Open Access Article
Open Access ArticleA simple copper(II) dppy-based receptor for sensing of L-cysteine and L-histidine in aqueous acetonitrile medium†
Dipankar
Das
 a,
Aritra
Roy‡
a,
Aritra
Roy‡
 b,
Sourav
Sutradhar
b,
Sourav
Sutradhar
 a,
Felipe
Fantuzzi
a,
Felipe
Fantuzzi
 *c and
Biswa Nath
Ghosh
*c and
Biswa Nath
Ghosh
 *a
*a
aDepartment of Chemistry, National Institute of Technology Silchar, Silchar-788010, Assam, India. E-mail: bnghosh@che.nits.ac.in; Tel: +91 801 812 3682
bDepartment of Chemistry, Pondicherry University, Pondicherry 605014, India
cSchool of Chemistry and Forensic Science, University of Kent, Park Wood Rd, Canterbury CT2 7NH, UK. E-mail: f.fantuzzi@kent.ac.uk
First published on 21st October 2023
Abstract
The development of simple yet efficient receptors that rapidly detect and monitor amino acids with high sensitivity and reliability is crucial for the early-stage identification of various diseases. In this work, we report the synthesis and characterisation of a copper(II) complex, CuCl2L, by employing a 2,6-dipyrazinylpyridine (dppy)-based ligand (L = 2,2′-(4-(3,4,5-trimethoxyphenyl)pyridine-2,6-diyl)dipyrazine). The in situ prepared CuCl2L receptor exhibits an instantaneous response to the presence of L-cysteine (Cys) and L-histidine (His) in aqueous acetonitrile (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer, pH 7.4). Furthermore, competitive experiments demonstrate the selectivity of CuCl2L towards Cys (1 equiv.) in the vicinity of other L-amino acids in the aforementioned solvent conditions. The detection limits for Cys and His are calculated as 0.33 μM and 1.40 μM, respectively. DFT calculations offer a plausible explanation for the observed selectivity of the CuCl2L receptor towards Cys and His. They reveal that the most stable conformer of Cu
1 v/v, 10 mM HEPES buffer, pH 7.4). Furthermore, competitive experiments demonstrate the selectivity of CuCl2L towards Cys (1 equiv.) in the vicinity of other L-amino acids in the aforementioned solvent conditions. The detection limits for Cys and His are calculated as 0.33 μM and 1.40 μM, respectively. DFT calculations offer a plausible explanation for the observed selectivity of the CuCl2L receptor towards Cys and His. They reveal that the most stable conformer of Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cys complex (1
Cys complex (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) is a five-membered ring formed through N,S-coordination mode (ΔG = −26.7 kcal mol−1) over various other possible coordination modes, while comparable ΔG values are only obtained for Cu
1) is a five-membered ring formed through N,S-coordination mode (ΔG = −26.7 kcal mol−1) over various other possible coordination modes, while comparable ΔG values are only obtained for Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) His complexes featuring two His moieties.
His complexes featuring two His moieties.
Introduction
L-Cysteine (Cys), one of the three naturally occurring biothiols, is a crucial amino acid that acts as an antioxidant to protect cells and tissues from oxidation by free radicals and reactive oxygen species.1,2 It is also the only amino acid that participates in peptide and protein biosynthesis and plays a key role in enzyme-active sites.2–5 The deficiency of Cys can lead to various health issues, such as liver damage,6 hair depigmentation,7 skin lesions,8 edema,9 lethargy,10 child growth retardation,11 and so on. In turn, an elevated level of Cys can result in neurological,12 and cardiovascular diseases.13,14L-Histidine (His), another essential amino acid, performs vital functions in the nervous system,15 including serving as a neurotransmitter,16 and promoting tissue growth and repair.17 His deficiency can lead to kidney disease,18 Parkinson's disease,19 epilepsy,20 and other disorders. On the other hand, an elevated level of His can result in liver cirrhosis,21 asthma,22 and other conditions.23Designing a receptor with a fast response, high sensitivity, and reliability for detecting and monitoring Cys and His concentrations can potentially aid in the early-stage recognition of various diseases.24–28 In this context, several receptor analogues have been proposed for Cys and His recognition, including Schiff base,29 1,8-naphthalimide,30 benzothiazole,31 coumarin,32 fluorescein,33 BODIPY,34 α,β-unsaturated ketone,35 rhodamine,36 imidazole,37 nanomaterials based receptors,38 metal–organic frameworks (MOFs),39etc. Various detection techniques have been employed for Cys and His detection, such as UV-visible spectroscopy,40,41 fluorescence spectroscopy,42 flow injection,43 capillary electrophoresis,44 Raman microspectroscopy,45 liquid chromatography,46 voltametric,47 mass spectrometry,48etc. Recently, nitrogen-based heterocyclic ligands such as 2,2′:6′,2′′-terpyridine, 2,6-dipyrazinylpyridine (dppy), and their transition metal complexes have garnered significant attention due to their easy one-pot synthesis, high stability, fascinating electrochemical and photophysical properties, and potential physiological activities.49–53 These ligands have been incorporated into various applications, including self-assembly,54 hydrogelation,55–57 halogen bonding,58,59 and anion sensing,60,61etc.
Most of the existing receptor analogues (non-metal complexes) for Cys sensing suffer from severe disadvantages, including laborious synthetic processes30,31,34–36,62,63 and high response time.33,64–66 Very few receptor systems (based on metal complexes, especially copper) reported Cys and His sensing over other L-amino acids, with a low detection limit and response time; however, the exact binding interaction mode of Cys with copper complexes has not been provided.1,9,104 Herein, we present a novel copper(II) complex (CuCl2L) featuring a one-pot, readily synthesisable dppy-based ligand (L = 2,2′-(4-(3,4,5-trimethoxyphenyl)pyridine-2,6-diyl)dipyrazine). This complex is designed for the immediate sensing of Cys and His over sixteen other L-amino acids (1 equiv.), seventeen anions (10 equiv.), and nine metal ions (1 equiv.). The detection is achieved at physiological pH (7.4) in aqueous acetonitrile (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer) with a comparatively low detection limit for Cys (0.33 μM) and His (1.40 μM), respectively, as determined through absorption spectral analysis. Additionally, we have also conducted DFT studies to i) assess the potential displacement of Cu(II) from CuCl2L for the sensing of Cys and His and ii) investigate the optimal binding modes of Cu(II) interaction with Cys and His in an aqueous acetonitrile medium. To the best of our knowledge, this is the first instance of a substituted 2,6-dipyrazinylpyridine (dppy) based receptor system reported for L-amino acid sensing.
1 v/v, 10 mM HEPES buffer) with a comparatively low detection limit for Cys (0.33 μM) and His (1.40 μM), respectively, as determined through absorption spectral analysis. Additionally, we have also conducted DFT studies to i) assess the potential displacement of Cu(II) from CuCl2L for the sensing of Cys and His and ii) investigate the optimal binding modes of Cu(II) interaction with Cys and His in an aqueous acetonitrile medium. To the best of our knowledge, this is the first instance of a substituted 2,6-dipyrazinylpyridine (dppy) based receptor system reported for L-amino acid sensing.
Experimental section
Materials and methods
The spectroscopic and analytical grade chemicals used in the synthesis and spectral analyses were procured commercially. The ligand 2,2′-(4-(3,4,5-trimethoxyphenyl)pyridine-2,6-diyl)dipyrazine L has been prepared following the literature method.67 3000 Hyperion FT-IR spectrometer (Bruker), ECZ500R/S1 (JEOL), and G2-XS QTOF mass spectrometer (XEVO), Thermo Electron Flash EA 1112 series were used to obtain FT-IR, 1H NMR and HRMS and CHN analysis respectively. A Motras Scientific UV plus MSGUI3.1.0 absorption spectrophotometer was used to record absorption spectra.Preparation of L
To an ethanolic solution (20 mL) of 2-acetylpyrazine (1.221 g, 10 mmol), KOH pellets (0.561 g, 10 mmol) were added and stirred, followed by the addition of 3,4,5-trimethoxybenzaldehyde (0.981 g, 5 mmol) and aqueous ammonia solution (15 mL). Stirring the resultant mixture at room temperature for 8 hours yielded a crude precipitate, which was separated by filtration and washed with ethanol (50 mL). The precipitate was dissolved in chloroform (10 mL), and an excess of n-hexane (80 mL) was added to it to obtain the white precipitate of L. The precipitate was filtered, washed with n-hexane, and dried. Yield: 0.802 g (2 mmol, 40%). 1H NMR (500 MHz, CDCl3) δ/ppm: 3.93 (s, 3H), 4.00 (s, 6H), 7.02 (s, 2H), 8.65–8.68 (m, 6H), 9.87 (s, 2H). 13C (125 MHz, CDCl3) δ/ppm: 56.5, 61.13, 104.65, 119.87, 133.77, 139.44, 143.65, 143.77, 144.92, 150.81, 151.02, 153.89, 154.42. ESI-MS [L + H]+m/z 402.20. Anal. calcd. C22H19N5O3 (401.426 g mol−1): C, 65.83; H, 4.77; N, 17.45. Found: C, 65.58; H, 4.72; N, 17.55.Preparation of CuCl2L
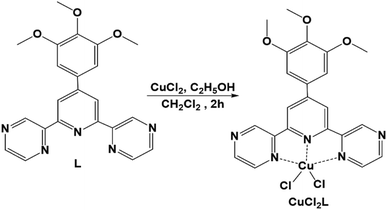
L (0.08 g, 0.2 mmol) was dissolved in dichloromethane (10 mL), and then 10 mL of ethanolic solution of copper chloride (0.027 g, 0.2 mmol) was added to it. Stirring the resultant mixture at room temperature for 2 hours afforded a green color precipitate. The precipitate was filtered off, washed with ethanol (20 mL) and diethyl ether (20 mL), and then dried to get the green-colored copper complex CuCl2L. Yield: 0.085 g (85%). ESI-MS [CuClL]+m/z 499.0720. Anal. calcd. C22H19Cl2CuN5O3 (535.872 g mol−1): C, 49.31; H, 3.57; N, 13.07. Found: C, 49.10 H, 3.49; N, 13.15.Preparation of analyte solutions for UV-vis absorption spectral study
An acetonitrile solution of L (200 μM, 25 mL), an aqueous solution of CuCl2 (1 mM, 10 mL), and aqueous solutions of glycine (Gly) and eighteen L-amino acids, namely alanine (Ala), aspartic acid (Asp), histidine (His), arginine (Arg), asparagine (Asn), cysteine (Cys), glutamic acid (Glu), methionine (Met), lysine (Lys), isoleucine (Ile), serine (Ser), proline (Pro), tryptophan (Trp), phenylalanine (Phe), valine (Val), leucine (Leu), threonine (Thr), and tyrosine (Tyr) (1 mM, 50 mL) were prepared separately. The CuCl2L receptor solution was prepared in situ by mixing L (200 μM, 6 mL) and CuCl2 (1 mM, 1.2 mL), followed by dilution to an aqueous acetonitrile HEPES buffer (10 mM, 32 mL, pH 7.4).Computational details
All quantum chemical calculations were conducted using the Gaussian 16, Revision C.01 program package.68 Geometry optimisations were carried out using the PBE069,70 functional in combination with the D3(BJ) method71,72 for dispersion corrections. The basis set employed for all atoms, except Cu, was def2-SVP, while the triple-zeta def2-TZVP basis set73 was used for Cu. This specific combination of basis sets is denoted as bs1; thus, the corresponding level of theory is referred to as PBE0-D3(BJ)/bs1. For open-shell systems, the unrestricted Kohn–Sham formalism was employed. For the calculation of the free energy values, Gibbs corrections at the PBE0-D3(BJ)/bs1 level were applied to single point energy calculations by utilising the same PBE0-D3(BJ) method but with a larger basis set. Specifically, the def2-TZVP basis set was used for all atoms, except for Cu, where the quadruple-zeta def2-QZVP basis set was employed. This combination of basis sets is denoted as bs2. Solvation effects were incorporated using the solvent model based on density (SMD),74 with a solvent mixture of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) water/acetonitrile considered in the calculations. The energy calculations were, therefore, performed at the SMD/PBE0-D3(BJ)/bs2 level of theory. A concentration correction of ΔG0→* = 1.89 kcal mol−1 was applied to the free energy values of all species to account for the change in standard states when transitioning from the gas phase (1 atm) to the condensed phase (1 M).75–77 This correction ensures an accurate description of associative and dissociative steps. All the optimised geometries were characterised as minima on the corresponding potential energy surfaces by performing vibrational frequency calculations, confirming only positive eigenvalues in the Hessian matrices. The selected levels of theory for geometry optimisation and free energy calculations were benchmarked against other DFT functionals and basis sets, consistently yielding similar results. To ensure the identification of the global minimum energy structures, different starting structures were considered for all geometries. Finally, time-dependent DFT (TD-DFT) calculations with 20 states were conducted to describe the electronic excitation features of CuCl2L. The resulting data were further analysed using the Multiwfn 3.8 program.78
1 (v/v) water/acetonitrile considered in the calculations. The energy calculations were, therefore, performed at the SMD/PBE0-D3(BJ)/bs2 level of theory. A concentration correction of ΔG0→* = 1.89 kcal mol−1 was applied to the free energy values of all species to account for the change in standard states when transitioning from the gas phase (1 atm) to the condensed phase (1 M).75–77 This correction ensures an accurate description of associative and dissociative steps. All the optimised geometries were characterised as minima on the corresponding potential energy surfaces by performing vibrational frequency calculations, confirming only positive eigenvalues in the Hessian matrices. The selected levels of theory for geometry optimisation and free energy calculations were benchmarked against other DFT functionals and basis sets, consistently yielding similar results. To ensure the identification of the global minimum energy structures, different starting structures were considered for all geometries. Finally, time-dependent DFT (TD-DFT) calculations with 20 states were conducted to describe the electronic excitation features of CuCl2L. The resulting data were further analysed using the Multiwfn 3.8 program.78
Results and discussion
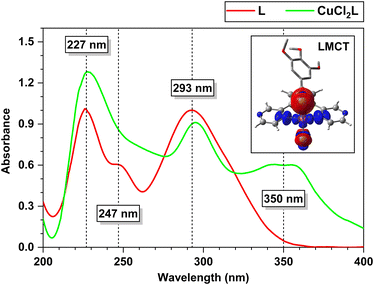
The experimental protocol of preparation of L and its Cu(II) complex CuCl2L are shown in Scheme S1 in the ESI† and Scheme 1, respectively. L and CuCl2L have been characterised using 1H NMR, HRMS, elemental analysis, ESR, FT-IR, and the corresponding spectra can be found in ESI† (Fig. S1–S8). While we could not obtain an X-ray crystal structure for CuCl2L, we have carried out DFT calculations which suggest favourable formation of CuCl2L from isolated L and CuCl2 moieties in aqueous acetonitrile solution (ΔG: −32.9 kcal mol−1). In CuC2L, the Cu(II) interacts with three donor nitrogen atoms of L, resulting in a penta-coordinate complex with a distorted square-pyramidal geometry (angular structural index parameter, τ = 0.08).79–82 Similar structures have been observed in the case of copper complexes with analogous ligands,79,81–83 many of which have been fully characterised by X-ray diffraction analysis.Compound L (200 μM, 300 μL) exhibits absorption maxima at 227 nm and 293 nm in aqueous acetonitrile (2 mL, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer, pH 7.4) (Fig. 1). The absorption maximum at 293 nm corresponds to an n → π* transition.67,84,85 The addition of one equiv. of aqueous CuCl2 (1 mM, 60 μL) to the solution of L (200 μM, 300 μL) in aqueous acetonitrile (2 mL, 4
1 v/v, 10 mM HEPES buffer, pH 7.4) (Fig. 1). The absorption maximum at 293 nm corresponds to an n → π* transition.67,84,85 The addition of one equiv. of aqueous CuCl2 (1 mM, 60 μL) to the solution of L (200 μM, 300 μL) in aqueous acetonitrile (2 mL, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
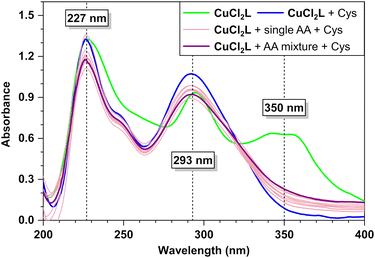
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer, pH 7.4) resulted in the appearance of a new absorption band at 350 nm (Fig. 1). The absorption band at 350 nm, attributed to a ligand-to-metal charge transfer (LMCT) transition,86–88 indicates the in situ formation of copper(II) complex CuCl2L. The stability constant of this complex is determined to be 7.285 × 104 M−1, calculated from B–H plot (Fig. S9 and S10 in the ESI†). The assignment of these transitions is in agreement with our TD-DFT calculations (see ESI† for more details). Specifically, the charge density difference (CDD)89–91 plot (Fig. 1) demonstrates that the band at 350 nm is primarily attributed to a Cl-to-Cu LMCT transition, accompanied by a minor contribution from ligand-to-ligand charge transfer. The findings from the interfragment charge transfer (IFCT) analysis78 further support these results, revealing an overall charge transfer character of 82% for the 350 nm band. Within this, the Cl ligands contribute to 87% of the hole density, while Cu and the dppy ligand contribute 68% and 18% of the electron density, respectively.
1 v/v, 10 mM HEPES buffer, pH 7.4) resulted in the appearance of a new absorption band at 350 nm (Fig. 1). The absorption band at 350 nm, attributed to a ligand-to-metal charge transfer (LMCT) transition,86–88 indicates the in situ formation of copper(II) complex CuCl2L. The stability constant of this complex is determined to be 7.285 × 104 M−1, calculated from B–H plot (Fig. S9 and S10 in the ESI†). The assignment of these transitions is in agreement with our TD-DFT calculations (see ESI† for more details). Specifically, the charge density difference (CDD)89–91 plot (Fig. 1) demonstrates that the band at 350 nm is primarily attributed to a Cl-to-Cu LMCT transition, accompanied by a minor contribution from ligand-to-ligand charge transfer. The findings from the interfragment charge transfer (IFCT) analysis78 further support these results, revealing an overall charge transfer character of 82% for the 350 nm band. Within this, the Cl ligands contribute to 87% of the hole density, while Cu and the dppy ligand contribute 68% and 18% of the electron density, respectively.
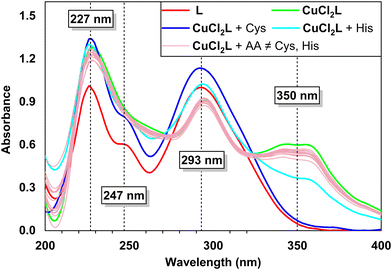
Aqueous solutions of Gly and various L-amino acids (Ala, Asp, His, Arg, Asn, Cys, Glu, Met, Lys, Ile, Ser, Pro, Trp, Val, Leu, Phe, Thr, and Tyr) at a concentration of 1 mM (60 μL) were individually added to solutions of the in situ prepared CuCl2L (30 μM, 2 mL) in aqueous acetonitrile (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer, pH 7.4), and the corresponding absorption spectra were recorded. It was observed that the absorption spectrum of the copper(II) complex (CuCl2L) remained unchanged upon the addition of most amino acid solutions, except for Cys and His (see Fig. 2). Specifically, when one equiv. of Cys was added to the copper complex solution, the 350 nm absorption band of CuCl2L disappeared, resulting in absorption spectra closely resembling that of the free ligand L (Fig. 2). This suggests that Cys displaced CuCl2 from the CuCl2L receptor previously formed upon adding L to the copper solution. Additionally, the 350 nm absorption band of CuCl2L underwent a hypochromic shift when one equiv. of His was added to the receptor solution (Fig. 2), indicating the sensitivity of CuCl2L towards His, along with Cys. Notably, almost 4 equiv. of His (1 mM, ∼200 μL) were required to perturb the 350 nm absorption band of CuCl2L completely. Furthermore, the detection study showed that in situ prepared CuCl2L is most effective in the pH range of 4 to 9 for Cys detection, while pH 6.5 to 9 is most effective for His (1 equiv.) sensing (see details in Fig. S13 in the ESI†).
1 v/v, 10 mM HEPES buffer, pH 7.4), and the corresponding absorption spectra were recorded. It was observed that the absorption spectrum of the copper(II) complex (CuCl2L) remained unchanged upon the addition of most amino acid solutions, except for Cys and His (see Fig. 2). Specifically, when one equiv. of Cys was added to the copper complex solution, the 350 nm absorption band of CuCl2L disappeared, resulting in absorption spectra closely resembling that of the free ligand L (Fig. 2). This suggests that Cys displaced CuCl2 from the CuCl2L receptor previously formed upon adding L to the copper solution. Additionally, the 350 nm absorption band of CuCl2L underwent a hypochromic shift when one equiv. of His was added to the receptor solution (Fig. 2), indicating the sensitivity of CuCl2L towards His, along with Cys. Notably, almost 4 equiv. of His (1 mM, ∼200 μL) were required to perturb the 350 nm absorption band of CuCl2L completely. Furthermore, the detection study showed that in situ prepared CuCl2L is most effective in the pH range of 4 to 9 for Cys detection, while pH 6.5 to 9 is most effective for His (1 equiv.) sensing (see details in Fig. S13 in the ESI†).
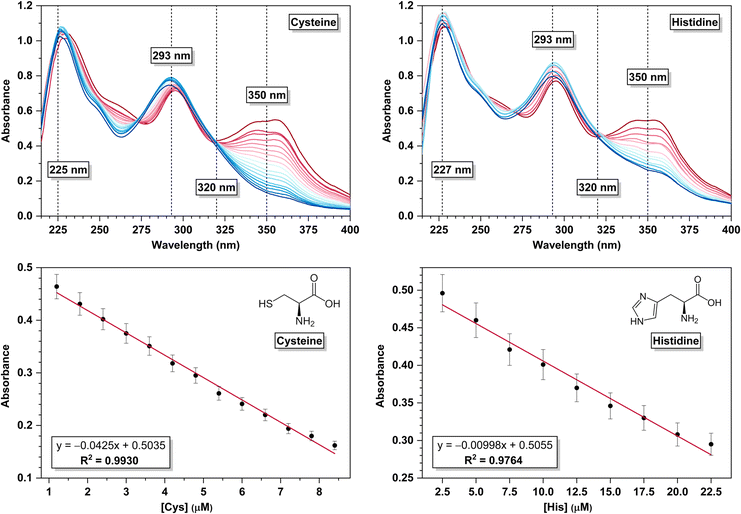
A UV-vis absorption spectral titration was carried out by gradually adding an aqueous solution of Cys (0.2 mM, 6 μL) to the in situ prepared CuCl2L receptor (30 μM, 2 mL) in aqueous acetonitrile (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES, pH 7.4). The absorption band at 293 nm of CuCl2L exhibited gradual hyperchromic shifts, and the 350 nm absorption band underwent gradual hypochromic shifts as Cys solution was added incrementally to the CuCl2L receptor (see Fig. 3a, top left). The presence of an isosbestic point at 320 nm for the above titration indicated the existence of an equilibrium between the CuCl2L receptor and free L along with the Cu
1 v/v, 10 mM HEPES, pH 7.4). The absorption band at 293 nm of CuCl2L exhibited gradual hyperchromic shifts, and the 350 nm absorption band underwent gradual hypochromic shifts as Cys solution was added incrementally to the CuCl2L receptor (see Fig. 3a, top left). The presence of an isosbestic point at 320 nm for the above titration indicated the existence of an equilibrium between the CuCl2L receptor and free L along with the Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cys moiety. Similar observations were made in a UV-vis absorption spectral titration conducted by gradually adding His (0.5 mM, 5 μL) to the CuCl2L receptor (30 μM, 2 mL) (see Fig. 3, top right). Both absorption spectral titrations demonstrate the sensitivity of the CuCl2L receptor towards minute changes in Cys and His concentrations. The detection limits92–94 for Cys and His were calculated to be 0.33 μM and 1.40 μM, respectively, indicating the high sensitivity of the CuCl2L receptor for these analytes. The linear relationships found between the absorbance and concentration of Cys and His are shown in Fig. 3, bottom left and right, respectively.
Cys moiety. Similar observations were made in a UV-vis absorption spectral titration conducted by gradually adding His (0.5 mM, 5 μL) to the CuCl2L receptor (30 μM, 2 mL) (see Fig. 3, top right). Both absorption spectral titrations demonstrate the sensitivity of the CuCl2L receptor towards minute changes in Cys and His concentrations. The detection limits92–94 for Cys and His were calculated to be 0.33 μM and 1.40 μM, respectively, indicating the high sensitivity of the CuCl2L receptor for these analytes. The linear relationships found between the absorbance and concentration of Cys and His are shown in Fig. 3, bottom left and right, respectively.
To assess the selectivity of our receptor towards Cys, we conducted competition experiments, whose results are shown in Fig. 4. Initially, aqueous solutions of various amino acids (excluding Cys) at a concentration of 1 mM (60 μL) were individually added to distinct CuCl2L solutions (30 μM, 2 mL) in aqueous acetonitrile (2 mL, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer, pH 7.4), followed by the addition of one equiv. of Cys (1 mM, 60 μL) to each of these above mixtures. Interestingly, the 350 nm absorption band of CuCl2L, which remained largely unaffected in the presence of other amino acids, disappeared upon addition of one equiv. of Cys. Similarly, when a mixture of different amino acids (60 μL) (excluding Cys) at a concentration of 1 mM was added collectively to the CuCl2L solution (30 μM, 2 mL), followed by the addition of one equiv. of Cys (1 mM, 60 μL), we observed a similar outcome (see Fig. 4). These findings provide compelling evidence that the CuCl2L receptor selectively detected Cys even in the presence of other L-amino acids.
1 v/v, 10 mM HEPES buffer, pH 7.4), followed by the addition of one equiv. of Cys (1 mM, 60 μL) to each of these above mixtures. Interestingly, the 350 nm absorption band of CuCl2L, which remained largely unaffected in the presence of other amino acids, disappeared upon addition of one equiv. of Cys. Similarly, when a mixture of different amino acids (60 μL) (excluding Cys) at a concentration of 1 mM was added collectively to the CuCl2L solution (30 μM, 2 mL), followed by the addition of one equiv. of Cys (1 mM, 60 μL), we observed a similar outcome (see Fig. 4). These findings provide compelling evidence that the CuCl2L receptor selectively detected Cys even in the presence of other L-amino acids.
To elucidate the selectivity mechanism of CuCl2L towards Cys and His binding, we conducted additional DFT calculations (vide supra) on distinct Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cys and Cu
Cys and Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) His complexes. The results are summarised here, with more details given in the ESI.† Aligned with our experimental results, a 1
His complexes. The results are summarised here, with more details given in the ESI.† Aligned with our experimental results, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio was used for Cu
1 stoichiometric ratio was used for Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cys complexes, while both 1
Cys complexes, while both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratios were explored for Cu
2 ratios were explored for Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) His complexes. In our calculations, we considered that the SH and NH3+ groups of Cys undergo deprotonation upon metal binding, as observed in other metal–Cys complexes,95 resulting in a doubly anionic [Cys]2− ligand. Indeed, our preliminary calculations considering the interaction of CuCl2 with Cys at distinct charge states revealed that the complexes with doubly anionic [Cys]2− ligands are those with the most negative binding free energies (see Table S2 in the ESI† for further details). The [Cys]2− structure was obtained by considering the most stable zwitterionic form of Cys as described by Fernández-Ramos et al.96 and removing the appropriate protons as the starting point for the geometry optimisation calculations. The most stable structure identified for [Cys]2− (I, see ESI†) was obtained through a systematic conformer search. This structure exhibits intramolecular N–H⋯O and N–H⋯S hydrogen bonds, which contribute to its stability. Notably, I is merely 0.4 kcal mol−1 more stable than that proposed by Foley and Enescu (II, see ESI†).95
His complexes. In our calculations, we considered that the SH and NH3+ groups of Cys undergo deprotonation upon metal binding, as observed in other metal–Cys complexes,95 resulting in a doubly anionic [Cys]2− ligand. Indeed, our preliminary calculations considering the interaction of CuCl2 with Cys at distinct charge states revealed that the complexes with doubly anionic [Cys]2− ligands are those with the most negative binding free energies (see Table S2 in the ESI† for further details). The [Cys]2− structure was obtained by considering the most stable zwitterionic form of Cys as described by Fernández-Ramos et al.96 and removing the appropriate protons as the starting point for the geometry optimisation calculations. The most stable structure identified for [Cys]2− (I, see ESI†) was obtained through a systematic conformer search. This structure exhibits intramolecular N–H⋯O and N–H⋯S hydrogen bonds, which contribute to its stability. Notably, I is merely 0.4 kcal mol−1 more stable than that proposed by Foley and Enescu (II, see ESI†).95
Regarding the metal site, we examined both the bare, open-shell Cu(II) ion and the neutral, open-shell CuCl2 moiety in our calculations. Consequently, we focused our investigations on the structure of [CuCys] and [CuCysCl2]2− complexes. Whenever appropriate, we compared our [CuCys] results with those of [CuCys]2+ by Belcastro and co-workers.97 Regarding His, we only considered the R–CH(NH2)–COO− anionic state. As a result, we thoroughly examined the characteristics of [CuHisCl2]− and [Cu(His)2] complexes. To explore the preference for chloride-bearing complexes in comparison to those without these ions, we conducted free energy calculations on a series of reactions (see ESI† for more details). Specifically, our results strongly indicate that the Cl groups remain bound to Cu following coordination with Cys, with the [CuCysCl2]2− structure featuring the N,S-coordination (Fig. 5A) being the most stable one. This aligns well with the experimental data, which also supports the presence of such coordination. In turn, unlike Cys, our findings suggest that His exhibits a preference for forming complexes with the bare Cu(II) ion rather than CuCl2. The most stable structure of [Cu(His)2] features a mixed configuration (Fig. 5B), where one His works as a tridentate ligand with N,N,O-coordination, and the other as a bidentate ligand through the amino and carboxyl groups (N,O-coordination). Our computational findings substantiate the observed differences in the stoichiometric ratio between copper cysteine and histidine complexes. Furthermore, they provide compelling evidence that the selectivity of CuCl2L in amino acid sensing is attributed to the exceptionally stable coordination modes formed between CuCl2 and the cysteine and histidine residues.
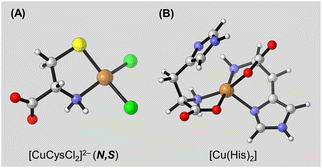 | ||
| Fig. 5 Most stable structures of (A) [CuCysCl2]2− and (B) [Cu(His)2] at the SMD/PBE0-D3(BJ)/bs2 level of theory. Geometries were optimised at the PBE0-D3(BJ)/bs1 level. For low-lying isomers, see Fig. S14 and S18 in the ESI.† Gray: carbon; white: hydrogen; red: oxygen; blue: nitrogen; green: chlorine; orange: copper. | ||
The detection limits and detection conditions achieved for Cys and His using the copper(II) complex in our work are comparable to those reported in previous studies involving different receptor systems, as summarised in Table 1. These results highlight the effectiveness of our receptor in detecting and monitoring Cys and His. Moreover, our sensing studies demonstrate an immediate response time, notably faster than most previously reported systems. This rapid response further emphasises the efficiency and reliability of our copper(II) complex as a sensing tool for the identification of Cys and His in various applications.
| Amino acid | Probe No. | Receptor probe | Conditions/detection medium | Detection limit (approx.) | Response time (approx.) | Ref. |
|---|---|---|---|---|---|---|
| L-Cysteine | 01 | Cu2+ based 4-(2-pyridylazo) resorcinol dye | H2O | 0.07 μM | 30 s | 1 |
| 02 | Cu2+ based zwitterionic chromophore dye | DMF | 4.07 μM | 10 s | 9 | |
| 03 | Chloropropionate-caged fluorescein probe | H2O/DMSO (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v, PBS, pH 7.4) 2, v/v, PBS, pH 7.4) |
12.8 μM | 10 min | 33 | |
| 04 | BODIPY-based receptor | CH3CN/HEPES buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, 20 mM, pH 7.4) 1, v/v, 20 mM, pH 7.4) |
78 nM | 20 min | 98 | |
| 05 | Two-photon fluorescent probe | DMF/PBS buffer (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, v/v, 10 mM, pH 7.4) 7, v/v, 10 mM, pH 7.4) |
— | 30 min | 99 | |
| 06 | Sulfonyl benzoxadiazole (SBD) dye having a chloride unit | DMF/PBS buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v, 10 mM, pH 7.4) 9, v/v, 10 mM, pH 7.4) |
12.21 nM | 20 min | 100 | |
| 07 | Imidazo[1,2-a]pyridine based probe having a acryloyl group | DMSO/HEPES buffer (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v, pH 7.4) 2, v/v, pH 7.4) |
0.33 μM | 12 min | 101 | |
| 08 | Silver-ion-mediated Mg2+-dependent DNAzyme | HEPES buffer (25 mM, pH 7.2) containing NaNO3 (100 mM) and Mg(NO3)2 (10 mM) | 2 nM | 30 min | 102 | |
| 09 | Two-photon ratiometric fluorescent probe (DNEPI) | DMSO–PBS buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, pH 7.4) 1, v/v, pH 7.4) |
0.29 μM | 2 min | 103 | |
| 10 | Imidazopyridine-based receptor | HEPES buffer (pH 7.34) | 99 nM | Immediate | 104 | |
| 11 | Acrylate group-based dye | HEPES/DMSO buffer (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, 10 mM, pH 7.4) 1, v/v, 10 mM, pH 7.4) |
0.16 μM | ∼3 min | 105 | |
| 12 | Boron dipyrromethene-based probe | CH3CN/HEPES buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, pH 7.4) 1, v/v, pH 7.4) |
0.857 μM | 30 min | 64 | |
| 13 | Flavone-based ESIPT ratiometric chemodosimeter | CH3CN/HEPES buffer (10 mM, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) 1, v/v) |
1 μM | 60 min | 65 | |
| 14 | Fluorescein and rhodamine B-based chemosensor | CH3CN/HEPES buffer (10 mM, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v, pH 7.4) 4, v/v, pH 7.4) |
88 nM | 20 min | 106 | |
| 15 | Nitroolefin-based coumarin | CH3CN/HEPES buffer (0.1 M, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v, pH 7.4) 1, v/v, pH 7.4) |
0.86 μM | 0.5 min | 107 | |
| 16 | Styryl quinolinium/G-quadruplex complex | Tris-HAc buffer solution (pH 5) | 2 nM | 10 min | 108 | |
| 17 | AIE-based receptor (Schiff base) | HEPES buffer (10 mM, pH 7.4) | 0.1 μM | 3 min | 109 | |
| 18 | Dppy-based Cu(II) receptor |
CH
3
CN/HEPES buffer (10 mM, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4, v/v, pH 7.4) 4, v/v, pH 7.4)
|
0.33 μM | Immediate | Present work | |
| L-Histidine | 01 | Oxidase-like activity of Cu(II) of O-phenylenediamine | CH3CN (15%)/Tris–HCl buffer (50 mM pH 7.4) | 0.33 μM | 60 min | 110 |
| 02 | Carbon quantum dots–Hg(II) system | PBS buffer (pH 6.0) | 0.15 μM | 2 min | 111 | |
| 03 | Copper nanoclusters | — | 0.28 μM | 4 min | 112 | |
| 05 | Doped zinc sulfide quantum dots | 0.10 M PBS (pH 9.0) | 0.74 μM | 13 min | 113 | |
| 06 | Terbium(III) coordination polymer–copper(II) ensemble | NEM (5 mM, 10 μL) and HEPES buffer (100 mM, 10 μL, pH 7.4) | 1.2 μM | 20 min | 114 | |
| 07 | (S)-BINOL based receptor |
iPrOH/MeOH 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v 1, v/v |
70.3 nM | 180 min | 115 | |
| 08 | Dppy-based Cu(II) receptor |
CH
3
CN/HEPES buffer (10 mM, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4, v/v, pH 7.4) 4, v/v, pH 7.4)
|
1.40 μM | Immediate | Present work |
Conclusions
In summary, we have synthesised and characterised a novel copper(II) complex, CuCl2L, utilising a dppy-based ligand (L = 2,2′-(4-(3,4,5-trimethoxyphenyl)pyridine-2,6-diyl)dipyrazine). The in situ prepared CuCl2L receptor exhibits rapid and sensitive detection of Cys and His amino acids in aqueous acetonitrile (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 10 mM HEPES buffer, pH 7.4). Competitive experiments have demonstrated the selectivity of the CuCl2L receptor towards Cys (1 equiv.) in the presence of other L-amino acids in the same solvent system. Notably, the detection limits for Cys and His were determined as 0.33 μM and 1.40 μM, respectively. These values are in line with those reported in previous studies utilising distinct receptors. Additionally, our sensing studies have demonstrated an exceptional response time, outperforming many existing systems. Our computational results strongly support the variations in the stoichiometric ratio observed in copper complexes with cysteine and histidine. Additionally, they strongly indicate that the remarkable selectivity of CuCl2L in amino acid sensing originates the formation of highly stable coordination modes between CuCl2 and the cysteine and histidine residues. These results underscore the promising potential of the CuCl2L receptor as an efficient and reliable tool for the early-stage identification and monitoring of Cys and His in various applications.
1 v/v, 10 mM HEPES buffer, pH 7.4). Competitive experiments have demonstrated the selectivity of the CuCl2L receptor towards Cys (1 equiv.) in the presence of other L-amino acids in the same solvent system. Notably, the detection limits for Cys and His were determined as 0.33 μM and 1.40 μM, respectively. These values are in line with those reported in previous studies utilising distinct receptors. Additionally, our sensing studies have demonstrated an exceptional response time, outperforming many existing systems. Our computational results strongly support the variations in the stoichiometric ratio observed in copper complexes with cysteine and histidine. Additionally, they strongly indicate that the remarkable selectivity of CuCl2L in amino acid sensing originates the formation of highly stable coordination modes between CuCl2 and the cysteine and histidine residues. These results underscore the promising potential of the CuCl2L receptor as an efficient and reliable tool for the early-stage identification and monitoring of Cys and His in various applications.
Author contributions
Dipankar Das: conceptualisation, investigation, formal analysis, data curation, writing – original draft. Aritra Roy: software, formal analysis, investigation. Sourav Sutradhar: methodology, data curation. Felipe Fantuzzi: conceptualisation, methodology, software, writing – original draft, writing – review & editing, supervision. Biswa Nath Ghosh: conceptualisation, validation, writing – original draft, writing – review & editing, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
F. F. acknowledges the University of Kent for financial and computational support. Special thanks are extended to Dr Timothy Kinnear for HPC assistance. D. D. acknowledges SAIF IIT Patna for providing NMR facility.Notes and references
- H. Tavallali, G. Deilamy-Rad, M. A. Karimi and E. Rahimy, Anal. Biochem., 2019, 583, 113376 CrossRef CAS PubMed.
- R. Zhang, J. Yong, J. Yuan and Z. Ping Xu, Coord. Chem. Rev., 2020, 408, 213182 CrossRef CAS.
- S. V. Mulay, Y. Kim, M. Choi, D. Y. Lee, J. Choi, Y. Lee, S. Jon and D. G. Churchill, Anal. Chem., 2018, 90, 2648–2654 CrossRef CAS PubMed.
- S. Muthusamy, L. Zhao, K. Rajalakshmi, D. Zhu, R. Soy, J. Mack, T. Nyokong, S. Wang, K. B. Lee and W. Zhu, Dyes Pigm., 2021, 193, 109556 CrossRef CAS.
- G. Zhao, W. Yang, F. Li, Z. Deng and Y. Hu, J. Lumin., 2020, 226, 117506 CrossRef CAS.
- U. Tamima, C. W. Song, M. Santra, Y. J. Reo, H. Banna, M. R. Islam and K. H. Ahn, Sens. Actuators, B, 2020, 322, 128588 CrossRef CAS.
- Y. N. Wei, B. Lin, Y. Shu and J. H. Wang, Analyst, 2021, 146, 4642–4648 RSC.
- X. Yang, Y. Guo and R. M. Strongin, Angew. Chem., Int. Ed., 2011, 50, 10690–10693 CrossRef CAS PubMed.
- W. Hao, A. McBride, S. McBride, J. P. Gao and Z. Y. Wang, J. Mater. Chem., 2011, 21, 1040–1048 RSC.
- Z. Li, Y. Zhang, Y. Jiang, H. Li, C. Chen and W. Liu, J. Mater. Chem. B, 2022, 10, 6207–6213 RSC.
- S. Shahrokhian, Anal. Chem., 2001, 73, 5972–5978 CrossRef CAS PubMed.
- J. P. Lomont and J. P. Smith, Spectrochim. Acta, Part A, 2022, 274, 121068 CrossRef CAS PubMed.
- H. Huang, X. Ji, Y. Jiang, C. Zhang, X. Kang, J. Zhu, L. Sun and L. Yi, Org. Biomol. Chem., 2020, 18, 4004–4008 RSC.
- L. El-Khairy, P. M. Ueland, H. Refsum, I. M. Graham and S. E. Vollset, Circulation, 2001, 103, 2544–2549 CrossRef CAS PubMed.
- X. Huang, K. Li, X. Wang and P. Xia, Spectrochim. Acta, Part A, 2018, 205, 287–291 CrossRef CAS PubMed.
- S. G. Eswaran, M. A. Ashkar, M. H. Mamat, S. Sahila, V. Mahalingam, H. V. S. R. M. Koppisetti and N. Vasimalai, J. Sci.: Adv. Mater. Devices, 2021, 6, 100–107 Search PubMed.
- P. Munjal and H. M. Chawla, J. Lumin., 2018, 203, 364–370 CrossRef CAS.
- M. Watanabe, M. E. Suliman, A. R. Qureshi, E. Garcia-Lopez, P. Bárány, O. Heimbürger, P. Stenvinkel and B. Lindholm, Am. J. Clin. Nutr., 2008, 87, 1860–1866 CrossRef CAS PubMed.
- Q. Zhang, P. Zhang, S. Li, C. Fu and C. Ding, Dyes Pigm., 2019, 171, 107697 CrossRef CAS.
- T. Nagae, S. Aikawa, K. Inoue and Y. Fukushima, Tetrahedron Lett., 2018, 59, 3988–3993 CrossRef CAS.
- M. Chakraborty, M. Mohanty, R. Dinda, S. Sengupta and S. K. Chattopadhyay, Polyhedron, 2022, 211, 115554 CrossRef CAS.
- G. Wei, F. Meng, Y. Wang, Y. Cheng and C. Zhu, Macromol. Rapid Commun., 2014, 35, 2077–2081 CrossRef CAS PubMed.
- P. Gunasekaran, C. I. David, S. Shanmugam, K. Ramanagul, R. Rajendran, V. Gothandapani, V. R. Kannan, J. Prabhu and R. Nandhakumar, J. Agric. Food Chem., 2023, 71, 802–814 CrossRef CAS PubMed.
- F. Yan, X. Sun, F. Zu, Z. Bai, Y. Jiang, K. Fan and J. Wang, Methods Appl. Fluoresc., 2018, 6, 42001 CrossRef CAS PubMed.
- Y. Wang, Q. Meng, Q. Han, G. He, Y. Hu, H. Feng, H. Jia, R. Zhang and Z. Zhang, New J. Chem., 2018, 42, 15839–15846 RSC.
- S. Tajik, Z. Dourandish, P. M. Jahani, I. Sheikhshoaie, H. Beitollahi, M. Shahedi Asl, H. W. Jang and M. Shokouhimehr, RSC Adv., 2021, 11, 5411–5425 RSC.
- Q. Meng, H. Jia, X. Gao, Y. Wang, R. Zhang, R. Wang and Z. Zhang, Chem. – Asian J., 2015, 10, 2411–2418 CrossRef CAS PubMed.
- Y. S. Kim, G. J. Park, S. A. Lee and C. Kim, RSC Adv., 2015, 5, 31179–31188 RSC.
- T. Anand, A. S. K. Kumar and S. K. Sahoo, Photochem. Photobiol. Sci., 2018, 17, 414–422 CrossRef CAS PubMed.
- R. Shen, J. J. Yang, H. Luo, B. Wang and Y. Jiang, Tetrahedron, 2017, 73, 373–377 CrossRef CAS.
- H. Li, L. Jin, Y. Kan and B. Yin, Sens. Actuators, B, 2014, 196, 546–554 CrossRef CAS.
- X. Dai, Q. H. Wu, P. C. Wang, J. Tian, Y. Xu, S. Q. Wang, J. Y. Miao and B. X. Zhao, Biosens. Bioelectron., 2014, 59, 35–39 CrossRef CAS PubMed.
- D. P. Murale, H. Kim, W. S. Choi and D. G. Churchill, RSC Adv., 2014, 4, 5289–5292 RSC.
- J. Shao, H. Guo, S. Ji and J. Zhao, Biosens. Bioelectron., 2011, 26, 3012–3017 CrossRef CAS PubMed.
- J. Li, Y. Yue, F. Huo and C. Yin, Dyes Pigm., 2019, 164, 335–340 CrossRef CAS.
- S. Y. Lim, D. H. Yoon, D. Y. Ha, J. M. Ahn, D. Il Kim, H. Kown, H. J. Ha and H. J. Kim, Sens. Actuators, B, 2013, 188, 111–116 CrossRef CAS.
- B. Saha, P. Saha, A. Mandal, J. P. Naskar, D. Maiti and S. Chowdhury, J. Chin. Chem. Soc., 2019, 66, 506–514 CrossRef CAS.
- S. Yang and F. Liao, Synth. Met., 2012, 162, 1343–1347 CrossRef CAS.
- E. Lee, H. Ju, J. H. Jung, M. Ikeda, Y. Habata and S. S. Lee, Inorg. Chem., 2019, 58, 1177–1183 CrossRef CAS PubMed.
- M. R. Hormozi-Nezhad, E. Seyedhosseini and H. Robatjazi, Sci. Iran., 2012, 19, 958–963 CrossRef CAS.
- X. Li, K. Fan, X. Zhang, L. Wang, B. Qu and L. Lu, Microchem. J., 2019, 146, 486–491 CrossRef CAS.
- S. C. Liang, H. Wang, Z. M. Zhang, X. Zhang and H. S. Zhang, Spectrochim. Acta, Part A, 2002, 58, 2605–2611 CrossRef PubMed.
- A. Waseem, M. Yaqoob and A. Nabi, Curr. Pharm. Anal., 2013, 9, 363–395 CrossRef CAS.
- J. S. Stamler and J. Loscalzo, Anal. Chem., 1992, 64, 779–785 CrossRef CAS PubMed.
- N. Cebi, C. E. Dogan, A. Develioglu, M. E. A. Yayla and O. Sagdic, Food Chem., 2017, 228, 116–124 CrossRef CAS PubMed.
- S. Wadud, M. M. Or-Rashid and R. Onodera, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2002, 767, 369–374 CrossRef CAS PubMed.
- A. Nezamzadeh-Ejhieh and H. S. Hashemi, Talanta, 2012, 88, 201–208 CrossRef CAS PubMed.
- M. Rafii, R. Elango, G. Courtney-Martin, J. D. House, L. Fisher and P. B. Pencharz, Anal. Biochem., 2007, 371, 71–81 CrossRef CAS PubMed.
- D. Das, R. M. Gomila, P. Sarkar, S. Sutradhar, A. Frontera and B. N. Ghosh, Polyhedron, 2022, 223, 115959 CrossRef CAS.
- B. N. Ghosh, F. Topić, P. K. Sahoo, P. Mal, J. Linnera, E. Kalenius, H. M. Tuononen and K. Rissanen, Dalton Trans., 2015, 44, 254–267 RSC.
- S. Myadaraboina, M. Alla, V. Saddanapu, V. R. Bommena and A. Addlagatta, Eur. J. Med. Chem., 2010, 45, 5208–5216 CrossRef CAS PubMed.
- G. Ramesh, N. M. S. Kumar, P. R. Kumar, P. A. Suchetan, S. Devaraja, F. Sabine and G. Nagaraju, J. Mol. Struct., 2020, 1200, 127040 CrossRef CAS.
- K. Q. Wu, J. Guo, J. F. Yan, L. L. Xie, F. B. Xu, S. Bai, P. Nockemann and Y. F. Yuan, Organometallics, 2011, 30, 3504–3511 CrossRef CAS.
- B. N. Ghosh, S. Bhowmik, P. Mal and K. Rissanen, Chem. Commun., 2014, 50, 734–736 RSC.
- S. Sutradhar, S. Basak, D. Das and B. N. Ghosh, Polyhedron, 2023, 236, 116344 CrossRef CAS.
- S. Bhowmik, B. N. Ghosh and K. Rissanen, Org. Biomol. Chem., 2014, 12, 8836–8839 RSC.
- S. Sutradhar, D. Das and B. N. Ghosh, J. Mol. Struct., 2022, 1265, 133442 CrossRef CAS.
- D. Das, S. Sutradhar, K. Rissanen and B. N. Ghosh, Z. Anorg. Allg. Chem., 2020, 646, 301–306 CrossRef CAS.
- B. N. Ghosh, M. Lahtinen, E. Kalenius, P. Mal and K. Rissanen, Cryst. Growth Des., 2016, 16, 2527–2534 CrossRef CAS.
- S. Bhowmik, B. N. Ghosh, V. Marjomäki and K. Rissanen, J. Am. Chem. Soc., 2014, 136, 5543–5546 CrossRef CAS PubMed.
- D. Das, S. Sutradhar, A. Singh and B. N. Ghosh, Z. Anorg. Allg. Chem., 2021, 647, 1234–1238 CrossRef CAS.
- S. Manna, P. Karmakar, S. S. Ali, U. N. Guria, R. Sarkar, P. Datta, D. Mandal and A. K. Mahapatra, New J. Chem., 2018, 42, 4951–4958 RSC.
- S. Manna, P. Karmakar, S. S. Ali, U. N. Guria, S. K. Samanta, R. Sarkar, P. Datta and A. K. Mahapatra, Anal. Methods, 2019, 11, 1192–1198 RSC.
- Q. Wu, Y. Wu, C. Yu, Z. Wang, E. Hao and L. Jiao, Sens. Actuators, B, 2017, 253, 1079–1086 CrossRef CAS.
- B. Liu, J. Wang, G. Zhang, R. Bai and Y. Pang, ACS Appl. Mater. Interfaces, 2014, 6, 4402–4407 CrossRef CAS PubMed.
- J. Wang, H. Wang, Y. Hao, S. Yang, H. Tian, B. Sun and Y. Liu, Food Chem., 2018, 262, 67–71 CrossRef CAS PubMed.
- R. Golla, P. R. Kumar, P. A. Suchethan, S. Foro and G. Nagaraju, J. Mol. Struct., 2020, 1201, 127118 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- C. Adamo and V. Barone, J. Chem. Phys., 1999, 110, 6158–6170 CrossRef CAS.
- M. Ernzerhof and G. E. Scuseria, J. Chem. Phys., 1999, 110, 5029–5036 CrossRef CAS.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed.
- S. Grimme, S. Ehrlich and L. Goerigk, J. Comput. Chem., 2011, 32, 1456–1465 CrossRef CAS PubMed.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed.
- R. L. Martin, P. J. Hay and L. R. Pratt, J. Phys. Chem. A, 1998, 102, 3565–3573 CrossRef CAS.
- M. Sparta, C. Riplinger and F. Neese, J. Chem. Theory Comput., 2014, 10, 1099–1108 CrossRef CAS PubMed.
- F. Fantuzzi, M. A. C. Nascimento, B. Ginovska, R. M. Bullock and S. Raugei, Dalton Trans., 2021, 50, 840–849 RSC.
- T. Lu and F. Chen, J. Comput. Chem., 2012, 33, 580–592 CrossRef CAS PubMed.
- K. Choroba, B. Machura, S. Kula, L. R. Raposo, A. R. Fernandes, R. Kruszynski, K. Erfurt, L. S. Shul'Pina, Y. N. Kozlov and G. B. Shul'Pin, Dalton Trans., 2019, 48, 12656–12673 RSC.
- A. W. Addison and T. N. Rao, Polyhedron, 1998, 17, 1349–1356 Search PubMed.
- H. R. Khavasi and M. Esmaeili, Cryst. Growth Des., 2019, 19, 4369–4377 CrossRef CAS.
- L. Li, Y. Z. Zhang, C. Yang, E. Liu, J. C. Fettinger and G. Zhang, J. Mol. Struct., 2016, 1110, 19–23 CrossRef CAS.
- H. R. Khavasi and M. Esmaeili, Langmuir, 2019, 35, 4660–4671 CrossRef CAS PubMed.
- F. A. Al-Mutlaq, P. G. Potvin, A. I. Philippopoulos and P. Falaras, Eur. J. Inorg. Chem., 2007, 2121–2128 CrossRef CAS.
- M. Małecka, B. Machura and A. Szlapa-Kula, Dyes Pigm., 2021, 188, 109168 CrossRef.
- X.-X. Han, X. Han, Y. Wang, D. Shang, Y.-H. Xing and F.-Y. Bai, Polyhedron, 2018, 151, 192–198 CrossRef CAS.
- R. Hao, L. Li, S. Zhu, Z.-H. Wang, X.-J. Zhao and E.-C. Yang, J. Mol. Struct., 2019, 1176, 376–385 CrossRef CAS.
- N. Zhang, J. Tang, Y. Ma, M. Liang, D. Zeng and G. Hefter, Phys. Chem. Chem. Phys., 2021, 23, 6807–6814 RSC.
- T. Le Bahers, C. Adamo and I. Ciofini, J. Chem. Theory Comput., 2011, 7, 2498–2506 CrossRef CAS PubMed.
- D. Jacquemin, T. Le Bahers, C. Adamo and I. Ciofini, Phys. Chem. Chem. Phys., 2012, 14, 5383 RSC.
- I. Ciofini, T. Le Bahers, C. Adamo, F. Odobel and D. Jacquemin, J. Phys. Chem. C, 2012, 116, 11946–11955 CrossRef CAS.
- J. Xu, H. Li, L. Li, J. Wang, F. Wang and L. He, J. Braz. Chem. Soc., 2020, 31, 1778–1786 CAS.
- D. Das, P. Sarkar, A. H. U. Kumar, S. Sutradhar, M. Kotakonda, N. K. Lokanath and B. N. Ghosh, J. Photochem. Photobiol., A, 2023, 441, 114726 CrossRef CAS.
- D. Das, S. Sutradhar, R. M. Gomila, K. Rissanen, A. Frontera and B. N. Ghosh, J. Mol. Struct., 2023, 1273, 134269 CrossRef CAS.
- S. Foley and M. Enescu, Vib. Spectrosc., 2007, 44, 256–265 CrossRef CAS.
- A. Fernández-Ramos, E. Cabaleiro-Lago, J. M. Hermida-Ramón, E. Martínez-Núñez and A. Peña-Gallego, J. Mol. Struct.: THEOCHEM, 2000, 498, 191–200 CrossRef.
- M. Belcastro, T. Marino, N. Russo and M. Toscano, J. Mass Spectrom., 2005, 40, 300–306 CrossRef CAS PubMed.
- F. Wang, Z. Guo, X. Li, X. Li and C. Zhao, Chem. – Eur. J., 2014, 20, 11471–11478 CrossRef CAS PubMed.
- S. Yang, C. Guo, Y. Li, J. Guo, J. Xiao, Z. Qing, J. Li and R. Yang, ACS Sens., 2018, 3, 2415–2422 CrossRef CAS PubMed.
- K. B. Li, W. B. Qu, D. M. Han, S. Zhang, W. Shi, C. X. Chen and X. X. Liang, Talanta, 2019, 194, 803–808 CrossRef CAS PubMed.
- M. Zhu, L. Wang, X. Wu, R. Na, Y. Wang, Q. X. Li and B. D. Hammock, Anal. Chim. Acta, 2019, 1058, 155–165 CrossRef CAS PubMed.
- X. H. Zhao, L. Z. Zhang, S. Y. Zhao, X. H. Cui, L. Gong, R. Zhao, B. F. Yu and J. Xie, Analyst, 2019, 144, 1982–1987 RSC.
- L. Fan, W. Zhang, X. Wang, W. Dong, Y. Tong, C. Dong and S. Shuang, Analyst, 2019, 144, 439–447 RSC.
- S. Priyanga, T. Khamrang, M. Velusamy, S. Karthi, B. Ashokkumar and R. Mayilmurugan, Dalton Trans., 2019, 48, 1489–1503 RSC.
- J. Zhang, J. Wang, J. Liu, L. Ning, X. Zhu, B. Yu, X. Liu, X. Yao and H. Zhang, Anal. Chem., 2015, 87, 4856–4863 CrossRef CAS PubMed.
- H. Chen, B. Zhou, R. Ye, J. Zhu and X. Bao, Sens. Actuators, B, 2017, 251, 481–489 CrossRef CAS.
- Y. Q. Sun, M. Chen, J. Liu, X. Lv, J. F. Li and W. Guo, Chem. Commun., 2011, 47, 11029–11031 RSC.
- Y. J. Lu, N. Ma, Y. J. Li, Z. Y. Lin, B. Qiu, G. N. Chen and K. Y. Wong, Sens. Actuators, B, 2012, 173, 295–299 CrossRef CAS.
- L. Yan, Z. Kong, W. Shen, W. Du, Y. Zhou and Z. Qi, RSC Adv., 2016, 6, 5636–5640 RSC.
- Y. Xu, X. Q. Wu, J. S. Shen and H. W. Zhang, RSC Adv., 2015, 5, 92114–92120 RSC.
- J. Hou, F. Zhang, X. Yan, L. Wang, J. Yan, H. Ding and L. Ding, Anal. Chim. Acta, 2015, 859, 72–78 CAS.
- Q. Tan, J. Qiao, R. Zhang and L. Qi, Microchem. J., 2020, 153, 2–7 CrossRef.
- W. Bian, F. Wang, Y. Wei, L. Wang, Q. Liu, W. Dong, S. Shuang and M. M. F. Choi, Anal. Chim. Acta, 2015, 856, 82–89 CrossRef CAS PubMed.
- S. F. Xue, L. F. Lu, Q. X. Wang, S. Zhang, M. Zhang and G. Shi, Talanta, 2016, 158, 208–213 CrossRef CAS PubMed.
- J. Tian, K. Lu, Y. Wang, Y. Chen, B. Huo, Y. Jiang, S. Yu, X. Yu and L. Pu, Tetrahedron, 2021, 95, 132366 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00183k |
| ‡ Current Address: Department of Chemical and Energy Engineering, London South Bank University, 103 Borough Road, London SE1 0AA, UK. |
| This journal is © The Royal Society of Chemistry 2023 |