L. Yuh, X. Sun, M. Rabinowitz, M. J. Vassar, S. R. Taylor, E. A. Winkler, R. C. Puffer, H. Deng, M. McCrea, M. B. Stein, C. S. Robertson, H. S. Levin, S. Dikmen, N. R. Temkin, J. T. Giacino, P. Mukherjee, K. K. W. Wang, D. O. Okonkwo, A. J. Markowitz, S. Jain, G. T. Manley and R. Diaz-Arrastia, J. Neurotrauma, 2021, 38, 918–927 CrossRef PubMed.
- T. Shetty, T. Cogsil, A. Dalal, E. Kim, K. Halvorsen, K. Cummings and J. T. Nguyen, J. Head Trauma Rehabil., 2019, 34, E28 CrossRef PubMed.
- K. Visser, M. Koggel, J. Blaauw, H. J. van der Horn, B. Jacobs and J. van der Naalt, Neurosci. Biobehav. Rev., 2022, 132, 154–168 CrossRef CAS PubMed.
- R. Sharma, A. Rosenberg, E. R. Bennett, D. T. Laskowitz and S. K. Acheson, PLoS One, 2017, 12, 1–18 Search PubMed.
- W. Miao, Chem. Rev., 2008, 108, 2506–2553 CrossRef CAS PubMed.
- Y. Zu and A. J. Bard, Anal. Chem., 2001, 73, 3960–3964 CrossRef CAS PubMed.
- J. J. Bazarian, P. Biberthaler, R. D. Welch, L. M. Lewis, P. Barzo, V. Bogner-Flatz, P. G. Brolinson, A. Büki, J. Y. Chen, R. H. Christenson, D. Hack, J. S. Huff, S. Johar, J. D. Jordan, B. A. Leidel, T. Lindner, E. Ludington, D. O. Okonkwo, J. Ornato, W. F. Peacock, K. Schmidt, J. A. Tyndall, A. Vossough and A. S. Jagoda, Lancet Neurol., 2018, 17, 782–789 CrossRef CAS PubMed.
- L. Lagerstedt, J. J. Egea-Guerrero, A. Bustamante, J. Montaner, A. Rodriguez-Rodriguez, A. El Rahal, N. Turck, M. Quintana, R. Garcia-Armengol, C. M. Prica, E. Andereggen, L. Rinaldi, A. Sarrafzadeh, K. Schaller and J.-C. Sanchez, PLoS One, 2017, 12, 1–11 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: SI-1: schematic representation of prototype design; SI-2: CAD drawings of the prototype and it's components; SI-3: software interface; SI-4: concept of the ECL microarray; SI-5: immunoassay conditions; SI-6: brief overview of recent publications reported on miniaturized ECL systems/devices. See DOI: https://doi.org/10.1039/d3sd00090g |
| This journal is © The Royal Society of Chemistry 2023 |